Abstract
Infectious fungi have been an increasing global concern in the present era. A promising approach to tackle this pressing concern involves utilizing Antifungal peptides (AFP) to develop an antifungal drug that can selectively eliminate fungal pathogens from a host with minimal toxicity to the host. Accordingly, identifying precise therapeutic antifungal peptides is crucial for developing effective drugs and treatments. This study proposed MLAFP-XN, a neural network-based strategy for accurately detecting active AFP in sequencing data to achieve this objective. In this work, eight feature extraction techniques and the XGB feature selection strategy are utilized together to present an enhanced methodology. A total of 24 classification models were evaluated, and the most effective four have been selected. Each of these models demonstrated superior accuracy on independent test sets, with respective scores of 97.93 %, 99.47 %, and 99.48 %. Our model outperforms current state of the art methods. In addition, we created a companion website to demonstrate our AFP recognition process and use SHAP to identify the most influential properties.
Keywords: Antifungal peptide, Neural network, Antifungal drug, Feature extraction, Feature selection, Drug discovery
1. Introduction
Fungal infection is a common disease that affects the body. This infection is spreading rapidly, and it is noted that billions of people are injured by a fungal infection [[1], [2], [3]]. The conventional therapies and treatment for fungal infection typically result in genotoxicity, skin problems, and nutrient deficiency [4,5]. Alternatively, researchers have discovered that novel antifungal drugs are an effective way to combat these significant problems with current efforts focused on evaluating peptide-based therapeutic strategies, which are more advantageous and patient-friendly than the traditional approaches [6,7]. Antifungal peptides (AFP) are natural molecules generated by microorganisms to protect themselves from fungi. They represent promising options for the treatment of fungal infections due to their effectiveness and preference. AFPs are classified into three types according to their mode of action: compounds that communicate with membranes, molecules that target the cell wall, nucleotide-inhibiting agents, and additional sequences with distinctive or unexplored modes of function. These peptides are involved in the innate immune system of many species, including humans, plants, and animals. They are crucial in the defense against fungal infections. Classifying many AFP candidates are usually a time-consuming and resource-demanding operation. As a result, suitable computational models are crucial to estimate AFPs rapidly and effectively [8].
In recent decades, most studies have discovered AFP using various machine-learning approaches. In 2018, Agrawal et al. proposed a support vector machine (SVM) method to detect the AFP with three feature extraction methods. While developing their proposed method, they did not accomplish a higher performance for accurate prediction [9]. In 2019, Fang et al. integrated the CNN-LSTM hybrid method with character feature embedding for detecting AFP [10]. In 2021, Ahmad et al. introduced the Deep-AntiFP model based on a neural network to predict the AFP. The sigmoid function is usually preferable for identifying binary classifications, but the authors used the SoftMax function [11]. In 2022, Ahmad et al. proposed an iAFPs-EnC-GA model based on ensemble methods with sequential and evolutionary information, minimum redundancy, and maximum relevance extraction approach. However, these manual feature properties involved dimensionality and redundancy challenges, leading to increased computational complexity [12]. In 2022, in another study, Sharma et al. proposed the DeepAFPpred model based on transfer learning and the 1D-CNN-BiLSTM method to detect the AFP classes. However, these methods use sequence-based features and do not have the peptide sequences' evolutionary information and physicochemical properties [13]. In 2023, Fang et al. developed the AFP-MFL framework with a multi-view features method to predict the AFP. The authors used the co-attention technique and SoftMax functions, where the sigmoid function is more appropriate for detecting binary-based classification [8]. In the same year, 2023, Yao et al. used CNN-BiLSTM architecture to predict AFP with three feature extraction methods. However, they used redundant data. As a result, their outcome showed a better performance than the other models [14]. Additionally, Asad et al. proposed the Antifp SRF model based on the random forest (RF) classifier method, where the authors obtained a better accuracy [15]. In another study, Fang et al. conducted a review of diagnostic tools for fungal infections [16]. Hassan et al. proposed a deep learning-based approach for the detection of AFP [17]. They employed a transfer learning approach. However, they did not mention any information how the applied transfer learning method was adapted to the target field. In this case, domain adaptation to the target field is not clarified and further development of the model for other dataset may face some domain adaptation issues.
Given the above limitations, there is still room for improvement to detect AFP more precisely. This study proposed a more robust and efficient model compared to the existing models. These desirable properties should be conducive to the timely and advantageous development of the proposed model. The high-level steps leading up to the MLAFP-XN model can be briefly summarized as follows:
-
1.
We first applied eight feature extraction methods with a larger dataset of 8743 sequences. To minimize the redundancy, we reduced the duplication of the datasets.
-
2.
We applied an extreme gradient boosting (XGB) feature selection process where we extracted 500 top-importance features.
-
3.
We applied a neural networks-based MLAFP-XN model, which enables the successful classification of AFP.
-
4.
We built a web server based on our proposed model for demonstration purposes. The link to the demo web server is accessible at https://immediate-rash-hire.anvil.app/.
2. Materials and methods
2.1. Datasets
To build a machine learning model, a relevant and high-quality dataset is a very important component. Consequently, we searched for publicly available datasets and found three available datasets that are relevant in the literature. So, to conduct this study, we curated all three benchmark datasets: Antifp_Main, Antifp_DS1, and Antifp_DS2 from the literature search [9]. Antifp_Main and Antifp_DS2 contain 1459 AFP and 1457 non-AFP samples, and Antifp_DS1 includes 1459 AFP and 1459 non-AFP samples. The three datasets are obtained from the Data Repository of Antimicrobial Peptides (DRAMP) because antimicrobials have various properties: anticancer, antiviral, antibacterial, and antifungal among others [18]. According to the study, the positive samples in the training datasets are the same in each dataset [9]. In particular, the negative samples originate from Swiss-prot databases [19]. The training datasets have no duplication data, but the testing sets have 7 sequences that are duplicated. Thus, to handle the redundancy of the datasets, we reduced the duplicate samples from the datasets and obtained 8743 samples in total. Table 1 shows the summary of the datasets of our study.
Table 1.
Different datasets overview of the study.
| Mode | Datasets | Train-Test | AFP | Non-AFP | Total |
|---|---|---|---|---|---|
| Original sets | Antifp_Main | Train | 1168 | 1167 | 2916 |
| Test | 291 | 290 | |||
| Antifp_DS2 | Train | 1168 | 1166 | 2916 | |
| Test | 291 | 291 | |||
| Antifp_DS1 | Train | 1168 | 1168 | 2918 | |
| Test | 291 | 291 | |||
| Pre-processed sets | Antifp_Main | Train | 1168 | 1167 | 2914 |
| Test | 291 | 288 | |||
| Antifp_DS2 | Train | 1168 | 1168 | 2911 | |
| Test | 291 | 286 | |||
| Antifp_DS1 | Train | 1168 | 1168 | 2918 | |
| Test | 291 | 291 |
2.2. Overall framework of MLAFP-XN
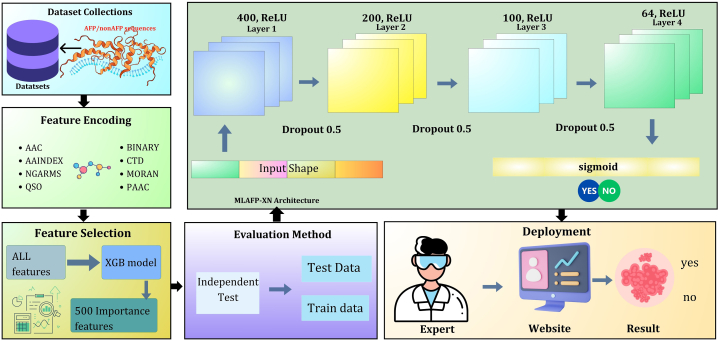
From the collected dataset, we leveraged several machine learning methods to develop the MLAFP-XN model to identify the AFP accurately. Fig. 1 shows the overall workflow of the analysis. To reduce the feature dimensions, we implemented the XGB method and selected the 500 features based on the feature importance of the XGB outcomes. Afterwards, we applied several ensemble learning models such as a light gradient boosting machine (LGBM), extra tree classifier (ETC), category boosting classifier (Catboost), and neural network approach called (MLAFP-XN). According to the evaluation metrics, the neural network-based approach performed better than the other models, and this model obtained a higher accuracy, sensitivity, and specificity than the other existing AFP predictor tools. Finally, we built a web server based on our proposed model for practical hands-on demonstration.
Fig. 1.
The overall workflow of the study: Data collection, feature encoding, feature selection process, applied evaluation method, development of the MLAFP-XN model, and deployment based on the proposed model.
Our dataset's size and internal heterogeneity led us to use an 80 % training and 20 % testing split. To achieve a balance between training and validation, machine learning frequently uses this ratio. The model can acquire knowledge from a significant amount of the information, recognizing a range of patterns and subtleties, with 80 % of the data set aside for training. A trustworthy estimate of the model's performance on a substantial amount of unobserved data is provided by the remaining 20 %, which is set aside for testing. To assess the generalization capabilities of the model, this technique keeps a representative test set and guarantees that the training set is sufficiently large to generate a robust model.
Eight feature extraction techniques were chosen because they have a track record of successfully collecting a variety of peptide sequence physicochemical, structural, and compositional properties: AAC, AAIndex, Ngrams, Binary, QSO, CTD, Moran, and PAAC. The combination of these techniques yields a thorough quantification of the peptides, which is essential for precisely determining their antifungal characteristics. By methodically removing unnecessary and uninformative characteristics and concentrating on the most pertinent ones, the XGB feature selection strategy helped to further improve computing efficiency. In addition to lowering the dataset's dimensionality, this procedure enhanced the performance and speed of the following classification models, guaranteeing reliable and precise predictions.
2.3. Feature extraction
Feature extraction is a highly significant process in the bioinformatics field. This study employed eight distinct feature extraction techniques, encompassing compositional, binary, autocorrelation, and physicochemical-based feature groups. Compositional-based features such as amino acid composition (AAC), pseudo amino acid composition (PAAC), n-gram composition (Ngrams), and conjoint triad descriptors (CTD). The binary-based feature is a binary profile pattern (Binary), the autocorrelation-based feature is the moran-autocorrelation (Moran), and physicochemical-based features are quasi-sequence-order (QSO) and amino acid index (AAIndex).
2.3.1. Amino acid composition (AAC)
AAC is one of the most popular feature extraction methods, where this feature brings protein information of each amino acid and delivers 20 nt features [20,21]. The formula of the AAC can be stated as:
| (1) |
where x means the residues of the amino acids, nx is the length of each residue of x, and T is the total number of residues.
2.3.2. Pseudo amino acid composition (PAAC)
In 2001, Kuo-Chen first proposed the PAAC feature extraction method, where the features were extracted the features with a d-tier-based correlation matrix that brings (20 + d) dimensions [22,23]. The calculation is as follows:
| (2) |
| (3) |
where, l denotes the weight vector of the sequences, fx means the normalized value, the value defines as a type which is A.
2.3.3. Quasi-sequence-order (QSO)
QSO-based features are splitting the distance function between the 20 amino acids [24,25]. A quasi-sequence-order descriptor for each amino acid can be determined as follows:
| (4) |
where, is the normalized occurrence of amino acid type of i, X denotes the weight vector. Where the value is 0.01. nlag means the highest number of lag factors, and Ad is the amino acid position.
2.3.4. N-gram composition (Ngrams)
This method describes that an amino acid sequence is di- or tripeptide. As a result, the functional measurement n can only be associated with decisions 2 and 3 [26]. The formula of this feature can be stated as:
| (5) |
| (6) |
where ndi means the di-peptide and ntri is the tri-peptide. N is the total number of sequence lengths.
2.3.5. Conjoint triad descriptors (CTD)
CTD descriptors were developed to express connections between proteins. The amino acids are organized into seven distinctive groups based on their dipoles and adjacent chain dimensions, which indicate their energetic and hydrophobic interactions [27,28]. The mathematical formula is defined as:
| (7) |
where, fi is the frequency of each divided group in the ith sequences, and i is the consisting of the CTD group.
2.3.6. Binary profile pattern (binary)
Binary profile patterns were designed for every amino acid sequence in the information set. When it is applied, the corresponding matrix with each amino acid is mapped to a 20-dimensional array having only a single non-zero element [29,30].
2.3.7. Moran-autocorrelation (moran)
Moran features are referred to as correlation-based amino acid dissemination. It was adopted for analyzing 233 adjacent metrics in a piece of spatial information [[31], [32], [33]]. The formula is as follows:
| (8) |
| (9) |
where, F’ means the property of the F sequence, T, k, Fi are in the correlation value.
2.3.8. Amino acid index (AAIndex)
The AAindex descriptor provides a collection of 20 quantities indicating differing amino acid physicochemical and ecological components. It includes 566 indexes 553 of which have no NaNs obtained for each amino acid in a sequence and are aggregated periodically [[34], [35], [36]]. The calculation is as follows:
| (10) |
where, i means the 566 AAIndex index's, aan is the amino acids location in n, and T is the total quantity of residues.
2.4. Feature selection process
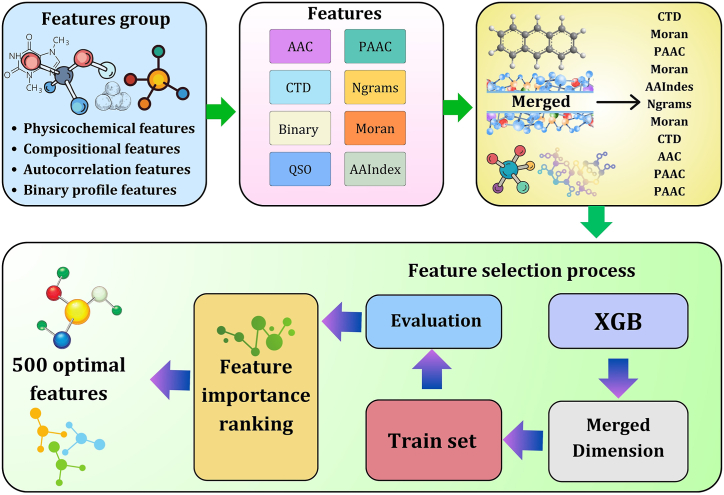
In this study, we have merged all the features of eight encodings. After combining the features, we took an XGB feature selection approach, where the model provides 500 crucial features. XGB possesses a feature ranking procedure that assists in identifying the most essential features in the dataset [37]. The large number of features increases the model's complexity. We aimed to reduce the number of features and include the most significant features so that we could build a model to identify AFP employing the minimum number of features with higher efficiency. This feature selection approach also minimizes the chances of overfitting and boosts the framework's adaptation capacities [38,39]. Fig. 2 shows the feature selection process of this study.
Fig. 2.
Feature selection process of the study. Features are selected based on feature importance scores and the selected features are combined accordingly. The XGB feature selection approach was applied to the merged dimension and delivered the 500 optimal features.
2.5. Construction of MLAFP-XN model
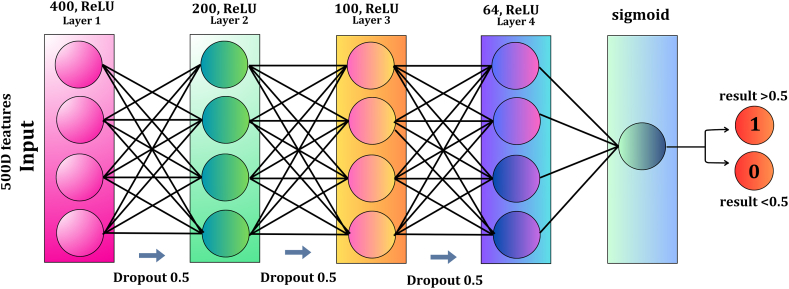
We opted to construct a fundamental structure that provides optimal efficiency while being as straightforward as feasible in Fig. 3. This deliberate strategy aims to facilitate potential enhancements and practical implementations in real-life situations.
Fig. 3.
MLAFP-XN model's architecture, input layer, an output layer, and four hidden layers with ReLU and sigmoid activation function for 500-dimensional features.
We used a neural network design for this study that consists of an input layer, four hidden layers, and an output layer. The requirement for a model that could preserve computational efficiency while capturing intricate patterns in the data, motivated the selection of this design [[40], [41], [42], [43]]. With 500 input features and 400, 200, 100, and 64 neurons in the hidden layers, this particular configuration was created to balance model capacity and avoid overfitting, using each hidden layer's 0.5 dropout rate to enhance generalization. The Adam Optimizer was picked due to its effectiveness in managing sparse gradients and adaptive learning rates. A learning rate of 0.001 was chosen based on best practices in comparable research and empirical tweaking. To fit the binary classification problem of predicting 0s and 1s, binary cross-entropy was employed as the loss function. Since the rectified linear unit (ReLU) activation function effectively introduces non-linearity and mitigates the vanishing gradient problem, it was used for every hidden layer. In the output layer, the sigmoid function was used to generate probabilistic binary results. In order to strike a balance between training stability and model complexity, traditional procedures in neural network design and optimization served as a guide for the selection of these architectural elements and hyperparameters. Equations 11–16 of our suggested model are detailed in the supplemental file, along with further information and explanations for the particular decisions made regarding the neural network design and hyperparameters.
3. Experimental results
Our method MLAFP-XN achieved superior performance on three different datasets based on the independent test approach, where the model more accurately predicts the AFP compared to other models. This study used six different evaluation criteria for the proper detection in the independent method to validate the models' performances. We applied accuracy (ACC), sensitivity (Sn), specificity (Sp), area under curve score (AUC), Matthews's coefficient correlation (MCC), and Cohen kappa scores (Kp) in every model. Table 2 describes all evaluation metrics performances. In addition, we applied 24 classification algorithms including ETC, LGBM, Catboost, and MLAFP-XN to the different descriptor generated datasets and the performance results are presented in Supplementary Tables 1–3. After applying 24 classification algorithms, we have represented the best-performing four algorithm outcomes in the result section. Analyzing all the results, we found that MLAFP-XN performed with higher accuracy, which proved that the proposed approach successfully detected the AFP on each feature extraction. All the results are provided in the supplementary file's Tables 1–3 This section will discuss the performances of the proposed method's results with the other classification algorithms in different settings to justify the efficiency of the proposed model.
Table 2.
Models’ performance in three datasets is based on an independent test approach.
| Datasets | Classifiers | ACC | Sn | Sp | MCC | Kp | AUC |
|---|---|---|---|---|---|---|---|
| Antifp_DS1 | ETC | 0.9518 | 0.9244 | 0.9794 | 0.9051 | 0.9038 | 0.9933 |
| LGBM | 0.9587 | 0.9381 | 0.9794 | 0.9183 | 0.9175 | 0.9903 | |
| Catboost | 0.9759 | 0.9622 | 0.9897 | 0.9522 | 0.9519 | 0.9788 | |
| MLAFP-XN | 0.9793 | 0.9622 | 0.9897 | 0.9596 | 0.9588 | 0.9999 | |
| Antifp_DS2 | ETC | 0.9791 | 0.9794 | 0.9789 | 0.9583 | 0.9583 | 0.9959 |
| LGBM | 0.9791 | 0.9759 | 0.9825 | 0.9583 | 0.9584 | 0.9951 | |
| Catboost | 0.9878 | 0.9863 | 0.9895 | 0.9757 | 0.9757 | 0.9914 | |
| MLAFP-XN | 0.9947 | 0.9897 | 0.9895 | 0.9896 | 0.9896 | 0.9989 | |
| Antifp_Main | ETC | 0.9446 | 0.9278 | 0.9617 | 0.8898 | 0.8893 | 0.9912 |
| LGBM | 0.9584 | 0.9622 | 0.9547 | 0.9170 | 0.9169 | 0.9919 | |
| Catboost | 0.9809 | 0.9725 | 0.9895 | 0.9621 | 0.9619 | 0.9859 | |
| MLAFP-XN | 0.9948 | 0.9725 | 0.9895 | 0.9897 | 0.9896 | 0.9999 |
According to Tables 2 and it is clearly stated that the MLAFP-XN model has the potential to predict the antifungal peptide sequences in all datasets. In the Antifp_DS1 dataset, MLAFP-XN delivered an excellent result in each evaluation metric, where in accuracy, this model provided 97.93 %, which denotes the model has predicted accurately between the positive and negative class of the fungal sequences. Moreover, in Sp, and Sn this model delivered 98.97 %, and 96.22 % which means the model identified the positive sequences more than 95 times and specifically detected the negative sequences more than 98 times. In addition, MCC, Kp, and AUC scores are also acceptable for this model, where this model distinguishes between positive and negative class 99.99 %. Although ETC, LGBM, and Catboost methods resulted in better performance, the MLAFP-XN model significantly outperforms the others. When we utilized the six metrics with an independent test method on Antifp_DS2 and Antift_Main, MLAFP-XN also outperformed the other models. In the Antifp_DS2 dataset, our proposed model generated 99.47 % accuracy with robust performances in other metrics. This model achieved 98.97 % of Sn, 98.95 % of Sp, 98.96 % in MCC, and Kp terms with 99.89 % AUC scores. In the Antifp_Main, this model provided 99.48 % accuracy with all other evaluation methods. Based on the overall performance of these three datasets, it is evident that MLAFP-XN is a promising development for AFP identifiers, as it is more efficient and faster with satisfactory results.
Supplementary Fig. 1 depicts a detailed overview of the performance of three datasets about several evaluated factors. Considering the emphasis on the accuracy subplot, all models outperformed the Antifp_DS2 dataset. However, in the context of the Antifp_Main dataset, the MLAFP-XN model exceeds the others, expressing significantly more accurate results. In the analyses of sensitivity and specificity subplots, it was observed that both the Antifp_Main and Antifp_DS2 datasets exhibited superior performance when subjected to the Catboost and MLAFP-XN frameworks. Meanwhile, the MLAFP-XN framework surpassed other MCC and Kp parameters models over all three datasets, indicating its comprehensive estimation capacities. Based on the Area Under the Curve (AUC) estimation, Catboost and MLAFP-XN consistently outscored other models among the three datasets, highlighting their usefulness in classification tasks. When all assessment criteria were considered together, the MLAFP-XN model emerged as the emphatic victor, proving its proficiency as a highly efficient predictor in precisely identifying categories.
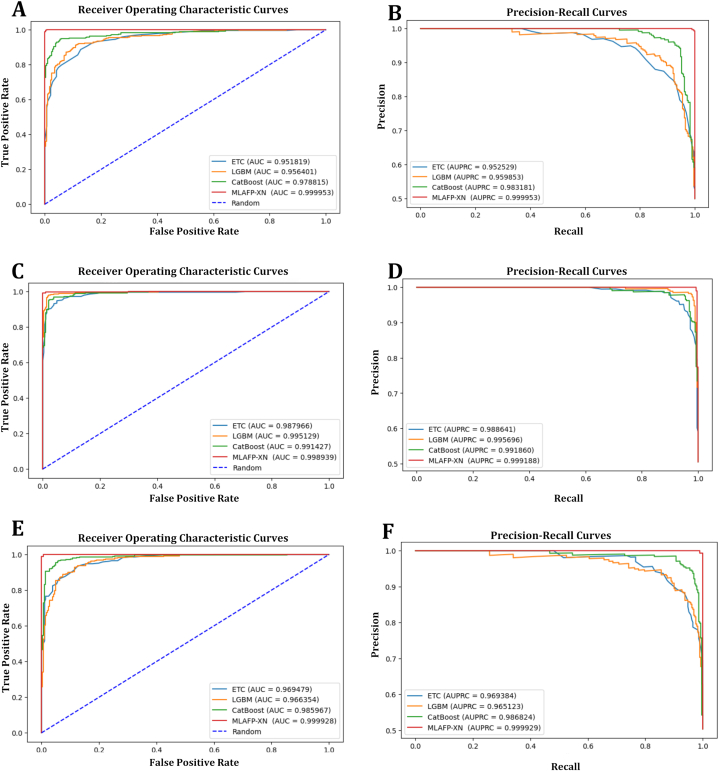
Our study also included receiver operating characteristics curves (ROC) and precision-recall curves (PR) for the models' abilities. Fig. 4 shows the overall ROC and PR curves, where A, C, E subplots denote the ROC curves and B, D, F are the PR analysis curves. In, A C, E subplots, MLAFP-XN obtained higher AUC scores with positive performance, which means the model successfully received the true positive rate more precisely. In addition, in B, D, and F subplots, the proposed model attained greater performances than others, where we can see that the red curve increased in every subplot. This indicates that the MLAFP-XN model can detect the antifungal peptide along with true positive rates, precision, and recall rates.
Fig. 4.
Receiver operating characteristics curves (ROC) and precision-recall curves (PR) analysis of all models with AUC scores, where the blue straight line is the random line. (A) Antifp_DS1 dataset's ROC curves (B) Antifp_DS1 dataset's PR curves, (C) Antifp_DS2 dataset's ROC curves (D) Antifp_DS2 dataset's PR curves, (E) Antifp_Main dataset's ROC curves (F) Antifp_Main dataset's PR curves. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
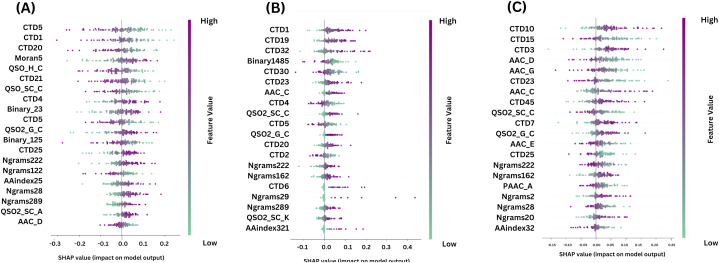
Fig. 5 demonstrates the MLAFP-XN model's outcome features based on SHAP [44], where 5(A) denotes the Antifp_DS1, 5(B) is the Antifp_DS2, and 5(C) indicates the Antifp_Main dataset.
Fig. 5.
SHAP feature analysis of ML-AFP-XN model, (A) Antifp_DS1 dataset's outcome, (B) Antifp_DS2 dataset's results, (C) Antifp_Main dataset's findings.
The three subplots exhibit similar CTD features. The presence of CTD, QSO, and Ngram features in Fig. 5(A) indicates an effective association for determining AFP. Additionally, Fig. 5(B) demonstrates that CTD, Ngrams, and QSO features have connections to the identification of AFP. The identification of AFP corresponds with CTD, AAC, QSO, and Ngrams features in Fig. 5(C). Overall, from the subplots, it is clear that CTD, QSO, and Ngram-based features are crucial features of this proposed model, which will benefit in the biological platform. According to the study, most of the researchers obtained a better result based on CTD, QSO, AAC based features [45,46]. Accordingly, we applied ETC, Catboost, LGBM, and MLAFP-XN models with these SHAP selected features (20D) in each dataset and without these SHAP selected features (480D) in the three datasets to validate our proposed approach. All the performance results are included in the Supplementary file Tables 4 and 5 respectively. With 20D features, our proposed method performed with 0.819, 0.928, and 0.861 accuracy for Antifp_DS1, Antifp_DS2, and Antifp_Main datasets respectively. Afterwards, with 480D features, this framework obtained 0.945, 0.803, and 0.977 accuracies for Antifp_DS1, Antifp_DS2, and Antifp_Main datasets respectively. The result indicates that all the features are crucial to detect the antifungal peptides with the proposed framework though these 20D features are crucial to detect the AFP from the sequences.
4. Discussion
The detection and development of antifungal peptides are vital for addressing the challenges associated with fungal infections, including drug resistance, side effects, and the need for effective broad-spectrum therapeutics. Continued research in this area holds promise for improving treatment options in the medical and biological sectors. Our study successfully detected the antifungal peptide from the various types of sequences based on three distinct datasets. The MLAFP-XN model was developed using a neural network technique, prioritizing sensible design that balances accuracy and affordability. According to the literature search, we have built our model with input, output, and four hidden layers, where we have fine-tuned several neurons, activation functions, and dropout rates. We focused on delivering a simple model with a high accuracy rate, and our model performed better than the other existing models. To demonstrate the model's performance and dependability, we implemented a web server that should serve as a useful starting point for future practical deployment. The study will contribute to the biological fields in different aspects, especially in drug discovery and precision medicine for fungal infections. In precision medicine, the identification of biological agents for specific disease is crucial to developing vaccines and other medications. The identification of AFP can assist biologists in cultivating AFP in laboratory to inject it into fungal-infected patients who have a deficiency of AFP. Additionally, the outcomes of the model have the potential to contribute enormously to bioinformatics and computational biology research. To check the overfitting and underfitting of the proposed model, we have applied cross-validation and obtained the ROC curve. The use of cross-validation ensures that the model generalizes well. The results of the cross-validation are included in the Supplementary Tables 6 and 7 respectively.
4.1. Performances comparison of state-of-the-art methods
In this subsection, we compare our proposed model with the existing AFP predictor tools. Many researchers proposed their model in different datasets and different strategies. Table 3 shows the overall performance comparison based on accuracy.
Table 3.
Performance comparison of MLAFP-XN with existing proposed methods on similar datasets.
| Dataset name | Proposed model | Accuracy (%) | References |
|---|---|---|---|
| Antifp_DS1 | AntiFP | 87.29 | [9] |
| AFPDeep | 90.21 | [10] | |
| iAMPCN | 69.59 | [41] | |
| AFPtransferPred | 48.97 | [42] | |
| AFP-MFL | 94.40 | [8] | |
| DeepAFP | 92.44 | [14] | |
| MLAFP-XN | 97.93 | This study | |
| Antifp_DS2 | AntiFP | 90.21 | [9] |
| AFPDeep | 94.67 | [10] | |
| iAMPCN | 84.19 | [41] | |
| AFPtransferPred | 66.32 | [42] | |
| AFP-MFL | 96.84 | [8] | |
| DeepAFP | 96.05 | [14] | |
| Deep-AFPpred | 56.49 | [47] | |
| MLAFP-XN | 99.47 | This study | |
| Antifp_Main | AntiFP | 84.98 | [9] |
| AFPDeep | 91.05 | [10] | |
| iAMPCN | 79.35 | [41] | |
| AFPtransferPred | 58.35 | [42] | |
| AFP-MFL | 95.84 | [8] | |
| DeepAFP | 93.29 | [14] | |
| Deep-AFPpred | 51.45 | [47] | |
| MLAFP-XN | 99.48 | This study | |
| Other datasets | Deep-AFPpred | 94.27 | [47] |
| PhytoAFP | 94.4 | [48] |
According to the results in Table 3, the MLAFP-XN model outscored all other models based on overall reliability. When validated on the DS1 dataset, our proposed approach outperformed more than 5 % compared to other methods. In contrast, the recent DeepAFP and AFP-MFL models utilized complex models and a limited set of features for detection on the same dataset. Our approach used several feature extraction methods and handled redundant information. We additionally evaluated our findings in the iAMPCN model, which focuses on antimicrobial property categorisation, including antifungal peptides. Despite using the same datasets, the accuracy of iAMPCN for AFP detection has not been sufficient. On Antifp_DS1datasets, our model outperformed the iAMPCN model by more than 20 % in accuracy. Besides, we compared our proposed model with other models which were developed using other datasets [47,48]. In this case, we observed that our proposed model is outperforming more than 5 % in terms of accuracy, delivering greater performance, when compared to existing models.
4.2. Web server implementation
We designed a website based on ANVIL software, which provides free hosting for Python-based projects [49]. Supplementary Fig. 2 presents an overview of the web pages for our MLAFP-XN framework. At the top-right corner, there is a "SERVER ACTIVATION" button. When viewers click on this button, the server has been configured to be active within 10 min. If the server activation doesn't work, users are urged to contact us by clicking the "EMAIL US" button. For the verification of models, we presented a few instances of antifungal peptide sequences. Users can view predetermined sequences in the input box by clicking the "EXAMPLES" button. After hitting the predict button, the model's result will appear in the output box. Additionally, necessary instructions are highlighted in red squares. Users should attentively read and follow these directions to ensure that the website operates correctly. The website is accessible at the following link: https://immediate-rash-hire.anvil.app/.
5. Conclusion
Identifying antifungal peptides (AFPs) is a significant step in developing future therapeutics. We designed an intuitive structure focusing on time, space, and cost savings. We used a neural network method with hidden layers, which is demonstrated to a higher accuracy. This approach will benefit future applications since previous investigations have used sophisticated models with poor outcomes. Compared to existing frameworks, our MLAFP-XN model identifies AFPs successfully and reliably. We used eight feature encoding approaches in conjunction with the XGB feature selection strategy to identify the most desirable features. Afterwards, we chose the MLAFP-XN model for its higher performance after analyzing other frameworks. With SHAP, we additionally discovered key features associated with AFP identification. Furthermore, we generated a practical online website based on our strategy. However, present paper has some shortcomings, namely the use of limited datasets and selective machine learning models. In the future, we will develop a more advanced model based on many antifungal resistances with a more diverse strategy to predict the AFP and explore the most optimal features directly associated with drug discoveries. Our proposed framework is a powerful prediction tool for AFP identification, providing significant benefits to computational biology, therapeutic studies, and medicinal upgrades.
Data availability
The data sets are available as supplementary files and to access the code please follow the link: Code availability: https://github.com/Shazzad-Shaon3404/MLAFP-XN.git. The website is accessible at the following link: https://immediate-rash-hire.anvil.app/.
Funding
This research is funded in part by the Natural Sciences and Engineering Research Council of Canada (NSERC).
Ethical consideration
Not Applicable.
CRediT authorship contribution statement
Md. Fahim Sultan: Writing – original draft, Visualization, Software, Resources, Investigation, Formal analysis. Md. Shazzad Hossain Shaon: Writing – original draft, Visualization, Software, Resources, Methodology, Investigation, Formal analysis. Tasmin Karim: Writing – original draft, Investigation, Formal analysis. Md. Mamun Ali: Writing – review & editing, Writing – original draft, Visualization, Validation, Supervision, Software, Resources, Project administration, Methodology, Investigation, Formal analysis, Data curation, Conceptualization. Md. Zahid Hasan: Validation, Supervision. Kawsar Ahmed: Writing – review & editing, Writing – original draft, Validation, Supervision, Project administration, Methodology, Formal analysis, Conceptualization. Francis M. Bui: Writing – review & editing, Supervision, Project administration, Funding acquisition. Li Chen: Writing – review & editing, Supervision, Project administration, Funding acquisition. Vigneswaran Dhasarathan: Writing – review & editing, Project administration. Mohammad Ali Moni: Writing – review & editing, Validation, Supervision, Resources, Project administration, Methodology.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2024.e37820.
Contributor Information
Md. Fahim Sultan, Email: fahim15-3416@diu.edu.bd.
Md. Shazzad Hossain Shaon, Email: shazzad15-3404@diu.edu.bd.
Tasmin Karim, Email: tasmin15-2920@diu.edu.bd.
Md. Mamun Ali, Email: m.ali@usask.ca.
Md. Zahid Hasan, Email: zahid.cse@diu.edu.bd.
Kawsar Ahmed, Email: k.ahmed.bd@ieee.org, kawsar.ict@mbstu.ac.bd, k.ahmed@usask.ca.
Francis M. Bui, Email: francis.bui@usask.ca.
Li Chen, Email: lic900@usask.ca.
Vigneswaran Dhasarathan, Email: dhasa.viki@gmail.com.
Mohammad Ali Moni, Email: mmoni@csu.edu.au.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Bongomin F., Gago S., Oladele R.O., Denning D.W. Global and multi-national prevalence of fungal diseases—estimate precision. Journal of fungi. 2017;3(4):57. doi: 10.3390/jof3040057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brown G.D., Denning D.W., Gow N.A., Levitz S.M., Netea M.G., White T.C. Hidden killers: human fungal infections. Sci. Transl. Med. 2012;4(165) doi: 10.1126/scitranslmed.3004404. 165rv13-165rv13. [DOI] [PubMed] [Google Scholar]
- 3.Richardson M.D. Changing patterns and trends in systemic fungal infections. J. Antimicrob. Chemother. 2005;56(suppl_1):i5–i11. doi: 10.1093/jac/dki218. [DOI] [PubMed] [Google Scholar]
- 4.Sanglard D. Emerging threats in antifungal-resistant fungal pathogens. Front. Med. 2016;3:11. doi: 10.3389/fmed.2016.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Capita R., Alonso-Calleja C. Antibiotic-resistant bacteria: a challenge for the food industry. Crit. Rev. Food Sci. Nutr. 2013;53(1):11–48. doi: 10.1080/10408398.2010.519837. [DOI] [PubMed] [Google Scholar]
- 6.Usmani S.S., Bedi G., Samuel J.S., Singh S., Kalra S., Kumar P., Ahuja A.A., Sharma M., Gautam A., Raghava G.P. THPdb: database of FDA-approved peptide and protein therapeutics. PLoS One. 2017;12(7) doi: 10.1371/journal.pone.0181748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Singh S., Chaudhary K., Dhanda S.K., Bhalla S., Usmani S.S., Gautam A., Tuknait A., Agrawal P., Mathur D., Raghava G.P. SATPdb: a database of structurally annotated therapeutic peptides. Nucleic Acids Res. 2016;44(D1):D1119–D1126. doi: 10.1093/nar/gkv1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fang Y., Xu F., Wei L., Jiang Y., Chen J., Wei L., Wei D.Q. AFP-MFL: accurate identification of antifungal peptides using multi-view feature learning. Briefings Bioinf. 2023;24(1):bbac606. doi: 10.1093/bib/bbac606. [DOI] [PubMed] [Google Scholar]
- 9.Agrawal P., Bhalla S., Chaudhary K., Kumar R., Sharma M., Raghava G.P. In silico approach for prediction of antifungal peptides. Front. Microbiol. 2018;9:323. doi: 10.3389/fmicb.2018.00323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fang C., Moriwaki Y., Li C., Shimizu K. Prediction of antifungal peptides by deep learning with character embedding. IPSJ Transactions on Bioinformatics. 2019;12:21–29. doi: 10.2197/ipsjtbio.12.21. [DOI] [Google Scholar]
- 11.Ahmad A., Akbar S., Khan S., Hayat M., Ali F., Ahmed A., Tahir M. Deep-AntiFP: prediction of antifungal peptides using distanct multi-informative features incorporating with deep neural networks. Chemometr. Intell. Lab. Syst. 2021;208 doi: 10.1016/j.chemolab.2020.104214. [DOI] [Google Scholar]
- 12.Ahmad A., Akbar S., Tahir M., Hayat M., Ali F. iAFPs-EnC-GA: identifying antifungal peptides using sequential and evolutionary descriptors based multi-information fusion and ensemble learning approach. Chemometr. Intell. Lab. Syst. 2022;222 doi: 10.1016/j.chemolab.2022.104516. [DOI] [Google Scholar]
- 13.Sharma R., Shrivastava S., Kumar Singh S., Kumar A., Saxena S., Kumar Singh R. Deep-AFPpred: identifying novel antifungal peptides using pretrained embeddings from seq2vec with 1DCNN-BiLSTM. Briefings Bioinf. 2022;23(1):bbab422. doi: 10.1093/bib/bbab422. [DOI] [PubMed] [Google Scholar]
- 14.Yao L., Zhang Y., Li W., Chung C.R., Guan J., Zhang W., Chiang Y.C., Lee T.Y. D eepAFP: an effective computational framework for identifying antifungal peptides based on deep learning. Protein Sci. 2023;32(10):e4758. doi: 10.1002/pro.4758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Asad H. Antifp SRF: identifying antifungal peptides by sequence statistical moments and random forest classifier. Journal of Innovative Research in Mathematical and Computational Sciences. 2023;2(2):109–125. doi: 10.62270/jirmcs.v2i2.23. [DOI] [Google Scholar]
- 16.Fang W., Wu J., Cheng M., Zhu X., Du M., Chen C., Liao W., Zhi K., Pan W. Diagnosis of invasive fungal infections: challenges and recent developments. J. Biomed. Sci. 2023;30(1):42. doi: 10.1186/s12929-023-00926-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hassan E., Talaat F.M., Adel S., Abdelrazek S., Aziz A., Nam Y., El-Rashidy N. Robust deep learning model for black fungus detection based on gabor filter and transfer learning. Comput. Syst. Sci. Eng. 2023;47(2) doi: 10.32604/csse.2023.037493. [DOI] [Google Scholar]
- 18.Fan L., Sun J., Zhou M., Zhou J., Lao X., Zheng H., Xu H. DRAMP: a comprehensive data repository of antimicrobial peptides. Sci. Rep. 2016;6(1) doi: 10.1038/srep24482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gasteiger E., Jung E., Bairoch A. SWISS-PROT: connecting biomolecular knowledge via a protein database. Curr. Issues Mol. Biol. 2001;3(3):47–55. doi: 10.21775/cimb.003.047. [DOI] [PubMed] [Google Scholar]
- 20.Zhang Z. 2018 IEEE/ACM 26th International Symposium on Quality of Service (IWQoS) Ieee; 2018, June. Improved adam optimizer for deep neural networks; pp. 1–2. [DOI] [Google Scholar]
- 21.Zeiler M.D. Adadelta: an adaptive learning rate method. 2012. arXiv preprint arXiv:1212.5701. [DOI]
- 22.Agarap A.F. Deep learning using rectified linear units (relu) arXiv preprint arXiv:1803.08375. 2018 doi: 10.48550/arXiv.1803.08375. [DOI] [Google Scholar]
- 23.Menon A., Mehrotra K., Mohan C.K., Ranka S. Characterization of a class of sigmoid functions with applications to neural networks. Neural Network. 1996;9(5):819–835. doi: 10.1016/0893-6080(95)00107-7. [DOI] [PubMed] [Google Scholar]
- 24.Zhang Y.F., Wang Y.H., Gu Z.F., Pan X.R., Li J., Ding H., Zhang Y., Deng K.J. Bitter-RF: a random forest machine model for recognizing bitter peptides. Front. Med. 2023;10 doi: 10.3389/fmed.2023.1052923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ahmed Z., Zulfiqar H., Tang L., Lin H. A statistical analysis of the sequence and structure of thermophilic and non-thermophilic proteins. Int. J. Mol. Sci. 2022;23(17) doi: 10.3390/ijms231710116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chou K.C. Prediction of protein cellular attributes using pseudo‐amino acid composition. Proteins: Struct., Funct., Bioinf. 2001;43(3):246–255. doi: 10.1002/prot.1035. [DOI] [PubMed] [Google Scholar]
- 27.Chou K.C. Pseudo amino acid composition and its applications in bioinformatics, proteomics and system biology. Curr. Proteonomics. 2009;6(4):262–274. doi: 10.2174/157016409789973707. [DOI] [Google Scholar]
- 28.Chou K.C. Prediction of protein subcellular locations by incorporating quasi-sequence-order effect. Biochemical and biophysical research communications. 2000;278(2):477–483. doi: 10.1006/bbrc.2000.3815. [DOI] [PubMed] [Google Scholar]
- 29.Chou K.C., Cai Y.D. Prediction of protein subcellular locations by GO–FunD–PseAA predictor. Biochem. Biophys. Res. Commun. 2004;320(4):1236–1239. doi: 10.1016/j.bbrc.2004.06.073. [DOI] [PubMed] [Google Scholar]
- 30.Roark B., Saraclar M., Collins M. Discriminative n-gram language modeling. Comput. Speech Lang. 2007;21(2):373–392. doi: 10.1016/j.csl.2006.06.006. [DOI] [Google Scholar]
- 31.Shen J., Zhang J., Luo X., Zhu W., Yu K., Chen K., Li Y., Jiang H. Predicting protein–protein interactions based only on sequences information. Proc. Natl. Acad. Sci. USA. 2007;104(11):4337–4341. doi: 10.1073/pnas.0607879104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Malik A., Shoombuatong W., Kim C.B., Manavalan B. GPApred: the first computational predictor for identifying proteins with LPXTG-like motif using sequence-based optimal features. Int. J. Biol. Macromol. 2023;229:529–538. doi: 10.1016/j.ijbiomac.2022.12.315. [DOI] [PubMed] [Google Scholar]
- 33.Ansari H.R., Raghava G.P. Identification of conformational B-cell Epitopes in an antigen from its primary sequence. Immunome Res. 2010;6(1):1–9. doi: 10.1186/1745-7580-6-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen Z., Chen Y.Z., Wang X.F., Wang C., Yan R.X., Zhang Z. Prediction of ubiquitination sites by using the composition of k-spaced amino acid pairs. PLoS One. 2011;6(7) doi: 10.1371/journal.pone.0022930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moran P.A. Notes on continuous stochastic phenomena. Biometrika. 1950;37(1/2):17–23. doi: 10.2307/2332142. [DOI] [PubMed] [Google Scholar]
- 36.Xiao N., Cao D.S., Zhu M.F., Xu Q.S. protr/ProtrWeb: R package and web server for generating various numerical representation schemes of protein sequences. Bioinformatics. 2015;31(11):1857–1859. doi: 10.1093/bioinformatics/btv042. [DOI] [PubMed] [Google Scholar]
- 37.Li H., Calder C.A., Cressie N. Beyond Moran's I: testing for spatial dependence based on the spatial autoregressive model. Geogr. Anal. 2007;39(4):357–375. doi: 10.1111/j.1538-4632.2007.00708.x. [DOI] [Google Scholar]
- 38.Kawashima S., Kanehisa M. AAindex: amino acid index database. Nucleic Acids Res. 2000;28(1) doi: 10.1093/nar/28.1.374. 374-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nakai K., Kidera A., Kanehisa M. Cluster analysis of amino acid indices for prediction of protein structure and function. Protein Eng. Des. Sel. 1988;2(2):93–100. doi: 10.1093/protein/2.2.93. [DOI] [PubMed] [Google Scholar]
- 40.Tomii K., Kanehisa M. Analysis of amino acid indices and mutation matrices for sequence comparison and structure prediction of proteins. Protein Eng. Des. Sel. 1996;9(1):27–36. doi: 10.1093/protein/9.1.27. [DOI] [PubMed] [Google Scholar]
- 41.Alsahaf A., Petkov N., Shenoy V., Azzopardi G. A framework for feature selection through boosting. Expert Syst. Appl. 2022;187 doi: 10.1016/j.eswa.2021.115895. [DOI] [Google Scholar]
- 42.Chen C., Zhang Q., Yu B., Yu Z., Lawrence P.J., Ma Q., Zhang Y. Improving protein-protein interactions prediction accuracy using XGBoost feature selection and stacked ensemble classifier. Comput. Biol. Med. 2020;123 doi: 10.1016/j.compbiomed.2020.103899. [DOI] [PubMed] [Google Scholar]
- 43.Chen T., He T., Benesty M., Khotilovich V., Tang Y., Cho H., Chen K., Mitchell R., Cano I., Zhou T. Xgboost: extreme gradient boosting. R package version 0.4-2. 2015;1(4):1–4. https://scholar.google.com/scholar?hl=en&as_sdt=0%2C5&q=extreme+gradient+boosting&btnG=&oq=extremely+gradient [Google Scholar]
- 44.García M.V., Aznarte J.L. Shapley additive explanations for NO2 forecasting. Ecol. Inf. 2020;56 doi: 10.1016/j.ecoinf.2019.101039. [DOI] [Google Scholar]
- 45.Xu J., Li F., Li C., Guo X., Landersdorfer C., Shen H.H., Peleg A.Y., Li J., Imoto S., Yao J., Akutsu T. iAMPCN: a deep-learning approach for identifying antimicrobial peptides and their functional activities. Briefings Bioinf. 2023;24(4):bbad240. doi: 10.1093/bib/bbad240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lobo F., González M.S., Boto A., Pérez de la Lastra J.M. Prediction of antifungal activity of antimicrobial peptides by transfer learning from protein pretrained models. Int. J. Mol. Sci. 2023;24(12) doi: 10.3390/ijms241210270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hasan M.M., Schaduangrat N., Basith S., Lee G., Shoombuatong W., Manavalan B. HLPpred-Fuse: improved and robust prediction of hemolytic peptide and its activity by fusing multiple feature representation. Bioinformatics. 2020;36(11):3350–3356. doi: 10.1093/bioinformatics/btaa160. [DOI] [PubMed] [Google Scholar]
- 48.Malik A., Shoombuatong W., Kim C.B., Manavalan B. GPApred: the first computational predictor for identifying proteins with LPXTG-like motif using sequence-based optimal features. Int. J. Biol. Macromol. 2023;229:529–538. doi: 10.1016/j.ijbiomac.2022.12.315. [DOI] [PubMed] [Google Scholar]
- 49.Anvil software for Python based project. 2023. https://anvil.works/ October.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data sets are available as supplementary files and to access the code please follow the link: Code availability: https://github.com/Shazzad-Shaon3404/MLAFP-XN.git. The website is accessible at the following link: https://immediate-rash-hire.anvil.app/.