Abstract
Spoligotyping was applied to 44 isolates of Mycobacterium bovis obtained from the Canadian province of Manitoba. Isolates were obtained from submissions of elk (n = 16), deer (n = 1), and cattle (n = 27) tissues spanning the period of 1990 to early 2003. Two spoligotype profiles were obtained differing only in the reaction with oligonucleotide number 12. Forty of the 44 isolates (90.9%) hybridized with oligonucleotide 12 (MB-1 type), while the remaining 4 of 44 (9.1%) did not show a signal at position 12 (MB-2 type). Octal codes for these 2 types are 656573377603600 and 656473377603600, respectively. These spoligotypes have not been reported as occurring elsewhere worldwide.
Résumé
Le spoligotypage a été utilisé pour caractériser 44 isolats de Mycobacterium bovis provenant du Manitoba, Canada. Les isolats provenaient d’échantillons de tissu de renne (n = 16), de cerf (n = 1) et de bovin (n = 27) soumis entre 1990 et 2003. Deux profiles de spoligotype ont été obtenus et ne différaient uniquement que par la réaction avec l’oligonucléotide 12. Quarante des 44 isolats (90,9 %) ont hybridé avec l’oligonucléotide 12 (type MB-1) alors que les 4 autres (9,1 %) ne présentaient aucun signal à la position 12 (type MB-2). Les codes pour ces deux types étaient respectivement 656573377603600 et 656473377603600. Ces deux spoligotypes n’ont pas été rapportés comme retrouvés ailleurs dans le monde.
(Traduit par Docteur Serge Messier)
Mycobacterium bovis, a causative agent of tuberculosis in livestock, is a member of the tuberculosis (TB) complex of mycobacteria including M. tuberculosis, bovis, microti, africanum, and canetti. Strain typing schemes have been developed for TB complex organisms, including restriction fragment length polymorpism (RFLP) analysis (1–3) using labelled probes for the insertion element IS6110 or the polymorphic GC-rich repeat sequence (PGRS). Spacer oligonucleotide typing or spoligotyping is a polymerase chain reaction (PCR) based method that exploits polymorphisms within the direct repeat region (DR) of the chromosome (4,5). The PCR products (with an incorporated label) are denatured and hybridized to immobilized oligonucleotides in a reverse line blot macroarray (4). The objective of this study was to conduct spoligotyping on isolates of M. bovis found in Manitoba.
Isolates of M. bovis from submissions from Manitoba, spanning the period 1990 through to May of 2003, were selected from the culture collection of the Mycobacterial Diseases Center of Expertise, Canadian Food Inspection Agency (CFIA), Ottawa, and represent all but one cattle outbreak and all wildlife isolates (elk and deer) during the period. Broth cultures were prepared by inoculation of 7H9 media with an aliquot of each isolate followed by incubation at 37°C for periods of 3 to 4 wk. Genomic DNA was extracted following the method of Skuce et al (1). The PCR assays were conducted as outlined in the ISOGEN Bioscience manual for the spoligotyping kit (ISOGEN, Maarssen, The Netherlands). Briefly, template DNA was amplified in the presence of 0.2 mM deoxynucleotide triphosphates (dNTP), 20 pmol of each primer (biotinylated DRa and DRb), 2.5 mM MgCl2, and 2.5 Units of AmpliTaq polymerase for a minimum of 20 cycles of 95°C for 1 min, 55°C for 1 min, and 72°C for 30 s. The PCR products were heat denatured and applied to the blot using an apparatus (Immunetics miniblotter 45; Immunetics, Cambridge, Massachusetts, USA). Hybridization signals were detected using a Biotin Detection System (Roche Diagnostics, Laval, Quebec) and film (Kodak X-Omat AR; Kodak, Ottawa, Ontario). Interpretation of spoligotype patterns and assignment of octal codes were done in accordance with the National Tuberculosis Genotyping and Surveillance Network (USA) recommendations as described by Crawford et al (6).
Typical spoligotype profiles for M. bovis isolates from Manitoba are shown in Figure 1. Of the 44 isolates tested, 40 were found to have the same profile (type MB-1). The remaining 4 isolates differed only by not having a hybridization signal for oligonucleotide 12 (MB-2). In an earlier report, Cousins et al (5) demonstrated that 2 spoligotypes were found in the limited (n = 3) number of isolates from Manitoba that were examined. They reported that spoligotype SP51 (equivalent to MB-2) was recovered from 2 cattle from the same location, while type SP52 (MB-1) was recovered from an elk. In the present expanded survey, all 4 examples of the MB-2 type were recovered from cattle submissions, all from different areas. Three locations were in the Rossburn area (1990 to 1991) and 1 location in the Grandview area in 2001 (Table I). Type MB-1 was recovered from cattle (n = 23), elk (n = 16), and the single deer isolate.
Figure 1.
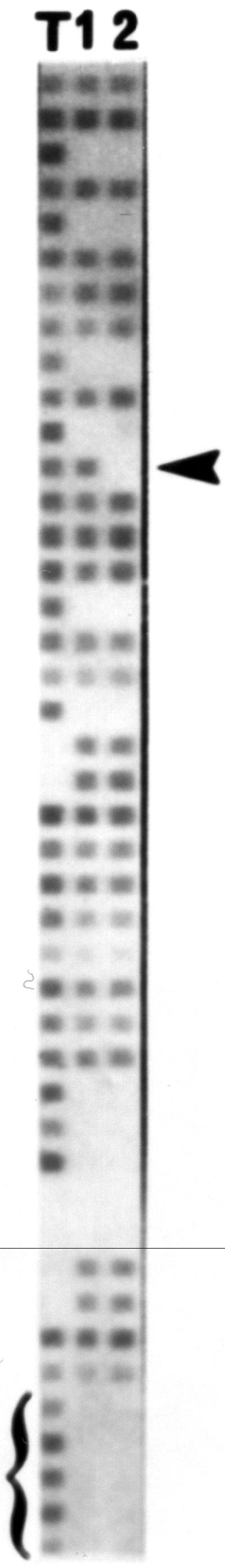
Spoligotype profiles for Manitoba types 1 and 2. Mycobacterium tuberculosis strain H37Rv (lane T) was used as a control. The arrowhead symbol is located at the position of oligonucleotide 12. A positive signal was found for MB-1 types (lane 1) while no signal was observed with type MB-2 (lane 2). Bracket symbol highlights oligonucleotides 39 to 43, which characteristically lack hybridization signals with M. bovis strains.
Table I.
Summary of spoligotyped isolates by year, species, and location
| Year submitted | Species | Spoligotype | Location | Number of Isolates |
|---|---|---|---|---|
| 1990 | Cattle | MB-2 | Rossburn | 1 |
| 1991 | Cattle | MB-2 | Rossburn | 2 |
| 1991 | Cattle | MB-1 | Erickson | 1 |
| 1992 | Elk | MB-1 | Rossburn | 1 |
| 1997 | Cattle | MB-1 | Rossburn | 6 |
| 1998 | Cattle | MB-1 | Rossburn | 10 |
| 1998 | Elk | MB-1 | RMNP | 1 |
| 1999 | Elk | MB-1 | RMNP | 1 |
| 2000 | Elk | MB-1 | RMNP | 3 |
| 2001 | Cattle | MB-2 | Grandview | 1 |
| 2001 | Elk | MB-1 | Rossburn | 1 |
| 2001 | Elk | MB-1 | RMNP | 1 |
| 2001 | Deer | MB-1 | Oakburn | 1 |
| 2002 | Cattle | MB-1 | Grandview | 5 |
| 2002 | Cattle | MB-1 | RMEA | 1 |
| 2003 | Cattle | MB-1 | Riding Mountain | 1 |
| 2003 | Elk | MB-1 | RMNP | 7 |
RMNP — Riding Mountain National Park; RMEA — Riding Mountain Eradication Area
Spoligotype patterns were converted to octal codes according to the method described by the National Tuberculosis Genotyping and Surveillance Network (6) for ease of comparison with other spoligotypes found in dataset SpolDB3.0 (7). This database contains 813 shared types of M. tuberculosis complex organisms, including M. bovis. Octal codes for the 2 Manitoba types are 656573377603600 (MB-1) and 656473377603600 (MB-2). These spoligotypes have not been reported as occurring elsewhere worldwide.
Spoligotyping is particularly useful for differentiating isolates harbouring only a single copy of IS6110. During the epidemiological investigation that follows a tuberculosis out-break, it is important to determine if there are links between infected areas. Strain characterization then becomes an important part of the investigation. Geographically within Canada, the strains of M. bovis described above have been isolated only from submissions of tissues from animals originating from Manitoba, exclusively from the area of Riding Mountain National Park and environs (Table I). All isolates examined in this study were found to originate from within the Riding Mountain Eradication Area of the province of Manitoba (8). Of the approximately 250 Canadian isolates of M. bovis examined by spoligotyping to date, originating from sporadic outbreaks of bovine tuberculosis since 1989 (9), all those originating from MB are distinct and MB-1 and 2 types have not been isolated from other jurisdictions either within Canada or worldwide (unpublished data). The area appears to represent a reservoir for these strain types.
Spoligotyping has generally been found to be very stable when repeated isolations have been derived from single or related outbreaks (10), despite some variation in the IS6110 RFLP pattern in M. tuberculosis strains. Determination of whether the MB-1 and MB-2 spoligotypes represent only minor variants of a single M. bovis strain or different strains will require full assessment by other complementary typing schemes. Limited RFLP (IS6110) analysis data confirms the profiles obtained by Cousins et al (5). The characteristic 1.9 kilo base pairs (kp) fragment is absent in the few isolates analyzed to date (unpublished data). The Manitoba strains examined by Cousins et al (5) generated different polymorphic GC-rich repeat sequence (PGRS) and direct repeat (DR) RFLP profiles, suggesting that there are greater differences between these 2 strains than would be expected for a single event affecting the spoligotype profile.
Acknowledgments
The authors gratefully acknowledge the contributions of L. Young, E. Rohonczy, and M. Koller-Jones of the CFIA for provision of information concerning the isolates used in this study and the technical assistance of D. Lloyd and G. McGinnis. The assistance of the CFIA field staff (B. Manns, D. Bates, B. Thompson) and Parks Canada Agency staff (D. Bergeson, P. Rousseau) for providing the information concerning cattle and wildlife samples was greatly appreciated.
References
- 1.Skuce RA, Brittain D, Hughes MS, Beck L-A, Neill SD. Genomic fingerprinting of Mycobacterium bovis from cattle by restriction fragment length polymorphism analysis. J Clin Microbiol. 1994;32:2387–2392. doi: 10.1128/jcm.32.10.2387-2392.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Skuce RA, Brittain D, Hughes MS, Neill SD. Differentiation of Mycobacterium bovis isolates from animals by DNA typing. J Clin Microbiol. 1996;34:2469–2474. doi: 10.1128/jcm.34.10.2469-2474.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Whipple DL, Clarke PR, Jarnagin JL, Payeur JB. Restriction fragment length polymorphism analysis of Mycobacterium bovis isolates from captive and free-ranging animals. J Vet Diagn Invest. 1997;9:381–386. doi: 10.1177/104063879700900407. [DOI] [PubMed] [Google Scholar]
- 4.Kamerbeek J, Schouls L, Kolk A, et al. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J Clin Microbiol. 1997;35:907–914. doi: 10.1128/jcm.35.4.907-914.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cousins D, Williams S, Liébana E, et al. Evaluation of four DNA typing techniques in epidemiological investigations of bovine tuberculosis. J Clin Microbiol. 1998;36:168–178. doi: 10.1128/jcm.36.1.168-178.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crawford JT, Braden CR, Schable BA, Onorato IM. National tuberculosis genotyping and surveillance network: design and methods. Emerg Infect Dis. 2002;8:1192–1196. doi: 10.3201/eid0811.020296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Filliol I, Driscoll JR, van Soolingen D, et al. Snapshot of moving and expanding clones of Mycobacterium tuberculosis and their global distribution assessed by spoligotyping in an international study. J Clin Microbiol. 2003;41:1963–1970. doi: 10.1128/JCM.41.5.1963-1970.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lees VW, Copeland S, Rousseau P. Bovine tuberculosis in elk (Cervus elaphus manitobensis) near Riding Mountain National Park, Manitoba, from 1992 to 2002. Can Vet J. 2003;44:830–831. [PMC free article] [PubMed] [Google Scholar]
- 9.Koller-Jones M. Bovine Tuberculosis Eradication Program — October 31, 2003. CAHNet Bulletin, 8th ed, 2004:33–36. Available at www.cahnet.org/bulletinsE/bulletin8ENG.pdf. Last accessed August 26, 2004.
- 10.Niemann S, Richter E, Rüsch-Gerdes S. Stability of Mycobacterium tuberculosis IS6110 restriction fragment length polymorphism patterns and spoligotypes determined by analyzing serial isolates from patients with drug-resistant tuberculosis. J Clin Microbiol. 1999;37:409–412. doi: 10.1128/jcm.37.2.409-412.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]


