Abstract
Glioma is a common invasive cancer with unfavorable prognosis in patients. Long non-coding RNAs have been reported to participate in modulating diverse cellular processes. Here, we focused on exploring the role of long intergenic non-protein coding RNA 1410 (LINC01410) in glioma and its underlying mechanism. The expression levels and protein levels of genes were analyzed by quantitative real-time PCR (RT-qPCR) analysis and western blot. Loss-of-function assays were performed to assess the function of LINC01410 in glioma cells. The interactions among MYC, LINC01410, microRNA-506-3p (miR-506-3p) and notch receptor 2 (NOTCH2) were validated through Chromatin immunoprecipitation (ChIP), RNA Binding Protein immunoprecipitation (RIP), RNA pull-down and luciferase reporter assays. Our data supported that LINC01410 was up-regulated in glioma cells. Bioinformatics predictions and the integrated experiments identified that MYC activated LINC01410 transcription and LINC01410 promoted the levels of NOTCH2 through sponging miR-506-3p and further motivated Notch signaling pathway. Rescue assays validated that LINC01410 exerted its influential functions on glioma cell proliferation and apoptosis via enhancing NOTCH2 expression. Our studies identified that LINC01410 accelerates the progression of glioma through acting as a ceRNA for miR-506-3p and elevating NOTCH2 expression to further activate the Notch signaling pathway, which indicated that LINC01410 might act as a novel regulator of glioma progression.
Keywords: Glioma, LINC01410, MYC, miR-506-3p, NOTCH2
Introduction
Glioma mainly originates from the central nervous system which accounts for almost 80% of malignant brain tumors (Ostrom et al. 2014). The most prominent hallmarks of glioma is invasive and aggressive behaviors, which makes it incurable and with frequent recurrence (Alfonso et al. 2017; Goodenberger and Jenkins 2012). Nevertheless, tumor etiology remains largely elusive. Besides, glioma patients still have a poor prognosis and a low overall survival rate. Therefore, identifying novel biomarkers is helpful for exploring effective strategies to decrease the incidence of glioma and improve the clinical treatment of glioma.
Long non-coding RNAs (lncRNAs) are a class of non-coding RNAs with no protein coding ability and they have more than 200 nucleotides. Although 60% of human transcripts are lncRNAs, their functions remain little known. In recent years, with the continuous development of biotechnology, the roles of lncRNAs in many cell phenotypes have been gradually recognized, such as metastasis, invasion, proliferation, apoptosis and so on (Sanchez Calle et al. 2018). In glioma, lncRNAs have been also found to be aberrant expressed and correlated with cancer development (Peng et al. 2018; Shi et al. 2017). For instance, LncRNA CASC11 promotes glioma progression via targeting miR-498/FOXK1 axis (Jin et al. 2019). LncRNA DNACR acts as an oncogene in glioma through activation of Wnt/β-catenin signaling pathway (Li and Zhou 2018). LncRNA LINC01857 participates in glioma tumorigenesis via sponging miR-1281 and modulating TRIM65 expression (Hu et al. 2019). Therefore, in order to understand the mechanism of glioma progression, it is urgent to constantly seek for new lncRNAs and investigate into their functions in cancers.
As a novel lncRNA, LINC01410 has been investigated in several cancers (Zhang et al. 2018; Wang et al. 2019; Jiang et al. 2020; Lu et al. 2020; Liu and Wen 2020; Luo et al. 2018; Cai et al. 2019). However, it has not been studied in glioma. In this paper, we concentrated on the roles and functions of LINC01410 In glioma and delving into the underlying mechanism.
Materials and Methods
Cell Lines and Culture
Human glioma cell lines (SHG44, T98G, LN229, A172), normal human glial cell line (HEB) and human embryonic kidney 293T (HEK-293T) cell line were chosen for this study. Among them, T98G, LN229, A172 and HEK-293T cells were obtained from American Type Culture Collection (ATCC; Manassas, VA, USA), while SHG44 and HEB cells were purchased from Huatuo Biotechnology Co., Ltd. (Shenzhen, China). All cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) except that T98G cell line was maintained in EMEM. All mediums were added with 10% fetal bovine serum (FBS) at 37 °C with 5% CO2 except that LN229 cell line was incubated in DMEM with 5% FBS.
Cell Transfection
The shRNAs targeting LINC01410 or MYC were designed by GenePharma (Shanghai, China) and then applied to reduce the expression of LINC01410 and MYC. MiR-506-3p mimics were used to overexpress miR-506-3p with NC mimics as the negative control. PcDNA3.1/MYC or pcDNA3.1/NOTCH2 was used to overexpressed MYC and NOTCH2, respectively. On the basis of the instruction of manufacturer, Lipofectamine 2000 was used to perform the transfection.
RNA Extraction and Quantitative Real-Time PCR (RT-qPCR) Analysis
TRIzol reagents were used to extract RNAs on the basis of the manufacturer’s instruction, and then the concentration and purity of RNAs were measured. The TaKaRa Reverse Transcription System was used to transcribe extracted RNA into cRNA. GoTaq qPCR Master Mix (Promega, Madison, WI, USA) was used to conduct RT-qPCR. U6 and GAPDH were used as the endogenous controls. StepOne Real-Time PCR System was used to performed reaction and the 2−△△Ct method was utilized to measure the relative fold differences and expression levels.
5-Ethynyl-20-deoxyuridine (EdU) Staining Assay
The SHG44 and LN229 cells were cultured in the cover glass in 24-well plates with EdU-labeled DMEM. The growth of cells was measured by EdU incorporation assay according to the product protocol. SHG44 and LN229 cells were fixed, permeated and stained by EdU and DAPI dye. At last, the treated cells were observed by the laser scanning microscope.
Colony Formation Assay
In brief, cultured cells were added into 6-well plates for incubation. After 14 days, cells were fixed in ethanol followed by staining with 0.1% crystal violet. The stained cell colonies were counted manually. The assay was independently carried out three times.
Flow Cytometry Analysis
After transfected for 48 h, SHG44 and LN229 cells were collected. Afterwards, cells were washed and fixed using 70% precooling ethanol at 4 °C for 12 h. At last, the cells were subjected to centrifugation. After washed with phosphate buffered saline, cells were added with 100 μl PI dye and 100 μl RNA enzymes, and went through incubation at 37 °C for 30 min before slowly and fully precipitation. A flow cytometer was used to observe cell apoptosis.
Chromatin Immunoprecipitation (ChIP)
SHG44 and LN229 cells were fixed in 1% formaldehyde for 30 min at room temperature. Sonication was used to cut the DNA to an average size. For chromatin immunoprecipitation, we used biotin-labeled MYC antibody, and anti-lgG was used as the negative control. Also, the RT-qPCR was conducted to quantify the purified chromatin.
Luciferase Reporter Assay
The LINC01410 promoter region containing the binding sites (wild type or mutant type) was constructed into the pGL3 vector (Promega, Madison, WI) and co-transfected along with pcDNA3.1/MYC or the empty vector into glioma cells. The sequence of LINC01410 or NOTCH2 mRNA containing wild-type (Wt) and mutant type (Mut) of miR-506-3p was inserted into pmirGLO dual-luciferase vector to form pmirGLO-LINC01410-Wt/Mut and pmirGLO-NOTCH2-Wt/Mut, respectively. Later, we co-transfected miR-506-3p mimics and NC mimics with the reporter gene into glioma cells. After transfected for 24 h, the cells were collected and lysed, and then the Luciferase Reporter Assay system was used to measure the luciferase activity.
Fluorescent In Situ Hybridization (FISH)
All the cells were fixed for 30 min in 4% PFA. The 0.5% TritonX-100 was used to permeabilize the cells for 15 min at 4 °C. Glioma cells were hybridized with LINC01410-specific FISH probe in buffer and counterstained with Hoechst solution. Images were obtained with a confocal laser microscope (Olympus).
Subcellular Fractionation
Subcellular fractionation assay was implemented with the employment of Cytoplasmic and Nuclear RNA Purification Kit (NORGEN, Thorold, ON, Canada). GAPDH or U6 was separately treated as the cytoplasmic control or the nuclear control.
RNA Pull Down
RNA pull down was carried out after Bio-miR-506-3p-Wt, Bio-miR-506-3p-Mut and negative control was constructed in glioma cells, and qRT-PCR was conducted to detect the enrichment. The cells were cultured in the cell lysate with streptavidin-coated magnetic beads.
RNA Binding Protein Immunoprecipitation (RIP)
On the basis of the manufacturer’s protocol, the Magna RIP RNA Binding Protein Immunoprecipitation Kit was used to perform the RNA immunoprecipitation experiment. Complete RIP lysis buffer was used to lyse the SHG44 and LN229 cells. At the same time, magnetic beads binding Ago2 or anti-IgG antibody were incubated. The negative control was IgG. And RT-qPCR was used to analyze the purified RNA.
Western Blot
The samples were added with RIPA buffer; after homogenization, the total protein was extracted. At room temperature, 5% skimmed dry milk was used to seal the PVDF membranes for 1 h. After incubation at 4 °C overnight, the membrane washed with TBST three times and the secondary antioxidant peroxides were incubated continuously. The antibodies against BCL2, Bax, cleaved caspase-3, total caspase-3, NOTCH2, HES1, HES6 and GAPDH were all bought from Abcam.
Bioinformatics Analysis
GEPIA (http://gepia.cancer-pku.cn/index.html) was used to predict the expression of LINC01410. Jaspar (http://jaspar.genereg.net/analysis) was used to predict the motif of MYC. HumanTFDB was used to predict transcription factors (http://bioinfo.life.hust.edu.cn/HumanTFDB#!/). DIANA (http://carolina.imis.athena-innovation.gr/diana_tools/web/index.php) was used to screen miRNAs. StarBase (http://starbase.sysu.edu.cn/) was used to screen mRNAs and predict the binding sites.
Statistical Analysis
All the experiments in vitro were performed three times. SPSS 18.0 software with Student’s t-test or one-way ANOVA was used to analyze the statistical difference. Mean ± standard deviation (SD) was used to present all the data. And when p < 0.05, it was considered to be statistically significant.
Results
LINC01410 is Up-regulated and Promotes the Development of Glioma
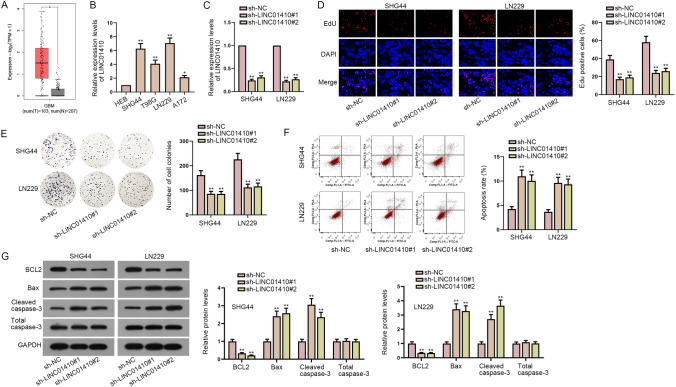
To disclose the role of LINC01410 in glioma, we firstly applied GEPIA 2 bioinformatics website (http://gepia2.cancer-pku.cn/#analysis) to analyze the expression of LINC01410 in 163 glioma tissues (tumor group) and 207 normal tissues (normal group). As shown in Fig. 1a, LINC01410 expression in tumor group was obviously higher than that in normal group. For further exploration, RT-qPCR was utilized to examine the expression levels of LINC01410 in glioma cells and the results showed that LINC01410 was highly expressed in glioma cells (SHG44, T98G, LN229, A172) compared with normal cell line (HEB) (Fig. 1b). We selected SHG44 and LN229 cells for subsequent experiments for that the expression levels of LINC01410 were higher in these two cells. Glioma cells were transfected with sh-LINC01410#1 and sh-LINC01410#2 to silence LINC01410 (Fig. 1c). To figure out the biological impacts of LINC01410 on glioma cell phenotypes, loss-of-function assays were performed. It was observed that the proliferative capacity of SHG44 and LN229 cells was inhibited by knockdown of LINC01410 in EdU and colony formation assays (Fig. 1d, e). On the contrary, flow cytometry analysis demonstrated that the apoptosis rate of SHG44 and LN229 cells was promoted by LINC01410 depletion (Fig. 1f). Besides, western blot analyzed that levels of apoptosis-related proteins (BCL2, Bax, cleaved caspase-3) were greatly changed due to LINC01410 inhibition (Fig. 1g). Taken together, LINC01410 is highly expressed in glioma and silencing of LINC01410 inhibits cell proliferation and promotes cell apoptosis in glioma.
Fig. 1.
LINC01410 is up-regulated and promotes the development of glioma. a GEPIA demonstrated the expression levels of LINC01410 in glioma tissues and normal tissues. b The expression levels of LINC01410 in glioma cell lines (SHG44, T98G, LN229, A172) and normal cell line (HEB) were detected by RT-qPCR. c RT-qPCR was used to detect the knockdown efficiency of LINC01410. d, e The proliferative capacity of glioma cells was measured by EdU and colony formation assays. f Cell apoptosis was examined by flow cytometry assay. g Levels of apoptosis-related proteins were detected by western blot. *P < 0.05, **P < 0.01
MYC Accelerates the Transcription of LINC01410 in Glioma
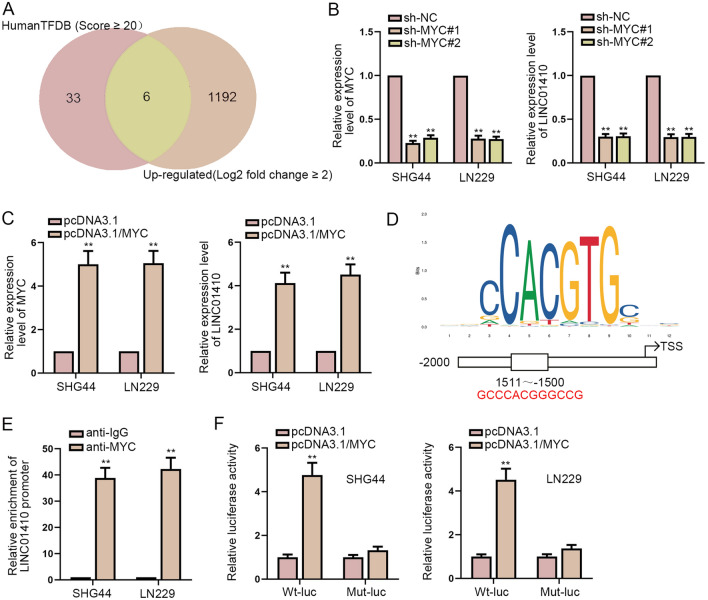
Through HumanTFDB and GEPIA database, we screened out 6 potential transcription factors of LINC01410 (EZH2, RNF2, ETV4, MYC, HIF1A, TP53) (Fig. 2a). Among them, MYC has been reported as a transcription factor in a variety of tumors. Therefore, we speculated that MYC could be used as a transcription factor of LINC01410. Firstly, we knocked down or overexpressed MYC, and then we observed that when MYC was knocked down, the expression of LINC01410 was also decreased. On the contrary, when MYC was overexpressed, the expression level of LINC01410 was increased (Fig. 2b, c). Then we used JASPAR to predict the motif of MYC (Fig. 2d). ChIP assay was carried out to uncover the high enrichment of MYC in the promoter of LINC01410 (Fig. 2e). Subsequently, we implemented luciferase reporter assay. The findings suggested that overexpression of MYC led to a significant increase in the luciferase activity of LINC01410 promoter (Fig. 2f). In summary, MYC acts as a transcription factor of LINC01410 in glioma and enhances the expression of LINC01410.
Fig. 2.
MYC accelerates the transcription of LINC01410 in glioma. a Transcription factors of LINC01410 were predicted by HumanTFDB and GEPIA database. b, c The expression levels of LINC01410 and MYC were detected after MYC expression was decreased or increased. d The motif of MYC predicted by JASPAR was demonstrated. e ChIP assay disclosed the enrichment of MYC in the LINC01410 promoter. f Luciferase reporter assay was conducted to verify the interaction between MYC and LINC01410 promoter at the predicted binding site. **P < 0.01
LINC01410 Sponges miR-506-3p
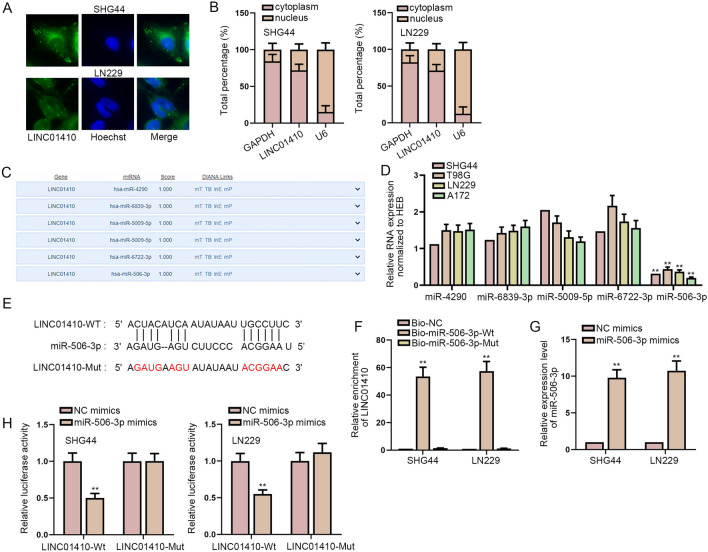
We intended to further study how LINC01410 plays a role in glioma. Firstly, the localization of LINC01410 was detected. FISH and subcellular fractionation experiments showed that LINC01410 was mainly accumulated in the cytoplasm of glioma cells (Fig. 3a, b). In the competing endogenous RNA (ceRNA) mechanism, lncRNAs can bind to miRNAs and regulate the target genes of miRNAs at the post-transcriptional level. Hence, DIANA tool was used to screen out six miRNAs (Fig. 3c). The results of RT-qPCR showed that only miR-506-3p was significantly down-regulated in glioma cells (Fig. 3d). The binding sites were predicted by DIANA (Fig. 3e). RNA pull-down assay demonstrated that LINC01410 could be effectively pulled down by biotinylated miR-506-3p-Wt probe instead of biotinylated miR-506-3p-Mut probe (Fig. 3f). The overexpression efficiency of miR-506-3p mimics was detected by RT-qPCR (Fig. 3g). The luciferase reporter assay confirmed that miR-506-3p had an interaction with LINC01410 (Fig. 3h). To be concluded, miR-506-3p is sponged by LINC01410.
Fig. 3.
LINC01410 sponges miR-506-3p. a, b FISH and subcellular fractionation assays were performed to the distribution of LINC01410 in glioma cells. c Six potential miRNAs were screened out by DIANA tool. d The expression levels of six miRNAs in glioma cells were tested. e Predicted binding site between LINC01410 and miR-506-3p was demonstrated. f The binding between LINC01410 and miR-506-3p was proved by RNA pull-down assay. g MiR-506-3p was overexpressed by transfection of miR-506-3p mimics. h Luciferase reporter assay further testified the relationship between LINC01410 and miR-506-3p. **P < 0.01
LINC01410 Competitively Sponges miR-506-3p to Increase the Expression of NOTCH2 and Activates Notch Signaling Pathway
Through luciferase reporter assay, we discovered that LINC01410 silencing obviously decreased the luciferase activity of Notch signaling pathway among six common signaling pathways (Fig. 4a). At the same time, we intriguingly found that NOTCH2, a key molecule in Notch signaling pathway, was a target gene of miR-506-3p. Hence, we conjectured that LINC01410 sponged miR-506-3p to modulate NOTCH2 expression, thereby being involved in Notch signaling pathway. Through RT-qPCR and western blot analysis, the expression levels and protein levels of key factors (NOTCH2, HES1 and HES6) in Notch signaling pathway were cut down due to the down-regulation of LINC01410 (Fig. 4b, c). Figure 4d shows the binding sites predicted by StarBase. In order to explore whether there was a binding force among LINC01410, miR-506-3p and NOTCH2, we carried out Ago2 RIP experiment. The results confirmed that they could co-exist in the RISC (RNA-induced silencing complex), providing a hypothesis that LINC01410, miR-506-3p and NOTCH2 might interact with each other (Fig. 4e). For further verification, the results of luciferase reporter assay exposed that after transfection of miR-506-3p mimics, the luciferase activity was reduced in the wild-type NOTCH2 group, while the luciferase activity of the mutant NOTCH2 group was barely changed (Fig. 4f). The expression level of miR-506-3p was decreased after transfection of miR-506-3p inhibitor into glioma cells (Fig. 4g). More surprisingly, we noticed that the depression of expression levels or protein levels of NOTCH2 caused by LINC01410 depletion was completely restored by miR-506-3p down-regulation (Fig. 4h, i). All these results support that LINC01410 competitively sponges miR-506-3p to increase the expression of NOTCH2, thereby activating Notch signaling pathway.
Fig. 4.
LINC01410 competitively sponges miR-506-3p to increase the expression of NOTCH2 and activates Notch signaling pathway. a Relative luciferase activities of six common signaling pathways were examined after LINC01410 expression was cut down. b, c After transfection of sh-LINC01410#1/2, the expression levels and protein levels of key factors in Notch signaling pathway were examined. d The binding site between miR-506-3p and NOTCH2 was predicted. e Ago2 RIP verified the interaction among LINC01410, miR-506-3p and NOTCH2. f The binding between miR-506-3p and NOTCH2 was disclosed by luciferase reporter assay. g The expression level of miR-506-3p was lessened after transfection of miR-506-3p inhibitor. h, i The expression levels and protein levels of NOTCH2 were examined in sh-NC, sh-LINC01410#1 and sh-LINC01410#1 + miR-506-3p inhibitor groups. **P < 0.01
LINC01410 Exerts its Influential Functions on Glioma Cell Proliferation and Apoptosis via Enhancing NOTCH2 Expression
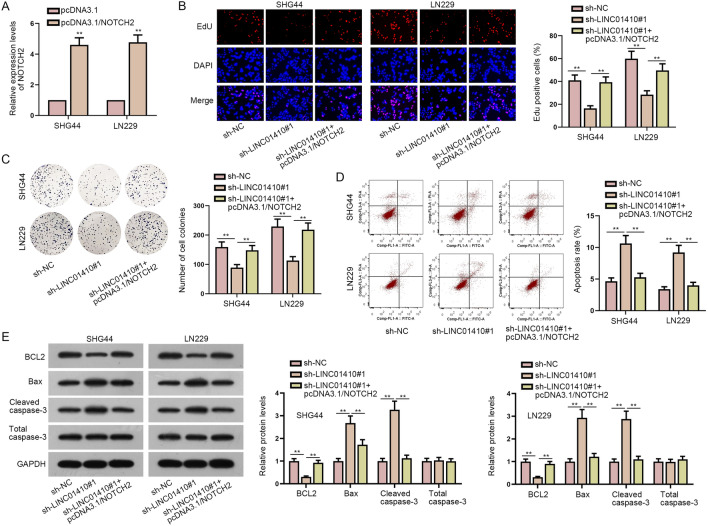
For further exploration, we conducted a series of rescue experiments. First, glioma cells were transfected with pcDNA3.1/NOTCH2 to increase the expression level of NOTCH2 (Fig. 5a). From the experimental results of EdU and colony formation assays, the reduction in cell proliferation caused by LINC01410 inhibition could be recovered by NOTCH2 up-regulation (Fig. 5b, c). Furthermore, LINC01410 down-regulation induced enhanced cell apoptosis was countervailed by NOTCH2 overexpression (Fig. 5d). Similarly, protein levels of BCL2, Bax and cleaved caspase-3 all had a significant change on account of LINC01410 knockdown while rescued by up-regulation of NOTCH2, indicating that LINC01410 hindered cell apoptosis in glioma via enhancing NOTCH2 expression (Fig. 5e). To sum up, LINC01410 affects the proliferation and apoptosis ability of glioma cell by enhancing the expression of NOTCH2.
Fig. 5.
LINC01410 exerts its influential functions on glioma cell proliferation and apoptosis via enhancing NOTCH2 expression. a PcDNA3.1/NOTCH2 was transfected to enhance the expression of NOTCH2. b, c EdU and colony formation assays were implemented to observe cell proliferation in sh-NC group, sh-LINC01410#1 group and sh-LINC01410#1 + pcDNA3.1/NOTCH2 group. d Flow cytometry analysis was utilized to observe cell apoptosis. e Apoptosis-related protein levels were detected in different groups by western blot. **P < 0.01
Discussion
Accumulating evidence has testified the importance of non-coding RNAs in glioma (Shi et al. 2017). LINC01410 has been identified to be an oncogene in gastric cancer, papillary thyroid carcinoma, colon cancer, pancreatic cancer, cervical cancer, endometrial cancer and cholangiocarcinoma (Zhang et al. 2018; Wang et al. 2019; Jiang et al. 2020; Lu et al. 2020; Liu and Wen 2020; Luo et al. 2018; Cai et al. 2019). Besides, LINC01410 has been reported to be concerned with angiogenesis, metastasis, proliferation, apoptosis and so on. Consistent with these findings, in our experiment, we found that LINC01410 had a high expression level in glioma tissues and cell lines. Moreover, knockdown of LINC01410 impeded cell proliferation and facilitated cell apoptosis in glioma. Furthermore, MYC was discovered to be a transcription activator of LINC01410 and stimulate the overexpression of LINC01410 in glioma. In a word, we demonstrated the oncogenic role of LINC01410 in glioma.
LncRNAs have been substantiated to be involved in targeting miRNAs and signaling pathways in glioma (Dang et al. 2018). In our research, we discovered that miR-506-3p was sponged by LINC01410. In fact, miR-506-3p has been proved to suppress glioma progression (Li et al. 2020). In addition, we also found miR-506-3p was down-regulated in glioma cells.
The functional roles of Notch signaling pathway have been elucidated in glioma and targeting Notch signaling pathway may improve malignant characteristics in glioma (Yan et al. 2018; Gersey et al. 2019). NOTCH2 has been defined to be an important member of the Notch signaling pathway (Penton et al. 2012). In addition, Chen et al. have certified that NOTCH2 is up-regulated in glioma (Chen et al. 2013). In this paper, we demonstrated the interaction among LINC01410, miR-506-3p and NOTCH2. Furthermore, rescue assays attested that LINC01410 enhanced NOTCH2 expression and activated Notch signaling pathway, thereby promoting cell proliferation and hindered cell apoptosis in glioma.
However, there are still many limitations that cannot be ignored in this study. In order to better understand how LINC01410 plays a regulatory role in glioma, we need to conduct further research on Notch signaling pathway. In addition, more experiments are needed to verify the effects of MYC/LINC01410/miR-506-3p/NOTCH/Notch signaling pathway on other malignant phenotypes in glioma.
In conclusion, we found that the expression of LINC01410 was high in glioma cells. Our study demonstrated that LINC01410 acted as a sponge of miR-506-3p, and then regulated NOTCH2 which activated Notch signaling pathway. At the same time, high expression of MYC in glioma could activate the transcription of LINC01410. Therefore, our study elucidated the molecular mechanism of LINC01410 in the occurrence of glioma, which might be an important target for clinical diagnosis and treatment of glioma in the future.
Acknowledgements
We are grateful to the help provided by all lab personnel in this research.
Author Contributions
XZ designed this study and performed experiments. XZ and FS contributed to data curation. BY wrote the manuscript. All authors reviewed and approved the final manuscript.
Compliance with Ethical Standards
Conflict of interest
No conflicts of interest were declared.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Alfonso JCL, Talkenberger K, Seifert M, Klink B, Hawkins-Daarud A, Swanson KR, Hatzikirou H, Deutsch A (2017) The biology and mathematical modelling of glioma invasion: a review. J Roy Soc, Interface 14(136):0490. 10.1098/rsif.2017.0490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai M, Xu L, Shen L, Zhang J (2019) The expression of long non-coding RNA-LINC01410 in pancreatic cancer and its effect on proliferation and migration of pancreatic cancer cells. Zhonghua yi xue za zhi 99(18):1406–1411. 10.3760/cma.j.issn.0376-2491.2019.18.010 [DOI] [PubMed] [Google Scholar]
- Chen L, Chen XR, Zhang R, Li P, Liu Y, Yan K, Jiang XD (2013) MicroRNA-107 inhibits glioma cell migration and invasion by modulating Notch2 expression. J Neurooncol 112(1):59–66. 10.1007/s11060-012-1037-7 [DOI] [PubMed] [Google Scholar]
- Dang Y, Wei X, Xue L, Wen F, Gu J, Zheng H (2018) Long non-coding RNA in glioma: target miRNA and signaling pathways. Clin Lab 64(6):887–894. 10.7754/Clin.Lab.2018.180107 [DOI] [PubMed] [Google Scholar]
- Gersey Z, Osiason AD, Bloom L, Shah S, Thompson JW, Bregy A, Agarwal N, Komotar RJ (2019) Therapeutic targeting of the notch pathway in glioblastoma multiforme. World Neurosurg 131:252-263.e252. 10.1016/j.wneu.2019.07.180 [DOI] [PubMed] [Google Scholar]
- Goodenberger ML, Jenkins RB (2012) Genetics of adult glioma. Cancer Genet 205(12):613–621. 10.1016/j.cancergen.2012.10.009 [DOI] [PubMed] [Google Scholar]
- Hu G, Liu N, Wang H, Wang Y, Guo Z (2019) LncRNA LINC01857 promotes growth, migration, and invasion of glioma by modulating miR-1281/TRIM65 axis. J Cell Physiol 234(12):22009–22016. 10.1002/jcp.28763 [DOI] [PubMed] [Google Scholar]
- Jiang T, Wang C, Zhu Y, Han H (2020) LINC01410 promotes cell proliferation and migration of cholangiocarcinoma through modulating miR-124-3p/SMAD5 axis. J Gene Med 22(6):e3162. 10.1002/jgm.3162 [DOI] [PubMed] [Google Scholar]
- Jin J, Zhang S, Hu Y, Zhang Y, Guo C, Feng F (2019) SP1 induced lncRNA CASC11 accelerates the glioma tumorigenesis through targeting FOXK1 via sponging miR-498. Biomed Pharmacother 116:108968. 10.1016/j.biopha.2019.108968 [DOI] [PubMed] [Google Scholar]
- Li J, Zhou L (2018) Overexpression of lncRNA DANCR positively affects progression of glioma via activating Wnt/β-catenin signaling. Biomed Pharmacother 102:602–607. 10.1016/j.biopha.2018.03.116 [DOI] [PubMed] [Google Scholar]
- Li H, Li T, Huang D, Zhang P (2020) Long noncoding RNA SNHG17 induced by YY1 facilitates the glioma progression through targeting miR-506-3p/CTNNB1 axis to activate Wnt/β-catenin signaling pathway. Cancer Cell Int 20:29. 10.1186/s12935-019-1088-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Wen C (2020) LINC01410 knockdown suppresses cervical cancer growth and invasion via targeting miR-2467-3p/VOPP1 axis. Cancer Manag Res 12:855–861. 10.2147/cmar.S236832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu M, Ding N, Zhuang S, Li Y (2020) LINC01410/miR-23c/CHD7 functions as a ceRNA network to affect the prognosis of patients with endometrial cancer and strengthen the malignant properties of endometrial cancer cells. Mol Cell Biochem 469(1–2):9–19. 10.1007/s11010-020-03723-9 [DOI] [PubMed] [Google Scholar]
- Luo J, Guo Y, Liu X, Yang X, Xiao F, Zhou M (2018) Long non-coding RNA LINC01410 promotes colon cancer cell proliferation and invasion by inhibiting miR-3128. Exp Ther Med 16(6):4824–4830. 10.3892/etm.2018.6806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrom QT, Bauchet L, Davis FG, Deltour I, Fisher JL, Langer CE, Pekmezci M, Schwartzbaum JA, Turner MC, Walsh KM, Wrensch MR, Barnholtz-Sloan JS (2014) The epidemiology of glioma in adults: a “state of the science” review. Neuro-oncology 16(7):896–913. 10.1093/neuonc/nou087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Z, Liu C, Wu M (2018) New insights into long noncoding RNAs and their roles in glioma. Mol Cancer 17(1):61. 10.1186/s12943-018-0812-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penton AL, Leonard LD, Spinner NB (2012) Notch signaling in human development and disease. Semin Cell Dev Biol 23(4):450–457. 10.1016/j.semcdb.2012.01.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez Calle A, Kawamura Y, Yamamoto Y, Takeshita F, Ochiya T (2018) Emerging roles of long non-coding RNA in cancer. Cancer Sci 109(7):2093–2100. 10.1111/cas.13642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Dong B, Cao J, Mao Y, Guan W, Peng Y, Wang S (2017) Long non-coding RNA in glioma: signaling pathways. Oncotarget 8(16):27582–27592. 10.18632/oncotarget.15175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Wang X, Jin Y (2019) LINC01410/miR-3619-5p/FOXM1 feedback loop regulates papillary thyroid carcinoma cell proliferation and apoptosis. Cancer Biother Radiopharm 34(9):572–580. 10.1089/cbr.2019.2854 [DOI] [PubMed] [Google Scholar]
- Yan D, Hao C, Xiao-Feng L, Yu-Chen L, Yu-Bin F, Lei Z (2018) Molecular mechanism of notch signaling with special emphasis on microRNAs: implications for glioma. J Cell Physiol 234(1):158–170. 10.1002/jcp.26775 [DOI] [PubMed] [Google Scholar]
- Zhang JX, Chen ZH, Chen DL, Tian XP, Wang CY, Zhou ZW, Gao Y, Xu Y, Chen C, Zheng ZS, Weng HW, Ye S, Kuang M, Xie D, Peng S (2018) LINC01410-miR-532-NCF2-NF-kB feedback loop promotes gastric cancer angiogenesis and metastasis. Oncogene 37(20):2660–2675. 10.1038/s41388-018-0162-y [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]