Abstract
While several cellular proteins are incorporated in the human immunodeficiency virus type 1 virion, cyclophilin (CyP) A is the only one whose absence has been demonstrated to impair infectivity. Incorporation of the cytosolic protein results from interaction with a highly exposed Pro-rich loop in the N-terminal region of the capsid (CA) domain of the precursor polyprotein, Pr55Gag. Even when prevented from interacting with CyP A, Pr55Gag still forms particles that proceed to mature into morphologically wild-type virions, suggesting that CyP A influences a postassembly event. The nature of this CyP A influence has yet to be elucidated. Here, we show that while CyP A binds both Gag and mature CA proteins, the two binding interactions are actually different. Tryptophan 121 (W121) in CyP A distinguished the two proteins: a phenylalanine substitution (W121F) impaired binding of mature CA protein but not of Gag. This indicates the occurrence of a maturation-dependent switch in the conformation of the Pro-rich loop. A structural consequence of Gag binding to CyP A was to block this maturational refolding, resulting in a 24-kDa CA protein retaining the immature Pro-rich loop conformation. Using trypsin as a structure probe, we demonstrate that the conformation of the C-terminal region in mature CA is also a product of maturational refolding. Binding to wild-type CyP A altered this conformation, as indicated by a reduction in the accessibility of Cys residue(s) in the region to chemical modification. Hence, the end result of binding to CyP A, whether the Pro-rich loop is in the context of Gag or mature CA protein, is a structurally modified mature CA protein. The postassembly role of CyP A may be mediated through these modified mature CA proteins.
The human immunodeficiency virus type 1 (HIV-1) virus incorporates a cytoplasmic form of cyclophilin (CyP) called CyP A. CyPs are a ubiquitous family of highly conserved cis-trans prolyl isomerases that assist protein folding (20) and also serve as targets for the immunosuppressive drug cyclosporin A (CsA) (25). Although several cellular proteins are incorporated in the virion (5, 40), only CyP A has been demonstrated to enhance virus infectivity (19, 47). Virus particles lacking CyP A assemble into morphologically wild-type (WT)-looking particles that are unable to productively replicate in target cells, which indicates that CyP A influences an early stage in the life cycle of the virus (8, 44). The exact step in the virus-target cell interaction for which CyP A is required has yet to be identified. That virion-incorporated CyP A may directly bind to receptors on the surface of target cells is suggested by observations that inhibitors of CyP A prevented efficient virus attachment (42, 43). That virion-incorporated CyP A influences functional uncoating to release the viral RNA and replicative enzymes is suggested by observations that CyP A-deficient virions are defective in the reverse transcription of viral RNA in target cells (8, 35, 44). That CyP A in target cells may also be involved is suggested by the observation that endocytic entry rescued replication of virus particles lacking CyP A when incorporation was abrogated by addition of CsA during virus production (4). Interestingly, endocytic entry did not rescue replication when incorporation was abrogated by a mutation in the binding site in HIV (4). Consistent with the suggestion that CyP A in the target cell may be utilized, virions devoid of CyP A by virtue of their production by cells in which both CyP A alleles have been knocked out exhibited delayed but nonetheless productive replication (D. Braaten and J. Luban, Abstr. 2000 Meet. Retroviruses, abstr. 156, 2000).
CyP A has the shape of a flattened beta barrel formed by eight antiparallel beta strands and two alpha helices that cap the top and bottom of the barrel (30, 31; reviewed in reference 46). Several of the beta strands and loop regions form a hydrophobic pocket on the surface of the protein which serve as the active site of CyP A. It is here that CsA and proline-containing proteins bind. The binding of a specific sequence within the capsid (CA) domain in Pr55Gag to this pocket is what permits the incorporation of CyP A in HIV-1 particles (19, 35, 47). Results of mutational analyses (19, 47, 50), cocrystallization (22, 48), and chimera construction followed by phenotypic analysis (12) identify the binding site in CA as residues 87 to 92 (HAGPIA). The binding of this sequence in the CyP A hydrophobic pocket is known in atomic detail (22, 48). The HAGPIA sequence by itself (48) or as part of a longer N-terminal CA fragment (22) lies in the pocket in an extended and slightly bowed position. Several CA and CyP A atoms are within van der Waals distance allowing stabilization of the binding through multiple hydrophobic interactions. Major contribution to binding stability, however, is made by seven direct and two water-mediated hydrogen bonding interactions between CA and CyP A atoms. Active site residues that are engaged in hydrogen bonding interaction are His54, Arg55, Asn71, Asn102, His126, and Trp121. Biochemical studies using engineered active site pocket mutants have demonstrated that these residues, except Trp121, are critical for incorporation of CyP A into HIV-1 particles (9, 14). The Trp121-to-Phe change (W121F) is particularly revealing. While virion incorporation, and hence Pr55Gag binding, remains at WT level (14), susceptibility to CsA is reduced ∼75-fold and peptidyl-prolyl isomerase activity is reduced ∼2-fold (34). These observations suggest differences in the binding of the CA sequence, cyclosporin A, and N-acetyl-Ala-Ala-Pro-Ala-amidomethylcoumarin (the model substrate used to assay prolyl isomerase activity [30]) within the hydrophobic pocket of CyP A.
The HAGPIA sequence is part of a Pro-rich region that forms a highly exposed loop in the N-terminal domain (NTD) of mature CA protein (7, 22–24, 38). This loop is also one of the most mobile or flexible region of CA (13, 24). Since CyP A is incorporated in HIV-1 by virtue of its association with Pr55Gag, presumably, the HAGPIA sequence in the context of the CA domain is likewise highly exposed. Presently, there is no available three-dimensional structure information to assess whether HAGPIA is presented differently in the precursor. However, results of two independent studies indicating differential binding of Gag and CA proteins to CyP A are suggestive. Endrich et al. (18), using fluorescence methods, demonstrated that mature CA was bound with higher affinity than an immature form of the protein. Bristow et al. (11), using a sensitive enzyme-linked immunosorbent assay, obtained higher affinities with immature forms of CA. Although conflicting results were obtained in these studies, their observations suggest a difference in the structure or presentation of the HAGPIA sequence in precursor and mature CA contexts. As part of the precursor polyprotein, the N- and C-terminal tails of the CA domain are tethered to matrix (MA) and the p2 spacer peptide, respectively. The prediction of extended conformations for these regions in the CA domain is consistent with the rod-like topography (21, 39) and multisector structure (21) of assembled Pr55Gag revealed by electron microscopy studies of immature particles. The β-hairpin structure of the N terminus that is stabilized by a salt bridge between Pro1 and Asp51 (22, 24) has been suggested to be a product of refolding (24). Evidence for the occurrence of analogous refolding by the C-terminal tail has yet to be obtained. The structure of this region, predicted to be helical when tethered (1), has eluded resolution in crystal structure studies (7, 23, 38).
The amount of CyP A that is incorporated in the assembling virion is influenced by the level of CyP A in the virus-producing cell (2, 10, 49). Yet even under the best of circumstances, the amount of incorporated CyP A remains a fraction of the amount of assembled Pr55Gag in the virion. In the subsequent maturation stage, proteolytic processing leads to release of the CA domain as a mature 24-kDa protein (45). As CyP A can also bind mature CA, based on the observed ability of CyP A to bind in vitro full-length CA protein (3, 36), an N-terminal CA fragment (22, 50) or an even shorter CA peptide containing the binding sequence (48), the expectation is that the situation leads to two populations of mature CA protein: unliganded and CyP A liganded. The morphogenic rearrangement that occurs next includes assembly of mature CA proteins into a shell around the genomic RNA and replicative enzymes. This shell disassembles (in a process called uncoating) during entry of the mature virion into a new host, allowing release and reverse transcription of the viral RNA.
In what manner do virion-associated CyP A-liganded CA proteins influence uncoating such that the end result is enhancement of infectivity? If, as some studies (4; Braaten and Luban, Abstr. 2000 meet. Retroviruses, 2000) suggest, infectivity is influenced by CyP A in the target cell, what postuncoating event do these newly formed CyP A-liganded mature CA proteins influence? To begin to address these questions, we examined the structural consequences of CyP A binding on Gag and the mature 24-kDa CA protein. In a previous study (3), we found that CyP A binding altered the trypsin sensitivity of the C-terminal domain (CTD) of the CA protein. In the present report, we demonstrate that (i) there are maturation-dependent structure changes in the Pro-rich loop and CTD of CA; (ii) maturational refolding of the Pro-rich loop is blocked when bound to CyP A prior to proteolytic processing of Gag by HIV-1 protease (PR); (iii) binding of mature CA protein to WT CyP A, but not to the W121F mutant, elicits a structural change in the CTD of the CA protein; and (iv) this structural change correlated with reduced accessibility of the Cys residue(s).
MATERIALS AND METHODS
Constructs and mutagenesis.
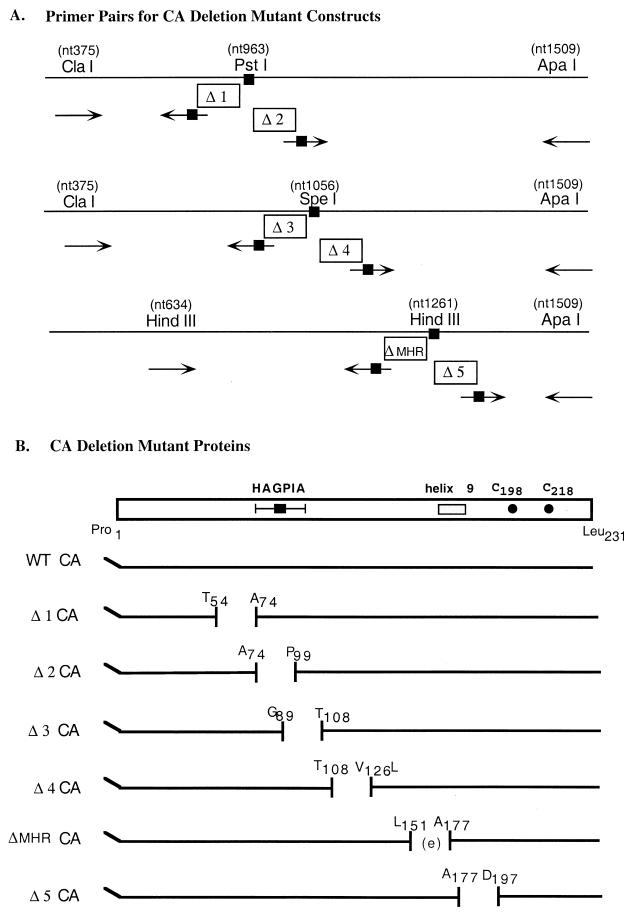
WT CyP A fused to glutathione S-transferase (GST-CyP A) and the single amino acid substitution mutants R55A GST-CyP A and W121F GST-CyP A were gifts from D. Braaten and J. Luban (9). CA, in both Gag and mature protein contexts, was expressed from constructs containing HIV-1 sequences derived from pBH10 (41). Constructs expressing N-terminally histidine (N-His)- and T7-tagged Gag and mature CA derived from in situ processing of a Gag-PR fusion protein have been described previously (15–17). N-His-tagged full-length CA protein and N-His-tagged CA mutants with deletions of ∼20 amino acids were constructed by PCR using convenient restriction sites in the gag gene. To engineer a deletion, a PCR product spanning two restriction sites but lacking ∼60 nucleotides (nt) was exchanged for a restriction fragment derived from the template plasmid (FS II; 17). Upstream-downstream primer pairs used to make each deletion mutant are summarized in Fig. 1A. To illustrate, deletion mutant Δ1 CA was made by synthesizing a PCR fragment using an upstream primer identical to nt 363 to 382 (thus including the endogenous ClaI site) and a downstream primer with an engineered PstI site that annealed to nt 892 to 904 (thus positioning the engineered site downstream of the endogenous PstI site). The PCR product obtained was purified and digested with restriction enzymes ClaI and PstI to obtain a ClaI/PstI PCR fragment containing the deletion. The template plasmid was likewise digested, and the longer of two ClaI/PstI restriction fragments generated was isolated and ligated with the ClaI/PstI PCR fragment. This general cloning strategy was used to engineer deletions. Each mutant required a unique pair of upstream and downstream primers (Fig. 1A). All mutations were subsequently confirmed by DNA sequencing. For ease of purifying the mutant CA proteins, the CA sequences in the FS II mutant plasmids were subcloned into pB6, a derivative of pET 11a that expresses N-His-tagged proteins (a gift from P. Tegtmeyer). A schematic of the full-length and six deletion mutant proteins in this context is shown in Fig. 1B.
FIG. 1.
Schematic of CA deletion mutants. (A) Upstream and downstream primer pairs (arrows) are aligned to CA sequences on the template plasmid (horizontal line) to which they anneal. One primer (plain arrow) in the pair includes an endogenous restriction site, while the other primer contains an engineered restriction site (arrow with a black solid box). The PCR product synthesized with these primer pairs was used in a cloning strategy described in Materials and Methods, resulting in deletion mutants with the following juncture sequences: Δ1, nt 904/nt 963; Δ2, nt 963/nt 1021; Δ3, nt 996/nt 1056; Δ4, nt 1056/nt 1108; ΔMHR, nt 1183/nt 1261; and Δ5, nt 1261/nt 1318. (B) Schematic of (WT CA) and the six mutant CA proteins. The CA polypeptide starts an N-terminal histidine tag (indicated by angled line) fused to the CA residues Pro1 to Leu231 (horizontal line). Positions of engineered deletions are indicated by a gap in the horizontal line. Pertinent landmarks in CA are shown at the top: CyP A binding sequence (HAGPIA), region that participates in forming helix 9, and cysteine residues 198 and 218.
Recombinant protein expression and purification.
All proteins used in this study were purified under native conditions. The HIV-1 T7-tagged and His-tagged Gag and mature CA derived from in situ processing of a Gag-PR fusion protein (CA1-231) were isolated as previously described (15–17). N-His-tagged HIV-1 CA proteins, full-length and deletion mutants, were expressed in Escherichia coli strain HMS174 and purified by chromatography on a Ni-agarose column under nondenaturing conditions as described by the manufacturer (Qiagen). Bound proteins were eluted by successive washing with 500 mM imidazole. WT, R55A, and W121F GST-CyP A proteins were expressed in C600 cells and purified by chromatography on a glutathione-agarose column also under nondenaturing conditions as described by the manufacturer (Sigma). Bound proteins were eluted by successive washing with 10 mM glutathione. Eluted proteins were analyzed for purity by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), dialyzed against 1,000 volumes of appropriate binding buffer, and measured for protein concentration using the Bio-Rad dye assay.
In vitro CyP A binding assays.
The binding of Gag or mature CA proteins to WT, R55A, or W121F mutant GST-CyP A was examined in two binding assay formats. In format A, Gag or CA and WT or W121F GST-CyP A proteins were mixed in TK buffer (20 mM Tris-HCl [pH 7.5], 100 mM KCI, 2 mM CaCl2, 2 mM MgCl2, 5 mM dithiothreitol, 0.5% Nonidet P-40, 0.5 mM phenylmethylsulfonyl fluoride, 5% glycerol [9]) and incubated at room temperature for 30 min. Glutathione-agarose beads were added, and the resulting slurry was incubated for 60 min in a rotary devise at 5°C to bind GST-CyP A and complexed CA or Gag proteins. In format B, Gag or CA protein was loaded on a column of packed glutathione-agarose beads with immobilized GST-CyP A protein. Flowthrough (FT) and successive washes with TK buffer were collected. GST-CyP A was eluted from the bead with TK buffer containing 10 mM glutathione. Proteins in FT, washes, and eluates were separated by SDS-PAGE and analyzed for the presence of Gag or CA protein by Coomassie blue staining or Western blotting.
Cell culture, transfection, and preparation of cell lysates.
CH-1, a cell line derived from Cos7 that stably expresses all of the HIV-1 genes except env (6), was maintained in Dulbecco modified Eagle medium (GIBCO) supplemented with 10% fetal calf serum and 1% streptomycin-penicillin. Cell lysates were prepared by allowing cells to swell for 10 to 15 min in hypotonic buffer (10 mM Tris-HCl) [pH 7.4], 0.2 mM MgCl2) and then disrupting them with 30 strokes in a Dounce homogenizer with a tight-fitting pestle type B. EDTA was added to a final concentration of 1 mM. Nuclei and unbroken cells were removed by centrifugation for 10 min at 1,000 × g to obtain the cytosolic fraction.
Limited tryptic digestion.
Recombinant HIV-1 CA and Gag proteins in 100 mM Tris-HCl (pH 8.5) were subjected to limited digestion by trypsin (Roche) at a trypsin/total protein ratio of 1:100 (wt/wt) for various periods at 37°C. Digestion was stopped by addition of SDS-PAGE sample loading buffer and heating to 100°C. Substrates and tryptic fragments were separated by SDS-PAGE and visualized by either Coomassie blue staining or Western analysis.
Thiol modification of CA protein by FM.
His-tagged CA protein and GST-CyP A protein were mixed at a molar ratio of 1:2.5 and incubated at room temperature for 30 min. Fluorescein maleimide (FM; Molecular Probes, Inc.) in N,N-dimethyl formamide (10%, wt/vol) was used as stock solution from which a working solution in 10 mM sodium phosphate (NP) buffer (pH 6.0) (1:10, vol/vol) was prepared. The latter was added at 100-fold molar excess to the protein solution, and the labeling reaction mixture was incubated at room temperature for 2 h in the dark. At the end of the incubation period, the reaction mixture was loaded on a Ni-agarose column to remove excess FM. The FT was collected, and the column was washed extensively with several aliquots of the buffer until the yellow chromophore (FM) was no longer detectable. Bound N-His-tagged CA protein was eluted from the bead with buffer containing 500 mM imidazole. Proteins in the FT washes, and eluates were separated by SDS-PAGE and visualized by Coomassie blue staining or Western analysis.
Coimmunoprecipitation assay.
Samples used in the assay contained nondenatured CA and WT or W121F GST-CyP A proteins. Protein A-bead slurry was added to each sample, and the mixture was rotated for 30 min at 5°C for preclearing. CA protein in precleared samples was immunoprecipitated by addition of CA-specific rabbit antiserum. The mixture was warmed briefly (for 5 min) at 37°C and then incubated for 2 h at 5°C. Immune complexes were bound to protein A-beads by adding fresh slurry to the mixture and incubating for another hour at 5°C, this time with rotation. After centrifugation of the mixture, the supernatant was removed and complexes bound to the pelleted beads were extracted with 50 μl of 2× SDS-PAGE sample loading buffer (100 mM Tris-HCl [pH 6.8], 1.7 M β-mercaptoethanol, 4% SDS, 0.2% bromophenol blue, 20% glycerol). The extract was analyzed by Western blotting for CA and GST-CyP A proteins using mouse primary monoclonal antibodies (MAbs) specific to CA (α-CA MAb NEA-9306; Dupont-NEN) and GST (α-GST; Sigma), respectively.
SDS-PAGE and Western blot analysis.
Samples prepared for analysis on SDS-polyacrylamide gels (33) were mixed with an equal volume of 2× sample loading buffer, boiled for 3 min, and then loaded onto 12.5% gels. For visualization of all proteins separated, the gel was stained with Coomassie blue. For visualization of specific proteins or protein fragments, proteins on the gel were transferred onto a nitrocellulose membrane (Schleicher & Schuell) on a Milliblot-Graphite Electroblotter II (Bio-Rad), and the resulting blot was probed with appropriate primary and horseradish peroxidase-conjugated secondary antibodies (Amersham) as described in the text. Reactive protein bands were visualized by enhanced chemiluminescence (Amersham). Where indicated in the text, blots were stripped of associated antibodies by incubation with 3% trichloroacetic acid and reprobed with a different set of primary and secondary antibodies.
RESULTS
CTD of CA in Gag and in mature CA protein have different conformations.
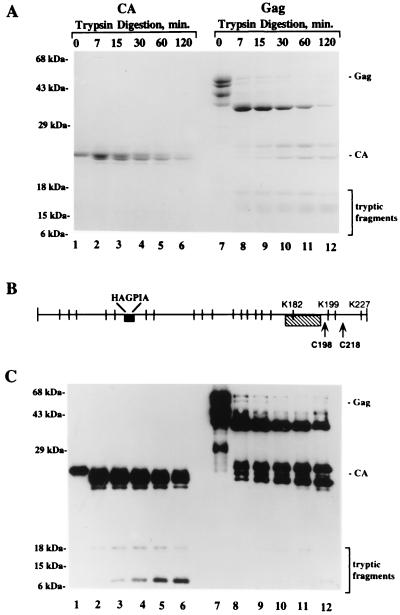
The accessibility of trypsin cleavage sites (Arg/X and Lys/X) was used to assess structure differences in the CTD of CA when in the context of Gag and when in the context of the mature protein. Both proteins were partially digested after incubation with the enzyme for 7 to 120 min (enzyme-to-substrate ratio = 1:100). Following limited trypsin digestion, residual Gag or CA protein and the newly generated fragments were separated by SDS-PAGE. Total protein was visualized by staining with Coomassie blue (Fig. 2A). The visually detectable yield of tryptic fragments from the Gag precursor indicated that the enzyme was not more limiting in the Gag than in the CA sample, even though the precursor contained more cleavage sites.
FIG. 2.
Limited trypsin digestion of Gag precursor and mature CA proteins. Equivalent amounts of purified recombinant Gag and CA proteins were subjected to partial proteolysis with trypsin as described in Material and Methods. The proteins in the digests were separated by SDS-PAGE and identified by Coomassie blue staining (A) or Western blotting with MAb 5-176, whose epitope resides within CA residues 178 to 196 (C). (B) Locations of Arg and Lys residues (vertical lines), the CyP A binding site (HAGPIA; solid box), cysteines 198 and 218 in CTD (arrows), and the region recognized by MAb 5-176 (hatched box). The brackets in panels A and C indicate the migration positions of tryptic fragments. Lanes 1 to 6, mature CA protein; lanes 7 to 12, Gag.
Tryptic fragments derived from the CTD of CA that were present in the digests were visualized by Western blotting with MAb 5-176. This MAb was generated using recombinant HIV-1 CA protein (17) as immunogen and recognizes both Gag and mature CA proteins. The epitope is within CA residues 178 to 196 as defined by mapping analysis using the CA deletion mutants described above (data not shown). As shown in Fig. 2B, this CA sequence in the CTD is flanked by several trypsin cleavage sites (K170, R173, K182, K199, K203, and K227). It is also adjacent to two highly conserved Cys residues (Cys198 and Cys218) which are both critical for HIV replication (37). As shown in Fig. 2C, fragments bearing the MAb 5-176 epitope were present in digests of the mature CA protein at the earliest time point (lane 2). In contrast, none were detected in digests of Gag even after 120 min of digestion (lane 12), although by this time residual substrate was barely detectable whereas tryptic fragments had accumulated in sufficient amounts to be stainable with Coomassie blue (Fig. 2A, lanes 8 to 12). This difference in accessibility of trypsin cleavage sites flanking the MAb 5-176 epitope indicates that the CTD assumes a different conformation in the context of the precursor and the mature CA protein.
Mature CTD conformation is contingent on removal of the p2 spacer peptide.
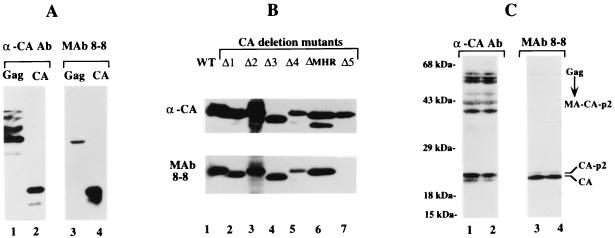
MAb 8-8 was generated using recombinant HIV-1 CA protein (17) as immunogen. This MAb recognizes the mature CA protein but, in contrast to MAb 5-176, not the full-length Pr55Gag precursor in a Western analysis (Fig. 3A, lanes 3 and 4). Analysis of mutants missing 20 amino acids from various regions in CA mapped the antibody's antigenic site to residues 178 to 196 (Fig. 3B, lane 7) in the CTD. This region coincides precisely with alpha helix 9, a region through which CA protein dimerization occurs (23). Note that MAb 8-8 also failed to recognize the faster migrating of two ΔMHR CA protein species (Fig. 3B, lane 6). These two CA species both have an N-terminal sequence that begins with Pro1 as revealed by Edman degradation analysis (data not shown), which indicates that the faster-migrating species (lane 6) is C-terminally truncated. Since the missing sequences are expected to be coincident with the assigned epitope of MAb 8-8, this nonrecognition of this species by the antibody corroborates the epitope assignment. However, it is also possible that the epitope exists elsewhere in the molecule and was masked by the mutation. Irrespective of where the epitope lies, the results in Fig. 3 demonstrate that the MAb 8-8 epitope is preferentially exposed in the mature CA protein rather than the Gag precursor.
FIG. 3.
Correlation of antigenic site 8-8 and cleavage of the p2 spacer peptide. (A) Specificity of MAb 8–8 for precursor versus mature protein. Recombinant Gag (lanes 1 and 3) and full-length mature CA1-231 (lanes 2 and 4) subjected to SDS-PAGE were analyzed by Western blotting using MAb 8-8 (lanes 3 and 4). The blot was stripped and reprobed with α-CA MAb NEA-9306 (lanes 1 and 2). (B) Mapping the antigenic site of MAb 8-8. Equal amounts of His-tagged full-length CA (lane 1) or mutant CA proteins containing 20-amino-acid deletions (lanes 2 to 7) were subjected to SDS-PAGE and analyzed by Western blotting with MAb 8-8 (bottom). The blot was stripped and reprobed with polyclonal α-CA antibody (top). (C) Western analysis of lysates prepared from CH-1 cells expressing HIV-1 proteins. Proteins in the cytoplasmic fraction of CH-1 cells were separated by SDS-PAGE on 15% gels and identified by Western blotting with MAb 8-8 (lanes 3 and 4). The blot was stripped and reprobed with α-CA MAb NEA-9306 (lanes 1 and 2). Lanes 1 and 3 and lanes 2 and 4 show two independent samples taken from adjacent sucrose density gradient fractions. Molecular weight markers are indicated on the left. The migration positions of Pr55Gag, CA, and cleavage intermediates are indicated on the right.
To delineate the processing event leading to exposure of the MAb 8-8 epitope, Gag processing intermediates and products from cytoplasmic extracts of CH-1 cells, which stably express the HIV-1 genome minus env (6), were separated by SDS-PAGE and assessed for recognition by MAb 8-8. As shown in Fig. 3C, Western analysis with MAb 8-8 revealed that this antibody recognized the mature CA protein efficiently but failed to detect the precursor or any of the cleavage intermediates (lanes 3 and 4). Stripping the nitrocellulose and reprobing with α-CA MAb NEA-9306 recognized the Pr55Gag precursor, mature CA protein, and bands that migrated at the molecular masses expected for the previously described CA-related cleavage intermediates, MA-CA-p2 (41 to 43 kDa), CA-p2-NC (nucleocapsid)-p1-p6 (∼39 kDa), and CA-p2 (25 kDa) (lanes 1 and 2) (6, 29). The identity of these bands was confirmed using antibodies against the MA, NC, and p6 domains in parallel experiments (data not shown). Thus, exposure of the epitope that is recognized by MAb 8-8 required removal of the p2 spacer peptide.
W121F CyP A binds Gag but not mature CA protein.
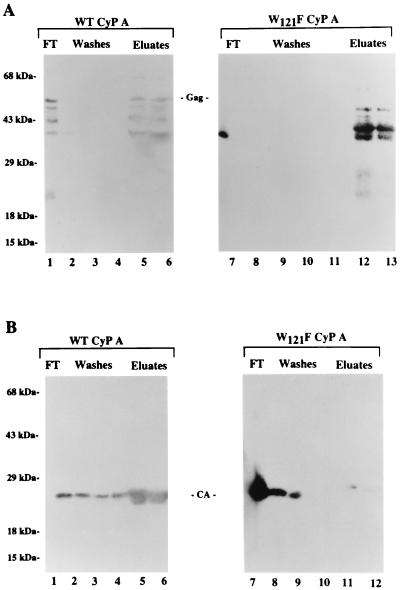
The differential interaction of W121F with CsA and Gag suggested that Trp121 plays a critical role in substrate recognition. To determine if this site could distinguish the Gag precursor and mature CA proteins, WT GST-CyP A or GST-CyP A-W121F was adsorbed to glutathione-agarose beads at 4°C as described previously (9). After removal of unbound protein, equivalent amounts of purified recombinant Gag or CA protein preparations were incubated with the CyP A-bead complex. As previously described (16), Pr55Gag expressed in E. coli is reproducibly isolated as full-length protein and three truncated fragments due to partial degradation by bacterial proteases (Fig. 2 and 3). The unbound proteins and the proteins retained by the beads were subjected to SDS-PAGE and identified by Western blotting using α-CA MAb NEA-9306.
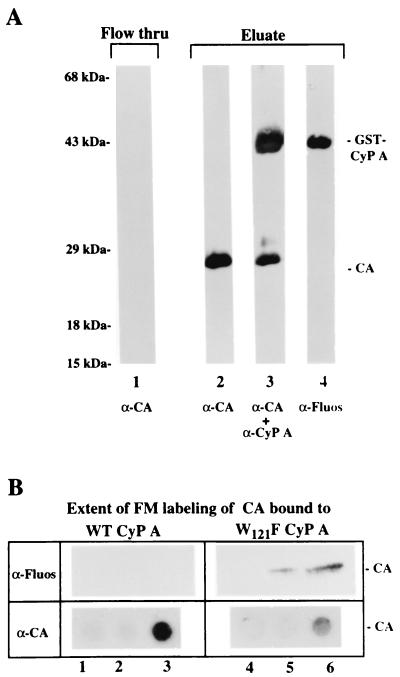
As expected, based on previous in vitro and in vivo studies (9, 14), the Pr55Gag precursor was retained by both WT CyP A (Fig. 4A, lanes 5 and 6) and the W121F mutant (lanes 12 and 13). Previous studies demonstrated that binding occurs through the CyP A domain of the GST fusion protein (9). In contrast to the WT protein, the W121F CyP A mutant bound the mature CA protein very inefficiently (Fig. 4B). A comparison of the amounts of bound CA protein in three independent experiments by matching signal intensities of different amounts of WT and mutant complexes indicated that WT GST-CyP A bound five-, five-, and fourfold more, respectively, mature CA protein than the W121F GST-CyP A mutant (data not shown). The results suggest that the W121 subsite in the substrate binding pocket determined the ability of CyP A to differentially recognize immature and mature forms of the CA protein.
FIG. 4.
In vitro binding of Gag and mature CA protein to WT and W121F CyP A. Recombinant Gag (A) or CA (B) protein was mixed with either WT (lanes 1 to 6) or W121F (lanes 7 to 13) GST-CyP A protein. Glutathione-agarose beads were added to each reaction mixture to bind GST-CyP A proteins and complexed Gag or CA protein. Following pelleting of the beads by centrifugation, the supernatant containing unbound protein (FT) was collected. The beads were washed three times with TK buffer (Washes) before successive elution with TK buffer containing glutathione (Eluates). Proteins in FT, washes, and eluates were subjected to SDS-PAGE followed by Western blotting. α-CA MAb NEA-9306 was used to detect the Gag and CA proteins. Molecular weight markers are on the left. The migration positions of full-length precursor (Gag) and 24-kDa mature CA protein (CA) are indicated.
W121F CyP A does not bind the truncated precursor.
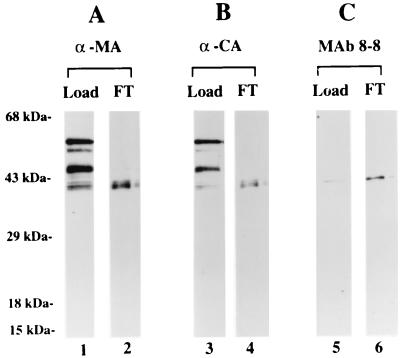
Closer examination of the elution profile of the W121F CyP A-Gag binding reaction shown in Fig. 4A revealed that in contrast to FT from the WT CyP A binding reaction, FT from the W121F CyP A binding reaction (lane 7) was enriched for the fastest-migrating Gag species. To determine if this was a technical aberration or an authentic feature of W121F CyP A, the experiment was repeated in a chromatography format where beads with immobilized W121F GST-CyP A protein were packed in a column. A solution of T7-tagged Gag protein was loaded onto the column under conditions where the ratio of total Gag protein to immobilized W121F CyP A protein was 1:2.5. The FT was collected, subjected to SDS-PAGE alongside the Gag sample used in the experiment, and analyzed by Western blotting (Fig. 5). As previously reported (16), the Gag sample contained several C-terminally truncated species, as indicated by their recognition by α-MA (α MA) (Fig. 5A, lane 1) and α-CA (Fig. 5B, lane 3) MAbs. The fastest-migrating Gag species was recognized by MAb 8-8 (Fig. 5C, lane 5), which indicates that the CTD in this truncated precursor already assumes the mature structure characteristic of the 24-kDa mature CA protein. Analysis of FT proteins demonstrated the near-exclusive presence of the fastest-migrating MA-CA-containing Gag species. The fact that the other Gag species were not represented in this fraction is consistent with our having done the binding experiment under conditions of excess column binding capacity. Thus, the presence of this Gag species in the FT is attributable to its inability to bind to the immobilized W121F CyP A. This corroborates the earlier observation shown in Fig. 4A. More importantly, that the MA-CA truncated Gag species was recognized by MAb 8-8 shows that acquisition of a mature CTD conformation does not require that CA be fully processed to the 24-kDa protein. Thus, there appears to be a causal relationship between two maturational refolding events, with the structural change originating in the CTD being relayed to the Pro-rich loop in the NTD.
FIG. 5.
Failure of truncated Gag precursor to bind to W121F CyP A. Recombinant HIV-1 Gag was loaded on a column of glutathione-agarose beads with immobilized W121F- GST CyP A protein under conditions of excess CyP A (CyP A:Gag = 2.5:1). Aliquots of the Gag preparation used for the binding reaction (Load) and the FT fraction were subjected to SDS-PAGE and analyzed by Western blotting. The blot was probed with α-MA (lanes 1 and 2), stripped, reprobed with α-CA MAb NEA-9306 (lanes 3 and 4), stripped, and reprobed with MAb 8-8 (lanes 5 and 6).
CyP A coimmunoprecipitates with CA protein derived from Gag complexed to WT and W121F CyP A.
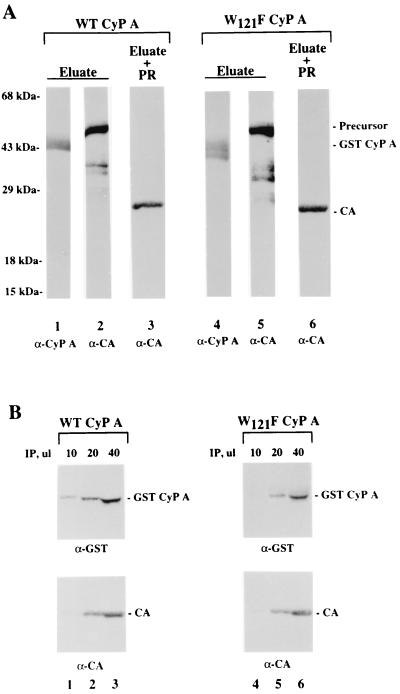
Taken together, the results above suggest that maturation-dependent changes initiating in the CTD of CA are transduced to the unliganded Pro-rich loop. In the natural setting, the ∼10% of assembled Pr55Gag is bound to CyP A. Are changes similarly transduced in this Pr55Gag population? To address this question, the Gag protein was complexed to bead-immobilized WT GST-CyP A or the W121F GST-CyP A mutant. These experiments used His-tagged Gag, as this could be isolated as a single predominant form of the precursor. Unbound Gag proteins were removed by extensive washing of the column with TK buffer. Bound Gag proteins were eluted with GST-CyP A proteins disengaged from the agarose matrix by washing with TK buffer containing 10 mM glutathione. An aliquot of both eluates was analyzed by Western blotting for GST-CyP A and Gag proteins using rabbit anti-human CyP A (BioMol) and anti-CA polyclonal antibodies, respectively. As shown in Fig. 6A, comparable amounts of the WT GST-CyP A (lane 1) and W121F GST-CyP A (lane 4) were in the column and comparable amounts of Gag precursor were complexed (lanes 2 and 5), which is consistent with the results in Fig. 4A. Next, eluates were dialyzed against MES (morpholineethanesulfonic acid) buffer (17), and HIV-1 PR was added to proteolytically cleave the Gag precursor proteins. As expected, the amount of Gag precursor diminished to undetectable levels, while mature 24-kDa CA protein was detected upon analysis of the digest by Western blotting (lanes 3 and 6). The ability WT GST-CyP A and W121F GST-CyP A proteins originally present in the eluates to bind the newly generated 24-kDa mature CA protein was tested by coimmunoprecipitation. The CA protein was reacted with rabbit serum containing CA-specific antibody. The immune precipitates were examined for the presence of WT and W121F GST-CyP A proteins by Western analysis. Since the blot is expected to contain the heavy (∼50-kDa) and light (∼25-kDa) chains from the rabbit immunoglobulin G (IgG) that was used in the immunoprecipitation assay, mouse MAbs were used as primary antibodies in the Western analysis (i.e., α-GST for detection of GST-CyP A and α-CA MAb NEA-9306 for detection of CA). With this Western regimen, horseradish peroxidase-conjugated sheep anti-mouse IgG antiserum was used as secondary antibody, which circumvented detection of the rabbit IgG components. As shown in Fig. 6B, almost comparable amounts of the WT and the W121F CyP A mutant were coimmunoprecipitated, indicating (i) that proteolytic processing of Gag with PR did not in itself cause release of CA protein from CyP A and (ii) that WT and W121F CyP A bound the newly formed CA protein to similar extents. Thus, binding of Gag to CyP A prior to proteolytic processing by the viral PR generated a mature CA protein that bound to the WT and the W121F mutant CyP A with comparable efficiency. This indicated that the CyP A-liganded Pro-rich loop is prevented from undergoing the maturation-dependent change seen in the unliganded Pro-rich loop (Fig. 4). Instead, the Pro-rich loop seemed to have been locked in the immature conformation of the precursor (i.e., it does not require the W121 site for stable binding to CyP A).
FIG. 6.
Coimmunoprecipitation of CyP A with CA protein derived from Gag complexed to WT and W121F CyP A. Gag was mixed with glutathione-agarose-immobilized WT or W121F GST-CyP A. The agarose beads were poured into a column and washed to remove unbound Gag proteins. GST-CyP A was eluted from the agarose matrix with buffer containing 10 mM glutathione. Eluates were analyzed for GST-CyP A and complexed Gag by Western blotting (A). The blot was probed with rabbit anti-CyP A polyclonal antibody to visualize eluted GST-CyP A (lanes 1 and 4), stripped, and reprobed with mouse α-CA MAb to visualize complexed Gag (lanes 2 and 5). Eluates were dialyzed against MES buffer and incubated with HIV-1 PR (17) at 37°C. Completeness of proteolytic processing of Gag was assessed by detection of mature 24-kDa CA protein following SDS-PAGE of the digest and analysis by Western blotting using mouse α-CA MAb (lanes 3 and 6). CA protein in the digest was immunoprecipitated using HIV-1 CA-specific rabbit antiserum and analyzed for coimmunoprecipitation of GST-CyP A by Western blotting (B). Immune precipitates from the digest of WT CyP A-Gag (lanes 1 to 3) and from the digest of W121F CyP A-Gag (lanes 4 to 6) were solubilized in SDS-PAGE loading buffer and loaded on SDS-polyacrylamide gels in three increments: 10 μl (lanes 1 and 4), 20 μl (lanes 2 and 5), and 40 μl (lanes 3 and 6). Separated proteins were transferred onto nitrocellulose, and the blot was probed with α-GST for GST-CyP A (top panels); the blot was stripped and reprobed with mouse α-CA MAb NEA-9306 for CA protein (bottom panels)
Binding to CyP A protected Cys residue(s) in mature CA protein CTD from thiol modification.
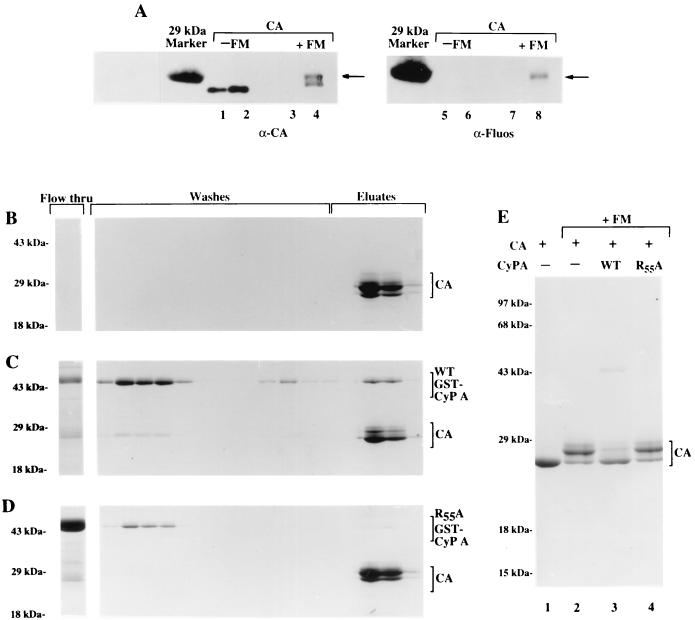
The results described above provide evidence that structure information is relayed from the CTD of CA to the Pro-rich loop. This observation suggests the following question: Does structure change originating from the Pro-rich loop also get relayed to the CTD of CA? A suggestion that this may be so was provided by our earlier observation that CyP A binding diminished disulfide bond formation by mature CA protein (3). To test this hypothesis directly, the effect of CyP A binding on the accessibility of the C-terminally located Cys residues to Cys-modifying reagents, fluorescein acetamide or FM, was examined using a gel shift assay (Fig. 7). Essentially identical results were obtained with fluorescein acetamide; only studies using FM, the more specific reagent, are presented below. All experiments were conducted at pH 6.0, where the maleimide specificity for thiol groups is highest. Figure 7A illustrates the extent to which modified CA proteins are retarded in their migration during SDS-PAGE. Two samples of CA1-231 were used in the experiment. The migration positions of the CA samples incubated with solvent buffer alone (Fig. 7A, lanes 1 and 2) confirm the relative amounts of CA protein in the samples. Incubation with FM resulted in the retarded electrophoretic migration of half of the original CA protein (lanes 3 and 4). The slower-migrating CA band was recognized by antifluorescein MAb (α-Fluos) (lane 8), indicating covalent attachment of the fluorescein moiety to the CA protein. Evidently, the distribution of the CA protein in two populations has lowered the protein-to-area ratio of the protein bands, accounting for the low signals obtained in lanes 3, 4, 7, and 8. Nonetheless, the study demonstrates that as reported for other FM-reactive proteins (28), there is correlation between gel shift and chemical modification of the CA protein.
FIG. 7.
Thiol modification of CA protein by FM in the presence of WT and R55A CyP A. (A) Retarded migration of FM-labeled CA1-231. Labeling with FM was done as described in the text. Samples incubated with solvent buffer only (−FM) or with the reagent (+FM) were analyzed by Western blotting. The blot was probed with α-CA MAb NEA-9306 for CA proteins (left), stripped, and reprobed with α-Fluos (right). Arrow indicates slower-migrating CA protein with covalently attached fluorescein moiety. (B to D) Coomassie blue-stained SDS-polyacrylamide gels of N-His-tagged CA protein eluted from Ni-agarose columns after labeling with FM. FM was added to N-His-tagged CA in buffer (B), N-His-tagged CA with WT GST-CyP A (C), and N-His-tagged CA with R55A GST-CyP A mutant (D); at the end of the labeling period, each reaction mixture was loaded onto a Ni-agarose column to separate the proteins from excess FM. Aliquots of FT washes, and eluates, obtained as described in Materials and Methods, were subjected to SDS-PAGE, and separated proteins were stained by Coomassie blue. (E) Eluate from each of the labeling reactions in panels B to D and unlabeled CA protein were loaded on an SDS-polyacrylamide gel, and the proteins were visualized by Coomassie blue staining. Lane 1, CA protein in buffer in the absence of FM; lanes 2 to 4, CA protein reacted with FM in the absence (lane 2) and presence of WT GST-CyP A (lane 3) or R55A GST-CyP A (lane 4).
The effect of CyP A binding on FM labeling was examined by incubating the CA protein with FM in the presence and absence of WT GST-CyP A (2.5-fold molar excess to the CA protein). Parallel studies were conducted with R55A GST-CyP A, a CyP A mutant impaired in stable binding to Pr55Gag (9, 14). The use of His-tagged CA facilitated the removal of unbound FM at the end of the reaction by chromatography of the samples on Ni-agarose columns. The scaled-up reaction mixtures (i.e., proteins in milligram quantities) allowed for visualization of proteins in the fractions by Coomassie blue staining. Figures 7B to D show Coomassie blue-stained SDS-polyacrylamide gels containing FT washes and eluates from columns loaded with FM labeling reactions containing CA protein alone, CA and WT GST-CyP A protein, and CA and R55A GST-CyP A mutant protein, respectively. In all three cases, both the yellow color and fluorescence attributable to FM were most intense in the FT fraction and were reduced to undetectable levels by washing (data not shown). The experiment represented in Fig. 7B was carried out as a control for labeling of the CA sample in the absence of GST-CyP A and for binding to the Ni-agarose column. Figure 7C shows that WT GST-CyP A was distributed among the FT (∼10%), 20 mM imidazole wash (∼80%), and CA protein-containing eluates (∼10%). In contrast, the R55A GST-CyP A mutant protein was distributed between the FT (∼80%) and the 20 mM imidazole washes (∼10%) and barely detectable in CA protein-containing eluates (Fig. 7D).
Figure 7E shows a Coomassie blue-stained gel with the CA protein used in the experiments and eluates from each of the labeling reactions described. Unmodified CA protein migrated as a single band at ∼24 kDa (lane 1). Incubation with FM diminished the intensity of this population and generated two additional species that migrated more slowly (lane 2). All three CA protein populations were detected in the sample incubated with WT GST-CyP A (lane 3), but the staining intensities of the slower-migrating species were considerably reduced and that of the fastest-migrating band was increased. In contrast, the CA protein preincubated with R55A GST-CyP A (lane 4) exhibited a pattern that was almost identical to that of CA protein modified by FM in the absence of CyP A (lane 2). Thus, CyP A induced a C-terminal structural change from one in which the Cys residues were highly exposed to one in which they were inaccessible.
A CA mutant missing the HAGPIA sequence is not protected by CyP A from thiol modification.
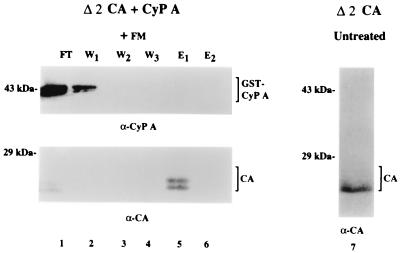
To ascertain that binding at Pro90 in the CA protein effected the CyP A-mediated conformational change, we determined the effect of deleting the N-terminal Pro-rich loop that contains the binding site. A CA mutant lacking residues 78 to 97 (Δ2 CA) (Fig. 8, lane 7) was incubated with FM and CyP A and examined for CyP A-induced changes in Cys accessibility (lanes 1 to 6). As expected, the mutant failed to bind CyP A, as indicated by the observation that CyP A was recovered in the FT and early washes (lanes 1 and 2) and not in the eluate fraction containing CA (lane 5). As described above for the WT CA protein incubated under conditions where CyP A does not bind (i.e., R55A GST-CyP A in Fig. 7), the addition of FM retarded the migration of a fraction of the Δ2 CA mutant protein (Fig. 8, lane 5). The results indicate that the observed changes in the CTD were mediated by binding of the Pro-rich loop in the NTD to CyP A.
FIG. 8.
Thiol modification of Δ2 CA mutant protein in the presence of WT CyP A. His-tagged CA protein lacking residues 78–97 (Δ2 CA) and WT GST-CyP A were mixed with 10 mM NP buffer at pH 6.0 at a ratio of 1 to 2.5 (CA to CyP A) and incubated at room temperature for 30 min. Labeling with FM, removal of unreacted FM by Ni-agarose chromatography, elution of bound CA protein, and gel analysis were as described in the text. Lanes 1 to 6, proteins in the FT, washes, and eluates analyzed by Western for GST-CyP A (α-CyP A; top) and for CA (α-CA; bottom). Lane 7, the N-His-tagged Δ2 CA mutant protein used in the experiment.
The W121F CyP A mutant does not protect mature CA protein from thiol modification.
The observed effect of CyP A on the C-terminal Cys residues might be mediated by the binding or isomerization functions of CyP A. The W121F CyP A mutant was used to address this question. The mutation does not affect binding to the site in Gag, as indicated by its incorporation in the virion at WT levels, but is 50% impaired for prolyl isomerase activity (14, 34). To dissociate binding and isomerization functions so that the role of the prolyl isomerase could be evaluated, the Cys modification of CA proteins that remained complexed to CyP A was examined (Fig. 9). The CA protein was mixed with glutathione-agarose beads to which W121F GST-CyP A protein (or WT GST-CyP A in control experiments) has been immobilized. After extensive washing to remove any uncomplexed CA proteins, the beads were resuspended in fresh buffer, FM was added, and the reaction mixture was incubated for 2 h at room temperature with constant rotation. At the end of the incubation period, excess FM was removed by pouring the slurry in a chromatography column and extensive washing of the packed beads. Proteins were eluted from the beads with buffer containing 10 mM glutathione. FT and eluates were analyzed for GST-CyPA and CA proteins by Western blotting (Fig. 9A). For both columns, practically all bound proteins were detected in the first eluate fraction.
FIG. 9.
Thiol modification of mature CA protein while bound to WT and W121F CyP A. Equivalent amounts of GST-CyP A and CyP A-W121F were immobilized to glutathione-agarose beads as described in the text, and the beads were packed in a column. N-His-tagged CA protein was loaded into each column, and unbound CA protein was removed by extensive washing with TK buffer. The washed beads were resuspended in NP buffer, FM was added, and the slurry was incubated in the dark for 2 h with constant rotation. At the end of the labeling period, the slurry was packed in a minicolumn, and excess FM was removed by extensive washing with buffer. CA protein-CyP A complexes that remained attached to the bead were eluted with 10 mM reduced glutathione, mixed with 0.2 volume of 5× SDS sample buffer, and analyzed by PAGE and Western blotting. (A) Analysis of the FT and eluate fractions from the sample containing CA protein bound to WT CyP A. Shown is a blot with FT (lane 1) and eluate (lane 2) probed with α-CA MAb NEA-9306 (α-CA). In lane 3, the eluate-containing blot shown in lane 2 was subsequently probed with CyP A-specific polyclonal antibody (α-CA + α-CyP A). In lane 4, the eluate-containing blot shown in lane 3 was stripped, checked, and then reprobed with α-Fluos. (B) Comparison of extent of FM modification in CA proteins bound to the WT CyP A (left) or to the W121F CyP A mutant (right). The top strip shows the 24-kDa region of the blot probed with α-Fluos; the bottom strip shows a dot blot of eluate samples probed with the α-CA MAb NEA-9306. Lanes 1 to 3, 5, 10, and 15 μl, respectively, of the eluate from the sample containing CA protein and WT CyP A; lanes 4 to 6, 20, 40, and 80 μl, respectively, of eluate from the sample containing CA and W121F CyP A mutant.
As expected (since the experiment was designed to have bead-immobilized GST-CyP A in excess of loaded CA protein), α-CA MAb failed to detect CA protein in the FT fraction, indicating the absence of unbound CA proteins (Fig. 9A, lane 1). In contrast, the α-CA MAb elicited a strong signal for CA in the eluate fraction obtained following disruption by glutathione of the interaction of GST with the bead matrix (lane 2). Subsequent probing without stripping of the same immunoblot with CyP A-specific antibody revealed an additional signal at 43 kDa representing GST-CyP A (lane 3). Results shown in Fig. 7 predicted that Cys residue(s) in CA protein complexed to WT GST-CyP A will not be labeled by FM. Consistent with this expectation, the GST-CyP A band, but not the CA protein band, was recognized after stripping of the blot and reprobing with α-Fluos (lane 4).
Figure 9B shows results of a comparison of the extent of modification of Cys residue(s) in CA proteins complexed to WT and to the W121F GST-CyP A. The amount of CA protein in the eluates were initially estimated in a Coomassie blue-stained SDS-polyacrylamide gel (data not shown). On the basis of the relative staining intensities of the 24-kDa CA protein band, the eluates were matched for CA protein content. For assessment of the level of CA-conjugated fluorescein (upper strip), 5, 10, and 15 μl of eluate from the reaction with WT (lanes 1 to 3) and from the reaction with the W121F mutant (lanes 4 to 6) GST-CyP A were analyzed by Western blotting using α-Fluos as the probe. The amount of CA in these eluates was again assessed, this time by Western dot blot analysis probed with α-CA MAb (lower strip). Consistent with earlier results (Fig. 7), the CA protein in the sample containing WT CyP A was not labeled, as indicated by the absence of detectable α-Fluos signal. In contrast, α-Fluos signal was detected in the 24-kDa CA protein band from the eluate with the W121F CyP A mutant. This result indicated that binding to the mutant CyP A did not induce the structure change mediated by the WT CyP A that made the Cys less accessible to FM modification. These results suggest participation of prolyl isomerase activity or the interaction between the W121 site in CyP A and the I91-A92-P93 residues in CA that it contacts.
DISCUSSION
In this report, we describe results indicating that while CyP A binds both Gag and mature CA protein, the two binding interactions are actually different. In GST pulldown experiments, WT CyP A bound both Gag and mature CA protein samples. However, the CyP A mutant W121F bound Gag but not mature CA protein. Molecular detail of the CyP A active site cleft based on the crystal structure of the protein shows W121 to be located near one end of the CyP A active site cleft (30, 31). Our observation that binding of mature CA was impaired by the mutation suggests that the CA binding sequence, HAGPIA, extends to this end of the cleft and requires direct interaction with W121 or with residues whose orientation in the cleft depended on W121. In this respect, the situation resembles the disposition of HAGPIA in the active site cleft as captured in the complex of CyP A with an N-terminal fragment of CA (CA1-151 [22]) and the short HAGPIA peptide (48). In both structure models, W121 is shown to form a direct hydrogen bond with the isoleucine residue in HAGPIA. Hydrogen bonding interaction is made by the backbone carbonyl oxygen atom of isoleucine with the hydrogen atom of the heterocyclic aromatic amine side chain of tryptophan. The indifference of Gag binding to the W121F mutation reflects a HAGPIA docking that does not require W121 for stabilization. This reflects a difference in the steric constraints imposed by residues flanking the Pro-rich loop. Hence, while the CyP A active site cleft can accommodate both immature (in Gag context) and mature (in mature CA context) Pro-rich loops, our studies show that the two conformations are distinguished by the W121 subsite (i.e., HAGPIA in mature Pro-rich loop requires interaction with W121 subsite). This maturation of the Pro-rich loop correlated with maturation of the CTD. Conceivably, structural changes initiated by maturational refolding in the CTD were transduced to the Pro-rich loop, leading to acquisition of a mature conformation by the latter.
Interestingly, we found that binding Gag to CyP A prior to proteolytic processing blocked the maturation-dependent change in the Pro-rich loop. The loop was locked in the immature conformation despite being in a 24-kDa CA protein. This finding predicts that natural immature HIV-1 particles formed by assembly of unliganded and CyP A-liganded Gag precursors will, upon maturation, release two populations of 24-kDa mature CA proteins that differ in Pro-rich loop conformation. The exact role of the Pro-rich loop in the virus life cycle is not known, making an assessment of the significance of having these two populations of mature CA proteins tentative. A role in core shell formation is unlikely or subtle at best, since cores with WT morphology are formed in the absence of incorporated CyP A (19, 44, 47). A role in core disassembly has been proposed (22, 36) where by if the Pro-rich loop is part of an associative interface, CyP A binding can be instrumental in dissolution of interface interaction, thereby influencing core disassembly.
Our finding that the HAGPIA sequences in the Pro-rich loop of Gag and of mature CA protein have different subsite requirements make them essentially distinct CyP A substrates. Given the importance of W121 to the prolyl isomerase activity of CyP A (34, 51), the action of CyP A on these two substrates can have different outcomes. Hence, the interaction of Pr55Gag with CyP A in producer cells and the interaction of mature CA proteins with CyP A (i.e., in the virion or in the cytosol of target cells) are distinct, with conceivably different influences on postassembly function of the CA protein. Relevant to this idea is that certain features on HIV-1 replication are not fully explained by CyP A-Pr55Gag interaction: (i) virions devoid of incorporated CyP A by virtue of being produced in cells where both CyP A alleles have been knocked out replicated with an initial lag but eventually displayed normal kinetics (Braaten and Luban, Abstr. 2000 Meet. Retroviruses, 2000); and (ii) forced endocytic entry rescued replication of virions that did not incorporate CyP A due to the presence of CsA during assembly but did not rescue replication of virions that did not incorporate CyP A as a result of a mutation in HAGPIA (4). Are these observations expainable by CyP A-mature CA protein interaction?
We examined the structural consequence of mature CA protein binding to CyP A using the C-terminally located Cys residues in CA as reporters of change in the region. Our examination focused on this region of the CTD, since an earlier result indicated that proclivity for disulfide bond formation was diminished in the presence of CyP A (3). We found that CyP A induced a structural change in the region that altered the accessibility of the Cys residue(s) to chemical modification. The structure change was not elicited when the CA protein was bound to W121F CyP A mutant, indicating that CyP A action required interaction of CA atoms with W121 itself or interaction with structural elements whose existence depended on W121. The structural change was also not elicited when CyP A was mixed with a CA protein mutant (Δ2 CA) that was missing HAGPIA and several flanking residues, indicating that the observed change was the consequence of CyP A action on HAGPIA. We can only speculate on how a change in HAGPIA located in the NTD can be transduced to a region in the CTD. It is noteworthy that Hong and Boulanger (26) found that the substitution of Pro for Leu136 in helix 7 of the NTD prevented virus-like particle formation. However, particle assembly was restored by complementation with another mutant in which Ser was substituted for Leu190 in helix 9 in the CTD. Le136 is also part of an interface in the crystal structure of the NTD (22). Leu190 is also part of an interface in the crystal structure of the CA CTD (23) and the full-length CA protein (7). Formation of an interface between helix 7 in the NTD of one subunit and helix 9 in the CTD of another subunit might place Cys198 and Pro90 in proximity. Such topology of the Pro-rich loop of one subunit and the CTD of another subunit is seen in the crystal structure of a tetrameric form of equine infectious anemia virus (EIAV) (27). Transduction of structure information within the CA polypeptide, while much more difficult to envision, cannot be eliminated.
Conceivably, the CyP A-induced structural change in the CTD has an influence on mature CA function. If the changes in the accessibility of the Cys residues (Cys198 and/or Cys218) reflect a global change in the CTD, the conformation of adjacent sequences that participate in CA dimerization may be changed as well, affecting CA protein-protein interaction. The CyP A-modified mature CA proteins might not coassemble with unliganded proteins. These may be sequestered elsewhere in the virion to provide a source for CyP A, CA protein, or CyP A-CA protein complex for yet undefined roles in productive entry of the virus in target cells (42, 43). Even if the CyP A-induced structural change is confined to the immediate region containing the Cys residues, an effect on CA function may still be obtainable. A relationship between oligomerization and the oxidation state of the Cys residues in the CTD has been proposed by Khorasanizadeh et al. (32). The authors made the observation that the structure of human T-cell leukemia virus type 1 (HTLV-1) CTD, which does not form dimers in solution, show the Cys to be in reduced form (32), whereas structures of HIV-1 CTD (23) and EIAV CA (27), both of which readily form dimers in solution, have Cys residues in oxidized form. It is possible that the CyP A-induced change in structure that made HIV-1 CA Cys residue(s) less accessible to chemical modification that we observed may also render them less accessible to oxidative agents. Based on the HTLV-1 model, reduced Cys in select CA subunits would create sites favorable for dissolution of subunit-subunit contacts and hence influence virus uncoating.
Taken together, results described in this report introduce the notion that the binding function that results in CyP A incorporation may be separate from a second, postassembly role for CyP A. We have shown that the end result of CyP A binding of the Pro-rich loop, whether the loop is in precursor or mature CA protein context, is a structurally modified mature CA protein. The functional consequence(s) of these structure changes may mediate the influence of CyP A on virus infectivity.
ACKNOWLEDGMENTS
We are grateful to Indra Jayatilaka and the Tissue Culture Facility, Department of Molecular Genetics and Microbiology, for excellent technical assistance. We thank Jeremy Luban, Douglas Braaten, Lilia Babe, and Charles Craik for generously providing plasmids and cells.
This work was supported by grant GM 48294 to C.C.
REFERENCES
- 1.Accola M A, Höglund S, Göttlinger H G. A putative α-helical structure which overlaps the capsid-p2 boundary in the human immunodeficiency virus type 1 Gag precursor is crucial for viral particle assembly. J Virol. 1998;72:2072–2078. doi: 10.1128/jvi.72.3.2072-2078.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ackerson B, Rey O, Canon J, Krogstad P. Cells with high cyclophilin A content support replication of human immunodeficiency virus type 1 Gag mutants with decreased ability to incorporate cyclophilin A. J Virol. 1998;72:303–308. doi: 10.1128/jvi.72.1.303-308.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Agresta B E, Carter C A. Cyclophilin A-induced alterations of human immunodeficiency virus type 1 CA protein in vitro. J Virol. 1997;71:6921–6927. doi: 10.1128/jvi.71.9.6921-6927.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aiken C. Pseudotyping human immunodeficiency virus type 1 (HIV-1) by the glycoprotein vesicular stomatitis virus targets HIV-1 entry to an endocytic pathway and suppresses both the requirement for Nef and the sensitivity to cyclosporin A. J Virol. 1997;71:5871–5877. doi: 10.1128/jvi.71.8.5871-5877.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arthur L O, Bess J W, Jr, Sowder II R C, Benveniste R E, Mann D L, Chermann J-C, Henderson L E. Cellular proteins bound to immunodeficiency viruses: implications for pathogenesis and vaccines. Science. 1992;258:1935–1938. doi: 10.1126/science.1470916. [DOI] [PubMed] [Google Scholar]
- 6.Babe L M, Craik C S. Constitutive production of nonenveloped human immunodeficiency virus type 1 particles by a mammalian cell line and effects of a protease inhibitor on particle maturation. Antimicrob Agents Chemother. 1994;38:2430–2439. doi: 10.1128/aac.38.10.2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berthet-Colominas C, Monaco S, Novelli A, Sibai G, Mallet F, Cusack S. Head-to-tail dimers and interdomain flexibility revealed by the crystal structure of HIV-1 capsid protein (p24) complexed with a monoclonal antibody Fab. EMBO J. 1999;18:1124–1136. doi: 10.1093/emboj/18.5.1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Braaten D, Franke E K, Luban J. Cyclophilin A is required for an early step in the life cycle of human immunodeficiency virus type 1 before the initiation of reverse transcription. J Virol. 1996;70:3551–3560. doi: 10.1128/jvi.70.6.3551-3560.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Braaten D, Ansari H, Luban J. The hydrophobic pocket of cyclophilin is the binding site for the human immunodeficiency virus type 1 Gag polyprotein. J Virol. 1997;71:2107–2113. doi: 10.1128/jvi.71.3.2107-2113.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Briggs C J, Tözser J, Oroszlan S. Effect of cyclosporin A on the replication cycle of human immunodeficiency virus type 1 derived from H9 and Molt-4 producer cells. J Gen Virol. 1996;77:2963–2967. doi: 10.1099/0022-1317-77-12-2963. [DOI] [PubMed] [Google Scholar]
- 11.Bristow R, Byrne J, Squirell J, Trencher H, Carter T, Rodgers B, Saman E, Duncan J. Human cyclophilin has a significantly higher affinity for HIV-1 recombinant p55 than p24. J Acquir Immune Defic Syndr Hum Retroviruses. 1999;20:334–336. doi: 10.1097/00042560-199904010-00002. [DOI] [PubMed] [Google Scholar]
- 12.Bukovsky A, Weimann A, Accola M, Göttlinger H G. Transfer of the HIV-1 cyclophilin-binding site to simian immunodeficiency virus from Macaca mulatta can confer both cyclosporin sensitivity and cyclosporin dependence. Proc Natl Acad Sci USA. 1997;94:10943–10948. doi: 10.1073/pnas.94.20.10943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Campos-Olivas R, Summers M F. Backbone dynamics of the N-terminal domain of the HIV-1 capsid protein and comparison with the G94D mutant conferring cyclosporin resistance/dependence. Biochemistry. 1999;38:10262–10271. doi: 10.1021/bi990991x. [DOI] [PubMed] [Google Scholar]
- 14.Dorfman T, Weimann A, Borsetti A, Walsh C T, Göttlinger H G. Active site residues of cyclophilin A are crucial for its incorporation into human immunodeficiency virus type 1 virions. J Virol. 1997;71:7110–7113. doi: 10.1128/jvi.71.9.7110-7113.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ebbets-Reed D, Scarlata S, Carter C A. The major homology region of the HIV-1 Gag precursor influences membrane affinity. Biochemistry. 1996;35:14268–14275. doi: 10.1021/bi9606399. [DOI] [PubMed] [Google Scholar]
- 16.Ehrlich L S, Fong S, Scarlata S, Zybarth G, Carter C. Partitioning of HIV-1 Gag and Gag-related proteins to membranes. Biochemistry. 1996;35:3933–3943. doi: 10.1021/bi952337x. [DOI] [PubMed] [Google Scholar]
- 17.Ehrlich L S, Kräusslich H-G, Wimmer E, Carter C A. Expression in Escherichia coli and purification of human immunodeficiency virus type 1 capsid protein (p24) AIDS Res Hum Retroviruses. 1990;6:1169–1175. doi: 10.1089/aid.1990.6.1169. [DOI] [PubMed] [Google Scholar]
- 18.Endrich M M, Gehrig P, Gehring H. Maturation-induced conformational changes of HIV-1 capsid protein and identification of two high affinity sites for cyclophilins in the C-terminal domain. J Biol Chem. 1999;274:5326–5332. doi: 10.1074/jbc.274.9.5326. [DOI] [PubMed] [Google Scholar]
- 19.Franke E K, Yuan H E H, Luban J. Specific incorporation of cyclophilin A into HIV-1 virions. Nature. 1994;372:359–362. doi: 10.1038/372359a0. [DOI] [PubMed] [Google Scholar]
- 20.Freskgard P-O, Bergenhem N, Jonsson B-H, Svensson M, Carlsson U. Isomerase and chaperone activity of prolyl isomerase in the folding of carbonic anhydrase. Science. 1992;258:466–468. doi: 10.1126/science.1357751. [DOI] [PubMed] [Google Scholar]
- 21.Fuller S D, Wilk T, Gowen B E, Kräusslich H-G, Vogt V. Cryo-electron microscopy reveals ordered domains in the immature HIV-1 particles. Curr Biol. 1997;7:729–738. doi: 10.1016/s0960-9822(06)00331-9. [DOI] [PubMed] [Google Scholar]
- 22.Gamble T R, Vajdos F F, Yoo S, Worthylake D K, Houseweart M, Sundquist W I, Hill C P. Crystal structure of human cyclophilin A bound to the amino-terminal domain of HIV-1 capsid. Cell. 1996;87:1285–1294. doi: 10.1016/s0092-8674(00)81823-1. [DOI] [PubMed] [Google Scholar]
- 23.Gamble T R, Yoo S, Vajdos F F, von Schwedler U K, Worthylake D K, Wang H, McCutcheon J P, Sundquist W I, Hill C P. Structure of the carboxyl-terminal dimerization domain of the HIV-1 capsid protein. Science. 1997;278:849–853. doi: 10.1126/science.278.5339.849. [DOI] [PubMed] [Google Scholar]
- 24.Gitti R K, Lee B M, Walker J, Summers M F, Yoo S, Sundquist W I. Structure of the amino-terminal core domain of the HIV-1 capsid protein. Science. 1996;273:231–235. doi: 10.1126/science.273.5272.231. [DOI] [PubMed] [Google Scholar]
- 25.Handschumacher R, Harding M, Rice J, Drugge R. Cyclophilin: a specific cytosolic binding protein for cyclosporin A. Science. 1984;226:544–547. doi: 10.1126/science.6238408. [DOI] [PubMed] [Google Scholar]
- 26.Hong S S, Boulanger P. Assembly-defective point mutants of the human immunodeficiency virus type 1 Gag precursor phenotypically expressed in recombinant baculovirus-infected cells. J Virol. 1993;67:2787–2798. doi: 10.1128/jvi.67.5.2787-2798.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jin Z, Jin L, Peterson D L, Lawson C L. Model for lentivirus capsid core assembly based on crystal dimers of EIAV p26. J Mol Biol. 1999;286:83–93. doi: 10.1006/jmbi.1998.2443. [DOI] [PubMed] [Google Scholar]
- 28.Jones P C, Sivaprasadarao A, Wray D, Findlay J B C. A method for determining transmembrane protein structure. Mol Membr Biol. 1996;13:53–60. doi: 10.3109/09687689609160575. [DOI] [PubMed] [Google Scholar]
- 29.Kaplan A H, Swanstrom R. Human immunodeficiency virus type 1 Gag proteins are processed in two cellular compartments. Proc Natl Acad Sci USA. 1991;88:4528–4532. doi: 10.1073/pnas.88.10.4528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kallen J, Walkinshaw M D. The X-ray structure of a tetrapeptide bound to the active site of human cyclophilin A. FEBS Lett. 1992;300:286–290. doi: 10.1016/0014-5793(92)80865-e. [DOI] [PubMed] [Google Scholar]
- 31.Ke H, Zydowsky L D, Lui J, Walsh C T. Crystal structure of recombinant human T-cell cyclophilin A at 2.5 Å resolution. Proc Natl Acad Sci USA. 1991;88:9483–9487. doi: 10.1073/pnas.88.21.9483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Khorasanizadeh S, Campos-Olivas R, Summers M F. Solution structure of the capsid protein from the human T-cell leukemia virus type 1. J Mol Biol. 1999;291:491–505. doi: 10.1006/jmbi.1999.2986. [DOI] [PubMed] [Google Scholar]
- 33.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 34.Liu J, Chen C-M, Walsh C T. Human and Escherichia coli cyclophilins: sensitivity to inhibition by the immunosuppressant cyclosporin A correlates with a specific tryptophan residue. Biochemistry. 1991;30:2306–2310. doi: 10.1021/bi00223a003. [DOI] [PubMed] [Google Scholar]
- 35.Luban J. Absconding with the chaperone: Essential cyclophilin-Gag interaction in HIV virions. Cell. 1996;87:1157–1159. doi: 10.1016/s0092-8674(00)81811-5. [DOI] [PubMed] [Google Scholar]
- 36.Luban J, Bossolt K A, Franke E K, Kalpana G V, Goff S P. Human immunodeficiency virus type 1 gag protein binds to cyclophilins A and B. Cell. 1993;73:1067–1078. doi: 10.1016/0092-8674(93)90637-6. [DOI] [PubMed] [Google Scholar]
- 37.McDermott J, Farrell L, Ross R, Barklis E. Structural analysis of human immunodeficiency virus type 1 Gag protein interactions, using cysteine-specific reagents. J Virol. 1996;70:5106–5114. doi: 10.1128/jvi.70.8.5106-5114.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Momany C, Kovari L C, Prongay A, Keller W, Gitti R K, Lee B M, Gorbalenya A E, Tong L, McClure J, Ehrlich L S, Summers M F, Carter C, Rossmann M G. Crystal structure of dimeric HIV-1 capsid protein. Nat Struct Biol. 1996;3:763–770. doi: 10.1038/nsb0996-763. [DOI] [PubMed] [Google Scholar]
- 39.Nermut M V, Hockley D J, Jowett J B M, Jones I M, Garreau M, Thomas D. Fullerene-like organization of the HIV-1 gag-protein shell in virus-like particles produced by recombinant baculovirus. Virology. 1994;198:288–296. doi: 10.1006/viro.1994.1032. [DOI] [PubMed] [Google Scholar]
- 40.Ott D E, Coren L V, Johnson D G, Kane B P, Sowder II R C, Kim Y D, Fisher R J, Zhou X Z, Lu K P, Henderson L E. Actin-binding cellular proteins inside human immunodeficiency virus type 1. Virology. 2000;266:42–51. doi: 10.1006/viro.1999.0075. [DOI] [PubMed] [Google Scholar]
- 41.Ratner L, Haseltine W, Patarca R, Livak K J, Starchich B, Josephs S F, Doran E R, Rafalski J A, Whitehorn E A, Baumeister K, Ivanoff L, Petteway S R, Pearson M L, Lautenberger J A, Papas T S, Ghrayeb J, Chang N T, Gallo R C, Wong-Staal F. Complete nucleotide sequence of the AIDS virus HTLV-III. Nature. 1985;313:277–284. doi: 10.1038/313277a0. [DOI] [PubMed] [Google Scholar]
- 42.Saphire A C S, Bobardt M D, Gallay P A. Host cyclophilin A mediates HIV-1 attachment to target cells via heparans. EMBO J. 1999;18:6771–6785. doi: 10.1093/emboj/18.23.6771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sherry B, Zybarth G, Alfano M, Dubrovsky L, Mitchell R, Rich D, Ulrich P, Bucala R, Cerami A, Bukrinsky M. Role of cyclophilin A in the uptake of HIV-1 by macrophages and T lymphocytes. Proc Natl Acad Sci USA. 1998;95:1758–1763. doi: 10.1073/pnas.95.4.1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Steinkasserer A, Harrison R, Billich A, Hammerschmid F, Werner G, Wolff B, Peichl P, Palfi G, Schnitzel W, Mlynar E, Rosenwirth B. Mode of action of SDZ NIM 811, a nonimmunosuppressive cyclosporin A analog with activity against human immunodeficiency virus type 1 (HIV-1): interference with early and late events in HIV-1 replication. J Virol. 1995;69:814–824. doi: 10.1128/jvi.69.2.814-824.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Swanstrom R, Wills J W. Synthesis, assembly and processing of viral proteins. In: Coffin J M, Hughes S H, Varmus H E, editors. Retroviruses. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1997. pp. 263–334. [PubMed] [Google Scholar]
- 46.Taylor P, Husi H, Kontopidis G, Walkinshaw M D. Structures of cyclophilin-ligand complexes. Prog Biophys Mol Biol. 1997;67:155–181. doi: 10.1016/s0079-6107(97)00014-x. [DOI] [PubMed] [Google Scholar]
- 47.Thali M, Bukovsky A, Kondo E, Rosenwirth B, Walsh C, Sodroski J, Göttlinger H G. Functional association of cyclophilin A with HIV-1 virions. Nature. 1994;372:363–365. doi: 10.1038/372363a0. [DOI] [PubMed] [Google Scholar]
- 48.Vajdos F F, Yoo S, Houseweart M, Sundquist W I, Hill C P. Crystal structure of cyclophilin A complexed with a binding site peptide from the HIV-1 capsid protein. Protein Sci. 1997;6:2297–2307. doi: 10.1002/pro.5560061103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yin L, Braaten D, Luban J. Human immunodeficiency virus type 1 replication is modulated by host cyclophilin A expression levels. J Virol. 1998;72:6430–6436. doi: 10.1128/jvi.72.8.6430-6436.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yoo S, Myszka D G, Yeh C-y, McMurray M, Hill C P, Sundquist W I. Molecular recognition in the HIV-1 capsid/cyclophilin A complex. J Mol Biol. 1997;269:780–795. doi: 10.1006/jmbi.1997.1051. [DOI] [PubMed] [Google Scholar]
- 51.Zydowsky L D, Etzkorn F A, Chang H Y, Ferguson S B, Stolz L A, Ho S I, Walsh C T. Active site mutants of human cyclophilin A separate peptidyl-prolyl isomerase activity from cyclosporin A binding and calcineurin inhibition. Protein Sci. 1992;1:1092–1099. doi: 10.1002/pro.5560010903. [DOI] [PMC free article] [PubMed] [Google Scholar]