Abstract
Nucleic acids have attracted widespread attention due to the simplicity with which they can be designed to form discrete structures and programmed to perform specific functions at the nanoscale. The advantages of DNA/RNA nanotechnology offer numerous opportunities for in-cell and in-vivo applications, and the technology holds great promise to advance the growing field of synthetic biology. Many elegant examples have revealed the potential in integrating nucleic acid nanostructures in cells and in vivo where they can perform important physiological functions. In this Review, we summarize the current abilities of DNA/RNA nanotechnology to realize applications in live cells and then discuss the key problems that must be solved to fully exploit the useful properties of nanostructures. Finally, we provide viewpoints on how to integrate the tools provided by DNA/RNA nanotechnology and related new technologies to construct nucleic acid nanostructure-based molecular circuitry for synthetic biology.
The intracellular environment is highly compartmentalized; biomolecules (nucleic acids, proteins, lipids and so on) are spatiotemporally organized to form functional modules, signal pathways and interacting networks. Inspired by nature, many researchers are interested in designing and/or repurposing artificial biomolecular assemblies to engineer living organisms with the goals of elucidating their molecular mechanisms and constructing novel biological computers or robots. These studies will have broad impacts in various areas ranging from fundamental biological studies to applications including synthetic biology, theranostics and biocomputing.
The past decade has witnessed the rapid emergence of new physical tools (for example, super-resolution and X-ray microscopy) and molecular biotechnologies (for example, DNA/RNA nanotechnology, CRISPR-Cas9) that have shifted our thinking and reshaped our ability to control biological systems. As an elegant example, DNA nanotechnology pioneered by Ned Seeman quickly evolved into a booming field1,2. By exploiting the unparalleled precision and programmability of DNA hybridization, researchers are able to construct virtually any prescribed DNA nanostructure in a bottom-up manner (Fig. 1). Importantly, the addressability of DNA nanostructures allows site-specific anchoring of biomolecules in vitro, which enables spatiotemporal control of biomolecular interactions resembling their intracellular counterparts. Despite the remarkable advances for studies in test tubes, the efficient and tailored fabrication and manipulation of circuit modules in cells and animals remains enormously challenging due to the extreme complexity of natural metabolic networks3,4. Here, we review the recent progress in this highly interdisciplinary field, with a focus on constructing molecular circuitry for synthetic biology by exploiting structural and computational DNA/RNA nanotechnology.
Figure 1 |. Representative examples of promising DNA nanostructures for synthetic biology.

a, 2D DNA crystalline arrays self-assembled from synthetic DNA double-crossover tiles155. Scale bar: 300 nm. b, 2D square lattice assembled with 4 × 4 DNA tiles156. Scale bar: 100 nm. c, DNA tetrahedral structure77. d, Self-assembled 3D DNA crystal from a tensegrity DNA triangle motif157. Scale bar: 200μm. e, Hierarchical polyhedral DNA structures158. f, 3D structures built with single-stranded tile DNA bricks159. g, DNA origami folded by a long single-stranded scaffold DNA and hundreds of short staple DNA oligos142. h, Wireframe DNA origami nanostructures with multi-arm junction vertices160. Scale bar: 100 nm. i, Hollow DNA box with a controllable lid18. j, 3D DNA origami built with multiple pleated layers115. Scale bar: 20 nm k, 3D DNA origami with complex curvatures161. l, Arbitrary 3D structure built with polyhedral meshes162. Scale bar: 50 nm
A toolbox for biomolecular engineering of living systems
The precision of DNA hybridization ensures the fidelity of DNA replication during the transmission of genetic information. DNA nanotechnology exploits the precise and predictable Watson–Crick base-pairing rules to construct a variety of self-assembled DNA nanostructures ranging from one-dimensional (1D) to three-dimensional (3D), from periodic to discrete, and from static to dynamic architectures1. Recent advances in the field have revealed the unprecedented power in using engineered nucleic acid nanostructures for various applications, especially biomolecular engineering both in vitro and in vivo. To demonstrate the potential for constructing modularly designed synthetic circuits in living organisms, we summarize DNA/RNA nanotechnology-enabled tools and related emerging techniques for on-demand nucleic acid manipulation (Fig. 2).
Figure 2 |. DNA/RNA nanotechnology-enabled toolbox for synthetic circuits.

A diverse set of useful tools have been available; for example, biomolecular scaffolds based on addressable DNA nanostructures8, logic units based on DNA strand displacement reactions15, DNA nanostructure cell-entry vehicle41, HCR-based isothermal construction of DNA structures44, targeted editing and error correction based on CRISPR systems60, signal readout based on fluorescent RNA motifs63 and triggers/switches based on siRNAs/microRNAs66 or riboregulators28. ORF, open reading frame.
Biomolecular scaffolds.
Self-assembled DNA nanostructures are fully addressable and can accommodate precise numbers of small molecules, DNA/RNA, proteins5,6, lipids7 and even nanoparticles at nearly any prescribed position8. This power is complemented by the commercial availability of a wide range of nucleotide modifications that offer great flexibility in choosing robust and nearly quantitative conjugation chemistry. Researchers have taken advantage of these attractive features to design various DNA nanostructure scaffolds to make functional modules such as enzyme cascades8–11, photonic coupling12 or electronic wiring13. The ability to spatially organize enzymes for enhanced substrate channelling has important implications for in-vivo applications8–11. Despite this progress, scaling-up these organized cascade reactions to large synthetic circuits mimicking intracellular signalling or metabolic pathways remains challenging. A very recent study demonstrating the assembly of DNA scaffolds in a ‘plug-and-play’ manner represents a plausible solution to this problem14.
Logic units.
Dynamic DNA nanostructures incorporating functional nucleic acids (FNAs; that is, aptamers, DNAzymes and RNAzymes) that are responsive to chemical/biochemical stimuli have been exploited to develop DNA logic gates that can be specifically triggered in physiological environments15–17. If integrated with strand-exchange reactions15, it is possible to transform input signals into the release of nucleic acids that can be relayed in cells to trigger subsequent logic-driven gene expression. In addition, tensioned or compressed DNA motifs have been used as ‘entropic springs’ that store stress energy that can be triggered to induce mechanical forces18,19.
A recent publication described the use of DNA as universal input/output (I/O) to construct multilayer logic gates and form scalable prototype molecular circuits20. In addition to using linear DNA sequences, the introduction of DNA nanostructures allows expansion to 2D and 3D and offers additional operations (for example, shape complementarity)21. Notably, new strategies including toehold exchange, strand displacement, hybridization chain reaction (HCR) and light-induced structural switching can dynamically and isothermally manipulate DNA hybridization, making biocomputing possible under physiological conditions22–26. In several recent reports, digital circuits regulating intracellular gene expression and mimicry of neural network computation have been realized using DNA- and RNA-based reactions27,28. FNAs provide new recognition and catalytic functions that nicely complement classic nucleic acid hybridization, which has been widely employed to process and execute biocomputation29,30. Given the ubiquity of natural ribozymes in cells, delivery of designed DNAzymes/RNAzymes into cells to regulate biological circuitry is a highly promising approach19,31. Nevertheless, the development of large-scale circuits in vivo remains challenging due to the need to avoid interference by the myriad biomolecules that are present32,33.
Reversibility is key to implementing continuous computation responsive to changing inputs, which is challenging for logic units based on kinetic-controlled non-equilibrium reactions. However, by using DNA structures that equilibrate between ON and OFF states34, or DNA nanodevices possessing good mechanical reversibility,35 resettable DNA logic units have also been implemented.
Cell-entry vehicles.
Cytoplasmic membranes are largely negatively charged and form a natural electrostatic barrier for polyanionic nucleic acids. Interestingly, self-assembled DNA nanostructures are readily internalized by mammalian cells via energy-dependent endocytosis36,37. Several groups have employed DNA nanostructures as cell-entry vehicles to deliver molecular payloads that can stimulate immunological responses and suppress tumour growth38–40. The use of biocompatible and degradable nucleic acids carriers largely circumvents the toxicological concerns of inorganic nanomaterial-based nanomedicine. Moreover, cargos can be covalently or non-covalently loaded on DNA nanostructures41. The addressability of DNA nanostructures allows precise control of the quantity and stoichiometric ratio of cargo molecules and cell-targeting ligands37, which could enable the targeted intracellular delivery and controlled release of various small molecules, nucleic acids and proteins40,41. The tailorable nature of DNA nanostructures also opens new opportunities for developing dynamic, responsive vehicles to circumvent many barriers encountered at different stages in cell entry42.
Isothermal construction.
DNA nanostructures are often self-assembled in test tubes by annealing at non-physiological temperatures, limiting their potential for in-vivo assembly under physiological conditions. A variety of isothermal strategies have recently been developed to fabricate DNA/RNA nanostructures, paving the way for intracellular DNA/RNA nanostructure replication. Several nanostructures with a few or even a single DNA/RNA strand have been successfully assembled using isothermal protocols at physiological temperatures using intramolecular self-cont-folding43 and HCR44 assembly approaches. Structures composed of dozens of strands have been assembled using single-stranded DNA tiles45. Anderson and colleagues46 designed an elegant approach to co-transcriptionally fold single-stranded RNAs into well-defined nanostructures with T7 RNA polymerase. Natural RNA motifs (for example, transfer RNAs [tRNAs] and packing RNAs [pRNAs]) have also been utilized to construct nano-assemblies under mild conditions47. These nanostructures can be cloned in cells by delivering plasmids carrying DNA templates into host cells and rely on the intracellular transcriptional machinery to perform replication48. Transcription from plasmids has also been successfully applied to the assembly of periodic RNA nanostructures within Escherichia coli49. Very recently, synthesis of single-stranded DNA motifs in living bacteria has been realized by Elbaz et al.50 via reverse transcription. Moreover, the Dietz group employed transcription activator-like (TAL) effector proteins as staples to isothermally fold dsDNAs into nanostructures at room temperature in near-physiological buffer conditions, which provides a potential path to fabrication of intracellular nanostructures using biosynthesized protein and nucleic acid components51. Although the designability of isothermally folded nanostructures is still limited, the development of computer-assisted algorithms might provide a feasible solution to isothermal construction in cells and in vivo46.
Targeting and editing.
Zinc finger nuclease (ZFN), transcription activator-like effector nuclease (TALEN), and clustered regularly interspaced short palindromic repeats (CRISPR) are three highly precise targeted gene editing tools52. The rapidly emerging RNA-guided CRISPR-Cas9 endonuclease system has revolutionized our ability to edit genomes in cells and even whole organisms. CRISPR is composed of two components: a Cas9 endonuclease and a guide RNA. Highly specific RNA–DNA hybridization guides Cas9 to targeted sites in the genome, where it precisely cuts gene fragments53–55. This approach opens a new door to efficiently rewrite naturally existing molecular circuitry and/or adapt artificial circuitry to molecular machinery in cells and animals56. We envision that the marriage of DNA nanotechnology with CRISPR might overcome some short-comings of the latter (for example, high off-target rates57), allow the former to better control biological circuits56,58,59, and serve to augment CRISPR capabilities by coupling its activity with other enzymes on a DNA scaffold60.
Signal readout.
Several conditionally fluorescent RNA motifs including ‘Spinach’ and ‘Broccoli’ are new tools for signal transduction and readout in cells. These live-cell fluorescent reporters mimic green fluorescent protein (GFP) to allow real-time monitoring of genetically associated processes and can be co-transcribed with other genetic modules in cells61–63. Hence, these fluorescent RNA motifs might be coupled with cloneable DNA/RNA nanostructures to signal the output of synthetic circuits. The fluorescent emission might be relayed to optogenetic control of cells and animals64. Nevertheless, in vivo signal reporting has long been a challenge due to strong background emission and limited imaging depth65. The development of fluorescent RNA motifs with near-infrared emission, and probably other advanced probes with two-photon excitation, magnetic resonance or photoacoustic properties, should greatly expand the potential of in-vivo biocomputing.
Triggers and switches.
Nucleic acids can perform various recognition and regulation functions aside from their genetic roles. Non-coding RNAs (for example, microRNA and small interfering RNA) are a class of naturally existing FNAs that dynamically regulate gene expression at the post-transcriptional level. RNA interference (RNAi) technology in particular has shown great potential in gene therapy and synthetic biology; it has been used to implement various synthetic logic gates functioning in live cells66. In addition, certain secondary structures at the 5’-untranslated region (UTR) of mRNAs (termed riboregulators) have been de-novo designed as prokaryotic translational regulators that prevent binding of ribosomes to the ribosomal binding site (RBS), thereby blocking translation initiation. When a trans-activating RNA is introduced, the repressed secondary structure is unfolded by this trigger RNA via strand displacement, which leads to target gene expression28,67. A wide assortment of other RNA-based systems have been developed to regulate at the transcriptional level68,69 and to repress gene expression in response to a trigger RNA70,71.
FNAs are readily obtained from genomic DNAs in natural organisms or systematic evolution of ligands by exponential enrichment (SELEX) to serve as gene regulation switches72. For example, endonuclease-like ribozymes are utilized to cleave mRNAs in a sequence-specific way to selectively repress their expression73. Notably, FNA catalytic activity often relies on certain ions or small molecules that are regarded as trans-factors for gene regulation73. Thus, a wide range of FNAs clearly enriches the toolbox for specific and tunable regulation and detection of intracellular events74,75.
Error correction and resilience.
The precision of DNA base pairing does not guarantee error-free signal transduction in either in-vivo replication or artificial molecular circuitry. In natural living systems, self-monitoring and repairing machineries (for example, the immune system and DNA damage repairing system) are indispensable for error correction and functional resilience of the system. These mechanisms provide inspiration for developing self-correction mechanisms within artificial biomolecular circuitry76. A straightforward approach to error correction is to make use of redundancy. In a typical DNA self-assembly reaction, over 1010 nanostructures can be fabricated in a 1-mL reaction system77. Hence, DNA nanotechnology enables massively parallel computing and the potential to design redundant units to reduce error rates24,78,79. In addition, multilayered DNA information storage systems (for example, realized by selective modification of DNAs)80 may also provide redundant space for error-suppression tasks. These simple-yet-powerful mechanisms are expected to be adapted to increase the resilience of complicated synthetic systems. Alternatively, targeted editing may provide a more intelligent way to implement error correction in synthetic biology, especially with the rise of the CRISPR technology81. These gene-editing tools provide unprecedented precision for cleaving and repairing error-containing segments via homologous recombination or homology-independent pathways in vivo.
Targeted applications
Exploiting the DNA/RNA nanotechnology-enabled toolbox makes it possible to develop novel synthetic circuitry that allows the rewiring of natural pathways for various applications. Here we summarize recent progress in drug delivery, cellular imaging, metabolic engineering and cellular-pathway investigation.
Drug delivery.
Targeted drug delivery holds the promise to improve drug efficacy and reduce side effects. In a typical protocol, drug molecules are conjugated with antibodies or aptamers that possess high affinity to the target. The Tan group82,83 demonstrated that DNA nanostructures appended with polyvalent aptamer motifs greatly improved targeted delivery. However, difficulty in finding highly specific ligand–receptor pairs largely restricts practical applications. Because DNA logic gates can process multiple inputs to make a pondered decision, it is possible to rely on DNA nanotechnology to integrate multiple factors subject to user-specified Boolean logic expressions to reliably distinguish targets. For example, the Church group19 developed a DNA origami-based logistic nanorobot triggered by two aptamers. Using AND logic, the nanocontainer can only be opened when both aptamers bind their corresponding targets. A recent study demonstrated successful application of this strategy in living cockroaches31.
Cellular imaging.
Given the flexibility and versatility in position-specific functionalization with various ligands and fluorophores, DNA nanostructures have demonstrated great potential for targeted imaging in cells and in vivo84,85. For example, Förster resonance energy transfer pairs can be site-specifically anchored on well-defined dynamic DNA nanostructures to construct structural switching probes that undergo stimuli-responsive fluorescence changes in cells16,83. The Krishnan group86 successfully performed spatiotemporal intracellular imaging of ions using responsive DNA nanostructures. The incorporation of the CRISPR-Cas9 system may increase targeting ability in live cells, thus enabling real-time imaging of chromatin structure dynamics67,68. Indeed, CRISPR-Cas9 has been employed for dynamic imaging of genomic loci in live cells87. However, this method proved suitable only for repetitive sequences or required tiling of guide RNAs along the target locus to yield detectable fluorescence signals. DNA nanostructures have been deployed in multiple previous reports to improve the sensitivity and resolution of cellular imaging. To image low-copy mRNAs in cells, Pierce and co-workers88 exploited HCR to amplify fluorescent signals. Jungmann et al.89,90 realized multiplexed 3D cellular super-resolution imaging by exploiting transient binding of short fluorescent-labelled oligonucleotides on synthetic DNA nanostructures, a method dubbed DNA-PAINT (point accumulation for imaging in nanoscale topography).
Metabolic engineering and cellular-pathway investigation.
Metabolic pathway efficiency largely depends on the spatiotemporal organization of the enzymes involved. Given the organizational capability of nucleic acid nanostructures, they are powerful tools for metabolic engineering in synthetic biology. Delebecque et al.49 used intracellularly assembled RNA structures as scaffolds to organize the interaction between hydrogenase and ferredoxin in bacteria. The coupling efficiency of these enzymes was improved through control of their relative positions, which led to dramatic increases in hydrogen production. Using a similar strategy, several enzyme cascade systems have been engineered to realize high-efficiency coupling5,9,11. The development of caged 3D DNA nanostructures enables further confinement of enzymes, reactants, and intermediates in 3D space, which could allow the construction of nanoscale reactors with high catalytic efficiency10,14.
DNA technology-mediated delivery of CRISPR-Cas9 and RNAi also provide new ways to probe cellular pathways by allowing specific knock-in or knock-out of target genes or regulatory elements. For example, with the use of CRISPR-targeted gene suppression, genetic logic circuits could be implemented to conditionally activate/deactivate genes in a pathway56,91. Nucleic acid strand exchange was also employed to conditionally activate RNA interference in live cells33,92. Conditional gene knock-in and knock-out tools are therefore very promising for future cellular-pathway investigations.
DNA/RNA nanotechnology-enabled computing kernel.
A cell is in principle an elaborate natural biocomputer that collects physical/chemical information from the environment, performs calculations via signal pathways or molecular circuitry, and uses this data to perform actions. Several types of live-cell logic gates and biocomputers have been developed by engineering gene regulatory networks in bacterial, yeast and mammalian cells28,56,66,93–96. So far, most designed biological circuits are highly case specific, making them difficult to adapt and transplant from one organism to another. Hence, it is highly desirable to develop a more general synthetic circuitry scheme. DNA and RNA are ideal tools for realizing such organism-independent circuitry given their ubiquity in all forms of life and their predictable base pairing interactions.
In an electronic computing kernel, rational compilation of basic elements (for example, transistors and wires) can reach arbitrary complexity. Given the proven ability of DNA to encode and solve combinatorial/high-dimensional mathematical problems97–100, it is envisioned that the computing kernel of a synthetic circuit can be similarly constructed by the rational assembly of a few basic DNA logic gates. Specifically, strand exchange of nucleic acids provides a route to wire DNA logic gates in cells and in vivo. The I/O in these reactions are nucleic acids that can seamlessly implement multilayer logic gates24. More importantly, strand exchange reaction kinetics can be finely tuned with rational sequence programming to fit the compilation of arbitrarily defined systems15. Hence, strand exchange appears to possess the universality and flexibility required for biocomputing.
Soloveichik et al.32 utilized strand exchange as a molecular primitive to implement the formalism of arbitrary chemical reaction networks. They demonstrated several fascinating examples including a limit cycle oscillator, a chaotic system, and feedback logic systems, all of which provide inspiration for constructing generic computing modules. Qian et al.20 transformed logic unit form and function by simply introducing rationally designed auxiliary strands. Without changing other components, they could switch the gates between AND and OR functions by tuning the concentration of a ‘threshold’ strand (Fig. 3a). Green et al.28 employed multiple toehold switch riboregulators in E. coli to form a layered logic circuit for evaluating 4-input AND expression using RNAs as inputs. Toehold switches were also used to detect the expression of endogenous RNAs in the cell, demonstrating the capacity for FNAs to directly interface with native cellular RNAs. Very recently, the Seelig group33 demonstrated the successful use of strand exchange in biological environments for biocomputing. By designing four-way strand exchange reactions, they activated functional siRNAs and realized subcellular co-localization of logic gate operation by exploiting native mRNA as a scaffold (Fig. 3b). To further increase the sophistication of intracellular DNA/RNA circuitry, the use of DNA nanostructures to precisely position DNA logic gates is predicted to substantially increase the computing power of strand exchange circuits through localization of reaction components (Fig. 3c)101. Such localization approaches can improve circuit performance in much the same way that substrate channelling is known to increase metabolic reaction rates.
Figure 3 |. Typical AND gate circuits.

a, A DNAzyme-enabled AND gate29. b, A CRISPR-Cas9 system91. c, A logic-gated DNA nanorobot that conditionally releases payload molecules19,31. d, A combination of riboregulators and recombinases104. Based on these principles, other kinds of logic gates (OR, XOR, NOR and so on) and logic-gated cascades can be readily implemented as well.
In addition to DNA/RNA, a range of enzymes including restriction endonucleases, ligases, recombinases and polymerases can serve as biocomputational operators by specifically cutting, ligating, recombining and replicating input nucleic acids under physiological conditions. These constitute programmable and autonomous computing machine components to manipulate gene expression in cells (Fig. 3d)102–104. The recently developed CRISPR-Cas9 technology is in fact a combination of DNA/RNA hybridization and the restriction endonuclease activity, which offers highly flexible and precise genome targeting. These molecular tools should be adapted with DNA nanotechnology to regulate cellular networks and implement live-cell biocomputation91.
Natural biological systems often enable continuous responses to environmental stimuli due to the analogue nature of biochemical reaction networks93. Hence, DNA-based analogue computation represents an alternative-yet-flexible approach for dealing with wide-ranges graded signals (for example, molecular concentrations) and producing output with rich information93. As an example, Qian et al.105 implemented analogue time-domain circuits by exploiting DNA strand-displacement reactions that are intrinsically analogue. More recently, by using a droplet-based microfluidic system, Genot et al.106 realized massively parallel analogue circuits for DNA-bistable switches and oscillators. Given the remarkably high ability of analogy circuits for computing non-linear functions107, they are especially useful for high-throughput analysis in drug discovery, systematic evolution and next-generation sequencing.
I/O and human-computer interfacing.
In modern electronic computers, a keyboard and mouse translate human actions to digital electronic signals, and the computed results are converted to visualizable output on the display. Recapitulating these capabilities using biomolecular components requires a diverse set of transducers for converting inputs into usable biomolecular signals and an array of outputs to modulate cell state and to return the results of the biocomputation. Regulatory proteins (for example, receptors, kinases and trans-regulatory factors) and nucleic acids (for example, siRNA and microRNA) provide a rich library of molecular signal transducers to trigger intracellular events. They effectively put cells in a predefined state (for example, protein phosphorylation and gene activation/deactivation), which is readily converted to optical output signals with the use of live-cell reporters (for example, GFPs). The readout device is often a fluorescence microscope that can image live cells and identify their specific states using coupled fluorescence. Although this approach has been widely employed to develop live-cell biocomputers, many of these systems involve highly specialized reactions under elaborate protocols.
DNA nanotechnology could provide a straightforward approach to modularly design user-friendly interfaces for signal I/O in which all signal transduction, information processing and signal generation tasks are carried out by DNA/RNA circuit elements (Fig. 4). First, since many DNA nanostructures are readily internalized by live cells36,84 and present numerous sites for binding, they can be employed as multifunctional delivery vehicles to carry regulatory proteins and DNA/RNA computing circuits into living organisms31,38. Furthermore, these complexes can be guided to targeted organelles or organs with small ligands, aptamers or peptide signals pre-loaded onto DNA nanostructures37,40,83. Such systems would make it possible to design a universal system for bringing genetic instructions into the cell using DNA nanostructure-based modules. Second, dynamic DNA nanostructures incorporating FNAs offer great flexibility in designing signal transducers due to the availability of a wide range of FNAs identified by SELEX or mined from natural genomes. These transducers can specifically transform an input signal (often a chemical/biochemical stimulus) into digital, binary states (ON/OFF) via structural switching108. Regulatory biomolecules released during structural switching can be relayed and rewired to intracellular signal pathways or circuits28,67,109. Third, DNA/RNA circuitry can act directly on the nucleic acids produced during input-signal transduction to process information and reach logical decisions. Fourth, once signals have been processed, DNA/RNA can be used to generate the output signals that are passed to an external observer or other cells. Such signals can take multiple forms; for instance, as a combination of new genes expressed by the cell. Alternatively, conditionally fluorescent RNA motifs (for example, Spinach, Broccoli) are GFP-like live-cell reporters that do not need to be translated20,52, making them desirable as fluorescent signal outputs for cell-based biocomputation. We also envision that DNA nanotechnology may enable high-resolution ‘soft lithographic’ patterns to present outputs with visualizable DNA origami that resemble display screens under atomic force microscopy110,111 or employ precise localization of fluorophores to provide highly multiplexed circuit readout112.
Figure 4 |. I/O interface scheme for synthetic circuits.

Three main types of components are employed to convert input signals into output signals. Signal transducers like aptamers, riboswitches and ribozymes, detect the inputs (in forms of pH value, UV light, metal ions, small molecules, proteins and so on) and convert them into nucleic acid signal molecules. These DNA/RNA signals undergo information processing using DNA/RNA-based circuitry. Following processing, signals are generated through transcription, translation or DNA lithography and output (in the form of fluorescence, protein products, a coded pattern and so on) from the circuit.
Given these new opportunities offered by DNA nanotechnology, it is possible to standardize the control of signal I/O and achieve multi-signal integration and processing in live cells. The ability to develop fan-in and fan-out cell-based circuits should have valuable applications in multi-maker solution, pattern recognition and neural network systems for intelligent disease diagnosis and therapy.
Information storage
DNA molecules are arguably the most important and powerful carrier of information in nature. Several recent studies have shown that using DNA molecules to store digital information leads to extremely high capacity and durability, far exceeding those of currently available storage media (Fig. 5a)113,114. These studies exploited the nucleotide sequence information of DNA alone to store data. DNA nanostructures, however, provide a number of unique features that could be harnessed to substantially increase DNA information storage density and to provide read/write/erase access of stored information for biocomputing.
Figure 5 |. Nucleic acid nanostructures as information storage media.

a, Comparison of different information storage media with regard to data density and capacity (Modified from ref. 114). b, Multidimensional information storage can be realized with DNA/RNA nanostructures with various geometrical/topological properties, chemical/biochemical modifications, and dynamic responses.
Similar to the chromatin structures found in eukaryotes, DNA nanostructures can condense DNA down to nearly the smallest possible volumes, yet without intervening proteins. 3D DNA origami, for instance, consists of tightly packed DNA double helices with inter-helix spaces down to only 0.1 to 0.4 nm115. Thus, storing information within a compact DNA nanostructure could yield immediate increases in the number of bits per unit volume. The structural control afforded by DNA nanostructures also provides additional means of encoding information beyond the basic DNA sequence specification. Just as the information contained in digital files is defined by their file format, information contained within DNA could be stored in 3D assemblies with a precisely defined structure in terms of the number of helices, turns, topology, and overall geometry (Fig. 5b). Knowing the ‘file format’ of this genetic information, sequencing results could be threaded into the predefined target structure. Thus, each element of sequence information can also be tied to its location within the DNA nanostructure, increasing the overall information content of the DNA itself. This additional information content could be used to store more data per unit volume or mass, or to establish additional error correction and redundancy measures.
Nucleic acid nanostructures formed within the cells could also serve as repositories for storing and presenting information concerning the past history of the cell. Nanostructures can be constructed with a programmable number of addressable capture or release sites (for example, DNA/RNA binding sites or aptamers) that can interact with different species produced by the cell, such as RNAs, proteins and small molecules. The binding and unbinding of these species effectively become the write/erase elements of the information storage device. The resulting intracellular recorders can then be read by DNA-based logic circuitry that directly interfaces with nucleic acid nanostructure to process the stored information. Beyond serving as breadboards for information storage, nanostructures also provide the opportunity to dynamically change their geometry in response to intracellular events, such as the binding of RNA molecules or interactions with a protein. Structure switching can be used to control access to the stored information in much the same way that chromatin structures in eukaryotes exert spatiotemporal control over the availability of genetic information116.
Application of DNA nanostructure-based storage media to biocomputing will also benefit from new mechanisms for reading, writing and copying data. The Sleiman group117 demonstrated the transfer of encoded DNA strands on different 3D DNA nanostructures to the surface of gold nanoparticles in a manner similar to lithography. This method provides a way to read multidimensional information from DNA storage media. To realize DNA writing in live cells, Farzadfard and Lu118 developed a genetically encoded approach to record arbitrary transcriptional signals in genomic DNA by generating targeted mutations. The stored information could be read out with fluorescence imaging of reporter genes. The CRISPR-Cas9 system has also been exploited to develop addressable tools for writing and reading information on genomic DNA, which have proven to be a robust data storage method119,120. Inspired by epigenetics, Mayer et al.80 recently demonstrated that multiple layers of information could be encoded in and retrieved from a single DNA template via selective chemical modifications of nucleobases. In addition, advances in autonomous self-replication and the cloneable fabrication of nucleic acid nanostructures are paving the way to realizing information duplication and signal amplification in cells and in vivo88,118,121.
Perspectives
Given their programmable and self-assembling nature, artificial DNA nanostructures can provide a library of modularly designed scaffolds for spatially organizing DNA/RNA-based logic gates into synthetic circuits reminiscent of electronic breadboards but with DNA/RNA as the principal signal carrier. Use of DNA/RNA ensures that there is substantial signal bandwidth for information processing and diverse options for input and output signals. This vision for DNA/RNA nanotechnology-based live-cell circuitry is outlined schematically in Fig. 6. With the assistance of cell-targeting aptamers and lipid-functionalized nanostructures, programmable DNA-based input signal transducers can be positioned at the cell membrane to monitor the presence of circulating input molecules via FNAs. Upon binding, input signals are converted into nucleic acid signal molecules that are passed on to intracellular DNA/RNA-based processor and memory elements that perform biocomputation and memory storage operations, respectively. Both processor and memory units can communicate with one another via nucleic acid signals. Once a decision is reached or more circuit elements are needed, the DNA/RNA circuitry can harness the cell as a biomolecular assembly plant to fabricate the required components. DNA or RNA synthesized by the cell can be programmed to self-assemble into new processor and memory storage modules. Outputs in the form of proteins, genetically encoded fluorescent reporters (for example, GFPs, Spinach) or DNA/RNA circuitry can be synthesized and passed as signals to other cells or used within the parent cell itself.
Figure 6 |. The scheme of an integrated live-cell circuit enabled by DNA/RNA nanotechnology.
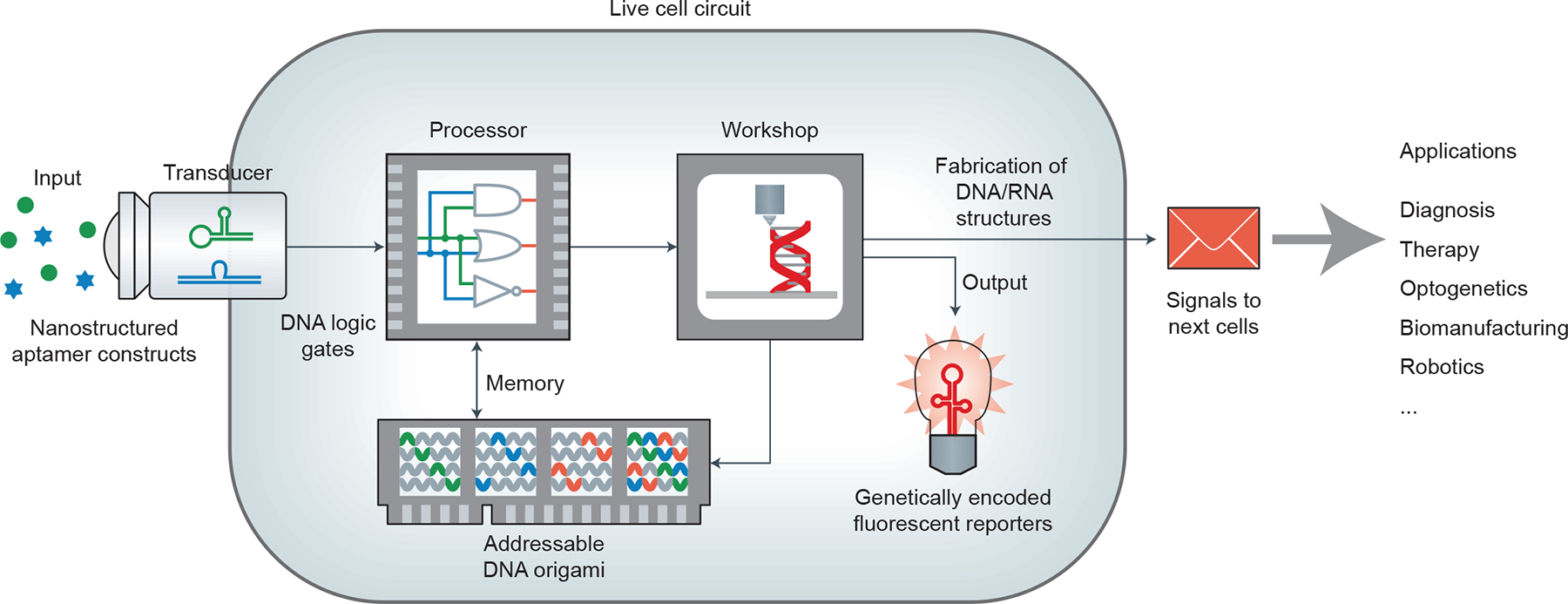
Inputs from outside the cell are converted via transducers into DNA/RNA signals that interface directly with intracellular DNA/RNA-based circuitry and memory storage elements. The biosynthetic capabilities of the cell itself are used to fabricate DNA/RNA nanostructures and to produce output signals for a human operator or the intended recipient cells. This live-cell circuit holds promise in applications like diagnosis, therapy, optogenetics, biomanufacturing and robotics.
Such DNA/RNA nanotechnology-based circuits can also make use of non-nucleic-acid functional elements such as endonucleases, recombinases, cell-targeting proteins and the CRISPR complex to expand their function. These elements along with FNAs may also be precisely incorporated in fixed copy numbers to receive input from environmental or cellular signals, specifically target organelles in cells or organs in vivo, and responsively trigger signal pathways in a predefined time-series. The outputs in the form of new biomolecules from such circuits can be used to correct a malignant cell state, induce apoptosis or initiate production and release of daughter DNA/RNA circuits. Hence, we expect that DNA nanotechnology will provide great opportunities to rewire intracellular signal pathways in a plug-and-play manner. Such engineered molecular circuitry could be repurposed for applications including diagnosis, cancer therapy, optogenetics, biomanufacturing and robotics.
DNA nanotechnology has played important roles in many facets of cell-based based biocomputation31,84,122. However, our fundamental understanding of how DNA nanostructures function in cells remains poor. Additional studies are needed to clarify their structural-functional relationship to optimize intracellular delivery; elucidate their endocytic pathways and intracellular traffic; and understand their circulation, distribution, metabolism, and excretion in vivo. Furthermore, it is imperative to refine DNA nanotechnology to allow its application in synthetic biology. Indeed, there have been several examples of DNA nanotechnology-based logical control in vivo31,123,124. For instance, DNA nanorobots implementing various logic calculations have been deployed in living cockroaches, which can release different drug molecules in response to protein cues31. Notably, in-vivo implementation of a half adder has also been realized, revealing the high potential of complex biocomputation in living systems. In other examples, by using DNA/RNA nanodevices, siRNAs have also been delivered and activated in living cells based on logic calculations, resulting in conditional knocking-down of target genes33,125. Yet, given the complex environment in living systems, grand challenges remain for in-vivo biocomputation.
Concerning the speed of a single logic operation, DNA/RNA-based computation is intrinsically limited by the kinetics of chemical reactions; a typical DNA hybridization reaction takes seconds, which is nine-to-ten orders of magnitude slower than an electronic logic switch. Nevertheless, given the massively parallel operation of DNA/RNA-based biocomputation, it is probably wise to employ it for solving combinatorial problems. More importantly, the potential of seamlessly integrating biocomputation within living organisms makes DNA/RNA circuitry appropriate for biomedical applications, where operations that take place over minutes are adequate.
The cellular environment is generally complex and hostile to foreign structures due to the presence of various ions, enzymes and endogenous nucleic acids. Although DNA nanostructures are more resistant to enzymatic degradation than single- or double-stranded DNA126,127, their stability remains a major concern when performing complicated computations in cells and in vivo. Previous studies have shown that DNA nanostructures remain intact in cell lysates and live cells for up to several hours, and these results form the basis for intracellular drug delivery and cellular imaging36,128,129. However, in-vivo studies revealed that the blood circulation time of DNA nanostructures in mice was at the minute scale130, which restricts their biomedical applications. Recent investigations also reported that DNA origami nanostructures tended to disintegrate in the presence of physiological concentrations of Mg2+, which could compromise their intracellular and in-vivo use130.
Several approaches have been developed to further stabilize DNA nanostructures in cells by ligating free ends that are vulnerable to nucleases or by designing closed and interlocked structures131,132. Niemeyer and co-workers133 reported that non-covalent modification with intercalators could rigidify DNA origami to prevent disintegration. Also, it might be possible to enhance intracellular DNA nanostructure stability by combining them with metal nanoparticles134,135 or lipids7. Seelig’s group33 recently presented an elegant example showing operation of nucleic acid-based computing in cells by optimizing various factors including composition, chemical modifications and transfection. Despite the progress, there is no simple and generic approach to engineer DNA nanostructures for reliable intracellular applications. The goal remains to design DNA nanostructures with long-term intracellular and in-vivo stability. At this stage, it might be wise to exploit their conditional stability to temporally trigger controlled intracellular release, as exemplified by several elegant designs and applications136–138. Alternatively, DNA/RNA nanostructures could be continually synthesized within the cell49 and reach a stable steady state where the rate of nanostructure repair matches the rate of degradation.
The advantages of DNA nanotechnology are often compromised by the high cost of large-scale DNA nanostructure synthesis, which greatly hampers its practical application and the ability to screen large numbers of potential structures for optimal function. Chip-based de-novo DNA synthesis provides a new avenue to produce large numbers (>20,000)139 of different DNA strands140. Shih and colleagues141 developed a multi-round rolling-circle amplification strategy to selectively amplify sequences from chip-synthesized libraries to produce DNA origami and brick structures. Use of these protocols could enable up to ~100 DNA origami designs to be tested from a single chip, facilitating the screening of nanostructure designs for desired properties, such as cellular uptake, endosomal release and in-vivo stability. Chip-based oligonucleotides can thus be used to identify nanostructures with a desirable combination of properties that can later be scaled. Although there have been several approaches to controllably assemble DNA nanostructures with nearly 100% yield77,142,143, the requirement for hundreds of oligonucleotides to form a single structure makes production beyond the μg-mg scale challenging. Cloneable DNA nanostructures that exploit natural DNA biosynthesis systems in cells are a potential solution to the scale problem43. RNA is also an intriguing alternative for large-scale nucleic acid nanostructure synthesis. RNA nanostructures have the potential to be produced intracellularly through co-transcriptional folding using approaches such as those described by Geary et al.46. In addition, previous studies have shown that non-structured recombinant RNA molecules can be produced at levels as high as 4.5 mg per litre of bacterial culture144, while structured RNAs can be obtained at up to 50 mg per litre of culture145. The bacterium Rhodovulum sulfidophilum — which naturally produces extracellular RNAs — has been harnessed to secrete recombinant RNA aptamers into its growth medium, suggesting potential uses for continuous RNA production in industrial bioreactors146. Hence, with continued improvements, a combination of the above RNA assembly and biosynthesis methods could enable expansion into gram-scale production levels.
Intracellular environments are highly heterogeneous in comparison to the homogenous solutions of test tubes. Because biomolecule motion and distribution are generally restricted in time and space by vesicular membranes and cellular trafficking, DNA nanostructures do not naturally locate themselves in the right place or time. For these reasons, artificial circuits that work well in test tubes may not be effective in live cells. To address this problem, several groups developed modular DNA nanostructures coupled with specific peptide sequences that are widely recognized as signals for transcellular/transmembrane transportation and intracellular localization37,147,148.
For example, cell-penetrating peptides improve the internalization efficiency of DNA origami nanostructures135, and nuclear localization sequences can guide the entry of tetrahedral DNA nanostructures into nuclei37. On a related note, it is challenging to couple peptides to DNA nanostructures with high efficiency, but several non-covalent coupling approaches were recently developed to overcome this problem. The researchers exploited naturally existing specific interactions between nucleic acids and proteins (for example, transcription factors149 and zinc fingers150) to facilitate quantitative coupling. This bioinspired wisdom is expected to endow DNA nanotechnology with desirable targeting abilities in cells and in vivo.
It is probably even more challenging to realize in-vivo biocomputation by rewiring existing signalling networks in a prescribed way at the multi-cellular/organism level. Tamsir et al.151 developed an elegant strategy to implement multi-cell computation by rewiring intercellular communication among cell colonies. Re-engineering the maternal genes on Drosophila has also been realized, which could lead to animal-scale population control123. Given the utility of CRISPR-Cas9 in genetically modifying embryos and organisms with high precision, it has been possible to directly translate designed synthetic networks from the cellular level to the tissue, organ or even organism level with unprecedented power52,152,153.
Despite these advances, the off-target rate and delivery efficiency of CRISPR-Cas9 are still two major concerns60. The use of elaborately designed DNA nanostructures to overcome these problems may open new doors for in-vivo computing in whole organisms154.
In summary, the recent progress in DNA/RNA nanotechnology has provided exciting opportunities to precisely manipulate naturally existing signal pathways and networks. By discovering and adopting new rules of nucleic-acid-based molecular design and programming, we are now in a position to rewire signal pathways in cells and even in vivo. Nevertheless, numerous challenges must be overcome before achieving the ultimate goal of reconstructing modularized, transplantable, and versatile integrated circuitry systems for synthetic biology.
Box 1 |. Glossary.
DNA origami.
A method for fabricating finite DNA nanostructures143. It employs hundreds of short oligonucleotides (staples) to fold a long ssDNA strand (scaffold, often the genomic DNA of M13 bacteriophage), resulting in high yields of predefined shapes or patterns with nanometre precision and addressability.
CRISPR-Cas.
Clustered regularly interspaced short palindromic repeats (CRISPR) are prokaryotic DNA segments spaced by foreign sequences. Their transcripts, known as CRISPR RNAs (crRNAs), can guide CRISPR associated (Cas) endonucleases to recognize and cut foreign DNAs. They together constitute the prokaryotic adaptive immune system known as the CRISPR-Cas system. CRISPR-Cas-based genome editing allows permanent modification of genes in eukaryotic organisms52,163.
Logic unit.
A computational unit that implements a fixed logic function (for example, AND, OR, NOT) with one, two or more binary inputs, and produces a binary output. Assembly of logic units yields digital circuits capable of evaluating complex calculations.
DNA computing.
A type of biocomputation using DNA and DNA-involved reactions. In the pioneering work reported by Adleman et al. in 1994164, a seven-point Hamiltonian path problem was solved by applying operations (including enzymatic ligation, PCR amplification and separation) on DNA strands that encoded input information. The output was visualized with gel electrophoresis.
Digital computing & Analogue computing.
Digital computing is based on logic units that deal with binary values (discrete states). Thus, for continuous signals commonly seen in nature, analogue-to-digital conversion is required. In contrast, analogue computing is based on gates that can directly compute using continuous values, and is thus fast and cost-efficient in solving problems like differential equations. So far, both modes of computing have been implemented using DNA-based models.
FNAs.
A class of nucleic acids that either possess cofactor-dependent catalytic activities toward their substrate (known as DNAzymes/RNAzymes, also termed deoxyribozymes/ribozymes, or catalytic DNAs/RNAs), or show high affinity to certain targets (known as aptamers) from metal ions to small molecules, drugs, proteins, DNA/RNA and even whole cells. Binding of FNAs to their targets often leads to changes in FNA structure, which can be used to generate output signals in DNA/RNA-based circuits. Stefanovic and Willner et al.30,165,166 implemented a series of logic gates using FNAs.
SDR.
The process through which an oligonucleotide can initially bind to a partially double-stranded complex by a single-stranded domain called a toehold, then displace and release the originally bound strand after branch migration occurs. A series of enzyme-free logic gates and related circuits have been developed based on SDR15,24. These gates also use oligonucleotides as inputs and outputs, enabling large scale circuits with thresholding and catalysis within every logical operation to perform digital signal restoration20.
Fluorescent RNAs.
RNA motifs that fluorescence when bound to dye molecules, which enables facile fluorescent tagging of endogenous RNAs through genetic fusion in a way similar to fluorescent proteins. Jaffrey et al. reported the ‘Spinach’ RNA aptamer which binds to the dye DFHBI to emit green fluorescence61.
Riboregulator.
An engineered RNA motif that can regulate the translation or transcription of a gene though a conformational change mediated by other nucleic acids. In a classic strategy developed by Collins et al.67, a motif is inserted into the 5’ untranslated region (UTR) of an mRNA, forming a secondary structure that sequesters the RBS and prevents the ribosome from initiating translation. Binding of a complementary nucleic acid releases the RBS from the repressing secondary structure and allows the ribosome to begin translation.
Acknowledgements
Financial support from the Ministry of Science and Technology of China (2013CB932803, 2013CB933802, 2016YFA0201200, 2016YFA0400900), the National Science Foundation of China (21390414, 21227804, 21329501, U1532119) and the Chinese Academy of Sciences (QYZDJ-SSW-SLH031, KJCX2-EW-N03) are acknowledged. Hao Yan also acknowledges financial support from the US National Science Foundation, the National Institutes of Health, the Army Research Office, the Office of Naval Research and funds from Arizona State University. A.A.G. acknowledges financial support from the US National Science Foundation, the Gates Foundation, the Arizona Biomedical Research Commission, an Alfred P. Sloan Research Fellowship (FG-2017-9108) and Arizona State University.
Footnotes
Competing financial interests
The authors declare no competing financial interests.
References
- 1.Jones MR, Seeman NC & Mirkin CA Programmable materials and the nature of the DNA bond. Science 347, 1260901 (2015). [DOI] [PubMed] [Google Scholar]
- 2.Gerling T, Wagenbauer KF, Neuner AM & Dietz H Dynamic DNA devices and assemblies formed by shape-complementary, non-base pairing 3D components. Science 347, 1446–1452 (2015). [DOI] [PubMed] [Google Scholar]
- 3.Benner SA & Sismour AM Synthetic biology. Nat. Rev. Genet 6, 533–543 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Church GM, Elowitz MB, Smolke CD, Voigt CA & Weiss R Realizing the potential of synthetic biology. Nat. Rev. Mol. Cell Biol 15, 289–294 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wilner OI et al. Enzyme cascades activated on topologically programmed DNA scaffolds. Nat. Nanotechnol 4, 249–254 (2009). [DOI] [PubMed] [Google Scholar]
- 6.Myhrvold C & Silver PA Using synthetic RNAs as scaffolds and regulators. Nat. Struct. Mol. Biol 22, 8–10 (2015). [DOI] [PubMed] [Google Scholar]
- 7.Perrault SD & Shih WM Virus-inspired membrane encapsulation of DNA nanostructures to achieve in vivo stability. ACS Nano 8, 5132–5140 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ye D, Zuo X & Fan C DNA Nanostructure-Based Engineering of the Biosensing Interface for Biomolecular Detection. Prog. Chem 29, 36–46 (2017). [Google Scholar]
- 9.Fu J et al. Multi-enzyme complexes on DNA scaffolds capable of substrate channelling with an artificial swinging arm. Nat. Nanotechnol 9, 531–536 (2014). [DOI] [PubMed] [Google Scholar]
- 10.Fu YM et al. Single-step rapid assembly of DNA origami nanostructures for addressable nanoscale bioreactors. J. Am. Chem. Soc 135, 696–702 (2013). [DOI] [PubMed] [Google Scholar]
- 11.Fu JL, Liu MH, Liu Y, Woodbury NW & Yan H Interenzyme substrate diffusion for an enzyme cascade organized on spatially addressable DNA nanostructures. J. Am. Chem. Soc 134, 5516–5519 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pal S et al. DNA directed self-assembly of anisotropic plasmonic nanostructures. J. Am. Chem. Soc 133, 17606–17609 (2011). [DOI] [PubMed] [Google Scholar]
- 13.Maune HT et al. Self-assembly of carbon nanotubes into two-dimensional geometries using DNA origami templates. Nat. Nanotechnol 5, 61–66 (2010). [DOI] [PubMed] [Google Scholar]
- 14.Zhao Z et al. Nano-caged enzymes with enhanced catalytic activity and increased stability against protease digestion. Nat. Commun 7, 10619 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang DY & Seelig G Dynamic DNA nanotechnology using strand-displacement reactions. Nat. Chem 3, 103–113 (2011). [DOI] [PubMed] [Google Scholar]
- 16.Pei H et al. Reconfigurable three-dimensional DNA nanostructures for the construction of intracellular logic sensors. Angew. Chem. Int. Ed 51, 9020–9024 (2012). [DOI] [PubMed] [Google Scholar]
- 17.Benenson Y Biomolecular computing systems: principles, progress and potential. Nat. Rev. Genet 13, 455–468 (2012). [DOI] [PubMed] [Google Scholar]
- 18.Andersen ES et al. Self-assembly of a nanoscale DNA box with a controllable lid. Nature 459, 73–76 (2009). [DOI] [PubMed] [Google Scholar]
- 19.Douglas SM, Bachelet I & Church GM A logic-gated nanorobot for targeted transport of molecular payloads. Science 335, 831–834 (2012). [DOI] [PubMed] [Google Scholar]
- 20.Qian L & Winfree E Scaling up digital circuit computation with DNA strand displacement cascades. Science 332, 1196–1201 (2011). [DOI] [PubMed] [Google Scholar]
- 21.Mao CD, LaBean TH, Reif JH & Seeman NC Logical computation using algorithmic self-assembly of DNA triple-crossover molecules. Nature 407, 493–496 (2000). [DOI] [PubMed] [Google Scholar]
- 22.Zhang DY & Winfree E Control of DNA strand displacement kinetics using toehold exchange. J. Am. Chem. Soc 131, 17303–17314 (2009). [DOI] [PubMed] [Google Scholar]
- 23.Ge Z et al. Hybridization chain reaction amplification of MicroRNA detection with a tetrahedral DNA nanostructure-based electrochemical biosensor. Anal. Chem 86, 2124–2130 (2014). [DOI] [PubMed] [Google Scholar]
- 24.Seelig G, Soloveichik D, Zhang DY & Winfree E Enzyme-free nucleic acid logic circuits. Science 314, 1585–1588 (2006). [DOI] [PubMed] [Google Scholar]
- 25.Zhou MG, Liang XG, Mochizuki T & Asanuma H A light-driven DNA nanomachine for the efficient photoswitching of RNA digestion. Angew. Chem. Int. Ed 49, 2167–2170 (2010). [DOI] [PubMed] [Google Scholar]
- 26.Lohmann F, Weigandt J, Valero J & Famulok M Logic gating by macrocycle displacement using a double-stranded DNA [3]rotaxane shuttle. Angew. Chem. Int. Ed 53, 10372–10376 (2014). [DOI] [PubMed] [Google Scholar]
- 27.Qian L, Winfree E & Bruck J Neural network computation with DNA strand displacement cascades. Nature 475, 368–372 (2011). [DOI] [PubMed] [Google Scholar]
- 28.Green AA, Silver PA, Collins JJ & Yin P Toehold switches: de-novo-designed regulators of gene expression. Cell 159, 925–939 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Elbaz J et al. DNA computing circuits using libraries of DNAzyme subunits. Nat. Nanotechnol 5, 417–422 (2010). [DOI] [PubMed] [Google Scholar]
- 30.Willner I, Shlyahovsky B, Zayats M & Willner B DNAzymes for sensing, nanobiotechnology and logic gate applications. Chem. Soc. Rev 37, 1153–1165 (2008). [DOI] [PubMed] [Google Scholar]
- 31.Amir Y et al. Universal computing by DNA origami robots in a living animal. Nat. Nanotechnol 9, 353–357 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Soloveichik D, Seelig G & Winfree E DNA as a universal substrate for chemical kinetics. Proc. Natl Acad. Sci. USA 107, 5393–5398 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Groves B et al. Computing in mammalian cells with nucleic acid strand exchange. Nat. Nanotechnol 11, 287–294 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Genot AJ, Bath J & Turberfield AJ Reversible logic circuits made of DNA. J. Am. Chem. Soc 133, 20080–20083 (2011). [DOI] [PubMed] [Google Scholar]
- 35.Li T, Lohmann F & Famulok M Interlocked DNA nanostructures controlled by a reversible logic circuit. Nat. Commun 5, 4940 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Walsh AS, Yin H, Erben CM, Wood MJ & Turberfield AJ DNA cage delivery to mammalian cells. ACS Nano 5, 5427–5432 (2011). [DOI] [PubMed] [Google Scholar]
- 37.Liang L et al. Single-particle tracking and modulation of cell entry pathways of a tetrahedral DNA nanostructure in live cells. Angew. Chem. Int. Ed 53, 7745–7750 (2014). [DOI] [PubMed] [Google Scholar]
- 38.Liu X et al. A DNA nanostructure platform for directed assembly of synthetic vaccines. Nano Lett. 12, 4254–4259 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jiang Q et al. DNA origami as a carrier for circumvention of drug resistance. J. Am. Chem. Soc 134, 13396–13403 (2012). [DOI] [PubMed] [Google Scholar]
- 40.Lee H et al. Molecularly self-assembled nucleic acid nanoparticles for targeted in vivo siRNA delivery. Nat. Nanotechnol 7, 389–393 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li J, Fan C, Pei H, Shi J & Huang Q Smart drug delivery nanocarriers with self-assembled DNA nanostructures. Adv. Mater 25, 4386–4396 (2013). [DOI] [PubMed] [Google Scholar]
- 42.Ohta S, Glancy D & Chan WC DNA-controlled dynamic colloidal nanoparticle systems for mediating cellular interaction. Science 351, 841–845 (2016). [DOI] [PubMed] [Google Scholar]
- 43.Lin C et al. In vivo cloning of artificial DNA nanostructures. Proc. Natl Acad. Sci. USA 105, 17626–17631 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dirks RM & Pierce NA Triggered amplification by hybridization chain reaction. Proc. Natl. Acad. Sci. USA 101, 15275–15278 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Myhrvold C, Dai M, Silver PA & Yin P Isothermal self-assembly of complex DNA structures under diverse and biocompatible conditions. Nano Lett. 13, 4242–4248 (2013). [DOI] [PubMed] [Google Scholar]
- 46.Geary C, Rothemund PWK & Andersen ES A single-stranded architecture for cotranscriptional folding of RNA nanostructures. Science 345, 799–804 (2014). [DOI] [PubMed] [Google Scholar]
- 47.Hao CH et al. Construction of RNA nanocages by re-engineering the packaging RNA of Phi29 bacteriophage. Nat. Commun 5, 3890 (2014). [DOI] [PubMed] [Google Scholar]
- 48.Grabow WW & Jaeger L RNA self-assembly and RNA nanotechnology. Acc. Chem. Res 47, 1871–1880 (2014). [DOI] [PubMed] [Google Scholar]
- 49.Delebecque CJ, Lindner AB, Silver PA & Aldaye FA Organization of intracellular reactions with rationally designed RNA assemblies. Science 333, 470–474 (2011). [DOI] [PubMed] [Google Scholar]
- 50.Elbaz J, Yin P & Voigt CA Genetic encoding of DNA nanostructures and their self-assembly in living bacteria. Nat. Commun 7, 11179 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Praetorius F & Dietz H Self-assembly of genetically encoded DNA–protein hybrid nanoscale shapes. Science 355, 1283 (2017). [DOI] [PubMed] [Google Scholar]
- 52.Gaj T, Gersbach CA & Barbas CF 3rd ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 31, 397–405 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cong L et al. Multiplex Genome Engineering Using CRISPR/Cas Systems. Science 339, 819–823 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mali P et al. RNA-guided human genome engineering via Cas9. Science 339, 823–826 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jinek M et al. RNA-programmed genome editing in human cells. Elife 2, e00471 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kiani S et al. CRISPR transcriptional repression devices and layered circuits in mammalian cells. Nat. Methods 11, 723–726 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tsai SQ et al. Dimeric CRISPR RNA-guided Fokl nucleases for highly specific genome editing. Nat. Biotechnol 32, 569–576 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gilbert LA et al. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell 154, 442–451 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Qi LS et al. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 152, 1173–1183 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wright AV, Nunez JK & Doudna JA Biology and applications of CRISPR systems: harnessing nature’s toolbox for genome engineering. Cell 164, 29–44 (2016). [DOI] [PubMed] [Google Scholar]
- 61.Paige JS, Wu KY & Jaffrey SR RNA mimics of green fluorescent protein. Science 333, 642–646 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Filonov GS, Moon JD, Svensen N & Jaffrey SR Broccoli: rapid selection of an RNA mimic of green fluorescent protein by fluorescence-based selection and directed evolution. J. Am. Chem. Soc 136, 16299–16308 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Paige JS, Nguyen-Duc T, Song W & Jaffrey SR Fluorescence imaging of cellular metabolites with RNA. Science 335, 1194 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tye KM & Deisseroth K Optogenetic investigation of neural circuits underlying brain disease in animal models. Nat. Rev. Neurosci 13, 251–266 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Antaris AL et al. A small-molecule dye for NIR-II imaging. Nat. Mater 15, 235–242 (2016). [DOI] [PubMed] [Google Scholar]
- 66.Rinaudo K et al. A universal RNAi-based logic evaluator that operates in mammalian cells. Nat. Biotechnol 25, 795–801 (2007). [DOI] [PubMed] [Google Scholar]
- 67.Isaacs FJ et al. Engineered riboregulators enable post-transcriptional control of gene expression. Nat. Biotechnol 22, 841–847 (2004). [DOI] [PubMed] [Google Scholar]
- 68.Lucks JB, Qi L, Mutalik VK, Wang D & Arkin AP Versatile RNA-sensing transcriptional regulators for engineering genetic networks. Proc. Natl. Acad. Sci. USA 108, 8617–8622 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chappell J, Takahashi MK & Lucks JB Creating small transcription activating RNAs. Nat. Chem. Biol 11, 214–220 (2015). [DOI] [PubMed] [Google Scholar]
- 70.Mutalik VK, Qi L, Guimaraes JC, Lucks JB & Arkin AP Rationally designed families of orthogonal RNA regulators of translation. Nat. Chem. Biol 8, 447–454 (2012). [DOI] [PubMed] [Google Scholar]
- 71.Takahashi MK & Lucks JB A modular strategy for engineering orthogonal chimeric RNA transcription regulators. Nucleic Acids Res. 41, 7577–7588 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bayer TS & Smolke CD Programmable ligand-controlled riboregulators of eukaryotic gene expression. Nat. Biotechnol 23, 337–343 (2005). [DOI] [PubMed] [Google Scholar]
- 73.Winkler WC, Nahvi A, Roth A, Collins JA & Breaker RR Control of gene expression by a natural metabolite-responsive ribozyme. Nature 428, 281–286 (2004). [DOI] [PubMed] [Google Scholar]
- 74.Isaacs FJ, Dwyer DJ & Collins JJ RNA synthetic biology. Nat. Biotechnol 24, 545–554 (2006). [DOI] [PubMed] [Google Scholar]
- 75.Callura JM, Cantor CR & Collins JJ Genetic switchboard for synthetic biology applications. Proc. Natl. Acad. Sci. USA 109, 5850–5855 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Heath JR, Kuekes PJ, Snider GS & Williams RS A defect-tolerant computer architecture: Opportunities for nanotechnology. Science 280, 1716–1721 (1998). [Google Scholar]
- 77.Goodman RP et al. Rapid chiral assembly of rigid DNA building blocks for molecular nanofabrication. Science 310, 1661–1665 (2005). [DOI] [PubMed] [Google Scholar]
- 78.Fujibayashi K, Zhang DY, Winfree E & Murata S Error suppression mechanisms for DNA tile self-assembly and their simulation. Nat. Comput 8, 589–612 (2008). [Google Scholar]
- 79.Schulman R, Wright C & Winfree E Increasing redundancy exponentially reduces error rates during algorithmic self-assembly. ACS Nano 9, 5760–5771 (2015). [DOI] [PubMed] [Google Scholar]
- 80.Mayer C, McInroy GR, Murat P, Van Delft P & Balasubramanian S An epigenetics-inspired DNA-based data storage system. Angew. Chem. Int. Ed 55, 11144–11148 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Brouns SJJ et al. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science 321, 960–964 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhu GZ et al. Noncanonical self-assembly of multifunctional DNA nanoflowers for biomedical applications. J. Am. Chem. Soc 135, 16438–16445 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hu R et al. DNA nanoflowers for multiplexed cellular imaging and traceable targeted drug delivery. Angew. Chem. Int. Ed 53, 5821–5826 (2014). [DOI] [PubMed] [Google Scholar]
- 84.Modi S et al. A DNA nanomachine that maps spatial and temporal pH changes inside living cells. Nat. Nanotechnol 4, 325–330 (2009). [DOI] [PubMed] [Google Scholar]
- 85.Bhatia D, Surana S, Chakraborty S, Koushika SP & Krishnan Y A synthetic icosahedral DNA-based host-cargo complex for functional in vivo imaging. Nat. Commun 2, 339 (2011). [DOI] [PubMed] [Google Scholar]
- 86.Modi S, Nizak C, Surana S, Halder S & Krishnan Y Two DNA nanomachines map pH changes along intersecting endocytic pathways inside the same cell. Nat. Nanotechnol 8, 459–467 (2013). [DOI] [PubMed] [Google Scholar]
- 87.Chen B et al. Dynamic imaging of genomic loci in living human cells by an optimized CRISPR/Cas system. Cell 155, 1479–1491 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Choi HM et al. Programmable in situ amplification for multiplexed imaging of mRNA expression. Nat. Biotechnol 28, 1208–1212 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Jungmann R et al. Single-molecule kinetics and super-resolution microscopy by fluorescence imaging of transient binding on DNA origami. Nano Lett. 10, 4756–4761 (2010). [DOI] [PubMed] [Google Scholar]
- 90.Jungmann R et al. Multiplexed 3D cellular super-resolution imaging with DNA-PAINT and Exchange-PAINT. Nat. Methods 11, 313–U292 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Liu Y et al. Synthesizing AND gate genetic circuits based on CRISPR-Cas9 for identification of bladder cancer cells. Nat. Commun 5, 5393 (2014). [DOI] [PubMed] [Google Scholar]
- 92.Bindewald E et al. Multistrand structure prediction of nucleic acid assemblies and design of RNA switches. Nano Lett. 16, 1726–1735 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Daniel R, Rubens JR, Sarpeshkar R & Lu TK Synthetic analog computation in living cells. Nature 497, 619–623 (2013). [DOI] [PubMed] [Google Scholar]
- 94.Damle SS & Davidson EH Synthetic in vivo validation of gene network circuitry. Proc. Natl. Acad. Sci. USA 109, 1548–1553 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Auslander S, Auslander D, Muller M, Wieland M & Fussenegger M Programmable single-cell mammalian biocomputers. Nature 487, 123–127 (2012). [DOI] [PubMed] [Google Scholar]
- 96.Nielsen AA et al. Genetic circuit design automation. Science 352, aac7341 (2016). [DOI] [PubMed] [Google Scholar]
- 97.Lipton RJ DNA solution of hard computational problems. Science 268, 542–545 (1995). [DOI] [PubMed] [Google Scholar]
- 98.Ouyang Q, Kaplan PD, Liu S & Libchaber A DNA solution of the maximal clique problem. Science 278, 446–449 (1997). [DOI] [PubMed] [Google Scholar]
- 99.Liu Q et al. DNA computing on surfaces. Nature 403, 175–179 (2000). [DOI] [PubMed] [Google Scholar]
- 100.Sakamoto K et al. Molecular computation by DNA hairpin formation. Science 288, 1223–1226 (2000). [DOI] [PubMed] [Google Scholar]
- 101.Chandran H, Gopalkrishnan N, Phillips A & Reif J In 17th Int. Conf. DNA Comput. Molecular Program (eds Cardelli L & Shih W) 64–83 (Springer, 2011). [Google Scholar]
- 102.Weizmann Y, Elnathan R, Lioubashevski O & Willner I Endonuclease-based logic gates and sensors using magnetic force-amplified readout of DNA scission on cantilevers. J. Am. Chem. Soc 127, 12666–12672 (2005). [DOI] [PubMed] [Google Scholar]
- 103.Bonnet J, Yin P, Ortiz ME, Subsoontorn P & Endy D Amplifying genetic logic gates. Science 340, 599–603 (2013). [DOI] [PubMed] [Google Scholar]
- 104.Siuti P, Yazbek J & Lu TK Synthetic circuits integrating logic and memory in living cells. Nat. Biotechnol 31, 448–452 (2013). [DOI] [PubMed] [Google Scholar]
- 105.Qian L & Winfree E A simple DNA gate motif for synthesizing large-scale circuits. J. R. Soc. Interface 8, 1281–1297 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Genot AJ et al. High-resolution mapping of bifurcations in nonlinear biochemical circuits. Nat. Chem 8, 760–767 (2016). [DOI] [PubMed] [Google Scholar]
- 107.Song TQ, Garg S, Mokhtar R, Bui H & Reif J Analog computation by DNA strand displacement circuits. ACS Synth. Biol 5, 898–912 (2016). [DOI] [PubMed] [Google Scholar]
- 108.Vinkenborg JL, Karnowski N & Famulok M Aptamers for allosteric regulation. Nat. Chem. Biol 7, 519–527 (2011). [DOI] [PubMed] [Google Scholar]
- 109.Pardee K et al. Paper-based synthetic gene networks. Cell 159, 940–954 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Zhang Z et al. A DNA-origami chip platform for label-free SNP genotyping using toehold-mediated strand displacement. Small 6, 1854–1858 (2010). [DOI] [PubMed] [Google Scholar]
- 111.Subramanian HKK, Chakraborty B, Sha R & Seeman NC The label-free unambiguous detection and symbolic display of single nucleotide polymorphisms on DNA origami. Nano Lett. 11, 910–913 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Lin C et al. Submicrometre geometrically encoded fluorescent barcodes self-assembled from DNA. Nat. Chem 4, 832–839 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Goldman N et al. Towards practical, high-capacity, low-maintenance information storage in synthesized DNA. Nature 494, 77–80 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Church GM, Gao Y & Kosuri S Next-generation digital information storage in DNA. Science 337, 1628 (2012). [DOI] [PubMed] [Google Scholar]
- 115.Douglas SM et al. Self-assembly of DNA into nanoscale three-dimensional shapes. Nature 459, 414–418 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Boettiger AN et al. Super-resolution imaging reveals distinct chromatin folding for different epigenetic states. Nature 529, 418–422 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Edwardson TGW, Lau KL, Bousmail D, Serpell CJ & Sleiman HF Transfer of molecular recognition information from DNA nanostructures to gold nanoparticles. Nat. Chem 8, 162–170 (2016). [DOI] [PubMed] [Google Scholar]
- 118.Farzadfard F & Lu TK Genomically encoded analog memory with precise in vivo DNA writing in living cell populations. Science 346, 1256272 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Kim KR et al. Sentinel lymph node imaging by a fluorescently labeled DNA tetrahedron. Biomaterials 34, 5226–5235 (2013). [DOI] [PubMed] [Google Scholar]
- 120.Mirkin CA, Letsinger RL, Mucic RC & Storhoff JJ A DNA-based method for rationally assembling nanoparticles into macroscopic materials. Nature 382, 607–609 (1996). [DOI] [PubMed] [Google Scholar]
- 121.Kim J, Lee J, Hamada S, Murata S & Ha Park S Self-replication of DNA rings. Nat. Nanotechnol 10, 528–533 (2015). [DOI] [PubMed] [Google Scholar]
- 122.Tam DY & Lo PK Multifunctional DNA nanomaterials for biomedical applications. J. Nanomater 2015, 765492 (2015). [Google Scholar]
- 123.Chen CH et al. A synthetic maternal-effect selfish genetic element drives population replacement in Drosophila. Science 316, 597–600 (2007). [DOI] [PubMed] [Google Scholar]
- 124.Kemmer C et al. Self-sufficient control of urate homeostasis in mice by a synthetic circuit. Nat. Biotechnol 28, 355–360 (2010). [DOI] [PubMed] [Google Scholar]
- 125.Ren K et al. A DNA dual lock-and-key strategy for cell-subtype-specific siRNA delivery. Nat. Commun 7, 13580 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Keum JW & Bermudez H Enhanced resistance of DNA nanostructures to enzymatic digestion. Chem. Commun, 7036–7038 (2009). [DOI] [PubMed] [Google Scholar]
- 127.Hamblin GD, Carneiro KMM, Fakhoury JF, Bujold KE & Sleiman HF Rolling circle amplification-templated DNA nanotubes show increased stability and cell penetration ability. J. Am. Chem. Soc 134, 2888–2891 (2012). [DOI] [PubMed] [Google Scholar]
- 128.Mei Q et al. Stability of DNA origami nanoarrays in cell lysate. Nano Lett. 11, 1477–1482 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Li J et al. Self-assembled multivalent DNA nanostructures for noninvasive intracellular delivery of immunostimulatory CpG oligonucleotides. ACS Nano 5, 8783–8789 (2011). [DOI] [PubMed] [Google Scholar]
- 130.Hahn J, Wickham SF, Shih WM & Perrault SD Addressing the instability of DNA nanostructures in tissue culture. ACS Nano 8, 8765–8775 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Hu L, Lu CH & Willner I Switchable catalytic DNA catenanes. Nano Lett. 15, 2099–2103 (2015). [DOI] [PubMed] [Google Scholar]
- 132.Cassinelli V et al. One-step formation of “chain-armor”-stabilized DNA nanostructures. Angew. Chem. Int. Ed 54, 7795–7798 (2015). [DOI] [PubMed] [Google Scholar]
- 133.Brglez J, Nikolov P, Angelin A & Niemeyer CM Designed intercalators for modification of DNA origami surface properties. Chem. Eur. J 21, 9440–9446 (2015). [DOI] [PubMed] [Google Scholar]
- 134.Cutler JI, Auyeung E & Mirkin CA Spherical nucleic acids. J. Am. Chem. Soc 134, 1376–1391 (2012). [DOI] [PubMed] [Google Scholar]
- 135.Yan J et al. Growth and origami folding of DNA on nanoparticles for high-efficiency molecular transport in cellular imaging and drug delivery. Angew. Chem. Int. Ed 54, 2431–2435 (2015). [DOI] [PubMed] [Google Scholar]
- 136.Sun WJ et al. Cocoon-like self-degradable DNA nanoclew for anticancer drug delivery. J. Am. Chem. Soc 136, 14722–14725 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Lu CH & Willner I Stimuli-responsive DNA-functionalized nano-/microcontainers for switchable and controlled release. Angew. Chem. Int. Ed 54, 12212–12235 (2015). [DOI] [PubMed] [Google Scholar]
- 138.Banerjee A et al. Controlled release of encapsulated cargo from a DNA icosahedron using a chemical trigger. Angew. Chem. Int. Ed 52, 6854–6857 (2013). [DOI] [PubMed] [Google Scholar]
- 139.LeProust EM et al. Synthesis of high-quality libraries of long (150mer) oligonucleotides by a novel depurination controlled process. Nucleic Acids Res. 38, 2522–2540 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Kosuri S & Church GM Large-scale de novo DNA synthesis: technologies and applications. Nat. Methods 11, 499–507 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Schmidt TL et al. Scalable amplification of strand subsets from chip-synthesized oligonucleotide libraries. Nat. Commun 6, 8634 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Rothemund PWK Folding DNA to create nanoscale shapes and patterns. Nature 440, 297–302 (2006). [DOI] [PubMed] [Google Scholar]
- 143.Zhao Z, Liu Y & Yan H Organizing DNA origami tiles into larger structures using preformed scaffold frames. Nano Lett. 11, 2997–3002 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Nelissen FH et al. Fast production of homogeneous recombinant RNA--towards large-scale production of RNA. Nucleic Acids Res. 40, e102 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Ponchon L & Dardel F Recombinant RNA technology: the tRNA scaffold. Nat. Methods 4, 571–576 (2007). [DOI] [PubMed] [Google Scholar]
- 146.Suzuki H, Ando T, Umekage S, Tanaka T & Kikuchi Y Extracellular production of an RNA aptamer by ribonuclease-free marine bacteria harboring engineered plasmids: a proposal for industrial RNA drug production. Appl. Environ. Microbiol 76, 786–793 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Chan MS & Lo PK Nanoneedle-assisted delivery of site-selective peptide-functionalized DNA nanocages for targeting mitochondria and nuclei. Small 10, 1255–1260 (2014). [DOI] [PubMed] [Google Scholar]
- 148.Patel PC, Giljohann DA, Seferos DS & Mirkin CA Peptide antisense nanoparticles. Proc. Natl. Acad. Sci. USA 105, 17222–17226 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Crawford R et al. Non-covalent single transcription factor encapsulation inside a DNA cage. Angew. Chem. Int. Ed 52, 2284–2288 (2013). [DOI] [PubMed] [Google Scholar]
- 150.Nakata E et al. Zinc-finger proteins for site-specific protein positioning on DNA-origami structures. Angew. Chem. Int. Ed 51, 2421–2424 (2012). [DOI] [PubMed] [Google Scholar]
- 151.Tamsir A, Tabor JJ & Voigt CA Robust multicellular computing using genetically encoded NOR gates and chemical ‘wires’. Nature 469, 212–215 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Gantz VM & Bier E The mutagenic chain reaction: a method for converting heterozygous to homozygous mutations. Science 348, 442–444 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Niu Y et al. Generation of gene-modified cynomolgus monkey via Cas9/RNA-mediated gene targeting in one-cell embryos. Cell 156, 836–843 (2014). [DOI] [PubMed] [Google Scholar]
- 154.Sun WJ et al. Self-assembled DNA nanoclews for the efficient delivery of CRISPR-Cas9 for genome editing. Angew. Chem. Int. Ed 54, 12029–12033 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Winfree E, Liu F, Wenzler LA & Seeman NC Design and self-assembly of two-dimensional DNA crystals. Nature 394, 539–544 (1998). [DOI] [PubMed] [Google Scholar]
- 156.Yan H, Park SH, Finkelstein G, Reif JH & LaBean TH DNA-templated self-assembly of protein arrays and highly conductive nanowires. Science 301, 1882–1884 (2003). [DOI] [PubMed] [Google Scholar]
- 157.Zheng JP et al. From molecular to macroscopic via the rational design of a self-assembled 3D DNA crystal. Nature 461, 74–77 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.He Y et al. Hierarchical self-assembly of DNA into symmetric supramolecular polyhedra. Nature 452, 198–201 (2008). [DOI] [PubMed] [Google Scholar]
- 159.Ke Y, Ong LL, Shih WM & Yin P Three-dimensional structures self-assembled from DNA bricks. Science 338, 1177–1183 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Zhang F et al. Complex wireframe DNA origami nanostructures with multi-arm junction vertices. Nat. Nanotechnol 10, 779–784 (2015). [DOI] [PubMed] [Google Scholar]
- 161.Han D et al. DNA origami with complex curvatures in three-dimensional space. Science 332, 342–346 (2011). [DOI] [PubMed] [Google Scholar]
- 162.Benson E et al. DNA rendering of polyhedral meshes at the nanoscale. Nature 523, 441–444 (2015). [DOI] [PubMed] [Google Scholar]
- 163.Jinek M et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Adleman LM Molecular computation of solutions to combinatorial problems. Science 266, 1021–1024 (1994). [DOI] [PubMed] [Google Scholar]
- 165.Stojanovic MN et al. Deoxyribozyme-based ligase logic gates and their initial circuits. J. Am. Chem. Soc 127, 6914–6915 (2005). [DOI] [PubMed] [Google Scholar]
- 166.Stojanovic MN, Mitchell TE & Stefanovic D Deoxyribozyme-based logic gates. J. Am. Chem. Soc 124, 3555–3561 (2002). [DOI] [PubMed] [Google Scholar]


