Abstract
It has previously been shown that the E7 protein from the cutaneous human papillomavirus type 1 (HPV1), which is associated with benign skin lesions, binds the product of the tumor suppressor gene retinoblastoma (pRb) with an efficiency similar to that of the E7 protein from the oncogenic HPV type 16. Despite this ability, HPV1 E7 does not display any activity in transforming primary cells. In addition, the two viral proteins differ in their mechanisms of targeting pRb. HPV16 E7 promotes pRb destabilization, while cells expressing HPV1 E7 do not show any decrease in pRb levels. In this study, we show that HPV1 E7, in contrast to HPV16 E7, has only a weak activity to neutralize the effect of cyclin-dependent kinase inhibitor p16INK4a. By generation of HPV1/16 E7 chimeric proteins, we have identified a central motif in the two E7 proteins, which determines their different abilities to overcome the p16INK4a-mediated cell cycle arrest. This motif is located downstream of the pRb-binding domain and comprises only three amino acids in HPV16 E7. Swapping this central motif in the two viral proteins causes an exchange of their activities involved in circumventing the inhibitory function of p16INK4a. Most importantly, our data show that the efficiency of the E7 proteins in neutralizing the inhibitory effect of p16INK4a correlates with their ability to promote pRb degradation.
Human papillomaviruses (HPVs) belong to a large family that comprises over 100 different genotypes (7). Based on their tissue tropism, HPVs are divided into two subgroups, cutaneous and mucosal genotypes, which infect the skin or the mucosae of the nasopharyngeal area, oral cavity, and genital tracts, respectively. All HPV types induce benign proliferative lesions. In addition, certain mucosal HPV types (e.g., HPV type 16 [HPV16]) have the ability to induce transformation of the benign lesion into invasive cancer (28). Thus, the mucosal HPVs are further subdivided into benign and malignant types (also termed “low risk” and “high risk”), according to the nature of the lesion induced. Several independent studies of the high-risk HPV types have demonstrated that the products of two early genes, E6 and E7, are the main transforming proteins of the virus and are directly involved in inducing benign proliferation and malignant transformation of the host cells (for a review, see reference 28). E7 from the mucosal high-risk HPV type 16, in cooperation with an activated cellular oncogene (e.g., ras), has the ability to induce full transformation of rodent and human primary cells, which are tumorigenic when injected into mice (15). In addition, HPV16 E7 together with E6 can efficiently immortalize human primary keratinocytes, the natural host of HPV (15). This is in part explained by the ability of HPV16 E7 to interact with and neutralize the functions of the so-called “pocket proteins,” pRb, p107, and p130 (6, 8, 9, 23). The pocket proteins have a central role in controlling the cell cycle. They negatively regulate, via direct association, the activity of several transcription factors, including members of the E2F family (reviewed in reference 12). In quiescent cells, pRb is hypophosphorylated and associates with E2F. When quiescent cells are exposed to mitogenic signals, the transcription of the genes encoding G1-specific D-type cyclins (D1, D2, and D3) is initiated. Subsequently, D-cyclins associate with and activate the cyclin-dependent kinases 4 and 6 (CDK4 and CDK6, respectively), which in turn phosphorylate pRb in mid-G1 phase, causing release of E2F (22). Ultimately, the free and active E2F promotes the transcription of a group of genes that encode proteins essential for cell cycle progression. Since the activation of CDK4 and CDK6 represents a key event for cell cycle entry, a number of cellular mechanisms regulate the activity of these kinases. In particular, members of a small protein family, p16INK4a, p15INK4b, p18INK4c and p19INK4d, associate with CDK4 and CDK6 and strongly inhibit their kinase activity (22). Ectopic expression of p16INK4a leads to accumulation of hypophosphorylated pRb, sequestration of E2F, and consequent G1 arrest (13). The interaction of HPV16 E7 with pRb, analogous to CDK-mediated phosphorylation, results in release of active E2Fs and stimulation of S-phase entry, even in the absence of active CDK4 and CDK6 complexes (25) and in the presence of high levels of p16INK4a (14). Studies of the low-risk HPV types 6 and 11, which are rarely associated with malignant lesions, have demonstrated that their E7s have reduced efficiency in binding pRb and lack in vitro transforming activity (17, 24). Thus, the potential oncogenicity of the different HPV types in vivo appears to correlate with the in vitro properties of the corresponding E7 protein. However, it has been shown that E7 from the cutaneous HPV1, which is normally associated with benign skin lesions, binds pRb with the same efficiency as E7 from the mucosal oncogenic HPV16 (5, 20). Despite this property, HPV1 E7, in contrast to HPV16 E7, fails to induce transformation of rodent or human primary cells in cooperation with the cellular oncogene ras (5, 20). Thus, the E7 association with pRb appears to be essential, but not sufficient, to induce transformation of the host cells. Together these findings indicate that HPV1 E7, in comparison with HPV16 E7, lacks additional activities that are essential to promote cellular transformation. Besides the ability to interact with pRb, HPV16 E7 induces degradation of the cellular protein (4, 10) via the proteasome pathway (4). Consistent with this property, HPV16 E7 has been found associated with the S4 subunit of the 26S proteasome (3). It has been shown recently that HPV1 E7 is not able to promote pRb degradation (1). Thus, the different mechanisms of HPV1 and -16 E7 proteins for targeting pRb may account for their different in vitro transforming activities and in vivo oncogenicities. However, a direct relationship between E7-induced pRb degradation and alteration of cell cycle regulation has not been demonstrated.
Here we show that the efficiency of HPV1 and -16 E7 proteins in circumventing the p16INK4a-mediated G1arrest is associated with their ability to degrade pRb. In addition, by using HPV1 and -16 E7 chimeric proteins, we have identified the domain that determines the different activities of the two viral proteins in destabilizing pRb.
MATERIALS AND METHODS
Retroviral expression vectors.
The retroviral vectors pBabe-puro and pBabe-neo were previously described (16). The open reading frames of HPV1 and -16 E7 were cloned in frame with the hemagglutinin (HA) tag sequence at the 5′ end. The pBabe-puroP16INK4a construct was kindly provided by Bruno Amati (DNAX Research Institute, Palo Alto, Calif.). The six chimeric HPV1/16 E7 genes and the HPV16 PP-E7 mutant were generated by overlapping PCR. The DNA sequence of the PCR products was verified by the dideoxy chain-termination method. The six chimeric proteins were obtained by fusing the following HPV1 and -16 E7 domains: HPV1 E7 conserved region 1 (CR1) amino acids 1 to 22, HPV1 E7 CR2 amino acids 23 to 32, HPV1 E7 CR3 amino acids 33 to 93, HPV16 E7 CR1 amino acids 1 to 20, HPV16 E7 CR2 amino acids 21 to 29, and HPV16 E7 CR3 amino acids 30 to 98. To avoid PCR artifacts due to the repetitive DNA sequence in the 3′ end of the E7 CR2s, the casein kinase II (CKII) phosphorylation site, which is normally considered to be part of CR2, was included in CR3.
Cell culture and retroviral infections.
NIH 3T3, HaCat, and Bosc23 cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% calf serum (NIH 3T3) or fetal calf serum. High-titer retroviral supernatants (>5 × 106 IU/ml) were generated by transient transfection of Bosc23 cells and used to infect NIH 3T3 cells as described previously (18). After infection, NIH 3T3 cells were selected in 1 mg of G418 per ml for 7 to 8 days and in 2.0 μg of puromycin per ml for 48 h.
Cell extract preparation.
Total cellular extracts were prepared from NIH 3T3 cells (10-cm-diameter plate, 80% confluence) by lysing the cells in 1 ml of lysing buffer (20 mM Tris-HCl [pH 8], 200 mM NaCl, 0.5% Nonidet P-40, 1 mM EDTA, 10 mM NaF, 0.1 mM Na3VO4, 1 mM phenylmethylsulfonyl fluoride, 1 μg of leupeptin per ml, 1 μg of aprotinin per ml) for 20 min at 4°C. After centrifugation (12,000 × g, 5 min) the supernatant was collected, and 100 μg of total extract was precipitated in acetone (9:1 [vol/vol]) for 20 min at −20°C, centrifuged (12,000 × g, 10 min), and resuspended in 20 μl of polyacrylamide gel loading buffer.
Immunoblot analysis and antibodies.
One hundred micrograms of total cell extract was fractionated by electrophoresis on a polyacrylamide gel containing 0.1% sodium dodecyl sulfate (SDS). Proteins were transferred onto a Polyscreen polyvinylidene difluoride (PVDF) membrane (NEN Life Sciences) in a Trans-Blot semidry electrophoretic transfer cell (Bio-Rad) (130 mA, 1 h 30 min), and the immunoblot analysis was performed as described in reference 1. The following antibodies were used: anti-HA epitope (MMS-101R; Babco; dilution of 1:1,000); anti-pRb (14001A; Pharmingen; dilution of 1:1,000); anti-β-tubulin (TUB2.1; Sigma; dilution of 1:1,000), and anti-p16INK4a (kindly provided by Gordon Peters, Imperial Cancer Research Fund, London, United Kingdom; dilution of 1:10).
Determination of the proliferative state of the different cell populations: colony formation.
As cells were split for selection with puromycin after p16INK4a or pBabe-puro infection, they were diluted 20, 200, or 2,000 times and allowed to grow for several days as described in reference 26. After this period, cells were washed in phosphate-buffered saline (PBS), and colonies were fixed and stained on the plates with crystal violet in 20% methanol. In order to determine the colony size, the number of cells in 10 randomly selected colonies was counted.
Time course.
As cells were split for selection, 100,000 cells were seeded in 60-mm-diameter plates and allowed to grow for 48 h. After this period of time, the growth was monitored daily for 3 days by determination of the protein concentration of each culture. Cells were lysed directly in lysis buffer, and the protein content was determined by a bicinchonic acid-based method. Triplicate determinations were performed in each independent experiment.
FACS analysis.
Fluorescence-activated cell sorter (FACS) analysis was performed as follows. After retroviral infection, cells were grown for 48 h, harvested by trypsinization, washed with PBS, and fixed in 80% methanol at −20°C for 30 min. A total of 105 cells were washed in PBS and resuspended in 500 μl of PBS containing RNase A (0.1 mg/ml). After incubation (30 min at 37°C), propidium iodide was added (5 μl of a 50-μg/ml solution). Analysis by flow cytometry was performed with a Becton Dickinson FACSort. Cell cycle profiles were determined by using the Modfit cell cycle analysis software package.
GST pull-down assay.
Glutathione S-transferase (GST) fusion protein synthesis in Escherichia coli BL 21 and protein purification were performed as described in the Pharmacia handbook (Pharmacia, Uppsala, Sweden). HaCat protein extracts were prepared as described previously (1). Approximately 0.6 mg of cell extract was incubated with GST-E7 proteins immobilized on glutathione beads (1 μg). After 1 h at 4°C, the beads were washed five times in a buffer containing 20 mM Tris-HCl (pH 8.0), 200 mM NaCl, 0.5% Nonidet P-40, 1 mM EDTA, and 10 mM NaF. The beads were resuspended in SDS-polyacrylamide gel electrophoresis sample buffer, and the eluted proteins were run on an SDS-polyacrylamide gel.
RESULTS
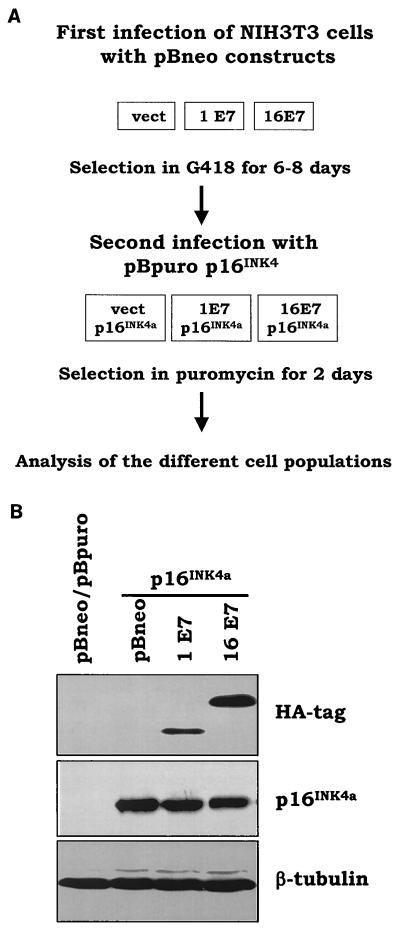
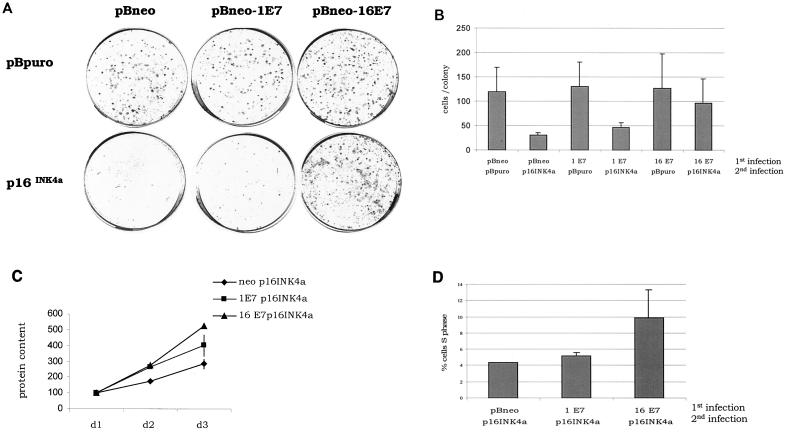
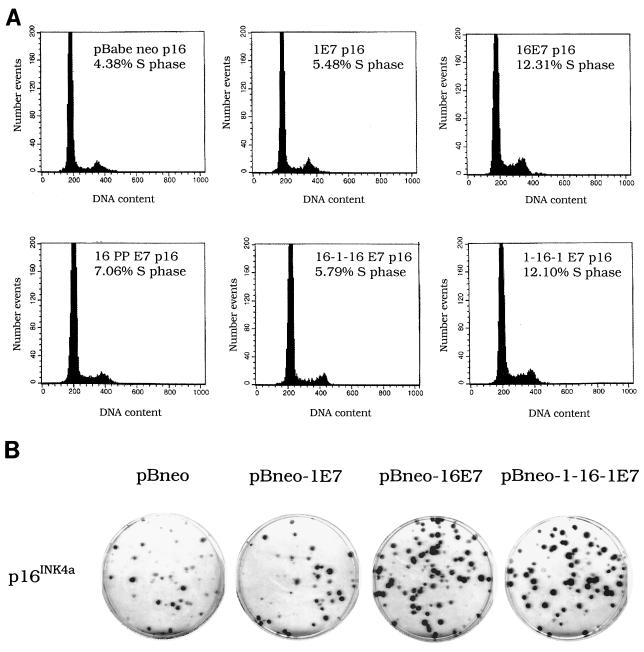
In order to understand the biological significance of E7-induced pRb degradation in deregulation of the G1 checkpoint, we compared the abilities of HPV1 and -16 E7 proteins to overcome the cell cycle block imposed by overexpression of p16INK4a, an inhibitor of specific G1-phase CDKs. We expressed E7 proteins and p16INK4a genes by using retroviral vectors containing the neomycin or puromycin resistance gene (16), respectively, as illustrated in Fig. 1A. Briefly, NIH 3T3 cells were infected with recombinant retrovirus expressing the HPV1 or -16 E7 gene. After neomycin selection, cells were reinfected with recombinant retrovirus expressing the p16INK4a gene and cultured in medium containing puromycin. Immunoblot analysis revealed that both proteins were expressed (Fig. 1B). To compare the level of expression of the viral proteins, the HA tag was fused in frame at the N terminus of both E7s. We have previously shown that the addition of an HA tag to the E7 proteins from HPV1 and -16 does not alter their biological activity (1). Figure 1B shows that HPV1 E7 is expressed at lower levels than HPV16 E7. Similar results were obtained in independent experiments in which different batches of recombinant retroviruses were used (data not shown). As expected, expression of p16INK4a alone resulted in a strong decrease in colony outgrowth (Fig. 2A, plate containing pBabe-neo and pBabe-puro-p16INK4a). HPV16 E7 efficiently overcomes the growth inhibition induced by p16INK4a, whereas HPV1 E7 appears to have no effect on reversion of the p16INK4a activity (Fig. 2A). However, we observed in several experiments that plates of HPV 1E7/p16INK4a-expressing cells consistently had a slightly higher number of colonies than plates of p16INK4a-expressing cells (data not shown). Indeed, by counting the number of cells per colony, we determined that HPV1 E7/p16INK4a-expressing cells form larger colonies than cells expressing p16INK4a alone (Fig. 2B). This observation suggests that HPV1 E7 has weak activity in sustaining proliferation in the presence of p16INK4a. To further confirm our initial results, we determined the proliferation states in all cell populations by alternative methods. The cell growth of the different cultures was monitored for the first 3 days after puromycin selection. Again, we observed that cells expressing HPV16 E7 and p16INK4a grow much faster than cells expressing p16INK4a alone, while HPV1 E7/p16INK4a cells have an intermediate phenotype (Fig. 2C). In addition, flow cytometry analysis revealed that the percentage of S-phase cells is lower in HPV1 E7/p16INK4a cells than in HPV16 E7/p16INK4a cells (Fig. 2D). Together these data show that HPV1 E7 has a much weaker activity than HPV16 E7 in neutralizing the p16INK4a-imposed cell cycle arrest.
FIG. 1.
Expression of HPV1 E7, HPV16 E7, and p16INK4a in NIH 3T3 cells. (A) Schematic representation of the double retroviral infections. The different recombinant retroviruses were generated by transient transfection of the Bosc23 cells as described in Materials and Methods. After the first infection with pBabe-neo (pBneo) retroviruses, NIH 3T3 cells were cultured in G418-containing medium for 6 to 8 days. The cells were then infected with pBabe-puro (pBpuro) retroviruses and cultured in puromycin-selective medium for 2 days. (B) Determination of E7s and p16INK4a expression levels by immunoblotting. One hundred micrograms of protein extracts of cells infected with different recombinant retroviruses as indicated was applied to a 15% polyacrylamide–SDS gel, transferred onto PVDF membrane, and incubated with an anti-HA tag (MMS-101R; Babco), p16INK4a (kindly provided by Gordon Peters, Imperial Cancer Research Fund, London, United Kingdom), or β-tubulin (TUB2.1; Sigma) antibody. β-Tubulin signal was used as a loading control.
FIG. 2.
HPV16 E7, but not HPV1 E7, efficiently neutralizes p16INK4a activity. (A) Colony formation assay. Double retroviral infections were performed as illustrated in Fig. 1. After 6 to 8 days of culture in puromycin-containing medium, colonies were fixed in 20% methanol and stained with crystal violet. (B) The size of colonies was determined by cell counting. The numbers in the figure are the means of 10 randomly selected colonies. (C) Growth time course. The growth of all cell populations was monitored for 3 days as described in Materials and Methods. The growth was assessed by determining the protein concentration at each point of the time course. The data represent the mean of three independent experiments, each performed in triplicate. (D) Percentage of S-phase cells in populations infected with different recombinant retroviruses as indicated. Double-infected cells were harvested after neomycin and puromycin selection. The cell cycle profile was analyzed by flow cytometry. The data represent the mean of two independent experiments.
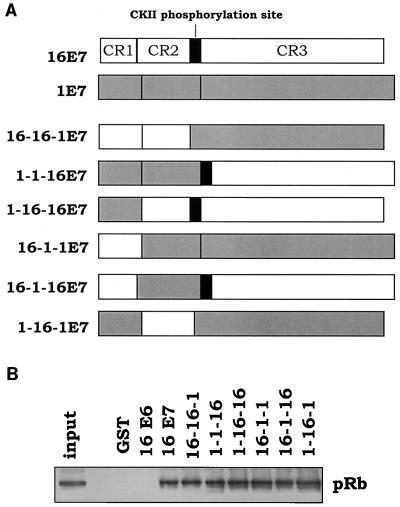
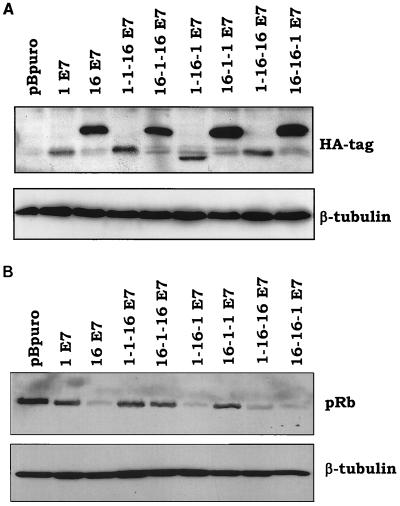
The different abilities of the two E7 molecules to counteract p16INK4a may be due to their different levels of expression or, alternatively, to their intrinsic biological properties, e.g., induction of pRb degradation. To discriminate between the two possibilities, we generated six chimeric E7 proteins, in which the three CRs of the two E7s were fused in all possible combinations (Fig. 3A). First, we determined the ability of the chimeric proteins to associate with pRb. For this purpose, the E7s were expressed in E. coli as GST fusion proteins. After purification, the bacterial recombinant proteins were immobilized on glutathione-Sepharose beads and incubated with human keratinocyte protein extracts to determine their ability to bind pRb. As shown in Fig. 3B, all chimeric E7 proteins associate with pRb with the same efficiency as HPV16 E7. Next, we determined the intracellular stability of the artificial E7 molecules. The E7 proteins were fused at the N terminus with the HA tag and expressed in NIH 3T3 cells by using the retroviral system described above. After retroviral infections, cellular extracts were prepared, and the intracellular levels of the E7 proteins were determined by immunoblot analysis. According to their expression levels and electrophoretic mobility, the six chimeric proteins can be divided into two different groups. Figure 4A shows that 1-1-16, 1-16-1, and 1-16-16 E7 proteins have features similar to those of HPV1 E7. In contrast, the 16-1-16, 16-1-1, and 16-16-1 E7 proteins behave like HPV16 E7. Since the common domain in the three proteins of each group is the CR1 of HPV1 or -16 E7, we reason that the amino acid sequence at the N terminus influences the intracellular level and the electrophoretic mobility of the E7 proteins. The fact that all HPV16 CR1-containing proteins have the same mobility is consistent with the results of a previous study, which showed that the aberrant electrophoretic mobility of HPV16 E7 protein is due to a stretch of negatively charged amino acids located in CR1 (2).
FIG. 3.
Generation of HPV1 and -16 E7 chimeric proteins. (A) Schematic representation of the HPV1/16 E7 chimeric proteins. The gray and white boxes represent the domains of HPV1 and -16 E7, respectively. The E7 proteins were fused at the N terminus with the HA tag. In the generation of the chimeric proteins, the CKII phosporylation site was included in CR3. For more details, see Materials and Methods. (B) HPV16 E7 and chimeric proteins have similar efficiencies in binding pRb in a GST pull-down assay. One microgram of the different GST-E7 proteins was immobilized on glutathione-Sepharose beads and incubated with 0.6 mg of HaCat protein extract. After 1 h, the beads were extensively washed and directly resuspended in SDS-sample buffer. The amount of pRb associated with the different GST-E7 proteins was determined by immunoblot analysis with a specific anti-pRb antibody (14001A; Pharmingen). One-tenth of the total cellular extract (60 μg) used in the GST pull-down assay (input) was applied to a polyacrylamide-SDS gel.
FIG. 4.
A central motif of HPV16 E7 CR2 located downstream of pRb-BD is essential for pRb destabilization. (A) Expression levels of the chimeric proteins. One hundred micrograms of protein extracts of cells expressing the different E7 proteins as indicated was applied to a 15% polyacrylamide–SDS gel, transferred onto a PVDF membrane, and incubated with an anti-HA tag (MMS-101R; Babco) or β-tubulin (TUB2.1; Sigma) antibody. β-Tubulin signal was used as a loading control. (B) pRb levels in cells expressing the different E7 proteins. One hundred micrograms of protein extracts of cells expressing the different E7 proteins as indicated was applied to an 8% polyacrylamide–SDS gel, transferred onto a PVDF membrane, and incubated with a pRb (14001A; Pharmingen) or β-tubulin (TUB2.1) antibody. β-Tubulin signal was used as a loading control.
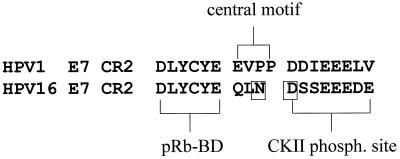
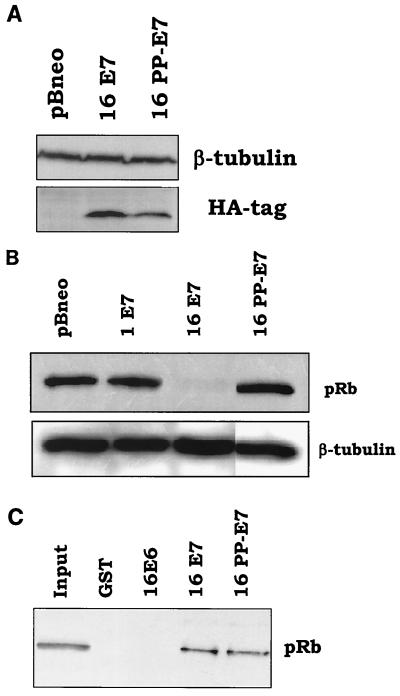
We next characterized the activity of the chimeric proteins to induce pRb degradation. Also in this case, the E7 proteins can be divided into two groups: weak and strong inducers of pRb degradation (Fig. 4B). Interestingly, the ability of the E7 proteins to promote pRb destabilization does not correlate with their expression levels but with the presence in the chimeric proteins of the HPV16 E7 CR2. Cells expressing the 1-16-1, 1-16-16, and 16-16-1 E7 proteins have a low level of pRb. In contrast, the remaining three E7 proteins, 1-1-16, 16-1-16, and 16-1-1, which contain the HPV1 E7 CR2, are less efficient in inducing pRb destabilization (Fig. 4B). Similar results were obtained in independent experiments, in which different batches of recombinant retroviruses were used to infect NIH 3T3 cells (data not shown). The central region of HPV16 E7 comprises two motifs, the pRb-binding domain (pRb-BD) and the CKII phosphorylation site, which are separated by a stretch of three amino acids (central motif) (Fig. 5). Comparison of the HPV1 and -16 E7 amino acid sequences reveals that both proteins contain an identical pRb-BD, but they differ in the downstream amino acid sequence (Fig. 5). Although HPV1 E7 has a stretch of negatively charged amino acids, characteristic of a CKII phosphorylation site, it lacks a serine or threonine, which are the substrates of CKII. In addition, the central motif in CR2 of HPV1 E7 comprises four amino acids, including two prolines, which are not present in HPV16 E7 (Fig. 5). In the chimeric proteins, the CKII phosphorylation site has been included in CR3. The data shown in Fig. 4B indicate that the presence of the CKII phosphorylation site does not influence E7's ability to induce pRb destabilization. Indeed, the 1-1-16 E7 (CKII phosphorylation site positive; Fig. 3A) and the 16-16-1 E7 (CKII phosphorylation site negative; Fig. 3A) are, respectively, weak and strong inducers of pRb degradation. These data are consistent with previous published data, which have shown that HPV16 E7 mutants lacking the CKII phosphorylation site retain most of the activity of the HPV16 E7 wild type to destabilize pRb (1, 11). Thus, it is likely that the difference in the central motif located immediately after pRb-BD (Fig. 5) is responsible for the different ability of the two E7 proteins to induce pRb degradation. To evaluate this possibility, we substituted the asparagine and aspartic acid at positions 29 and 30 in HPV16 for the two prolines of HPV1 E7 (Fig. 5). Immunoblot analysis revealed that the mutant HPV16 PP-E7 is expressed in NIH 3T3 cells at levels similar to those of wild-type HPV16 E7 (Fig. 6A). However, it is not able to promote pRb degradation (Fig. 6B), despite the fact that it binds the pocket protein with the same efficiency as the wild-type HPV16 E7 in a GST pull-down assay (Fig. 6C). Together these data demonstrate that the amino acid motif between the pRb-BD and the CKII phosphorylation site (amino acids QLN at positions 27 to 29 in HPV16 E7) has an essential role in E7-mediated pRb destabilization.
FIG. 5.
Schematic representation of HPV1 and -16 E7 CR2. The two amino acids (at positions 29 and 30) in the central motif of HPV16 CR2, which have been substituted for with two prolines, are boxed. The positions of the pRb-BD and CKII phosphorylation site (phosph.) are also indicated.
FIG. 6.
Characterization of the properties of the HPV16 PP-E7 mutant. (A) HPV16 PP-E7 is expressed in NIH 3T3 cells at levels similar to those of the HPV16 E7 wild type. One hundred micrograms of protein extracts of cells infected with different recombinant retroviruses as indicated in the figure was applied to a 15% polyacrylamide–SDS gel, transferred onto a PVDF membrane, and incubated with an anti-HA tag (MMS-101R; Babco) or β-tubulin (TUB2.1; Sigma) antibody. β-Tubulin signal was used as a loading control. (B) HPV16 PP-E7 does not promote pRb degradation. One hundred micrograms of protein extracts of cells infected with different recombinant retroviruses as indicated in the figure was applied to an 8% polyacrylamide–SDS gel, transferred onto a PVDF membrane, and incubated with an anti-pRb (14001A; Pharmingen) or anti-β-tubulin (TUB2.1) antibody. β-Tubulin signal was used as a loading control. (C) HPV16 PP-E7 and the HPV16 E7 wild type have similar efficiencies in binding pRb in a GST pull-down assay. One microgram of the different GST/E7 proteins was immobilized on glutathione-Sepharose beads and incubated with 0.6 mg of HaCat protein extract. The GST pull-down assay was performed as described in the legend to Fig. 3. One-tenth of the total cellular extract (60 μg) used in the GST pull-down assay (input) was applied to a polyacrylamide-SDS gel.
Next, we determined whether the HPV16 E7 activity in overcoming the p16INK4a-imposed G1 block is associated with its pRb degradation activity. For this purpose, the p16INK4a neutralization efficiencies of two inducers (HPV16 E7 wild type and 1-16-1 E7) and two noninducers (HPV16 PP-E7 mutant and 16-1-16 E7) of pRb degradation were directly compared. Table 1 and Fig. 7A show that only the two inducers of pRb degradation are able to efficiently neutralize the inhibitory function of p16INK4a. To further confirm that 1-16-1 E7, which contains the central motif of HPV16 E7 (QLN), is able to overcome the p16INK4a-induced G1 arrest, we performed the colony formation assay as described above. Figure 7B shows that, also in this assay, HPV16 E7 and 1-16-1 E7 have similar efficiencies in driving G1-arrested cells into the cell cycle. Thus, the two events, pRb degradation and efficient circumvention of p16INK4a cell cycle inhibition, are tightly linked.
TABLE 1.
Percentage of S-phase NIH 3T3 cells expressing p16INK4a alone or in combination with E7 proteinsa
| Expt | pBabe-puro p16INK4a
|
|||||
|---|---|---|---|---|---|---|
| pBabe-neo | HPV1 E7 | HPV16 E7 | HPV16 PP-E7 | HPV16-1-16 E7 | HPV1-16-1 E7 | |
| 1 | 4.38 | 5.48 | 12.31 | 7.06 | 5.79 | 12.10 |
| 2 | 7.63 | 8.50 | 13.10 | 8.90 | 8.80 | 11.80 |
| 3 | 7.21 | 7.10 | 12.20 | NDb | ND | 11.00 |
For flow cytometry, the different cell populations were prepared as described in the legend to Fig. 7.
ND, not determined.
FIG. 7.
Efficiency of the E7 proteins in overcoming p16INK4a-induced arrest correlates with their ability to promote pRb degradation. (A) Cell cycle profile. Cells infected with different recombinant retroviruses as indicated in the figure were harvested after the neomycin and puromycin selection. The cell cycle profile was analyzed by flow cytometry. (B) Colony formation assay. Cells were infected with two recombinant retroviruses as indicated in the figure. After 14 days of culture in puromycin-containing medium, colonies were fixed in 20% methanol and stained with crystal violet. pBneo, pBabe-neo.
DISCUSSION
E7 is one of the major transforming proteins of HPV (28). The neutralization of pocket protein functions by E7 and consequent activation of E2F-dependent transcription appear to play a key role in cell cycle deregulation (25). Indeed, mutations in pRb-BD LXCXE (amino acids 22 to 26 in HPV16 E7) lead to a loss of its ability to associate with pRb and to induce cellular transformation (19). Furthermore, E7 proteins from the low-risk HPV types weakly associate with pocket proteins and do not display any in vitro transforming activity (17, 24). However, studies of the cutaneous HPV1 E7 suggest that, in addition to the E7-pRb interaction, other E7-mediated events contribute to promote cellular transformation (5, 20). HPV1 E7 binds pRb with an affinity similar to that of HPV16 E7, but it does not cooperate with ras to induce transformation of rodent or human primary cells (5, 20). It has been shown that HPV16 E7 has the additional property of inducing pRb degradation (4, 10) via the proteasome pathway (4), while HPV1 E7 lacks this activity (1). In this study, we show that HPV1 E7 is less efficient than HPV16 E7 in overcoming G1 cell cycle arrest imposed by overexpression of the CDK inhibitor p16INK4a. We have generated HPV1/16 E7 chimeric proteins and identified a domain in HPV16 E7 that is essential to promote pRb degradation. This motif comprises three amino acids (QLN) and is located immediately after pRb-BD (Fig. 5). Exchanging these central motifs in HPV1 and -16 E7 results in an inversion of their activity in inducing pRb degradation and in efficiently overcoming the p16INK4a-mediated G1 block. The most significant difference between the central motifs of the two viral proteins is the presence of two proline residues in HPV1 E7. Introduction of two prolines in the central motif of HPV16 E7 CR2 results in a loss of ability to promote pRb destabilization and to efficiently stimulate cell cycle progression. It is possible that the introduction of two amino acids with a rigid structure, such as proline, into the HPV16 E7 protein induces a conformational change, which results in loss of some of the viral protein properties without affecting its ability to bind pRb. Since published data (1, 11) and the findings of this study show that the integrity of the CKII phosphorylation site is not required for efficient degradation of pRb, it is unlikely that the substitution of the aspartic acid at position 30 in the HPV16 E7 CKII phosphorylation site (Fig. 5) influences E7's ability to destabilize pRb.
It has previously been reported that HPV 16 E7 CR1 contributes to promote pRb degradation (1, 11). In this study, we show that the HPV1 E7 CR1 is functionally related to CR1 of HPV16 E7. Indeed, all chimeric proteins containing HPV1 E7 CR1 fused to HPV16 CR2 are able to induce pRb degradation. These data are consistent with the level of amino acid similarity between the two CR1s, which is approximately 55%.
Berezutskaya and Bagchi have reported that HPV16 E7 associates with the subunit S4 of the 26S proteasome with consequent stimulation of its ATPase activity (3). This interaction requires the integrity of the HPV16 E7 CR3 and is independent of the pRb-BD. Based on these data, the authors suggested that HPV16 E7 induces pRb degradation by targeting the pocket protein to the proteasomes. Thus, it would be interesting to determine whether the efficiency of the E7 proteins included in this study in inducing pRb degradation and overcoming p16INK4a-mediated G1 arrest correlates with their ability to associate with the S4 protein. Despite the fact that CR3 is the least conserved domain in the E7 proteins from different HPV types, HPV1 and -16 E7 have 44% identity in the last 50 amino acids. Therefore, it is likely that HPV1 E7, like HPV16 E7, is able to interact with the S4 protein, but the QLN domain of HPV16 E7 is required to promote pRb degradation.
The low activity of HPV1 E7 in neutralizing the p16INK4a inhibitory function is consistent with previous published data, which show that HPV1 E7 is not able to efficiently transform primary rodent or human cells in cooperation with the activated ras oncogene (5, 20). It has been shown that overexpression of ras alone leads to an accumulation of p16INK4a and consequent G1 cell cycle arrest (21). Thus, HPV16 E7, but not HPV1 E7, cooperates with ras in the induction of cellular transformation being able to efficiently neutralize p16INK4a inhibitory function. Together these findings provide a possible explanation for the nononcogenicity of HPV1 in vivo.
Our study demonstrates an association between the HPV16 E7-mediated events, pRb degradation, and abrogation of p16INK4a-induced cell cycle arrest. However, it does not address why pRb degradation is required for efficient stimulation of the cell cycle. Two different explanations can be envisaged. One possibility is that pRb degradation represents a more efficient way to neutralize pRb function. In this model, E7, after induction of pRb degradation, can be recycled to target a new pRb molecule. Thus, HPV16 E7 can efficiently antagonize pRb, even if present in the cell at a much lower level than the pocket protein. Alternatively, the E7-mediated pRb degradation may be required in order to release other pRb-associated factors, which are not displaced by E7 binding. For instance, it has been reported that the c-Abl tyrosine kinase interacts with the C-terminal domain of pRb. HPV16 E7 binding to the pocket domain of pRb does not lead to a disruption of the pRb–c-Abl complex (27). In this scenario, we could imagine that the cellular proteins released upon pRb degradation contribute to efficient deregulation of G1 checkpoints.
In conclusion, in this study, we have shown that E7-induced pRb degradation plays a key role in the neutralization of p16INK4a function and have identified an HPV16 E7 domain essential for this activity.
ACKNOWLEDGMENTS
We are grateful to Harald zur Hausen and Lutz Gissmann for continuous support and interest in our work. We also thank all of the members of our laboratory for their cooperation, Konstantinos Alevizopoulos and Bruno Amati for the pBabepuro-p16INK4a construct and technical advice, Gordon Peters for the p16INK4a antibody, and Leonie Ringrose for critical reading of the manuscript.
S.C. was supported by a PRAXIS XXI doctoral fellowship (Sub Programa Ciencia e Tecnologia do 2° Quadro Comunitario de Apoio).
REFERENCES
- 1.Alunni-Fabbroni M, Littlewood T, Deleu L, Caldeira S, Giarre M, Dell' Orco M, Tommasino M. Induction of S phase and apoptosis by the human papillomavirus type 16 E7 protein are separable events in immortalized rodent fibroblasts. Oncogene. 2000;19:2277–2285. doi: 10.1038/sj.onc.1203570. [DOI] [PubMed] [Google Scholar]
- 2.Armstrong D J, Roman A. The anomalous electrophoretic behavior of the human papillomavirus type 16 E7 protein is due to the high content of acidic amino acid residues. Biochem Biophys Res Commun. 1993;192:1380–1387. doi: 10.1006/bbrc.1993.1569. [DOI] [PubMed] [Google Scholar]
- 3.Berezutskaya E, Bagchi S. The human papillomavirus E7 oncoprotein functionally interacts with the S4 subunit of the 26 S proteasome. J Biol Chem. 1997;272:30135–30140. doi: 10.1074/jbc.272.48.30135. [DOI] [PubMed] [Google Scholar]
- 4.Boyer S N, Wazer D E, Band V. E7 protein of human papilloma virus-16 induces degradation of retinoblastoma protein through the ubiquitin-proteasome pathway. Cancer Res. 1996;56:4620–4624. [PubMed] [Google Scholar]
- 5.Ciccolini F, Di Pasquale G, Carlotti F, Crawford L, Tommasino M. Functional studies of E7 proteins from different HPV types. Oncogene. 1994;9:2342–2348. [PubMed] [Google Scholar]
- 6.Davies R, Hicks R, Crook T, Morris J, Vousden K. Human papillomavirus type 16 E7 associates with a histone H1 kinase and with p107 through sequences necessary for transformation. J Virol. 1993;67:2521–2528. doi: 10.1128/jvi.67.5.2521-2528.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Villiers E M. Papillomavirus and HPV typing. Clin Dermatol. 1997;15:199–206. doi: 10.1016/s0738-081x(96)00164-2. [DOI] [PubMed] [Google Scholar]
- 8.Dyson N, Howley P M, Munger K, Harlow E. The human papilloma virus-16 E7 oncoprotein is able to bind to the retinoblastoma gene product. Science. 1989;243:934–937. doi: 10.1126/science.2537532. [DOI] [PubMed] [Google Scholar]
- 9.Hu T H, Ferril S C, Snider A M, Barbosa M S. In vivo analysis of HPV E7 protein association with pRb, p107 and p130. Int J Oncol. 1995;6:167–174. doi: 10.3892/ijo.6.1.167. [DOI] [PubMed] [Google Scholar]
- 10.Jones D L, Münger K. Analysis of the p53-mediated G1 growth arrest pathway in cells expressing the human papillomavirus type 16 E7 oncoprotein. J Virol. 1997;71:2905–2912. doi: 10.1128/jvi.71.4.2905-2912.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jones D L, Thompson D A, Munger K. Destabilization of the RB tumor suppressor protein and stabilization of p53 contribute to HPV type 16 E7-induced apoptosis. Virology. 1997;239:97–107. doi: 10.1006/viro.1997.8851. [DOI] [PubMed] [Google Scholar]
- 12.Kouzarides T. Transcriptional control by the retinoblastoma protein. Semin Cancer Biol. 1995;6:91–98. doi: 10.1006/scbi.1995.0012. [DOI] [PubMed] [Google Scholar]
- 13.Lukas J, Parry D, Aagaard L, Mann D J, Bartkova J, Strauss M, Peters G, Bartek J. Retinoblastoma-protein-dependent cell-cycle inhibition by the tumour suppressor p16. Nature. 1995;375:503–506. doi: 10.1038/375503a0. [DOI] [PubMed] [Google Scholar]
- 14.Mann D J, Jones N C. E2F-1 but not E2F-4 can overcome p16-induced G1 cell-cycle arrest. Curr Biol. 1996;6:474–483. doi: 10.1016/s0960-9822(02)00515-8. [DOI] [PubMed] [Google Scholar]
- 15.Mansur C P, Androphy E J. Cellular transformation by papillomavirus oncoproteins. Biochim Biophys Acta. 1993;1155:323–345. doi: 10.1016/0304-419x(93)90013-3. [DOI] [PubMed] [Google Scholar]
- 16.Morgenstern J P, Land H. Advanced mammalian gene transfer: high titre retroviral vectors with multiple drug selection markers and a complementary helper-free packaging cell line. Nucleic Acids Res. 1990;18:3587–3596. doi: 10.1093/nar/18.12.3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Munger K, Werness B A, Dyson N, Phelps W C, Harlow E, Howley P M. Complex formation of human papillomavirus E7 proteins with the retinoblastoma tumor suppressor gene product. EMBO J. 1989;8:4099–4105. doi: 10.1002/j.1460-2075.1989.tb08594.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pear W S, Nolan G P, Scott M L, Baltimore D. Production of high-titer helper-free retroviruses by transient transfection. Proc Natl Acad Sci USA. 1993;90:8392–8396. doi: 10.1073/pnas.90.18.8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Phelps W C, Münger K, Yee C L, Barnes J A, Howley P M. Structure-function analysis of the human papillomavirus type 16 E7 oncoprotein. J Virol. 1992;66:2418–2427. doi: 10.1128/jvi.66.4.2418-2427.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schmitt A, Harry J B, Rapp B, Wettstein F O, Iftner T. Comparison of the properties of the E6 and E7 genes of low- and high-risk cutaneous papillomaviruses reveals strongly transforming and high Rb-binding activity for the E7 protein of the low-risk human papillomavirus type 1. J Virol. 1994;68:7051–7059. doi: 10.1128/jvi.68.11.7051-7059.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Serrano M, Lin A W, McCurrach M E, Beach D, Lowe S W. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell. 1997;88:593–602. doi: 10.1016/s0092-8674(00)81902-9. [DOI] [PubMed] [Google Scholar]
- 22.Sherr C J, Roberts J M. CDK inhibitors: positive and negative regulators of G1-phase progression. Genes Dev. 1999;13:1501–1512. doi: 10.1101/gad.13.12.1501. [DOI] [PubMed] [Google Scholar]
- 23.Smith-McCune K, Kalman D, Robbins C, Shivakumar S, Yuschenkoff L, Bishop J M. Intranuclear localization of human papillomavirus 16 E7 during transformation and preferential binding of E7 to the Rb family member p130. Proc Natl Acad Sci USA. 1999;96:6999–7004. doi: 10.1073/pnas.96.12.6999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Storey A, Pim D, Murray A, Osborn K, Banks L, Crawford L. Comparison of the in vitro transforming activities of human papillomavirus types. EMBO J. 1988;7:1815–1820. doi: 10.1002/j.1460-2075.1988.tb03013.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tommasino M, Crawford L. Human papillomavirus E6 and E7: proteins which deregulate the cell cycle. Bioessays. 1995;17:509–518. doi: 10.1002/bies.950170607. [DOI] [PubMed] [Google Scholar]
- 26.Vlach J, Hennecke S, Alevizopoulos K, Conti D, Amati B. Growth arrest by the cyclin-dependent kinase inhibitor p27Kip1 is abrogated by c-Myc. EMBO J. 1996;15:6595–6604. [PMC free article] [PubMed] [Google Scholar]
- 27.Welch P J, Wang J Y J. A C-terminal protein-binding domain in the retinoblastoma protein regulates nuclear c-Abl tyrosine kinase in the cell cycle. Cell. 1993;75:779–790. doi: 10.1016/0092-8674(93)90497-e. [DOI] [PubMed] [Google Scholar]
- 28.zur Hausen H. Papillomaviruses causing cancer: evasion from host-cell control in early events in carcinogenesis. J Natl Cancer Inst. 2000;92:690–698. doi: 10.1093/jnci/92.9.690. [DOI] [PubMed] [Google Scholar]