Abstract
Genetic studies of blood pressure (BP) traits to date have been performed on conventional measures by brachial cuff sphygmomanometer for systolic BP (SBP) and diastolic BP, integrating several physiologic occurrences. Genetic associations with central SBP (cSBP) have not been well-studied. Genetic discovery studies of BP have been most often performed in European-ancestry samples. Here, we investigated genetic associations with cSBP in a Chinese population and functionally validated the impact of a novel associated coiled-coil domain containing 93 (CCDC93) gene on BP regulation. An exome-wide association study (EWAS) was performed using a mixed linear model of non-invasive cSBP and peripheral BP traits in a Han Chinese population (N = 5,954) from Beijing, China genotyped with a customized Illumina ExomeChip array. We identified four SNP-trait associations with three SNPs, including two novel associations (rs2165468-SBP and rs33975708-cSBP). rs33975708 is a coding variant in the CCDC93 gene, c.535C>T, p.Arg179Cys (MAF = 0.15%), and was associated with increased cSBP (β = 29.3 mmHg, P = 1.23x10-7). CRISPR/Cas9 genome editing was used to model the effect of Ccdc93 loss in mice. Homozygous Ccdc93 deletion was lethal prior to day 10.5 of embryonic development. Ccdc93+/- heterozygous mice were viable and morphologically normal, with 1.3-fold lower aortic Ccdc93 protein expression (P = 0.0041) and elevated SBP as compared to littermate Ccdc93+/+ controls (110±8 mmHg vs 125±10 mmHg, P = 0.016). Wire myography of Ccdc93+/- aortae showed impaired acetylcholine-induced relaxation and enhanced phenylephrine-induced contraction. RNA-Seq transcriptome analysis of Ccdc93+/- mouse thoracic aortae identified significantly enriched pathways altered in fatty acid metabolism and mitochondrial metabolism. Plasma free fatty acid levels were elevated in Ccdc93+/- mice (96±7mM vs 124±13mM, P = 0.0031) and aortic mitochondrial dysfunction was observed through aberrant Parkin and Nix protein expression. Together, our genetic and functional studies support a novel role of CCDC93 in the regulation of BP through its effects on vascular mitochondrial function and endothelial function.
Author summary
More than 1000 loci have been reported in previous gene discovery efforts for blood pressure and hypertension. cSBP is a unique blood pressure trait that has not been as extensively studied as peripheral BP, and correlates of cSBP may provide a better indication of future cardiovascular events. In this study we replicated several BP associations in a Han Chinese cohort and found a novel association with a coding variant in CCDC93 (rs33975708, c.535C>T, p.Arg179Cys, MAF 0.15%) which was strongly associated with higher cSBP. To model loss of Ccdc93 and functionally validate the human genetic association, a new transgenic mouse was created. Ccdc93 homozygous deletion was embryonic lethal, and mice with heterozygous loss of Ccdc93 exhibited impaired arterial relaxation and enhanced contractile responses. Aortic RNA-seq analysis of Ccdc93+/- heterozygous mice identified metabolic dysregulation and mitochondrial dysfunction, which was validated by features of elevated plasma free fatty acids and increased aortic Parkin and Nix protein expression. Improved knowledge of the genes involved in BP regulation is needed to develop effective therapies to manage elevated BP, and this study identified the gene CCDC93 as a novel regulator of blood pressure and vascular function.
Introduction
Elevated blood pressure (BP) is the leading modifiable risk factor for cardiovascular disease and stroke [1,2]. BP is influenced by genetic, demographic, and environmental factors, and their interactions. Previous genome-wide association studies (GWAS) have identified and validated variants at more than 1000 loci with modest effects on population BP [3–6], explaining in aggregate ~6% of the trait variance [7]. Differences according to ancestry in the genetic architecture of BP and hypertension in the population have been identified [8] and highlight the importance of genetic studies in diverse ancestry populations [9].
Conventionally, BP is measured at the brachial artery by automatic BP monitors or auscultation of Korotkoff sounds. Thus, measured BP is an integrated readout of both the forward wave of BP according to cardiac output and arterial elasticity as well as a reflected wave mediated by total peripheral vascular resistance [10]. Although central systolic BP (cSBP) and peripheral SBP (pSBP) may share similar mean values, there are significant differences between these two measures. cSBP is determined by the interactions between the left ventricle and the vasculature. cSBP is elevated due to increased forward and reflected wave amplitudes and earlier return of the reflected wave to the proximal aorta and it is greatly influenced by arterial stiffness [11,12]. Additionally, BP amplification from the central aorta to peripheral arteries occurs. These factors together lead to discrepancies between central BP and peripheral BP, with underlying physiologic etiologies that are not defined through conventional BP measurement [13–15]. Although peripheral BP is strongly associated with central BP [16], cSBP responds differently to certain BP-lowering drugs [17] and cSBP may be more closely correlated with future cardiovascular events as compared to peripheral BP measured at the brachial artery [18,19], as well as measures of related cardiovascular disease risks [12,17,19–26].
cSBP is heritable, with estimated heritability 0.18 in a twin study [27]; conventionally measured SBP and diastolic BP (DBP) has estimated heritability 0.17–0.52 [28]. Despite numerous GWAS of peripheral BP, few studies have focused on genetic associations with central BP [29,30]. Furthermore, most BP GWAS studies have been conducted in European ancestry samples, and relatively fewer have been conducted in East Asian samples [31].
In the current study, we performed an exome-wide association study (EWAS) using the Illumina ExomeChip genotyping array for discovery associations with cSBP, pSBP, pDBP, and peripheral mean arterial pressure (pMAP) in Han Chinese populations, followed by replication experiments. We generated a novel transgenic mouse based upon the EWAS findings with heterozygous loss of Ccdc93 and conducted functional validation experiments.
Results
PUUMA cohort discovery study samples
The PUUMA Study included 5,954 subjects with peripheral BP measurements (pSBP, pDBP, and pMAP) and 4,938 subjects with cSBP. In the discovery PUUMA cohort, 63.3% (n = 3,769) of participants were female. Average cSBP was 133.1±18.6 mmHg and average pSBP was 133.7±16.7 mmHg. Other traits and covariates are summarized in Table 1.
Table 1. Characteristics of participants in the discovery stage EWAS.
| Characteristics | N (%) or Mean±S.d. |
|---|---|
| N | 5954 |
| Women | 3769 (63.3) |
| Age | 56.9±9.0 |
| Body mass index | 26.1±3.4 |
| Systolic blood pressure | 133.7±16.7 |
| Diastolic blood pressure | 75.0±10.0 |
| Mean arterial pressure | 94.6±10.8 |
| Central systolic blood pressure | 133.1±18.6 |
| Anti-hypertensive medication | 1890 (31.9) |
PUUMA cohort discovery stage EWAS
We conducted an EWAS for each BP trait, using mixed linear models, adjusted for age, age2, BMI, and sex, with resulting λGC values ranging from 0.997 to 1.008. The Manhattan plot, quantile-quantile plot (QQ), and regional association plot for cSBP are shown in Fig 1A–1C. Results for pSBP, pDBP and pMAP are shown in S1–S3 Figs. We identified four locus-trait combinations that reached the Bonferroni corrected genome wide significance threshold of P-value<5.8 × 10−7 in the discovery analysis, including 2 novel SNP-trait associations and 2 previously reported associations (Table 2).
Fig 1. Exome-wide association study (EWAS) for central systolic blood pressure (cSBP) and eQTL plot.
ExomeChip SNPs meeting quality control were analyzed in the EWAS. Results are shown in a (A) Manhattan plot and (B) Quantile-Quantile plot; λGC value was 1.0. (C) Regional association plots with gene annotation for the chromosome 2q14.1 region associated with cSBP is shown with the index SNP rs33975708 and additional SNPs within 500 kbp in each direction. Top panel: Regional cSBP association plot for the CCDC93 variant. LD (linkage disequilibrium) was calculated from our samples. Non-synonymous variants were annotated using ANNOVAR. The genetic recombination rate is based on Hapmap release 22. Middle Panel: Standardized varLD scores illustrate LD variations between populations (CEU vs. JPT+CHB, CEU vs. YRI, YRI vs. JPT+CHB) using genome positions from the Hapmap 3 reference. The red line represents the comparison between CEU (European ancestry) and JPT+CHB (East Asian ancestry), the purple line indicates CEU versus YRI (African ancestry), and the green line represents YRI versus JPT+CHB. Bottom Panel: Gene/transcript annotations are sourced from the reference downloaded from the UCSC database (EST, mRNA, uniGene, Encode, and RefGene). (D) eQTL plots for rs33975708 (chr2_117986054_G_A_b38 eQTL) and ENSG00000125633.10 (CCDC93), for human arterial tissues in GTEx data. Significant eQTLs were queried from the GTEx V8 portal for arterial tissues, which showed that the tibial artery had significant eQTL for rs33975708 and CCDC93 gene, based on GTEx defined permutation-based multiple test correction significance threshold. The eQTL violin plot demonstrated that the risk allele or alternative allele, A, for rs33975708, decreases the expression of CCDC93. A cross-tissue meta-analysis of 49 GTEx tissues using METASOFT tested the posterior probability that an eQTL effect exists in each tissue was also reported (meta-analysis RE2 P-value: 1.96E-15). A large m-value (>0.9) indicated the tissue was predicted to have an eQTL effect, whereas a small m-value (<0.1) indicated that it was predicted not to have an eQTL effect. Both the tibial artery and aorta artery had a larger m-value>0.9, indicating a strong eQTL effect. The risk allele of rs33975708, A, was associated with lower expression of CCDC93, in all the three arterial tissues (NES<0).
Table 2. EWAS SNP-trait associations.
SNP-trait associations meeting genome-wide significance are shown.
| Trait | SNP | Locus | Chr | Position1 | A1 | A2 | MAF | β | SE | P | Gene |
|---|---|---|---|---|---|---|---|---|---|---|---|
| pSBP | rs2165468 | p15.2 | 10 | 3516105 | A | C | 0.458 | 1.77 | 0.34 | 1.55x10-07 |
PITRM1(dist = 301072), KLF6(dist = 302083) |
| pDBP | rs13149993 | q21.21 | 4 | 81158545 | A | G | 0.423 | 1.06 | 0.21 | 3.16x10-07 |
PRDM8(dist = 33063), FGF5(dist = 29197) |
| pMAP | rs13149993 | q21.21 | 4 | 81158545 | A | G | 0.423 | 1.24 | 0.24 | 1.67x10-07 |
PRDM8(dist = 33063), FGF5(dist = 29197) |
| cSBP | rs33975708 | q14.1 | 2 | 118743630 | A | G | 0.002 | 29.31 | 5.53 | 1.23x10-07 | CCDC93 |
1Assembly version: GRCh37 (hg19)
The associations of rs13149993 with pDBP and rs13149993 with pMAP reported in previous studies [32] were replicated in our East-Asian population. rs13149993 on chromosome 4q21.21 locus within FGF5 was associated with pDBP (risk allele: A; risk allele frequency: 0.42; β: 1.06; SE: 0.21; P: 3.16×10−7) and pMAP (risk allele: A; risk allele frequency: 0.42; β: 1.24, SE: 0.24; P: 1.67×10−7). The two novel locus-trait combinations we identified were rs2165468 located on chromosome 10p15.2 and between PITRM1 (dist = 301,072 bp from GRCh37 genome assembly) and KLF6, was associated with pSBP (risk allele: A; risk allele frequency: 0.46; β = 1.77; SE = 0.34; P = 1.55×10−7), and rs33975708 located on chromosome 2q14.1 in CCDC93 was associated with cSBP (risk allele: A; risk allele frequency = 0.002; β = 29.31; SE = 5.53; P = 1.23×10−7).
To confirm major published BP association signals in our cohort, the association of a genetic risk score (GRS) based upon published SBP and DBP GWAS results, which included more than one million samples from the International Consortium for Blood Pressure (ICBP) and UK Biobank (UKB) cohorts, and the corresponding BP traits in our BP EWAS cohort samples were performed. GRS were weighted using effect sizes from the published GWAS of the same BP trait (5). Though subjects of the current study and previously published studies differed in ancestry, significant associations were identified with SBP (GRS-association of 611 SNPs β = 0.68, SE = 0.06, P = 3.42×10−30) and DBP (GRS-association of 734 SNPs β = 0.59, SE = 0.06, P = 2.5×10−24) in UKB with adjustment for age, age2, gender, BMI, and the top 10 PCs, and in ICBP (S1 Table).
Replication of novel SNP-trait associations
We conducted replication analyses of the 2 novel SNP-trait associations from the PUUMA EWAS. Given the rarity of rs33975708 (MAF in East Asians: 0.15%) and the lack of available cohorts with existing cSBP measurement, as well as that BP measurements may not be static across time for each sample, we attempted replication in two manners. First, we brought back subjects for an additional visit and collected an additional cSBP measurement. From our discovery stage cohort, 3448 individuals (84% samples of the discovery stage subjects) were evaluated at a mean follow-up of 2.35±0.08 years after the first visit and BP measurements. The association with rs33975708 for subjects with both first-visit and second-visit measurements (N = 2897), utilizing only the second visit cSBP measurement was significant, with a similar effect size as the discovery stage association of rs33975708 and cSBP (β = 23.63, SE = 7.84, P = 2.58×10−3). In a secondary analysis of the union set of samples (N = 4938) from these two visits, cSBP was averaged across the visits to yield a long-term averaged cSBP (LTA-cSBP) value [33]. LTA-cSBP was associated with rs33975708, with a stronger effect estimate and lower p-value (β = 24.63, SE = 6.62, P = 9.95×10−8) as compared to the initial discovered association result (S2 Table). An independent replication analysis of rs33975708 with cSBP measured by the same technique was performed in the Ji cohort (N = 76) [34], although underpowered for detection of association, with 9 individuals carrying the rs33975708-A CCDC93 risk allele and 67 individuals who did not have this risk allele; the association result was not statistically significant, but the effect direction was consistent with the discovery stage result (β = 11.18, SE = 9.64, P = 0.25). Inspection of the cSBP values showed that individuals in the Ji cohort with the rs33975708-A CCDC93 risk allele had a mean cSBP of 175.1 mmHg (s.d. = 30.7 mmHg) whereas individuals who did not carry the risk allele had a mean cSBP of 162.5 mmHg (s.d. = 26.2 mmHg) with t-test P-value = 0.19. Meta-analysis of discovery stage and Ji County data showed consistent directions of effect: β = 24.82, SE = 4.80, P = 2.29x10-7 utilizing inverse-variance-weighted method, and P = 7.03x10-8 by employing a weighted sum of Z-scores method [35]. We tested replication of the association of rs2165468 with pSBP using two independent resources (Peking University First Hospital and Third Hospital). This association was replicated in 565 participants from Peking University First Hospital (β = 2.45, SE = 1.15, P = 0.034), but not replicated in 127 participants from third hospital (β = 1.7, SE = 2.38, P = 0.48), although under-powered analyses; in the meta-analysis of the two replication results, a positive association with consistent effect direction was observed (β = 2.3, SE = 1.04, P = 0.026) (S2 Table).
In PUUMA, CCDC93-rs33975708 was evaluated for pleiotropic associations with peripheral BP traits, pulse wave velocity and blood lipid levels, as major risk factors for atherosclerotic coronary artery disease. rs33975708 was significantly associated with several BP traits including pSBP and Ba-PWV. (Table 3 and S4 Fig).
Table 3. Pleiotropy analysis of rs33975708.
For participants taking anti-hypertensive medications, pSBP and pDBP measurements were adjusted +15 mmHg and +10 mmHg, respectively. For blood lipid, participants taking lipid-lowering drugs were excluded.
| Trait | N | Sex and age adjusted model | |
|---|---|---|---|
| β(95%CI) | P | ||
| pSBP | |||
| GG | 5936 | ref | |
| AG | 18 | 14.47(5.87,23.07) | 0.001 |
| pDBP | |||
| GG | 5936 | ref | |
| AG | 18 | 6.55(1.37,11.74) | 0.013 |
| pPP | |||
| GG | 5936 | ref | |
| AG | 18 | 7.92(2.09,13.74) | 0.008 |
| pMAP | |||
| GG | 5936 | ref | |
| AG | 18 | 9.19(3.27,15.11) | 0.002 |
| Ba-PWV | |||
| GG | 5654 | ref | |
| AG | 18 | 285.94(143.44,428.44) | 8.51x10-5 |
| LDL cholesterol | |||
| GG | 5295 | ref | |
| AG | 15 | 0.41(0.01,0.82) | 0.047 |
| HDL cholesterol | |||
| GG | 5296 | ref | |
| AG | 15 | -0.11(-0.32,0.11) | 0.328 |
| Total cholesterol | |||
| GG | 5294 | ref | |
| AG | 15 | 0.37(-0.36,1.09) | 0.318 |
| Triglycerides | |||
| GG | 5308 | ref | |
| AG | 15 | 0.24(-0.35,0.84) | 0.425 |
Generation and characteristics of Ccdc93 mutant mice and eQTL analysis
Given the challenges of replicating the effect of a low-frequency variant, we hypothesized that gene loss of function in a mouse deficient in Ccdc93 expression would demonstrate higher BP and be a new model for functional studies of this effect. Supporting this direction, rs33975708 showed an expression QTL association with lower CCDC93 expression in tibial artery in the GTEx database (P = 0.000014, Fig 1D), with the caveat that most GTEx tissues were from European ancestry individuals. In GTEx, eQTLs for each tissue have been precalculated through linear regression employing programs like fastQTL or MatrixQTL. GTEx calculated permutation-based empirical p-values and multiple test corrections to assess the significance of variant-gene pairs. After multiple test correction (Storey’s q value), we identified a significant association between rs33975708 and the cis-eQTL gene CCDC93, in tibial artery (P = 1.4x10-5, normalized effect size = -0.2, Fig 1D).
Using GTEx v8 data, we identified 275 expression-associated SNPs (eSNPs) for CCDC93 in the aorta and 270 eSNPs in the tibial artery. Of these, 92 eSNPs were in linkage disequilibrium (LD) with rs33975708 (R2>0.2, +/- 500Kb) as per 1000G CEU population. All 92 proxy eSNPs showed the same effect as rs33975708 and CCDC93, with the minor allele linked to reduced CCDC93 expression, indirectly supporting our study’s signals. Neither the specific SNPs nor any proxy SNP, when available, identified any eQTL associations for the additional variants, rs2165468-pSBP and rs13149993-pDBP/pMAP, using the updated GTEx v8 portal and other eQTL databases (such as the Human Genetic Variation Database). Only rs33975708-cSBP exhibited a significant association with CCDC93 in tibial artery in GTEx v8.
Using CRISPR/Cas9 genome editing in mouse embryos, we created a novel transgenic mouse to model heterozygous deletion of Ccdc93. Ccdc93 heterozygous (Ccdc93+/-) mice have a 21-nucleotide deletion in a region spanning intron 6/exon 7 (2 nucleotides in intron 6 and 19 nucleotides in exon 7, S5 Fig), and significantly reduced aortic Ccdc93 mRNA transcript and protein expression (Fig 2C–2E). Ccdc93+/- mice were viable and morphologically normal. In contrast, Ccdc93 null (Ccdc93-/-) mice were embryonic lethal and died prior to embryonic day E10.5 (S6 Fig), supporting that Ccdc93 is indispensable for embryonic development. Timed mating was performed in Ccdc93+/- X Ccdc93+/- mice, and pregnant females were euthanized at E10.5 to assess embryonic abnormality in Ccdc93-/-. 5 fetoplacental units (FPUs) of total 56 were Ccdc93-/- (9%, S6A Fig). Histological analysis at E10.5 of a single uterine horn revealed 2 resorption sites and 6 healthy FPUs. FPUs of Ccdc93+/- heterozygote contained developing embryos that appeared viable and had no evidence of necrosis, inflammation, or hemorrhage (S6B Fig). Resorption sites of Ccdc93-/- homozygous FPUs contained hemorrhage and necrotic debris accompanied by neutrophilic and lymphocytic inflammation within and around chorioallantoic membranes on the fetal side. There were no embryos or embryonic tissues present (S6C Fig), suggesting that Ccdc93-/- embryos either did not develop or died very early in gestation.
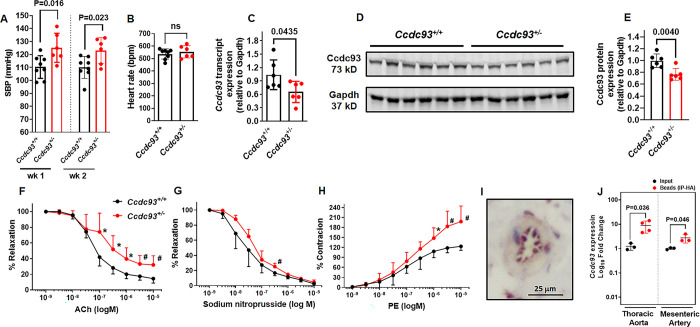
Fig 2. Ccdc93+/- influences systolic blood pressure and arterial relaxation in mice.
(A) Ccdc93+/- mice displayed higher systolic blood pressure as compared to Ccdc93+/+ littermate controls. (B) Heart rate (bpm, beats per minute) was not different in Ccdc93+/- mice as compared to Ccdc93+/+ littermate controls. Both male and female mice (N = 3–4 per genotype) were used in our study and since we identified no sex-specific signals (S11 Fig), further results are reported for the combined-sex analyses of all mice studied. (C) Ccdc93 transcript expression and protein expression (D-E) were significantly lower in the descending thoracic aortae in Ccdc93+/- mice as compared to the Ccdc93+/+ littermate control mice (N = 6–8 in each group). (F-G) In the descending thoracic aortae, Ccdc93+/- (red) displayed significant impairment in acetylcholine (ACh)-induced endothelial relaxation (endothelial-dependent) and a mild impairment in sodium nitroprusside-induced relaxation (endothelium-independent) as compared to the littermate controls (black). Dose response curve of Ach and sodium nitroprusside were performed in aortic rings precontracted with phenylephrine (10 μM). (H) Phenylephrine (PE)-induced contraction was significantly higher in Ccdc93+/- as compared to the littermate controls (Ccdc93+/+ N = 5, Ccdc93+/- N = 6). The dose-response curve of PE is expressed as % maximal response to high KCl buffer (60 mM). Data were analyzed by two-sided unpaired t-test. P<0.05 was defined to be statistically significant (* P<0.05, # P<0.03). (I) Immunohistochemistry showed Ccdc93 protein expression was enriched in the endothelium of mesenteric artery of Ccdc93+/+ wild-type mouse (129/Sv). (J) Endothelial cell-specific translating ribosome affinity purification (EC-TRAP) confirmed that Ccdc93 transcript expression was indeed enriched in the endothelial cells (immunoprecipitated-hemagglutinin IP-HA fraction) of thoracic aortae and mesenteric arteries of C57BL/6 wild-type mice (N = 3–4 in each group). All data are expressed as mean±s.d.
Total body weight, and lean mass measured by echo magnetic resonance imaging (Echo MRI) were higher in Ccdc93+/- heterozygous mice as compared to the littermate controls (S12A Fig). Heart weight, kidney weight and spleen weight normalized over body weight were not significantly different in Ccdc93+/- as compared to littermate controls (S7A–S7C Fig). Ccdc93+/- heterozygous mice displayed normal aortic histology on H&E and picrosirius red staining (S8 Fig) and normal peripheral blood counts (S3 Table).
Mice deficient in Ccdc93 demonstrated higher SBP and impaired arterial relaxation
Baseline SBP was significantly higher in Ccdc93+/- mice measured at two time-points one week apart (week 1: 125.15±10.24 mmHg, week 2: 123.01±9.19 mmHg, Fig 2A) as compared to littermate Ccdc93+/+ mice (week 1: 110.31±8.17 mmHg, P = 0.016, week 2: 110.03±8.87 mmHg, P = 0.023). Heart rate was similar in Ccdc93+/- and Ccdc93+/+ littermate control mice (Fig 2B). Both male and female mice (N = 3–4 per genotype) were studied, and secondary analyses were performed stratified by sex that identified no sex-specific signals (S11 Fig). Further results are reported for the combined-sex analyses of all mice studied. Vascular reactivity of aortae was evaluated ex vivo using wire myography. Descending thoracic aortic rings of Ccdc93+/- showed impaired acetylcholine (ACh)-induced relaxation starting at ACh dose 10−7 M (P = 0.043) and relaxation remained impaired for all increasing ACh doses tested up to a maximal ACh response, corresponding to 86% vs 68% (Ccdc93+/+ vs Ccdc93+/-, P = 0.029 at Ach 10−5 M, Fig 2F). Endothelium-independent relaxation induced by sodium nitroprusside was impaired in Ccdc93+/- aortic rings as compared to Ccdc93+/+ control aortic rings at 3.16×10−7 M sodium nitroprusside (P = 0.013, Fig 2G), although to an overall milder degree as compared to ACh-induced arterial relaxation.
Phenylephrine (PE)-induced contractile responses of Ccdc93+/- aorta to a high concentration of PE starting at 10−6 M was significantly higher in Ccdc93+/- when compared with Ccdc93+/+ littermate controls (P = 0.048) and remained highly contractile at all increasing PE doses, as compared to Ccdc93+/+ controls (maximal PE response at 10−5 M was 123% vs 198% normalized to maximal response to high KCl buffer (Ccdc93+/+ vs Ccdc93+/-, P = 0.015 Fig 2H).
Arterial transcript expression studies to localize Ccdc93 expression and identify mechanisms of Ccdc93 deficiency
Based upon the findings of endothelial dysfunction in Ccdc93+/- mice, we hypothesized that vascular Ccdc93 expression is enriched in the endothelium. We thus evaluated endothelial cell expression of Ccdc93 protein in a wild-type mouse (129/Sv) by immunohistochemistry which demonstrated endothelial expression in the mesenteric artery (Fig 2I). Quantification of Ccdc93 protein expression showed 1.3 fold higher expression in the endothelium as compared to the non-endothelial cells in mouse aorta (S15 Fig); however due to high dispersion and some degree of background staining, we employed cell-type targeted mRNA quantification by performing endothelial cell-specific translating ribosome affinity purification (EC-TRAP) in thoracic aortae and mesenteric arteries of Rpl22fl/fl, Tek-Cre+/0 C57BL/6 wild-type mice, which showed that Ccdc93 transcript expression was in fact enriched in endothelial cells (Fig 2J). As positive controls for the experiment, the expected enrichment of EC-specific transcript expression (Tek, Cdh5 and Vwf) and depletion of SMC transcripts (Tagln, Acta2 and Cnn1) were observed (S9A and S9B Fig) as well as immunostaining of EC-specific marker platelet and endothelial cell adhesion molecule-1 (PECAM-1) that showed expected staining of ECs in mouse aorta (S16A and S16B Fig). Immunostaining of PECAM-1 and Ccdc93 showed strong brown staining in the positive control (antibody present), which was not observed in the negative control of mouse aortic tissues (antibody absent) (S16A–S16D Fig). We queried the mouse and human single-cell RNA-seq database at Tabula Muris and GTEx respectively, which demonstrated Ccdc93 expression in endothelial cells (S14A–S14C Fig).
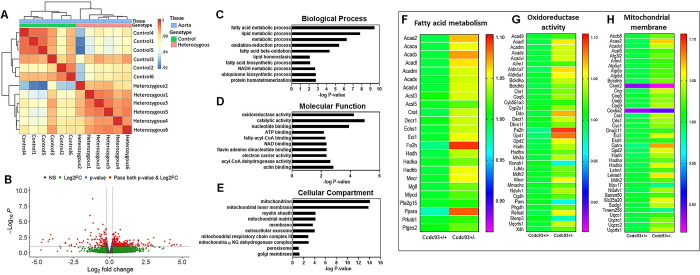
To gain insights into the underlying molecular mechanisms of impaired arterial relaxation and high SBP in Ccdc93+/- using unbiased transcriptome analysis, thoracic aortic RNA transcript expression was evaluated by bulk RNA-Seq. A heat map (Fig 3A) showed correlation between the samples of top differentially expressed genes according to genotype. PCA plot (S10A Fig) demonstrated no clear clustering of transcriptomes by mouse genotype, which may be partly explained by allelic compensation and high degree of global correlation in transcription expression profile between heterozygous and controls. Differential expression analysis comparing Ccdc93+/- to Ccdc93+/+ littermate control aortae identified 368 significantly upregulated and 131 downregulated transcripts (FDR<0.1, log2FC>0.5, Fig 3B), with reduced aortic Ccdc93 transcript expression observed as expected in Ccdc93+/- mice, consistent with qPCR data (S10B Fig). Gene ontology and pathway analyses showed a strong enrichment of pathways of mitochondrial function including fatty acid metabolism, oxidoreductase pathway, mitochondrial fatty acid beta oxidation, and reactive oxygen species production (Figs 3C–3H and S10C). Transcripts involved in the mitochondrial reactive oxygen species (ROS) cascade including malate dehydrogenase 2 (Mdh2), xanthine dehydrogenase (Xdh), cytochrome c1 (Cyc1), aldehyde dehydrogenase 5 family member A1 (Aldh5a1), and aldehyde dehydrogenase family 1, subfamily A7 (Aldh1a7) were significantly higher in Ccdc93+/- as compared to controls. Cyc1 and Aldh5a1, Aldh1a7 are elevated under oxidative stress and function as ROS scavenger and detoxification of highly reactive aldehydes, respectively [36,37]. Acetyl-Coenzyme A acyltransferase 2 (Acaa2), Acyl coenzyme-A dehydrogenase 9 (Acad9) and cytochrome P450, family 2, subfamily u, polypeptide 1 (Cyp2u1) transcript expression were higher in Ccdc93+/- and their functions are reported in fatty acid beta-oxidation and lipid metabolism, with increased activity associated with alteration of mitochondrial architecture, bioenergetics and oxidative stress [38–41].
Fig 3. RNA-seq analysis of Ccdc93+/- mouse aortae.
Descending thoracic aortae of male and female Ccdc93+/+ littermate controls and Ccdc93+/- heterozygous mice (N = 3 in each group) were analyzed by bulk RNA-Seq analysis. (A) A heat map represents the correlation between the samples of top differentially expressed genes by the genotype. Correlations were calculated using the Pearson correlation coefficient from the log-transformed raw reads counting per gene. (B) Volcano plot showing significantly upregulated genes (368) and downregulated genes (131) in thoracic aortae of Ccdc93+/- compared to Ccdc93+/+ mice. (C-E) Gene ontology (GO) analysis of the differentially expressed genes of descending thoracic aortae of Ccdc93+/- mice as compared to the littermate controls. GO is classified based on the (C) biological process, (D) molecular function and (E) cellular compartment and plotted against Benjamini adjusted P-value on the x-axis. (F-H) Significantly altered gene expression of fatty acid metabolic process, oxidoreductase activity and mitochondrial membrane affiliated gene regulators, are represented by heat maps. The normalized rlog transformed counts were used to generate heat maps (FDR<0.1).
Based upon these results, we hypothesized that concordant energetic abnormalities would be observed in Ccdc93+/- mice. Fat mass and weights of adipose tissues from three different anatomic locations (gonadal and inguinal white adipose tissue, and interscapular brown adipose tissue) were not statistically different between Ccdc93+/- mice compared to Ccdc93+/+ littermate controls (S12 Fig) Circulating plasma free fatty acids (FFA) levels but not total cholesterol was significantly higher in Ccdc93+/- mice as compared to littermate controls (Fig 4A and 4B). Concordant with the mouse data, human CCDC93 rs33975708 A carriers displayed higher FFA levels (1.67-fold increase, P = 0.35, S4 Table) that was not statistically significant in this underpowered analysis, owing to the rarity of this genetic variant.
Fig 4. Plasma free fatty acid, aortic Parkin, Nix and Ppar-α protein expression were increased in Ccdc93+/- mice.
(A) Plasma free fatty acid was significantly increased in Ccdc93+/- mice as compared to Ccdc93+/+ littermate controls. (B) Total plasma cholesterol was not different between the genotypes. (C-F) Aortic Parkin and Nix protein expression, markers of mitochondrial dysfunction, and (G-H) Aortic Ppar-α protein expression were significantly higher in Ccdc93+/- mice as compared to Ccdc93+/+ littermate controls. (G-I) Aortic Lrp1 protein expression was higher in Ccdc93+/- mice but did not reach statistical significance. P<0.05 was defined to be statistically significant. N = 5–6 in each group. Data are expressed as mean±s.d.
Liver weight relative to body weight was examined since the liver contributes to FFA pools in tissues and plasma [42], and this was mildly increased in Ccdc93+/-mice (S12B Fig). Analysis of liver histology by H&E showed no histologically evident lesions to explain the increased liver weight in Ccdc93+/- mice (S13 Fig). Plasma glucose was similar in Ccdc93+/- heterozygous mice as compared to the littermate Ccdc93+/+ control mice (S12C Fig), consistent with our observation where CCDC93 rs33975708 A carriers showed similar blood glucose levels as compared to non-carriers (S4 Table). Aortic Parkin and Nix protein, both of which are markers of mitochondrial dysfunction were significantly elevated in Ccdc93+/- mice (Fig 4C–4F). Aortic peroxisome proliferator-activated receptor-alpha (Ppar-α) was significantly higher in Ccdc93+/- mice as compared to littermate controls (1.28-fold higher, P = 0.018, Fig 4G and 4H), and low-density lipoprotein receptor-related protein (Lrp1) protein expression was higher but did not reach statistical significance (1.17-fold higher, P = 0.083, Fig 4G and 4I).
Discussion
In an association study of exome-wide variants and BP traits in 5,954 Han Chinese individuals, we identified 4 trait-locus associations with 4 BP traits, including traditionally measured BP traits and cSBP, with cSBP suggested to be a more accurate predictor of cardiovascular diseases. We replicated previously reported associations with BP traits, and identified 2 novel associations, including the association of cSBP with rs33975708, a nonsynonymous variant in CCDC93 (c.535C>T, p.Arg179Cys). This variant demonstrated pleiotropic associations with peripheral BP traits, Ba-PWV and LDL cholesterol, consistent with a recently described association of CCDC93 genetic variation with cellular LDL receptor trafficking [43]. Replication studies of specialized measurements, in this case cSBP, and rare genetic variation are challenged by sample size requirements of the replication resources. Although we were able to leverage a few resources to define robust human genetic evidence supporting the association of rs33975708 and cSBP, we undertook studies of a genetic mouse model of CCDC93 loss, as the variant in humans was predicted to lead to genetic deficiency of this gene. In functional validation experiments, mice with heterozygous loss of Ccdc93 indeed demonstrated higher SBP than littermate control mice. In contrast, Ccdc93-/- in its homozygous form was embryonic lethal, supporting its role in the embryonic development. Ccdc93 expression was enriched in the vascular endothelium, and Ccdc93+/- mice demonstrated endothelial dysfunction, evidenced by impaired acetylcholine mediated relaxation, as well as enhanced arterial contractility. Unbiased transcriptomic analysis demonstrated mitochondrial dysfunction, an effect that was validated in the elevated expression of Parkin and Nix, markers of mitochondrial dysfunction.
cSBP is associated with future cardiovascular events, and associations independent of pSBP have been defined [19,23,24,44]. In comparisons of the relationship of cSBP and pSBP measures with cardiovascular events, cSBP was superior to pSBP for the prediction of future cardiovascular events [45]. In addition to the CAFÉ study which reported differential effects on central pressure for different BP-lowering drugs despite similar effects on brachial pressure [17], a meta-analysis showed antihypertensive agents reduced pSBP more than cSBP (weighted mean difference [WMD] 2.52 mmHg, 95% CI 1.35 to 3.69) and β-Blockers posed a significantly greater reduction in pSBP as compared to cSBP (WMD 5.19 mmHg, 95% CI 3.21 to 7.18) [26]. cSBP measurement requires specialized techniques and has thus not been widely studied; we have thus studied cSBP in addition to traditional peripheral BP traits.
The coiled-coil domain containing 93 (CCDC93) gene encodes for a protein that is part of the “CCC” protein complex comprised of COMMD, CCDC22, and CCDC93. The recycling to the cell surface of Notch is dependent on the CCC complex [46,47]. Absence of COMMD9 resulted in reduced Notch-dependent signaling, and deletion of Commd9 led to embryonic lethality in mice due to abnormal cardiovascular development [47]. Similar to Commd9 deletion [47], embryonic lethality of Ccdc93 homozygous mice in our study supports an essential role in development at an early stage [47]. Both CCDC93 and the CCC complex mediate endosomal sorting of the low-density lipoprotein receptor (LDLR) [43,48,49]. Transcriptome analysis of Ccdc93+/- mice in our study notably identified alterations of adipogenesis and fatty acid metabolism, possibly linking the role of Ccdc93 in endosomal trafficking of Ldlr and the low-density lipoprotein receptor-related protein (Lrp1). In a previous genetic study of individuals of European ancestry, CCDC93 (rs17512204; C>T, p.Pro228Leu) was associated with lower LDL-C levels, lower cardiovascular mortality, and lower risk of myocardial infarction [43]. Additional CCDC93 variants have been associated with venous thromboembolism, triglyceride levels, and total cholesterol levels [50–52]. In the current study, another coding variant in CCDC93 (rs33975708; c.535C>T, p.Arg179Cys), was associated with higher LDL-C and supports potential regulatory function of CCDC93 in CVDs.
The cSBP-associated variant, rs33975708, has an overall MAF in the 1000 genomes reference of 4.07% but varies between populations, with the lowest in East Asians (0.00%) and the highest in Europeans (8.55%). Similarly, in the Exome Aggregation Consortium (ExAC) database, the frequency of this variant was 0.2% in individuals of East Asian ancestry and 7.43% in European-ancestry individuals.
In Ccdc93+/- heterozygous mice, arterial Ccdc93 transcript expression was reduced approximately 50% as expected, while protein expression was reduced by 22%, indicating that Ccdc93 protein expression is somewhat stabilized in these mice. Importantly for the objective of the current study, SBP elevation was observed in Ccdc93+/- mice as compared to their Ccdc93+/+ littermate controls, confirming the direction of the discovered human genetic effect. Concordant endothelial-dependent and endothelial-independent relaxation impairment in Ccdc93+/- heterozygous mice, along with increased contractile responses to vasoconstrictive challenges, suggested both endothelial and vascular smooth muscle defects in arterial function that result from Ccdc93 deficiency. Transcriptome analysis defined mitochondrial dysfunction and expression of key reactive oxygen species (ROS) transcripts including Mdh2, Xdh, Cyc1, Aldh5a1, Aldh1a7 increased in Ccdc93+/- mice suggest mitochondrial-derived ROS production may mediate the observed hypertensive effects, as ROS has been implicated in the mechanisms of hypertension in other settings [53,54]. Mdh2 is the final enzyme in the mitochondrial tricarboxylic acid cycle and its activity is associated with increased oxidative stress in the brain of Alzheimer disease [55]. Xdh is a source of superoxide ion, hydrogen peroxide, and nitric oxide, which can function as second messengers in various downstream vascular pathways [56]. Xdh-derived ROS are proinflammatory in the vasculature as they increase vascular permeability and arteriolar tone [57] and thus promote hypertension, cardiovascular diseases, and atherosclerosis [58,59].
Peroxisome proliferator-activated receptors-α (Ppar-α) is a member of the superfamily of ligand-activated nuclear transcription factors and its expression was significantly higher in Ccdc93+/- mice, both at transcript- and protein-levels. Ppar-α has been implicated in the regulation of genes involved in mitochondrial fatty acid β-oxidation. The promoters of medium-chain acyl-CoA dehydrogenase and mitochondrial HMG-CoA synthase are Ppar-α responsive [60]. PPAR agonists are also involved in the production of reactive oxygen species [61]. Concordant with the transcriptome data of the current study of abnormal mitochondrial fatty acid metabolism, plasma FFA levels were significantly higher in Ccdc93+/- mice, and a higher liver weight may suggest contribution to free fatty acid pools in tissues and plasma [42]. A positive correlation of mouse FFA levels was observed in our human samples. Human correlates of this finding include that elevated plasma FFA levels have been positively associated with SBP in nondiabetic subjects in a cross-sectional study [62]. Experimental FFA elevation achieved by intralipid infusion in non-obese women resulted in impaired capillary recruitment and acetylcholine-induced vasodilation in the skin microvasculature, as well as increased SBP [63]. Accumulating evidence suggests that FFAs contribute to hypertension by altering several pathways including alpha-adrenergic stimulation, increase in oxidative stress, stimulation of vascular cell growth, and endothelial dysfunction [64].
Mitochondrial dysfunction was validated through the examination of protein marker expression in Ccdc93+/- mice as compared to controls. The protein Parkin is a E3-ubiquitous ligase that accumulates on the outer membrane of damaged mitochondria which ubiquitinates mitochondrial outer membrane proteins [65,66], and activates mitochondria for autophagy and degradation in lysosomes [67]. Nix is a mitochondrial protein and a member of the Bcl-2 family of apoptotic regulators and is implicated in the pathogenesis of cancer and heart disease by regulating mitophagy and cell death [68]. Nix also plays a critical role in Parkin-mediated mitochondrial autophagy by controlling the mitochondrial translocation of Parkin [69]. Evidence suggests that alterations in mitochondrial function and ROS production have been associated with endothelial dysfunction, development of hypertension, and cardiac hypertrophy [70]. The current study’s findings suggest that fatty acid metabolism and mitochondrial dysfunction play key roles in regulating BP and vascular function in Ccdc93+/- mice.
The limitations of this study include a modest sample size, particularly compared to larger GWAS that have been carried out for peripheral BP traits. By using a custom exome chip array including Asian-specific nonsynonymous SNPs from sequenced samples in East Asian individuals, we identified a novel exonic variant association with cSBP that ideally would be replicated in large, independent samples. Due to the limited availability of ancestry-matched cohorts with cSBP measurement and the rarity of the discovered variant, replication experiments were under-powered, although using GRS, the complex genetic architecture of BP regulation in our cohort was aligned with that of previously reported BP GWAS. In order to further validate the effect of the variant identified, we undertook the generation and characterization of a novel transgenic mouse model deficiency in Ccdc93, which identified a similar phenotype and prioritized vascular mechanisms of BP regulation by Ccdc93 for further study.
Methods
Ethics statement
All animal procedures were approved by University of Michigan Institutional Animal Care and Use Committee (IACUC), and mice were housed and cared for in accordance with the Guide for the Care and Use of Laboratory Animals.
Discovery samples
In this study, subjects were recruited into the Peking University-University of Michigan Study of Atherosclerosis (PUUMA) in China, which is a large-scale project designed to study cardiovascular traits in China [22]. In total, 5,959 study participants of Chinese ancestry were genotyped in our BP study. Study protocols were approved by the Institutional Review Board (IRB) of Peking University First Hospital (IRB2014[816]) and Peking University Health Science center (IRB00001052-11086), and the study was approved by the University of Michigan IRB (HUM00074756). Written informed consent was collected from all subjects. Fasting venous blood samples were gathered for DNA genotyping and biochemical analyses. Medical and drug histories were accessed by standard questionnaire for each subject. Seated peripheral BP was measured for each participant with the standard method of calibration and appropriately sized cuffs after a 5-minute rest using an Omron HEM-7117 electronic sphygmomanometer. Triplicate measurements for each participant were taken with at least 1 minute between successive readings [22]. Noninvasive cSBP measurements were performed in a seated position using radial artery tonometry with an Omron HEM-9000AI device, as we have described and validated previously [22]. Brachial-ankle pulse wave velocity (Ba-PWV) was measured in the supine position after resting for at least 5 min (Omron Colin BP-203RPEIII; Omron Healthcare) by trained staffs. Total cholesterol (TC), triglycerides (TG), high-density lipoprotein cholesterol (HDL-C), low-density lipoprotein (LDL-C) and fasting blood glucose (FBG) were measured using Roche (Basel, Switzerland) C8000 Automatic Analyzer.
Replication and validation study samples
Replication analyses of the discovered associations were carried out using multiple replication resources. For pSBP, we accessed customized Illumina ExomeChip genotyped SNPs from patients in a study of myocardial infarction [71] at Peking University First Hospital (N = 565) and Peking University Third Hospital (N = 127) in China. For cSBP, we evaluated associations in all available data from our discovery cohort resource. cSBP measurement was repeated 2.3 years after the first visit (N = 2,897 among 3,448 were included in the discovery stage), labeled “visit 2” cSBP. The baseline cSBP and visit 2 cSBP measurements were then averaged to generate a long-term averaged (LTA) [33] cSBP trait (N = 4,938). The additional LTA and Visit 2 data were analyzed for association with cSBP. In an independent cohort from Ji County in Tianjin, China, available subjects harboring the cSBP candidate variant were selected for cSBP measurement (N = 9), and their cSBP values were compared with age- and sex-matched individuals without the variant (N = 67 invited to participate in the study). In total, there were nine subjects with heterozygous CCDC93 rs33975708-A allele. We used Student’s t-test and linear regression adjusting for age and sex to assess the differences of cSBP values between these two groups, and association of rs33975708-A with cSBP.
Genotyping
Genotyping was conducted using a custom Illumina ExomeChip array with additional variants included in the design of the genotyping array based upon sequencing data from East Asian individuals, as has been described and used in other EWAS studies in China [71]. This exome array was designed including custom content representing 58,317 variants detected through genome sequencing of Asian individuals, in addition to the standard Infinium Human Exome Bead Chip (Illumina, CA) [72], which accessed a total of 302,218 variants. Genotype calling was performed using GenTrain version 2.0 in GenomeStudio V2011.1 (Illumina) independently for both cohorts, followed by cohort-specific quality control (QC).
Data and sample quality controls
Sample and variant QC was performed as previously described [71], for evidence of bias or low-quality genotypes. Variants with low cluster score, Hardy-Weinberg equilibrium deviations P < 0.0001 and call rate < 99% were excluded. Variants with a minor allele count larger than 3 were included. For the sample QC, we excluded duplicates based on whole genome genotyping data IBD (Identical by descent) analysis, gender miss-matched samples, and samples with missing phenotypes. All samples had missing call rate < 1%. None of them failed QC in inbreeding coefficient check. Finally, 5,954 samples (2,185 males, 3,768 females) and 86,148 SNPs were retained after the QC in this BP EWAS. The correlation r2 of the allele frequencies between PUUMA Chinese samples and 1000 Genomes East Asian samples was 0.98.
Phenotypes and transformation
Our study phenotypes focused on BP measurements: pSBP, pDBP, pMAP and cSBP. Three repetitive measurements were recorded for pSBP (ex. SBP1, SBP2, SBP3) and pDBP (ex. DBP1, DBP2, DBP3), and the average number was used in the further analysis. Approximately 33% of individuals in the PUUMA cohort were taking anti-hypertensive medications, and their pSBP and pDBP measurements were adjusted +15 mm Hg and +10 mm Hg, respectively, before further analysis. pMAP was from (1/3 × pSBP) + (2/3 × pDBP). Distributions of pSBP, pDBP, pMAP, and cSBP were inspected, and outliers were detected as >3 standard deviations (s.d.) from the mean for each trait. For pSBP, pDBP, pMAP, and cSBP, the phenotypic values were substituted by the residual values using technique of linear regression adjustments: Trait ~ age + age2+ body mass index (BMI) sex.
Generation of Ccdc93 heterozygous mice
The loss of CCDC93 was modeled in a new transgenic mouse after CRIPSR/Cas9 targeting leading to an out-of-frame deletion in the Ccdc93 gene. Ccdc93 heterozygous mice (Ccdc93+/-) carrying a 21-nucleotide deletion were generated by injecting Cas9 mRNA, a guide RNA, and a donor DNA (Biomatrik, Ontario, Canada) into the embryo obtained by mating (C57BL/6 X SJL) F1 or C57BL/6 female mice with (C57BL/6 X SJL) F1 male mice. Pronuclear microinjection was performed as previously described [73]. Ccdc93+/- mice were mated with C57BL/6 mice and 129/Sv mice (Taconic Biosciences, NY, USA) for at least 10 generation to confirm germline transmission. Genotyping of Ccdc93+/- mice was performed by PCR amplifying the region using forward (5’GGAAGGGTGGGAGCGAGGAG3’) and reverse primer (5’ GTTGGCTTGCTTGCTTCTTTCTTTC 3’) and confirmed by sanger sequencing (Genewiz, NJ), S5 Fig.
BP measurement in mice
Male and female Ccdc93+/- (129/Sv) mice and their littermate controls at 12–16 weeks of age underwent SBP measurement by non-invasive tail-cuff plethysmography (BP-2000, Visitech Systems, NC) as previously described [74]. BP was measured after keeping trained mice quietly on a temperature-controlled restrainer for 10 minutes. Then, ten preliminary cycles were performed to allow the mice to acclimate and warm up to increase the blood-flow to the tail for optimal BP signals followed by 10 more cycles for the actual measurement. BP measurements were performed between 10 am and 12 noon. Mice received regular chow and water ad libitum.
Endothelial cell-specific translating ribosome affinity purification (EC-TRAP)
EC-TRAP was performed as we have previously described [75]. Briefly, Rpl22fl/fl, Tek+/0 C57BL/6 wild-type mice were perfused with cycloheximide (100 μg/ml, Sigma-Aldrich) to stop protein translation and stabilize ribosomal complexes, and tissues were snap frozen until further use. Frozen tissues were homogenized using a Dounce homogenizer in lysis buffer [75] (50 mM Tris pH 7.4, 100 mM KCl, 12 mM MgCl2, 1% Igepal CA-630, Protease inhibitor, 200 U/ml RNAse OUT, 1 mg/ml heparin, 1 mM dithiothreitol and 100 μg/mL cycloheximide). Cellular debris was removed by centrifugation and clear lysates were incubated with a purified mouse monoclonal antibody against the HA epitope tag (HA.11 clone 16B12, BioLegend, CA; 3μg/400μl lysate) for 1 hour at 4°C. Protein G magnetic beads (New England BioLabs, MA; 100μl/400μl lysate) equilibrated in polysome buffer were then added for an additional 30-minute incubation. The magnetic beads were subsequently washed 3 times with high salt buffer (50 mM Tris pH 7.4, 300 mM KCl, 12 mM MgCl2, 1% Igepal CA-630, 1 mM dithiothreitol and 100 μg/mL cycloheximide). RA1 buffer (NucleoSpin RNA, Takara Bio USA, CA) supplemented with 2-mercapto-ethanol (1% vol/vol) was added to the beads and vigorously vortexed to dissociate the mRNA from the HA-tagged polysomes. Total RNA from tissue lysate and TRAP polysomal complexes were isolated using NuceloSpin RNA kit (Takara Bio USA, CA), including an on-column DNase digestion step and RNA was quantified by Quant-it RiboGreen RNA assay (Thermo Fisher Scientific, MA). Ccdc93 transcript expression was queried in the mouse single-cell RNA-seq database at Tabula Muris with Tek used to identify aortic endothelial cells (https://tabula-muris.ds.czbiohub.org/) and human single-cell RNA-seq GTEx database at Protein Atlas (https://www.proteinatlas.org/).
RNA-Seq library preparation, sequencing, and qRT-PCR
To assess aortic transcriptional changes in Ccdc93+/- transgenic mice, RNA-Seq was performed on descending thoracic aortae from male and female Ccdc93+/- heterozygous mice (N = 3 per sex) and their Ccdc93+/+ littermate controls (N = 3 per sex). Total RNA was isolated using Trizol and chloroform extraction, followed by RNA purification as described above. RNA samples meeting RIN score >7 was included in the RNA-Seq. All libraries were prepared with 10 ng of RNA using SMART-Seq V4 kit for low RNA input (Takara Bio USA, CA). After quality control, all samples were pair-end sequenced by NovaSeq 6000 in MedGenome (Foster City, CA), with a read length of 100 bp and 40 million read depth per sample. Aortic Ccdc93 RNA expression in Ccdc93+/- heterozygous and Ccdc93+/+ littermate controls mice was measured as described previously [76]. Briefly, total aortic RNA was extracted and reverse transcription of 200 ng of total RNA was performed using SuperScript-III first-strand synthesis system (ThermoFisher, MA, USA) in a total volume of 20 μL. The exon spanning primers of Ccdc93 and Gapdh were designed (primer 3 software, S5 Table) and synthesized (Invitrogen, MA). cDNA was amplified using PCR master mix with SYBR-Green (Applied Biosystems, MA) and data were calculated by 2- ΔΔCT method [77] and presented as fold change of transcripts of Ccdc93 in mouse aortae and normalized over housekeeping Gapdh gene, as compared to control samples.
RNA-Seq analysis of differential gene expression
All data were aligned to the reference genome GRCh37 (hg19) by STAR aligner (2.7.0) with the annotation from GENCODE (GRCm39) [78,79]. Raw counts were extracted and then normalized with DESeq2 [80]. After quality assessment, analyses were performed in DESeq2 to determine differential transcript expression between Ccdc93+/- and Ccdc93+/+ littermate control groups. Gene ontology enrichment analysis and Gene set enrichment analysis were performed using DAVID Bioinformatic Resources, and by Hallmark pathways set files from the molecular signatures database, respectively [81,82]. The significant threshold was defined as false discovery rate (FDR) < 0.1.
Histological examination of fetal placental units, adult liver, and arterial section
Uterine horns of Ccdc93+/- X Ccdc93+/- mice subjected to timed mating were stored at E10.5 in 4% neutral buffered formalin until histologic analysis. The uterus was not opened but was cut into sections with 2 fetoplacental units (FPU) or resorption sites per section. Each section of two FPU or resorption sites were processed on an automated histology processor (Tissue Tek VIP, Sakura, Torrance, CA). Sections were embedded in paraffin with sections oriented laterally to enable visualization of both mesometrial and antimesometrial sides of the tissue. Liver tissues of adult Ccdc93+/- and littermate controls were paraffin embedded. Aorta and mesenteric artery of wild-type 129/Sv mice were paraffin embedded for immunohistochemical analysis by Ccdc93 antibody (1:200 dilution, 20861-1-AP, Proteintech) and endothelial specific markers platelet and endothelial cell adhesion molecule-1 (PECAM-1, 1:50 dilution, DIA-310, Dianova, Switzerland). Detection of Ccdc93 and PECAM-1 protein expression is based on horseradish peroxidase (HRP) catalysis of a 3,3′-Diaminobenzidine (DAB, brown) chromogenic reaction, with a hematoxylin (blue) nuclear counterstain. Sections of adult liver, artery and FPUs were cut from each block at 4 μm thickness. Pixel quantification of Ccdc93 protein expression was performed using ImageJ (NIH) and pixel intensity was normalized by arterial area encompassing endothelial cells and non-endothelial cells. Slides were evaluated by a board-certified veterinary pathologist using a BX45 Olympus light microscope. Slides were digitized to pyramidal Tiff files on a digital slide scanner (Leica Aperio AT2, Leica Biosystems) and representative images were taken from digitized slide files using freely available manufacturer-provided software (Leica Aperio ImageScope). Composite images were assembled in Adobe Photoshop CC (v 19.0, Adobe Systems Inc). Photo processing was confined to global adjustment of image size, white balance, brightness, or contrast that did not materially alter the interpretation of the image.
Vascular reactivity analysis using wire myography
Vascular reactivity assessed by wire myography was performed as previously described [83]. Briefly, descending thoracic aorta of Ccdc93+/- and Ccdc93+/+ mice were carefully dissected to remove perivascular fat and cut into 2 mm rings. Rings were transferred into physiological saline solution (PSS, 37°C gassed with 95% air and 5% CO2, PSS solution: 130 mM NaCl, 4.7 mM KCl, 1.17 mM MgSO4.7H2O, 14.9 mM NaHCO3, 1.18 mM KH2PO4, 0.026 mM EDTA, 1.6 mM CaCl2, and 5 mM glucose, pH 7.4) and mounted on pins in multi-wire myograph connected to a force transducer (DMT 620M, MI) to measure wall tension developed by the rings. Prior to assessing vascular function, aortic rings were pre-treated with high KCl buffer (60 mM) to induce contraction and to determine maximal contractility, and this was used for normalizing force generated by dose response of phenylephrine. Vascular function was evaluated by contraction induced by phenylephrine (10 μM) and vasodilation induced by acetylcholine (Ach, 10 μM) in aorta pre-contracted with phenylephrine (10 μM). Dose responses of acetylcholine and sodium-nitroprusside (1 nM to 10 μM) of aorta pre-contracted by phenylephrine were then recorded and analyzed. Rings were washed three times with physiological saline solution and equilibrated for 30 minutes at 37°C between each successive vasoactive treatment.
Plasma free fatty acid, glucose measurement, and body composition
Blood was collected from Ccdc93+/- and Ccdc93+/+ mice from the abdominal vena cava into citrate containing tubes (0.4% w/v) and centrifuged at 2,500 rpm for 15 minutes at 4°C to obtain plasma which was stored at -80°C until further use. Plasma free fatty acid level (FFA) in mouse and human samples was measured using commercially available sensitive enzyme-based assay (Abcam 65341) in which fatty acids are converted to their CoA derivatives in the presence of added acyl-CoA synthetase. Acyl-CoA products were then subsequently oxidized leading to the formation of color which was measured colorimetrically at 570 nm. Total plasma cholesterol in mice was measured by colorimetric detection at 450 nm by enzyme-based assay (Abcam 285242).
Fasting blood glucose level in human samples was measured and analyzed at the central laboratory of Tianjin Medical University General Hospital as previously described [34]. Mouse plasma glucose was measured using commercially available enzyme-based assay (K039-H1, Arbor Assays, MI). Glucose oxidase reacts with glucose to produce hydrogen peroxide, which, in the presence of HRP, reacts with the substrate to generate a colored product which was measured at 560 nm. Body composition scans in Ccdc93+/- and littermate controls were performed at the University of Michigan Mouse Metabolic and Phenotypic Center (MMPC). Body fat and lean mass were measured using a nuclear magnetic resonance (NMR)-based analyzer (EchoMRI, 4in1-500, Houston, TX). The measurements were performed in under 2 minutes while conscious mice were placed individually in the measuring tube.
Western blot
Aortic tissue protein (20 μg) from Ccdc93+/- and Ccdc93+/+ mice were separated by sodium dodecyl sulfate-polyacrylamide gel (12% SDS-PAGE) and transferred on to a nitrocellulose membrane. The blotted membranes were incubated with rabbit anti-Ccdc93 antibody (1:1000, 20861-1-AP, Proteintech), mouse anti-parkin (1:2000, ab77924, Abcam), rabbit anti-Nix/BNIP3L (1:1000, #12396S, CST), rabbit anti-Ppar-α (1:1000, ab126285, Abcam) and rabbit anti-Lrp1 (1:1000, # 64099, CST). The housekeeping gene product Gapdh (1:10,000, # 97166, CST) served as a loading control. Protein bands were visualized using a fluorescent-conjugated secondary antibody (anti-rabbit IRDye 680RD, anti-mouse IRDye 800CW, LI-COR Biosciences, Lincoln, NE) and imaged using LI-COR Odyssey CLx imaging system. Densitometric analysis of protein bands were quantified using NIH ImageJ software.
Statistical analyses
A mixed linear model considering kinship was performed using the EMMAX program [84], which is a statistical test for large scale human or model organism association mapping accounting for the sample structure. As described above, we adjusted for age, age2, BMI, and sex for our phenotypes first and run mixed linear model based on the residuals and the exome-wide SNPs. 5,954 samples and 86,148 SNPs were tested in the discovery stage and two novel identified SNPs with P <5.8 × 10−7 (0.05/86,148) were re-examined in replication stage. Manhattan plots were generated from exome-wide SNPs, with marked of known BP GWAS ± 500 Kb region as positive controls and to filter the newly discovered loci. Quantile-quantile plots were depicted for SNPs in different minor allele frequency groups. λGC was evaluated for evidence of population stratification.
A published BP GWAS finding that included more than one million people was used to construct BP weighted genetic risk score (GRS), where SNPs were weighted according to BP coefficients from the GWAS discovery results(5). Genome-wide summary statistics of SBP and DBP were downloaded from the GWAS Catalog. Variants that achieved a P-value less than 5×10−8 from GWAS of the same trait were selected and LD-pruned (R2>0.2 in a 500Kb window). Their beta estimates were then aligned with our study’s summary statistic based on the identical effect alleles. The GTX package (https://www.rdocumentation.org/packages/gtx/versions/0.0.8) in R was applied to test the association between PRS of known BP and our BP analysis using GWAS summary statistics adjusted for age, age2, sex, BMI, and principal components (PCs).
Associations of novel BP-related SNPs with risk factors of coronary artery disease (including blood lipids, BP traits, pulse wave velocity, and body mass index) were tested using multivariate linear regression, with models adjusting for sex and age, using R program. For the analyses of blood lipids, those who were taking lipid-lowering drugs were excluded.
For experiments involving mice, all data were reported as mean±s.d. Student’s t tests (unpaired, two-tailed) were used to compare the significance between the two groups. P<0.05 was defined to be statistically significant.
Supporting information
pSBP, peripheral systolic blood pressure; pDBP, peripheral diastolic blood pressure; pMAP, peripheral mean arterial pressure; cSBP, central systolic blood pressure.
(XLSX)
(XLSX)
WBC, white blood cell; NE, neutrophils; LY, lymphocyte; MO, monocyte; EO, eosinophil; BA, basophil; RBC, red blood cell; Hb, hemoglobin; HCT, hematocrit; MCV, mean corpuscular volume; MCH, mean corpuscular hemoglobin; MCHC, mean corpuscular hemoglobin concentration; RDW, red cell distribution width; PLT, platelet; MPV, mean platelet volume; K, thousand; M, million; g, gram; μL, microliter, dL, deciliter; fL, femtoliter.
(XLSX)
In human, CCDC93 rs33975708 A carriers displayed higher FFA levels as compared to non-carriers. Fasting blood glucose level was similar between CCDC93 rs33975708 A carriers and non-carriers.
(XLSX)
(XLSX)
ExomeChip SNPs meeting quality control were analyzed in the EWAS. Results are shown in a (A) Manhattan plot and (B) Quantile-quantile (QQ) plot; the λGC value was 1.0. (C) Regional association plots with gene annotation for the chromosomes 10p15.2 region associated with pSBP is shown with the index SNP rs2165468 and additional SNPs within 500 kbp in each direction. Top panel: Regional pSBP association plot for the PITRM1-KLF6 locus. LD (linkage disequilibrium) was calculated from our samples. Non-synonymous variants were annotated using ANNOVAR. The genetic recombination rate is based on Hapmap release 22. Middle Panel: Standardized varLD scores illustrate LD variations between populations (CEU vs. JPT+CHB, CEU vs. YRI, YRI vs. JPT+CHB) using genome positions from the Hapmap 3 reference. The red line represents the comparison between CEU (European ancestry) and JPT+CHB (East Asian ancestry), the purple line indicates CEU versus YRI (African ancestry), and the green line represents YRI versus JPT+CHB. Bottom Panel: Gene/transcript annotations are sourced from the reference downloaded from the UCSC database (EST, mRNA, uniGene, Encode, and RefGene).
(TIF)
ExomeChip SNPs meeting quality control were analyzed in the EWAS. Results are shown in a (A) Manhattan plot and (B) QQ plot; the λGC value was 1.0. (C) Regional association plots with gene annotation for the chromosomes 4q21.21 region associated with pDBP is shown with the index SNP rs13149993 and additional SNPs within 500 kbp in each direction. Top panel: Regional pDBP association plot for the PRDM8-FGF5 locus. LD was calculated from our samples. Non-synonymous variants were annotated using ANNOVAR. The genetic recombination rate is based on Hapmap release 22. Middle Panel: Standardized varLD scores illustrate LD variations between populations (CEU vs. JPT+CHB, CEU vs. YRI, YRI vs. JPT+CHB) using genome positions from the Hapmap 3 reference. The red line represents the comparison between CEU (European ancestry) and JPT+CHB (East Asian ancestry), the purple line indicates CEU versus YRI (African ancestry), and the green line represents YRI versus JPT+CHB. Bottom Panel: Gene/transcript annotations are sourced from the reference downloaded from the UCSC database (EST, mRNA, uniGene, Encode, and RefGene).
(TIF)
ExomeChip SNPs meeting quality control were analyzed in the EWAS. Results are shown in a (A) Manhattan plot and (B) QQ plot; the λGC value was 1.0. (C) Regional association plots with gene annotation for the chromosomes 4q21.21 region associated with pMAP is shown with the index SNP rs13149993 and additional SNPs within 500 kbp in each direction. Top panel: Regional pMAP association plot for the PRDM8-FGF5 locus. LD was calculated from our samples. Non-synonymous variants were annotated using ANNOVAR. The genetic recombination rate is based on Hapmap release 22. Middle Panel: Standardized varLD scores illustrate LD variations between populations (CEU vs. JPT+CHB, CEU vs. YRI, YRI vs. JPT+CHB) using genome positions from the Hapmap 3 reference. The red line represents the comparison between CEU (European ancestry) and JPT+CHB (East Asian ancestry), the purple line indicates CEU versus YRI (African ancestry), and the green line represents YRI versus JPT+CHB. Bottom Panel: Gene/transcript annotations are sourced from the reference downloaded from the UCSC database (EST, mRNA, uniGene, Encode, and RefGene).
(TIF)
(A) Peripheral systolic blood pressure (pSBP); (B) Peripheral diastolic blood pressure (pDBP); (C) Peripheral pulse pressure (pPP); (D) Peripheral mean arterial pressure (pMAP); (E) Brachial-ankle pulse wave velocity (Ba-PWV); and (F) Low-density lipoprotein-cholesterol (LDL-C). All these traits were significantly higher in CCDC93 risk allele A carriers as compared to non-carriers.
(TIF)
(A) Ccdc93 heterozygous mice were generated using the CRISPR/Cas9 system. (B) Ccdc93+/- carried a 21-nucleotide deletion. The molecular weight of DNA bands after PCR amplification of the Ccdc93 gene was not distinguishable by agarose gel electrophoresis. (C) Sanger sequencing of genomic DNA of Ccdc93 showed difference between littermate control and heterozygous of Ccdc93 with expected peak-on-peak DNA chromatogram observed in Ccdc93+/- at the transgenic site (yellow highlight).
(TIF)
(A) Genotype frequencies at birth and at E10.5 of Ccdc93+/- X Ccdc93+/- mating. At birth, homozygous Ccdc93-/- mice were not viable and time mating at E10.5 of embryos produced from Ccdc93+/- X Ccdc93+/- mating showed Ccdc93 homozygosity were embryonic lethal and died before E10.5 of gestation. (B) Histology of fetoplacental units showed uterine horns from a Ccdc93+/- (heterozygous) pregnant female mouse at day E10.5 of gestation. This pregnancy was the product of Ccdc93+/- X Ccdc93+/- timed mating. Ccdc93+/- heterozygous embryos appeared viable (no evidence of necrosis, inflammation, or hemorrhage). (C) 2 resorption sites of Ccdc93-/- homozygous contained maternal hemorrhage and necrotic debris accompanied by neutrophilic and lymphocytic inflammation within and around chorioallantoic membranes on the antimesenterial (fetal) side. There were no embryos or embryo tissues present.
(TIF)
Tissue weight (A-heart weight, B-kidney weight, C-spleen weight) normalized over body weight were not different between the groups (N = 6–8 in each group). All data are shown as mean±s.d.
(TIF)
(A-C) Picrosirius red staining (PSR) for total collagen content of the descending thoracic aorta of littermate Ccdc93+/+ and (D-F) Ccdc93+/- mice demonstrate no significant differences. Hematoxylin and eosin (H&E) staining of the descending thoracic aorta of (G-I) littermate Ccdc93+/+ and (J-L) Ccdc93+/- mice did not demonstrate abnormal vascular morphology. Each section was examined under 4x objective microscope (Nikon’s Eclipse E600), photographed with a digital camera (DS-Ri1, Nikons Instrument), and evaluated by the ImageJ analysis system (NIH). PSR staining was used to facilitate automatic detection in pixel intensity by the image processing macro in ImageJ. N = 6 in each group. Scale bars 200 μm. All data are shown as mean±s.d.
(TIF)
(A-B) Endothelial cell (EC)-specific enrichment and vascular smooth muscle-specific (VSMC) depletion of transcript markers by EC-TRAP in C57BL/6 wild-type mice in (A) thoracic aortae and (B) mesenteric arteries in vivo. Total RNA from was isolated from input and anti-HA beads (IP-HA) from descending thoracic aortae and mesenteric arteries of Rpl22fl/fl, Tie2-Cre+/0 (Tie2-RiboTag mouse) and showed expected enrichment of EC-specific transcripts (Tek, Cdh5 and Vwf) and depletion of vascular smooth muscle-specific markers (Cnn1, Acta2 and Tagln) in the IP-HA fraction as compared to input. Ccdc93 transcript expression in the Tek subpopulations (EC-specific) was enriched (Fig 2J), confirming predominantly EC expression. N = 3–4 in each group. All data are shown as mean±s.d.
(TIF)
(A) Principal component analysis (PCA) plot showed no clear clustering by the genotype. The first and second principal components (PC1 and PC2) accounted for 61% and 19% variability in the RNA-Seq dataset. (B) Aortic Ccdc93 transcript expression was significantly reduced in Ccdc93+/- as compared to Ccdc93+/+ in bulk RNA-seq analysis, validating qPCR findings (Fig 2C). (C) Gene set enrichment analysis showed hallmark pathway associations with altered adipogenesis and fatty acid metabolism that were significantly upregulated in the thoracic aortae of Ccdc93+/- mice as compared to Ccdc93+/+ littermate controls, false discovery rate (FDR<0.1).
(TIF)
A similar increase in SBP was observed in male and female Ccdc93+/- heterozygous mice as compared to littermate controls. All data are shown as mean±s.d.
(TIF)
(A) Body weight, and lean mass measured by EchoMRI were significantly higher in Ccdc93+/- mice as compared to Ccdc93+/+ control mice. (B) A mild increase in the liver weight of Ccdc93+/- heterozygous mice was observed, however adipose tissue weight (gonadal and inguinal white adipose tissue, and interscapular brown adipose tissue) were not different between the genotype groups. (C) Plasma glucose level was equivalent in Ccdc93+/- heterozygous mice as compared to littermate control mice. Equal numbers of male and female mice (N = 4 per sex per genotype) were analyzed. White adipose tissue (WAT), brown adipose tissue (BAT). All data are shown as mean±s.d.
(TIF)
Hematoxylin and eosin stain (H&E) stained images are shown. No histological lesions were observed in the liver of Ccdc93+/- mice as compared to the littermate controls. (A) Male Ccdc93+/+ wild-type control liver samples (inset-male wild-type biological replicates, N = 3). (B) Male Ccdc93+/- heterozygous liver samples (inset-male Ccdc93+/- heterozygous biological replicates, N = 3). (C) Female Ccdc93+/+ wild-type control liver samples (inset-female wild-type biological replicates, N = 3). (D) Female Ccdc93+/- heterozygous liver samples (inset-female Ccdc93+/- heterozygous biological replicates, N = 3). Scale bars 200 μm, insets 2 mm.
(TIF)
(A-B) Ccdc93 expression was queried in the mouse scRNA-seq database Tabula Muris (https://tabula-muris.ds.czbiohub.org/), and in (C) human scRNA-seq GTEx database protein atlas (https://www.proteinatlas.org/), which showed low tissue specificity of Ccdc93, and validated Ccdc93 expression in mouse aortic endothelial cells (blue dots in left figure panel A), shown in the tSNE plots, and enrichment of CCDC93 expression in human liver vascular endothelial cells (colored dot in panel C).
(TIF)
(A-C) Ccdc93 protein expression was observed in both endothelial and non-endothelial cells in wild-type mouse aortae (129/Sv). Three biological replicates of mouse thoracic aorta showed Ccdc93 expression (brown stained). (D) Pixel quantification showed 1.33-fold higher Ccdc93 protein expression in the endothelium (normalized by area) as compared to non-endothelial cells (P = NS). Scale bar = 200μm.
(TIF)
(A) Mouse aorta immunostaining of platelet and endothelial cell adhesion molecule-1 (PECAM-1), (B) Negative control (no antibody) immunostaining of mouse aorta serial section. (C) Positive control of mouse aorta immunostaining of Ccdc93. (D) Negative control of mouse aorta where Ccdc93 antibody was absent. Detection of PECAM-1 (black arrows) and Ccdc93 protein expression (black arrows represents EC expression and blue arrows represents medial expression) is based on horseradish peroxidase (HRP) catalysis of a 3,3′-Diaminobenzidine (DAB, brown) chromogenic reaction, with a hematoxylin (blue) nuclear counterstain, scale bar = 200μm.
(TIF)
Acknowledgments
We thank Dr. Daniel Goldstein (University of Michigan) for kindly providing the parkin antibody. We thank Wanda Filipiak & Galina Gavrilina for preparation of transgenic mice and In Vivo Animal Core (University of Michigan) for assistance in mouse histological examination.
Data Availability
All data associated with this manuscript is available in the supplementary files, summary statistics or under GCP IDs GCST90310248, GCST90310249, GCST90310250, GCST90310251, GCST90310252 on GWAS Catalog https://www.ebi.ac.uk/gwas/home.
Funding Statement
The PUUMA cohort genotyping and analyses were supported by the UM-PUHSC Joint Institute for Translational and Clinical Research (grant No: BMU20110177 and BMU20160530, to YZ and SKG). YZ was supported by grants from National High Level Hospital Clinical Research Funding (High Quality Clinical Research Project of Peking University First Hospital), No. 2022CR71; National Key Research and Development Program of China (No 2021YFC2500500, 2021YFC2500503), and Key Laboratory of Molecular Cardiovascular Sciences (Peking University), Ministry of Education; National Health Commission Key Laboratory of Cardiovascular Molecular Biology and Regulatory Peptides. SKG was supported by National Institutes of Health (R01HL139672, R01HL122684, R01HL086694, R35HL161016), Department of Defense, and the University of Michigan A. Alfred Taubman Institute. NK was supported by the National Center for Advancing Translational Sciences (UL1TR002240) and the University of Michigan Medical School Office of Research. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Collaborators GBDRF. Global, regional, and national comparative risk assessment of 84 behavioural, environmental and occupational, and metabolic risks or clusters of risks, 1990–2016: a systematic analysis for the Global Burden of Disease Study 2016. Lancet. 2017;390(10100):1345–422. doi: 10.1016/S0140-6736(17)32366-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Forouzanfar MH, Liu P, Roth GA, Ng M, Biryukov S, Marczak L, et al. Global Burden of Hypertension and Systolic Blood Pressure of at Least 110 to 115 mm Hg, 1990–2015. JAMA. 2017;317(2):165–82. doi: 10.1001/jama.2016.19043 [DOI] [PubMed] [Google Scholar]
- 3.Newton-Cheh C, Johnson T, Gateva V, Tobin MD, Bochud M, Coin L, et al. Genome-wide association study identifies eight loci associated with blood pressure. Nat Genet. 2009;41(6):666–76. doi: 10.1038/ng.361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Warren HR, Evangelou E, Cabrera CP, Gao H, Ren M, Mifsud B, et al. Genome-wide association analysis identifies novel blood pressure loci and offers biological insights into cardiovascular risk. Nat Genet. 2017;49(3):403–15. doi: 10.1038/ng.3768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Evangelou E, Warren HR, Mosen-Ansorena D, Mifsud B, Pazoki R, Gao H, et al. Genetic analysis of over 1 million people identifies 535 new loci associated with blood pressure traits. Nat Genet. 2018;50(10):1412–25. doi: 10.1038/s41588-018-0205-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ehret GB, Ferreira T, Chasman DI, Jackson AU, Schmidt EM, Johnson T, et al. The genetics of blood pressure regulation and its target organs from association studies in 342,415 individuals. Nat Genet. 2016;48(10):1171–84. doi: 10.1038/ng.3667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cabrera CP, Ng FL, Nicholls HL, Gupta A, Barnes MR, Munroe PB, et al. Over 1000 genetic loci influencing blood pressure with multiple systems and tissues implicated. Hum Mol Genet. 2019;28(R2):R151–R61. doi: 10.1093/hmg/ddz197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Takeuchi F, Akiyama M, Matoba N, Katsuya T, Nakatochi M, Tabara Y, et al. Interethnic analyses of blood pressure loci in populations of East Asian and European descent. Nat Commun. 2018;9(1):5052. doi: 10.1038/s41467-018-07345-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sung YJ, Winkler TW, de Las Fuentes L, Bentley AR, Brown MR, Kraja AT, et al. A Large-Scale Multi-ancestry Genome-wide Study Accounting for Smoking Behavior Identifies Multiple Significant Loci for Blood Pressure. Am J Hum Genet. 2018;102(3):375–400. doi: 10.1016/j.ajhg.2018.01.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang S, Zhou Z, Fan F, Qi L, Jia J, Sun P, et al. Joint Effect of Non-invasive Central Systolic Blood Pressure and Peripheral Systolic Blood Pressure on Incident Hypertension in a Chinese Community-based Population. Sci Rep. 2018;8(1):3229. doi: 10.1038/s41598-018-21023-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheng HM, Chuang SY, Wang TD, Kario K, Buranakitjaroen P, Chia YC, et al. Central blood pressure for the management of hypertension: Is it a practical clinical tool in current practice? J Clin Hypertens (Greenwich). 2020;22(3):391–406. doi: 10.1111/jch.13758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Safar ME, Jankowski P. Central blood pressure and hypertension: role in cardiovascular risk assessment. Clin Sci (Lond). 2009;116(4):273–82. doi: 10.1042/CS20080072 [DOI] [PubMed] [Google Scholar]
- 13.Segers P, Mahieu D, Kips J, Rietzschel E, De Buyzere M, De Bacquer D, et al. Amplification of the pressure pulse in the upper limb in healthy, middle-aged men and women. Hypertension. 2009;54(2):414–20. doi: 10.1161/HYPERTENSIONAHA.109.133009 [DOI] [PubMed] [Google Scholar]
- 14.Avolio AP, Van Bortel LM, Boutouyrie P, Cockcroft JR, McEniery CM, Protogerou AD, et al. Role of pulse pressure amplification in arterial hypertension: experts’ opinion and review of the data. Hypertension. 2009;54(2):375–83. doi: 10.1161/HYPERTENSIONAHA.109.134379 [DOI] [PubMed] [Google Scholar]
- 15.Benetos A, Thomas F, Joly L, Blacher J, Pannier B, Labat C, et al. Pulse pressure amplification a mechanical biomarker of cardiovascular risk. J Am Coll Cardiol. 2010;55(10):1032–7. doi: 10.1016/j.jacc.2009.09.061 [DOI] [PubMed] [Google Scholar]
- 16.Tarumi T, Sugawara J, Tanaka H. Association between ankle blood pressure and central arterial wave reflection. J Hum Hypertens. 2011;25(9):539–44. doi: 10.1038/jhh.2010.100 [DOI] [PubMed] [Google Scholar]
- 17.Williams B, Lacy PS, Thom SM, Cruickshank K, Stanton A, Collier D, et al. Differential impact of blood pressure-lowering drugs on central aortic pressure and clinical outcomes: principal results of the Conduit Artery Function Evaluation (CAFE) study. Circulation. 2006;113(9):1213–25. doi: 10.1161/CIRCULATIONAHA.105.595496 [DOI] [PubMed] [Google Scholar]
- 18.Roman MJ, Devereux RB, Kizer JR, Lee ET, Galloway JM, Ali T, et al. Central pressure more strongly relates to vascular disease and outcome than does brachial pressure: the Strong Heart Study. Hypertension. 2007;50(1):197–203. doi: 10.1161/HYPERTENSIONAHA.107.089078 [DOI] [PubMed] [Google Scholar]
- 19.Pini R, Cavallini MC, Palmieri V, Marchionni N, Di Bari M, Devereux RB, et al. Central but not brachial blood pressure predicts cardiovascular events in an unselected geriatric population: the ICARe Dicomano Study. J Am Coll Cardiol. 2008;51(25):2432–9. doi: 10.1016/j.jacc.2008.03.031 [DOI] [PubMed] [Google Scholar]
- 20.Kollias A, Lagou S, Zeniodi ME, Boubouchairopoulou N, Stergiou GS. Association of central versus brachial blood pressure with target-organ damage: systematic review and meta-analysis. Hypertension. 2016;67(1):183–90. doi: 10.1161/HYPERTENSIONAHA.115.06066 [DOI] [PubMed] [Google Scholar]
- 21.Sun P, Yang Y, Cheng G, Fan F, Qi L, Gao L, et al. Noninvasive central systolic blood pressure, not peripheral systolic blood pressure, independently predicts the progression of carotid intima-media thickness in a Chinese community-based population. Hypertens Res. 2019;42(3):392–9. doi: 10.1038/s41440-018-0175-5 [DOI] [PubMed] [Google Scholar]
- 22.Fan F, Qi L, Jia J, Xu X, Liu Y, Yang Y, et al. Noninvasive Central Systolic Blood Pressure Is More Strongly Related to Kidney Function Decline Than Peripheral Systolic Blood Pressure in a Chinese Community-Based Population. Hypertension. 2016;67(6):1166–72. doi: 10.1161/HYPERTENSIONAHA.115.07019 [DOI] [PubMed] [Google Scholar]
- 23.Cheng HM, Chuang SY, Sung SH, Yu WC, Pearson A, Lakatta EG, et al. Derivation and validation of diagnostic thresholds for central blood pressure measurements based on long-term cardiovascular risks. J Am Coll Cardiol. 2013;62(19):1780–7. doi: 10.1016/j.jacc.2013.06.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang KL, Cheng HM, Chuang SY, Spurgeon HA, Ting CT, Lakatta EG, et al. Central or peripheral systolic or pulse pressure: which best relates to target organs and future mortality? J Hypertens. 2009;27(3):461–7. doi: 10.1097/hjh.0b013e3283220ea4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rahman M, Hsu JY, Desai N, Hsu CY, Anderson AH, Appel LJ, et al. Central Blood Pressure and Cardiovascular Outcomes in Chronic Kidney Disease. Clin J Am Soc Nephrol. 2018;13(4):585–95. doi: 10.2215/CJN.08620817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McGaughey TJ, Fletcher EA, Shah SA. Impact of Antihypertensive Agents on Central Systolic Blood Pressure and Augmentation Index: A Meta-Analysis. Am J Hypertens. 2016;29(4):448–57. doi: 10.1093/ajh/hpv134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Snieder H, Hayward CS, Perks U, Kelly RP, Kelly PJ, Spector TD. Heritability of central systolic pressure augmentation: a twin study. Hypertension. 2000;35(2):574–9. doi: 10.1161/01.hyp.35.2.574 [DOI] [PubMed] [Google Scholar]
- 28.Kolifarhood G, Daneshpour M, Hadaegh F, Sabour S, Mozafar Saadati H, Akbar Haghdoust A, et al. Heritability of blood pressure traits in diverse populations: a systematic review and meta-analysis. J Hum Hypertens. 2019;33(11):775–85. doi: 10.1038/s41371-019-0253-4 [DOI] [PubMed] [Google Scholar]
- 29.Patel RS, Morris AA, Ahmed Y, Kavtaradze N, Sher S, Su S, et al. A genetic risk variant for myocardial infarction on chromosome 6p24 is associated with impaired central hemodynamic indexes. Am J Hypertens. 2012;25(7):797–803. doi: 10.1038/ajh.2012.41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cecelja M, Keehn L, Ye L, Spector TD, Hughes AD, Chowienczyk P. Genetic aetiology of blood pressure relates to aortic stiffness with bi-directional causality: evidence from heritability, blood pressure polymorphisms, and Mendelian randomization. Eur Heart J. 2020;41(35):3314–22. doi: 10.1093/eurheartj/ehaa238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Giri A, Hellwege JN, Keaton JM, Park J, Qiu C, Warren HR, et al. Trans-ethnic association study of blood pressure determinants in over 750,000 individuals. Nat Genet. 2019;51(1):51–62. doi: 10.1038/s41588-018-0303-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kelly TN, Takeuchi F, Tabara Y, Edwards TL, Kim YJ, Chen P, et al. Genome-wide association study meta-analysis reveals transethnic replication of mean arterial and pulse pressure loci. Hypertension. 2013;62(5):853–9. doi: 10.1161/HYPERTENSIONAHA.113.01148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ganesh SK, Chasman DI, Larson MG, Guo X, Verwoert G, Bis JC, et al. Effects of long-term averaging of quantitative blood pressure traits on the detection of genetic associations. Am J Hum Genet. 2014;95(1):49–65. doi: 10.1016/j.ajhg.2014.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhan C, Shi M, Yang Y, Pang H, Fei S, Bai L, et al. Prevalence and Risk Factors of Carotid Plaque Among Middle-aged and Elderly Adults in Rural Tianjin, China. Sci Rep. 2016;6:23870. doi: 10.1038/srep23870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee CH, Cook S, Lee JS, Han B. Comparison of Two Meta-Analysis Methods: Inverse-Variance-Weighted Average and Weighted Sum of Z-Scores. Genomics Inform. 2016;14(4):173–80. doi: 10.5808/GI.2016.14.4.173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Atlante A, Calissano P, Bobba A, Azzariti A, Marra E, Passarella S. Cytochrome c is released from mitochondria in a reactive oxygen species (ROS)-dependent fashion and can operate as a ROS scavenger and as a respiratory substrate in cerebellar neurons undergoing excitotoxic death. J Biol Chem. 2000;275(47):37159–66. doi: 10.1074/jbc.M002361200 [DOI] [PubMed] [Google Scholar]
- 37.Singh S, Brocker C, Koppaka V, Chen Y, Jackson BC, Matsumoto A, et al. Aldehyde dehydrogenases in cellular responses to oxidative/electrophilic stress. Free Radic Biol Med. 2013;56:89–101. doi: 10.1016/j.freeradbiomed.2012.11.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shen LF, Chen YJ, Liu KM, Haddad ANS, Song IW, Roan HY, et al. Role of S-Palmitoylation by ZDHHC13 in Mitochondrial function and Metabolism in Liver. Sci Rep. 2017;7(1):2182. doi: 10.1038/s41598-017-02159-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kwon DN, Park WJ, Choi YJ, Gurunathan S, Kim JH. Oxidative stress and ROS metabolism via down-regulation of sirtuin 3 expression in Cmah-null mice affect hearing loss. Aging (Albany NY). 2015;7(8):579–94. doi: 10.18632/aging.100800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tesson C, Nawara M, Salih MA, Rossignol R, Zaki MS, Al Balwi M, et al. Alteration of fatty-acid-metabolizing enzymes affects mitochondrial form and function in hereditary spastic paraplegia. Am J Hum Genet. 2012;91(6):1051–64. doi: 10.1016/j.ajhg.2012.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nouws J, Nijtmans L, Houten SM, van den Brand M, Huynen M, Venselaar H, et al. Acyl-CoA dehydrogenase 9 is required for the biogenesis of oxidative phosphorylation complex I. Cell Metab. 2010;12(3):283–94. doi: 10.1016/j.cmet.2010.08.002 [DOI] [PubMed] [Google Scholar]
- 42.Teusink B, Voshol PJ, Dahlmans VE, Rensen PC, Pijl H, Romijn JA, et al. Contribution of fatty acids released from lipolysis of plasma triglycerides to total plasma fatty acid flux and tissue-specific fatty acid uptake. Diabetes. 2003;52(3):614–20. doi: 10.2337/diabetes.52.3.614 [DOI] [PubMed] [Google Scholar]
- 43.Rimbert A, Dalila N, Wolters JC, Huijkman N, Smit M, Kloosterhuis N, et al. A common variant in CCDC93 protects against myocardial infarction and cardiovascular mortality by regulating endosomal trafficking of low-density lipoprotein receptor. Eur Heart J. 2020;41(9):1040–53. doi: 10.1093/eurheartj/ehz727 [DOI] [PubMed] [Google Scholar]
- 44.Dong Y, Jiang L, Wang X, Chen Z, Zhang L, Zhang Z, et al. Central rather than brachial pressures are stronger predictors of cardiovascular outcomes: a longitudinal prospective study in a Chinese population. J Clin Hypertens (Greenwich). 2020;22(4):623–30. doi: 10.1111/jch.13838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lamarche F, Agharazii M, Nadeau-Fredette AC, Madore F, Goupil R. Central and brachial blood pressures, statins, and low-density lipoprotein cholesterol: a mediation analysis. Hypertension. 2018;71(3):415–21. doi: 10.1161/HYPERTENSIONAHA.117.10476 [DOI] [PubMed] [Google Scholar]
- 46.Kopan R. Notch signaling. Cold Spring Harb Perspect Biol. 2012;4(10). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li H, Koo Y, Mao X, Sifuentes-Dominguez L, Morris LL, Jia D, et al. Endosomal sorting of Notch receptors through COMMD9-dependent pathways modulates Notch signaling. J Cell Biol. 2015;211(3):605–17. doi: 10.1083/jcb.201505108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fedoseienko A, Wijers M, Wolters JC, Dekker D, Smit M, Huijkman N, et al. The COMMD Family Regulates Plasma LDL Levels and Attenuates Atherosclerosis Through Stabilizing the CCC Complex in Endosomal LDLR Trafficking. Circ Res. 2018;122(12):1648–60. doi: 10.1161/CIRCRESAHA.117.312004 [DOI] [PubMed] [Google Scholar]
- 49.Bartuzi P, Billadeau DD, Favier R, Rong S, Dekker D, Fedoseienko A, et al. CCC- and WASH-mediated endosomal sorting of LDLR is required for normal clearance of circulating LDL. Nat Commun. 2016;7:10961. doi: 10.1038/ncomms10961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sinnott-Armstrong N, Tanigawa Y, Amar D, Mars N, Benner C, Aguirre M, et al. Genetics of 35 blood and urine biomarkers in the UK Biobank. Nat Genet. 2021;53(2):185–94. doi: 10.1038/s41588-020-00757-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Deguchi H, Shukla M, Hayat M, Torkamani A, Elias DJ, Griffin JH. Novel exomic rare variants associated with venous thrombosis. Br J Haematol. 2020;190(5):783–6. doi: 10.1111/bjh.16613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Klarin D, Damrauer SM, Cho K, Sun YV, Teslovich TM, Honerlaw J, et al. Genetics of blood lipids among ~300,000 multi-ethnic participants of the Million Veteran Program. Nat Genet. 2018;50(11):1514–23. doi: 10.1038/s41588-018-0222-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dikalov SI, Nazarewicz RR. Angiotensin II-induced production of mitochondrial reactive oxygen species: potential mechanisms and relevance for cardiovascular disease. Antioxid Redox Signal. 2013;19(10):1085–94. doi: 10.1089/ars.2012.4604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dikalov S. Cross talk between mitochondria and NADPH oxidases. Free Radic Biol Med. 2011.; 51(7):1289–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bubber P, Haroutunian V, Fisch G, Blass JP, Gibson GE. Mitochondrial abnormalities in Alzheimer brain: mechanistic implications. Ann Neurol. 2005;57(5):695–703. doi: 10.1002/ana.20474 [DOI] [PubMed] [Google Scholar]
- 56.Battelli MG, Polito L, Bortolotti M, Bolognesi A. Xanthine Oxidoreductase-Derived Reactive Species: Physiological and Pathological Effects. Oxid Med Cell Longev. 2016;2016:3527579. doi: 10.1155/2016/3527579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cai H, Harrison DG. Endothelial dysfunction in cardiovascular diseases: the role of oxidant stress. Circ Res. 2000;87(10):840–4. doi: 10.1161/01.res.87.10.840 [DOI] [PubMed] [Google Scholar]
- 58.Battelli MG, Polito L, Bolognesi A. Xanthine oxidoreductase in atherosclerosis pathogenesis: not only oxidative stress. Atherosclerosis. 2014;237(2):562–7. doi: 10.1016/j.atherosclerosis.2014.10.006 [DOI] [PubMed] [Google Scholar]
- 59.Ghosh SM, Kapil V, Fuentes-Calvo I, Bubb KJ, Pearl V, Milsom AB, et al. Enhanced vasodilator activity of nitrite in hypertension: critical role for erythrocytic xanthine oxidoreductase and translational potential. Hypertension. 2013;61(5):1091–102. doi: 10.1161/HYPERTENSIONAHA.111.00933 [DOI] [PubMed] [Google Scholar]
- 60.Grygiel-Gorniak B. Peroxisome proliferator-activated receptors and their ligands: nutritional and clinical implications—a review. Nutr J. 2014;13:17. doi: 10.1186/1475-2891-13-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Teissier E, Nohara A, Chinetti G, Paumelle R, Cariou B, Fruchart JC, et al. Peroxisome proliferator-activated receptor alpha induces NADPH oxidase activity in macrophages, leading to the generation of LDL with PPAR-alpha activation properties. Circ Res. 2004;95(12):1174–82. doi: 10.1161/01.RES.0000150594.95988.45 [DOI] [PubMed] [Google Scholar]
- 62.Carlsson M, Wessman Y, Almgren P, Groop L. High levels of nonesterified fatty acids are associated with increased familial risk of cardiovascular disease. Arterioscler Thromb Vasc Biol. 2000;20(6):1588–94. doi: 10.1161/01.atv.20.6.1588 [DOI] [PubMed] [Google Scholar]
- 63.de Jongh RT, Serne EH, Ijzerman RG, de Vries G, Stehouwer CD. Free fatty acid levels modulate microvascular function: relevance for obesity-associated insulin resistance, hypertension, and microangiopathy. Diabetes. 2004;53(11):2873–82. doi: 10.2337/diabetes.53.11.2873 [DOI] [PubMed] [Google Scholar]
- 64.Sarafidis PA, Bakris GL. Non-esterified fatty acids and blood pressure elevation: a mechanism for hypertension in subjects with obesity/insulin resistance? J Hum Hypertens. 2007;21(1):12–9. doi: 10.1038/sj.jhh.1002103 [DOI] [PubMed] [Google Scholar]
- 65.Ge P, Dawson VL, Dawson TM. PINK1 and Parkin mitochondrial quality control: a source of regional vulnerability in Parkinson’s disease. Mol Neurodegener. 2020;15(1):20. doi: 10.1186/s13024-020-00367-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tyrrell DJ, Blin MG, Song J, Wood SC, Goldstein DR. Aging Impairs Mitochondrial Function and Mitophagy and Elevates Interleukin 6 Within the Cerebral Vasculature. J Am Heart Assoc. 2020;9(23):e017820. doi: 10.1161/JAHA.120.017820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Narendra D, Tanaka A, Suen DF, Youle RJ. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J Cell Biol. 2008;183(5):795–803. doi: 10.1083/jcb.200809125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhang J, Ney PA. Role of BNIP3 and NIX in cell death, autophagy, and mitophagy. Cell Death Differ. 2009;16(7):939–46. doi: 10.1038/cdd.2009.16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ding WX, Ni HM, Li M, Liao Y, Chen X, Stolz DB, et al. Nix is critical to two distinct phases of mitophagy, reactive oxygen species-mediated autophagy induction and Parkin-ubiquitin-p62-mediated mitochondrial priming. J Biol Chem. 2010;285(36):27879–90. doi: 10.1074/jbc.M110.119537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lahera V, de Las Heras N, Lopez-Farre A, Manucha W, Ferder L. Role of Mitochondrial Dysfunction in Hypertension and Obesity. Curr Hypertens Rep. 2017;19(2):11. doi: 10.1007/s11906-017-0710-9 [DOI] [PubMed] [Google Scholar]
- 71.Tang CS, Zhang H, Cheung CY, Xu M, Ho JC, Zhou W, et al. Exome-wide association analysis reveals novel coding sequence variants associated with lipid traits in Chinese. Nat Commun. 2015;6:10206. doi: 10.1038/ncomms10206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhang Y, Long J, Lu W, Shu XO, Cai Q, Zheng Y, et al. Rare coding variants and breast cancer risk: evaluation of susceptibility Loci identified in genome-wide association studies. Cancer Epidemiol Biomarkers Prev. 2014;23(4):622–8. doi: 10.1158/1055-9965.EPI-13-1043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Katja Becker BJ. Generation of Transgenic Mice by Pronuclear Microinjection. Springer Advanced Protocols for Animal Transgenesis. 2011:99–115. [Google Scholar]
- 74.Kumar N, Liao TD, Romero CA, Maheshwari M, Peterson EL, Carretero OA. Thymosin beta4 Deficiency Exacerbates Renal and Cardiac Injury in Angiotensin-II-Induced Hypertension. Hypertension. 2018;71(6):1133–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cleuren ACA, van der Ent MA, Jiang H, Hunker KL, Yee A, Siemieniak DR, et al. The in vivo endothelial cell translatome is highly heterogeneous across vascular beds. Proc Natl Acad Sci U S A. 2019;116(47):23618–24. doi: 10.1073/pnas.1912409116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kumar N, Zuo Y, Yalavarthi S, Hunker KL, Knight JS, Kanthi Y, et al. SARS-CoV-2 Spike Protein S1-Mediated Endothelial Injury and Pro-Inflammatory State Is Amplified by Dihydrotestosterone and Prevented by Mineralocorticoid Antagonism. Viruses. 2021;13(11). doi: 10.3390/v13112209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–8. doi: 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- 78.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. doi: 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Frankish A, Diekhans M, Ferreira AM, Johnson R, Jungreis I, Loveland J, et al. GENCODE reference annotation for the human and mouse genomes. Nucleic Acids Res. 2019;47(D1):D766–D73. doi: 10.1093/nar/gky955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- 82.Liberzon A, Subramanian A, Pinchback R, Thorvaldsdottir H, Tamayo P, Mesirov JP. Molecular signatures database (MSigDB) 3.0. Bioinformatics. 2011;27(12):1739–40. doi: 10.1093/bioinformatics/btr260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Stevenson MD, Vendrov AE, Yang X, Chen Y, Navarro HA, Moss N, et al. Reactivity of renal and mesenteric resistance vessels to angiotensin II is mediated by NOXA1/NOX1 and superoxide signaling. Am J Physiol Renal Physiol. 2023. doi: 10.1152/ajprenal.00236.2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kang HM, Sul JH, Service SK, Zaitlen NA, Kong SY, Freimer NB, et al. Variance component model to account for sample structure in genome-wide association studies. Nat Genet. 2010;42(4):348–54. doi: 10.1038/ng.548 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
pSBP, peripheral systolic blood pressure; pDBP, peripheral diastolic blood pressure; pMAP, peripheral mean arterial pressure; cSBP, central systolic blood pressure.
(XLSX)
(XLSX)
WBC, white blood cell; NE, neutrophils; LY, lymphocyte; MO, monocyte; EO, eosinophil; BA, basophil; RBC, red blood cell; Hb, hemoglobin; HCT, hematocrit; MCV, mean corpuscular volume; MCH, mean corpuscular hemoglobin; MCHC, mean corpuscular hemoglobin concentration; RDW, red cell distribution width; PLT, platelet; MPV, mean platelet volume; K, thousand; M, million; g, gram; μL, microliter, dL, deciliter; fL, femtoliter.
(XLSX)
In human, CCDC93 rs33975708 A carriers displayed higher FFA levels as compared to non-carriers. Fasting blood glucose level was similar between CCDC93 rs33975708 A carriers and non-carriers.
(XLSX)
(XLSX)
ExomeChip SNPs meeting quality control were analyzed in the EWAS. Results are shown in a (A) Manhattan plot and (B) Quantile-quantile (QQ) plot; the λGC value was 1.0. (C) Regional association plots with gene annotation for the chromosomes 10p15.2 region associated with pSBP is shown with the index SNP rs2165468 and additional SNPs within 500 kbp in each direction. Top panel: Regional pSBP association plot for the PITRM1-KLF6 locus. LD (linkage disequilibrium) was calculated from our samples. Non-synonymous variants were annotated using ANNOVAR. The genetic recombination rate is based on Hapmap release 22. Middle Panel: Standardized varLD scores illustrate LD variations between populations (CEU vs. JPT+CHB, CEU vs. YRI, YRI vs. JPT+CHB) using genome positions from the Hapmap 3 reference. The red line represents the comparison between CEU (European ancestry) and JPT+CHB (East Asian ancestry), the purple line indicates CEU versus YRI (African ancestry), and the green line represents YRI versus JPT+CHB. Bottom Panel: Gene/transcript annotations are sourced from the reference downloaded from the UCSC database (EST, mRNA, uniGene, Encode, and RefGene).
(TIF)
ExomeChip SNPs meeting quality control were analyzed in the EWAS. Results are shown in a (A) Manhattan plot and (B) QQ plot; the λGC value was 1.0. (C) Regional association plots with gene annotation for the chromosomes 4q21.21 region associated with pDBP is shown with the index SNP rs13149993 and additional SNPs within 500 kbp in each direction. Top panel: Regional pDBP association plot for the PRDM8-FGF5 locus. LD was calculated from our samples. Non-synonymous variants were annotated using ANNOVAR. The genetic recombination rate is based on Hapmap release 22. Middle Panel: Standardized varLD scores illustrate LD variations between populations (CEU vs. JPT+CHB, CEU vs. YRI, YRI vs. JPT+CHB) using genome positions from the Hapmap 3 reference. The red line represents the comparison between CEU (European ancestry) and JPT+CHB (East Asian ancestry), the purple line indicates CEU versus YRI (African ancestry), and the green line represents YRI versus JPT+CHB. Bottom Panel: Gene/transcript annotations are sourced from the reference downloaded from the UCSC database (EST, mRNA, uniGene, Encode, and RefGene).
(TIF)
ExomeChip SNPs meeting quality control were analyzed in the EWAS. Results are shown in a (A) Manhattan plot and (B) QQ plot; the λGC value was 1.0. (C) Regional association plots with gene annotation for the chromosomes 4q21.21 region associated with pMAP is shown with the index SNP rs13149993 and additional SNPs within 500 kbp in each direction. Top panel: Regional pMAP association plot for the PRDM8-FGF5 locus. LD was calculated from our samples. Non-synonymous variants were annotated using ANNOVAR. The genetic recombination rate is based on Hapmap release 22. Middle Panel: Standardized varLD scores illustrate LD variations between populations (CEU vs. JPT+CHB, CEU vs. YRI, YRI vs. JPT+CHB) using genome positions from the Hapmap 3 reference. The red line represents the comparison between CEU (European ancestry) and JPT+CHB (East Asian ancestry), the purple line indicates CEU versus YRI (African ancestry), and the green line represents YRI versus JPT+CHB. Bottom Panel: Gene/transcript annotations are sourced from the reference downloaded from the UCSC database (EST, mRNA, uniGene, Encode, and RefGene).
(TIF)
(A) Peripheral systolic blood pressure (pSBP); (B) Peripheral diastolic blood pressure (pDBP); (C) Peripheral pulse pressure (pPP); (D) Peripheral mean arterial pressure (pMAP); (E) Brachial-ankle pulse wave velocity (Ba-PWV); and (F) Low-density lipoprotein-cholesterol (LDL-C). All these traits were significantly higher in CCDC93 risk allele A carriers as compared to non-carriers.
(TIF)
(A) Ccdc93 heterozygous mice were generated using the CRISPR/Cas9 system. (B) Ccdc93+/- carried a 21-nucleotide deletion. The molecular weight of DNA bands after PCR amplification of the Ccdc93 gene was not distinguishable by agarose gel electrophoresis. (C) Sanger sequencing of genomic DNA of Ccdc93 showed difference between littermate control and heterozygous of Ccdc93 with expected peak-on-peak DNA chromatogram observed in Ccdc93+/- at the transgenic site (yellow highlight).
(TIF)
(A) Genotype frequencies at birth and at E10.5 of Ccdc93+/- X Ccdc93+/- mating. At birth, homozygous Ccdc93-/- mice were not viable and time mating at E10.5 of embryos produced from Ccdc93+/- X Ccdc93+/- mating showed Ccdc93 homozygosity were embryonic lethal and died before E10.5 of gestation. (B) Histology of fetoplacental units showed uterine horns from a Ccdc93+/- (heterozygous) pregnant female mouse at day E10.5 of gestation. This pregnancy was the product of Ccdc93+/- X Ccdc93+/- timed mating. Ccdc93+/- heterozygous embryos appeared viable (no evidence of necrosis, inflammation, or hemorrhage). (C) 2 resorption sites of Ccdc93-/- homozygous contained maternal hemorrhage and necrotic debris accompanied by neutrophilic and lymphocytic inflammation within and around chorioallantoic membranes on the antimesenterial (fetal) side. There were no embryos or embryo tissues present.
(TIF)
Tissue weight (A-heart weight, B-kidney weight, C-spleen weight) normalized over body weight were not different between the groups (N = 6–8 in each group). All data are shown as mean±s.d.
(TIF)
(A-C) Picrosirius red staining (PSR) for total collagen content of the descending thoracic aorta of littermate Ccdc93+/+ and (D-F) Ccdc93+/- mice demonstrate no significant differences. Hematoxylin and eosin (H&E) staining of the descending thoracic aorta of (G-I) littermate Ccdc93+/+ and (J-L) Ccdc93+/- mice did not demonstrate abnormal vascular morphology. Each section was examined under 4x objective microscope (Nikon’s Eclipse E600), photographed with a digital camera (DS-Ri1, Nikons Instrument), and evaluated by the ImageJ analysis system (NIH). PSR staining was used to facilitate automatic detection in pixel intensity by the image processing macro in ImageJ. N = 6 in each group. Scale bars 200 μm. All data are shown as mean±s.d.
(TIF)
(A-B) Endothelial cell (EC)-specific enrichment and vascular smooth muscle-specific (VSMC) depletion of transcript markers by EC-TRAP in C57BL/6 wild-type mice in (A) thoracic aortae and (B) mesenteric arteries in vivo. Total RNA from was isolated from input and anti-HA beads (IP-HA) from descending thoracic aortae and mesenteric arteries of Rpl22fl/fl, Tie2-Cre+/0 (Tie2-RiboTag mouse) and showed expected enrichment of EC-specific transcripts (Tek, Cdh5 and Vwf) and depletion of vascular smooth muscle-specific markers (Cnn1, Acta2 and Tagln) in the IP-HA fraction as compared to input. Ccdc93 transcript expression in the Tek subpopulations (EC-specific) was enriched (Fig 2J), confirming predominantly EC expression. N = 3–4 in each group. All data are shown as mean±s.d.
(TIF)
(A) Principal component analysis (PCA) plot showed no clear clustering by the genotype. The first and second principal components (PC1 and PC2) accounted for 61% and 19% variability in the RNA-Seq dataset. (B) Aortic Ccdc93 transcript expression was significantly reduced in Ccdc93+/- as compared to Ccdc93+/+ in bulk RNA-seq analysis, validating qPCR findings (Fig 2C). (C) Gene set enrichment analysis showed hallmark pathway associations with altered adipogenesis and fatty acid metabolism that were significantly upregulated in the thoracic aortae of Ccdc93+/- mice as compared to Ccdc93+/+ littermate controls, false discovery rate (FDR<0.1).
(TIF)
A similar increase in SBP was observed in male and female Ccdc93+/- heterozygous mice as compared to littermate controls. All data are shown as mean±s.d.
(TIF)
(A) Body weight, and lean mass measured by EchoMRI were significantly higher in Ccdc93+/- mice as compared to Ccdc93+/+ control mice. (B) A mild increase in the liver weight of Ccdc93+/- heterozygous mice was observed, however adipose tissue weight (gonadal and inguinal white adipose tissue, and interscapular brown adipose tissue) were not different between the genotype groups. (C) Plasma glucose level was equivalent in Ccdc93+/- heterozygous mice as compared to littermate control mice. Equal numbers of male and female mice (N = 4 per sex per genotype) were analyzed. White adipose tissue (WAT), brown adipose tissue (BAT). All data are shown as mean±s.d.
(TIF)
Hematoxylin and eosin stain (H&E) stained images are shown. No histological lesions were observed in the liver of Ccdc93+/- mice as compared to the littermate controls. (A) Male Ccdc93+/+ wild-type control liver samples (inset-male wild-type biological replicates, N = 3). (B) Male Ccdc93+/- heterozygous liver samples (inset-male Ccdc93+/- heterozygous biological replicates, N = 3). (C) Female Ccdc93+/+ wild-type control liver samples (inset-female wild-type biological replicates, N = 3). (D) Female Ccdc93+/- heterozygous liver samples (inset-female Ccdc93+/- heterozygous biological replicates, N = 3). Scale bars 200 μm, insets 2 mm.
(TIF)
(A-B) Ccdc93 expression was queried in the mouse scRNA-seq database Tabula Muris (https://tabula-muris.ds.czbiohub.org/), and in (C) human scRNA-seq GTEx database protein atlas (https://www.proteinatlas.org/), which showed low tissue specificity of Ccdc93, and validated Ccdc93 expression in mouse aortic endothelial cells (blue dots in left figure panel A), shown in the tSNE plots, and enrichment of CCDC93 expression in human liver vascular endothelial cells (colored dot in panel C).
(TIF)
(A-C) Ccdc93 protein expression was observed in both endothelial and non-endothelial cells in wild-type mouse aortae (129/Sv). Three biological replicates of mouse thoracic aorta showed Ccdc93 expression (brown stained). (D) Pixel quantification showed 1.33-fold higher Ccdc93 protein expression in the endothelium (normalized by area) as compared to non-endothelial cells (P = NS). Scale bar = 200μm.
(TIF)
(A) Mouse aorta immunostaining of platelet and endothelial cell adhesion molecule-1 (PECAM-1), (B) Negative control (no antibody) immunostaining of mouse aorta serial section. (C) Positive control of mouse aorta immunostaining of Ccdc93. (D) Negative control of mouse aorta where Ccdc93 antibody was absent. Detection of PECAM-1 (black arrows) and Ccdc93 protein expression (black arrows represents EC expression and blue arrows represents medial expression) is based on horseradish peroxidase (HRP) catalysis of a 3,3′-Diaminobenzidine (DAB, brown) chromogenic reaction, with a hematoxylin (blue) nuclear counterstain, scale bar = 200μm.
(TIF)
Data Availability Statement
All data associated with this manuscript is available in the supplementary files, summary statistics or under GCP IDs GCST90310248, GCST90310249, GCST90310250, GCST90310251, GCST90310252 on GWAS Catalog https://www.ebi.ac.uk/gwas/home.