Abstract
Background
Early detection and proper management of maternal sepsis caused by carbapenem-resistant Klebsiella pneumoniae (CRKP) can significantly reduce severe complications and maternal mortality. This study aimed to describe the epidemiology, antimicrobial resistance profile, and management of carbapenem-resistant K. pneumoniae among sepsis-suspected maternal cases in Ethiopia.
Methods
A prospective cross-sectional study was conducted in five tertiary hospitals from June 2021 to December 2023. Isolation, identification, and antimicrobial susceptibility testing of the isolates were carried out following standard microbiological procedures as stated in the CLSI guidelines. Data on socio-demographics, risk factors, and management strategies were collected with structured questionnaires. Associations between variables were determined using logistic regression analysis in STATA-21. A p-value of less than 0.05 was statistically significant.
Results
Of the 5613 total women suspected of having maternal sepsis, 609 (10.8%) of them were infected with K. pneumoniae. The prevalence rates of MDR, XDR, and PDR K. pneumoniae strains were 93.9%, 24.3%, and 10.9%, respectively. The resistance rates for the last-resort antibiotics; amikacin, tigecycline, carbapenem, and third-generation cephalosporin were 16.4%, 29.1%, 31.9%, and 93.0%, respectively. The combination of carbapenem with tigecycline or amikacin therapy was used to manage maternal sepsis caused by cephalosporin-and carbapenem-resistant strains. Sepsis associated risk factors, including septic abortion [AOR = 5.3; 95%CI:2.2–14.4]; extended hospitalization [AOR = 3.7; 95%CI: 1.6–19.4]; dilatation and curettage [AOR = 2.2; 95%CI:1.3–13.4]; cesarean wound infection [AOR = 4.1; 95%CI:2.0–9.2]; indwelling catheterization [AOR = 2.1;95%CI: 1.4–6.2]; ICU admission [AOR = 4.3; 95%CI:2.4–11.2]; post abortion [AOR = 9.8; 95%CI:5.7–16.3], and recurrent UTI [AOR = 3.3; 95%CI: 1.6–13.2] were significantly associated with maternal sepsis caused by K. pneumoniae.
Conclusions
The prevalence of maternal sepsis caused by carbapenem- resistant K. pneumoniae is high and serious attention needs to be given to combat transmission. Therefore, improving awareness, early diagnosis, IPC, integrated maternal surveillance, improved sanitation and efficient antimicrobial stewardship are crucial to combating bacterial maternal sepsis.
Keywords: Antimicrobial resistance, Maternal sepsis, Klebsiella pneumoniae, Pan-drug resistant, Extreme drug-resistance
Background
During pregnancy, the body undergoes significant changes in the immune system and metabolism [1], which can increase the risk of bacterial infections, including bacterial sepsis [2]. Globally, there are approximately 48.9 million cases of maternal sepsis each year, 17 million of which are reported in Africa [3]. Annually, 5.7 million women develop severe sepsis during pregnancy, childbirth, postabortion, or the postpartum period. According to the WHO, the number of deaths from sepsis is greater in Sub-Saharan Africa and southern Asia, accounting for 253,000 (87%) maternal sepsis-related deaths [4]. Identifying and diagnosing maternal sepsis early can be difficult, as maternal sepsis is often similar to other serious obstetric conditions, according to previous studies [4, 5]. Thus, most sepsis-infected mothers experience severe morbidity when they arrive at health facilities [6]. Maternal sepsis is responsible for 11% of maternal mortality. On the other hand, mothers can also contract sepsis through bacterial contamination at health facilities. Up to 50% of sepsis survivors may experience lasting disabilities, including myopathy, neuropathy, chronic pelvic pain, infertility, and psychological distress [7, 8].
Emerging carbapenemase- and extended-spectrum β-lactamase producing K. pneumoniae (ESBL-Kp) threaten maternal and neonatal health despite progress in the treatment and management of bacterial sepsis [9, 10]. These strains can survive in various environments, such as water, soil, and the gastrointestinal tract [9–11]. Its ubiquitous nature leads to spontaneous mutations and acquired antimicrobial resistance genes that undergo gene transfer by mobile genetic elements (MGEs). Once they acquire resistance genes, they can transfer them horizontally and vertically within and between species [9–12]. This bacterium has acquired antimicrobial resistance genes (ARGs) (blaLAP-2, blaTEM-1B, and blaSHV-106) from the ecosystem [13], and the primary source genes blaKPC-2, NDM-1, and KPC-2 [14, 15]. As a result, they were shown to be non-susceptible to carbapenem, colistin, carbapenem, polymyxin, fluoroquinolone, aminopenicillin, third-generation cephalosporin, and tigecycline [13–18]. Klebsiella pneumoniae is estimated to cause 124,000 deaths annually, accounting for 20% of AMR-related fatalities and 18% of AMR-related deaths, resulting in healthcare costs of $9 billion per year [2, 18, 19].
Currently, MDR (multidrug resistant), XDR (extensively drug-resistant), and PDR (pan-drug resistant) K. pneumoniae strains are known to possess antimicrobial resistance genes (blaKPC, blaCTX, blaTEM, blaOXA, and blaSHV). Additionally, they also harbor virulence genes such as enterobactin, aerobactin, yersiniabactin, and other genes (irp1, fyuA, iutA, ST29, PAI, ehxA, toxB, eae, sfa, pai, fim H, aggR, hly, pap, hyl A, tra, Tpai, cnf-1, afa, and rmpA/A). These genes are fundamental for the development and spread of hypervirulent carbapenem resistant strains [13–15, 20–22]. Infections caused by this superbug, such as urinary tract infections, skin infections, respiratory tract infections, sepsis, meningitis, and pyogenic liver abscesses become major health problems in pregnant women, immunecompromised patients, and elderly adults [20, 22]. This bacterium possesses various virulence factors namely capsules, adhesions, fimbriae, type II secretion systems, siderophores, lipopolysaccharides, and biofilms that activate cellular immune responses mainly macrophages [15, 18, 23, 24]. In the absence of proper antibiotic treatment, hyperactivated macrophages have the potential to release large amounts of proinflammatory cytokines and interleukins, leading to sepsis which adversely affects maternal and fetal health [25, 26].
Maternal sepsis can be worsened by various factors such as socioeconomic status, stress, lack of education, poverty, poor sanitation, family responsibilities, limited financial resources, and an unhealthy diet [27, 28]. Many studies have shown that various contributing factors include a medical history of chronic diseases such as gestational hypertension, diabetes, cancer, hospitalization, ventilator use, catheterization, kidney or lung disease, preeclampsia, abortion, and a history of antibiotic use during gestation [29]. Habits, such as smoking, alcoholism, and drug abuse, can also play a role in the development of these bacterial infections [30]. Educational level and intravenous drug administration can also be contributing factors [28, 30]. In developing countries, most cases of maternal sepsis are treated with a combination of two or more antibiotics such as cephalosporins, carbapenems, aminoglycosides, and polymyxins [31–33]. If these antimicrobial agents fail, the recommended treatment for maternal sepsis is tigecycline combination therapy with avibactam/ceftazidime, or meropenem caused by MDR-/XDR-/PDR-K. pneumoniae [34–37].
The effectiveness of antibiotics for treating maternal sepsis in carbapenem-/ third generation cephalosporin-resistant K. pneumoniae strains has been compromised [35, 38]. Recent studies indicate that Klebsiella is responsible for 65.7% of antibiotic-resistant bacteria that cause healthcare-associated infections [39, 40]. Among these bacteria, half are ESBL-producing K. pneumoniae isolates. The primary source of these infections was multiple sites, for a prevalence rate of 76.8%. MDR K. pneumoniae was the most common type of bloodstream infection, with a prevalence rate of 62.9%. The rate of MDR infections was greater for hospital-acquired infections (72.1%) than for hospital- or community-acquired infections [19, 27, 33, 37, 41].
Despite efforts to reduce maternal mortality and IPC and AMS programs at health facilities, maternal sepsis caused by K. pneumoniae remains the leading HAI causing maternal morbidity and mortality [2, 4, 16, 42]. However, the full extent of the impact of maternal sepsis caused by carbapenem-/third-generation cephalosporin-resistant K. pneumoniae strains has not been thoroughly established [43–45]. Currently, paying attention to unseen maternal sepsis caused by carbapenem-/cephalosporin-resistant K. pneumoniae is highly important for reducing preventable maternal morbidity and mortality [3, 6, 38, 44, 46]. This research will help establish a baseline, measure interventions, and ensure that patients receive the most effective care. We performed a nationwide study to generate evidence-based data and comprehensive information on the burden, risk factors, diagnosis, and treatment of maternal sepsis to develop better strategies to combat carbapenem-resistant strains and improve the quality of maternal care.
Materials and methods
Study area and period
This study was conducted on women who were admitted for maternal sepsis in five tertiary hospitals between June 2021 and December 2023. It covers more than three-quarters of the national geographic areas. Tikur Anbessa Specialized Hospital (TASH) and Comprehensive Specialized Hospital (ALERT) were chosen from central Ethiopia. TASH has a capacity of 1600 beds, and ALERT has a capacity of 850 beds. Hiwot Fana Compressive Specialized Hospital (HFSH) is a specialized university hospital in eastern Ethiopia that is located 526 km from Addis Ababa and has 550 beds. It serves as a referral hospital from the eastern part of the country for three regional governments (Harar, Somalia, and Oromia). Jimma University Medical Center (JUMC) is located 350 km from AA in western Ethiopia and has 834 beds. It serves as a referral hospital for five regional governments (Oromia, the Central Ethiopia Regional State, Gambela, the South Ethiopia Regional State, and the Southwest Ethiopia Peoples’ Region), which cater to 20 million people from the western part of the country. Whereas, Hawassa University Specialized Hospital (HUSH) is located 370 km away from AA in the southern part of the country and has over 750 beds. It operates as a referral hospital for three regional governments (Oromia, South Ethiopia Regional State, and South West Ethiopia Peoples’ Region), serving a catchment area of more than 25 million people.
Study design
A prospective cross-sectional study was conducted on women who were suspected of having maternal sepsis and admitted to five tertiary hospitals between June 2021 and December 2023. The women were randomly selected from various departments, such as ICUs, emergency rooms, gynecology, surgery, emergence OPD, and obstetric wards.
Source of population and study population
Our study population consisted of all women seeking treatment at five tertiary hospitals during the study period. Moreover, all women suspected of having maternal sepsis and admitted to these hospitals were randomly recruited and followed from the day of admission until the day of discharge from the hospital.
Sampling technique and sample size
During the study period, 5613 women admitted for maternal cases suspected of having bacterial sepsis and meeting the WHO criteria for sepsis were randomly selected from five tertiary hospitals in four clusters of the country. To our knowledge, there are no prior epidemiological data on maternal sepsis caused by this bacterium in multicenter studies. Hence, the sample size was calculated using the single population proportion formula, which assumes a 95% confidence interval, 3% margin of error, P = 50%, and 10% non-response rate.
The sample size was calculated as follow: 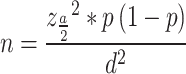
; then,
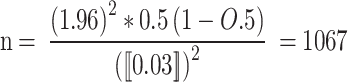
Non-responden t= 107
One tertiary hospital = 1067 + 107 = 1174.
Where n = sample size z = confidence level.
d = margin of error p = proportion.
The sample was obtained by multiplying 1174 by five tertiary hospitals (5*(1067 + 107)) = 5870) in the four clusters to estimate the prevalence of maternal bacterial sepsis, which is representative of nationwide data. However, 256 mothers suspected of having sepsis were excluded on the basis of national and WHO guidelines [2, 4] and the final sample size was 5613.
Sample collection, transportation and processing
The methods used for collecting, processing, and transporting samples were based on the guidelines provided by the CLSI for identifying Enterobacteriaceae and performing AST. Trained health professionals collected clinical samples such as blood, CSF, urine, wound exudates, and other samples aseptically from women admitted with maternal cases. Within an hour, the samples were transported to the bacteriology laboratory. The samples were subsequently processed and inoculated on agar plates with 5% sheep blood agar and MacConkey agar and incubated at 35–36 °C for 18–24 h. Biochemical tests were performed to identify K. pneumoniae. The identified K. pneumoniae isolates were inoculated in nutrient broth for subculture on nutrient agar and incubated aerobically for 18–24 h. AST was subsequently performed according to CLSI guidelines for Enterobacteriaceae [47]. Isolates resistant to carbapenem antibiotics (ertapenem, imipenem, and meropenem) were subsequently subjected to confirmation for carbapenemase production. Similarly, those that were resistantto cefotaxime and ceftazidime were subjected to the ESBL confirmatory test as per CLSI guidelines. The isolates were stored in 20% glycerol at -80 °C for further investigation for whole genome sequencing (WGS) [47] at the EPHI.
Antimicrobial susceptibility testing
Three to five colons with similar culture morphologies were transferred with a sterile inoculation loop to the nutrient broth until it was matched with a McFarland value of 0.5. The bacterial suspension was then streaked on MHA using a sterile swab to uniformly thicken the lawn of growth following incubation and the antibiotic discs were placed with sterile forceps as described in the disc diffusion method [47]. Following overnight incubation at 37 °C, the inhibitory zone diameters were measured and evaluated according to CLSI guidelines. Amoxicillin/clavulanic acid (20/10µg), gentamicin (10 µg), tobramycin (10 µg), ampicillin-sulbactam (10/10 µg), cefepime (30 µg), cefotaxime (30 µg), cefotetan (30 µg), cefuroxime (30 µg), ertapenem (10 µg), imipenem (10 µg), meropenem (10 µg), amikacin (30 µg), ciprofloxacin (5 µg), ceftriaxone (30 µg), tetracycline (30 µg), ceftazidime (30 µg), piperacillin-tazobactam (100/10 µg), trimethoprim-sulfamethoxazole (1.25/23.75 µg), and cefazolin (30 µg) are some of the Oxoid England products used for this study. Following CLSI guidelines, the quality of the media, biochemical reagents, and antibiotics was assessed using standard strains [47].
Phenotypic characterization
The identification of carbapenem resistance was conducted using the phenotypic-based disc diffusion method with meropenem, ertapenem, and imipenem as the primary approach to detect carbapenemase. The carbapenem-resistant strains were tested using minimum inhibitory concentration breakpoints to confirm their resistance using the carbapenem inactivation method (CIM). The K. pneumoniae isolates underwent testing with 1 antimicrobial 20 panels to identify their resistance patterns. If they showed resistance to cefotaxime, ceftazidime, and cefpodoxime, they were considered to be potential ESBL producers. Confirmatory tests using a double-disc synergy test (DDST) were then carried out according to CLSI guidelines [47].
Data analysis and interpretation
The data were subsequently entered into Epi Data Version 3.1 and exported for analysis using STATA-21. The prevalence of carbapenemase-producing and ESBL-producing K. pneumoniae, antimicrobial susceptibility testing, and risk factor identification were determined using descriptive analysis. Bivariate and multivariate logistic regressions were performed to evaluate associations between variables. A P-value less than 0.05 was considered to statistically significance.
Data quality assurance
Data regarding sociodemographic information, risk factors, and treatment and management were collected using predesigned questionnaires. The investigators at each selected hospital ensured patient data consistency and quality. Likewise, antibiotics, media, and procedures were ensured at preanalytical, analytical, and postanalytical phase qualities as per CLSI guidelines and adhered to national IPC guidelines [47].
Inclusion and exclusion criteria
Inclusion criteria
Maternal sepsis patients aged 15–49 years, 2) patients admitted to selected hospital departments such as ICUs, emergency rooms, gynecological, surgical, EOPD, and obstetric wards with infections-related maternal cases, 3) Patients who can able to provided samples and 4) patients who were not treated with carbapenem within three weeks during the study.
Exclusion criteria
(1) Patients with already known sepsis etiology such as malaria and viral; (2) patients treated with carbapenem antibiotics within two weeks of sample collection; (3) all male patients; and (4) women who were not suspected of having sepsis .
Ethics clearance
Approval was obtained from the Institutional Review Board (IRB) of the Institute of Biotechnology (RIB/001/22/IOB), ensuring that the study was conducted ethically. The hospitals involved in the study permitted this study to be carried out.
Results
Sociodemographic characteristics
The present study revealed that maternal bacterial sepsis-related hospitalization is caused mainly by K.pneumoniae sepsis. Of the 5,613 mothers who met the criteria for maternal sepsis, 609 (10.8%) mothers had carbapenem-resistant K. pneumoniae. The age of the women ranged from 15 to 49 years. 58% of the infected mothers had an education above the primary level. Almost all (97.7%) respondents were unaware of bacterial sepsis. Nearly half (43.8%) of the maternal sepsis cases caused by CRKP were identified from ICU wards, followed by GYN/OB (24.2%), surgical (16.4%), medical (11%), and emergency OPD (4.6%). Almost three-fourths (74.3%) of the participants were from rural areas (Table 1). The occurrence of maternal sepsis caused by CRKP differed among hospitals on the basis of their geographic location. In central Ethiopia, the prevalence of maternal sepsis caused by CRKP was11.3% at ALERT and 10.4% at TASH during the same study period. The prevalence rates of maternal sepsis caused by CRKP at HFSH, JUMC, and HUSH were 9.2%, 12.8%, and 10.8%, respectively (Table 1).
Table 1.
Sociodemographic and prevalence of K. pneumoniaeisolated from maternal sepsis casesin five tertiary hospitals in Ethiopia, 2024
| Covariates | Crude OR (95%CI) | P Value | Adjusted OR (95% CI) | P Value | |
|---|---|---|---|---|---|
| Onset of infection | Present on admissions | 1.00 | 1.00 | 1.00 | - |
| Nosocomial/hospital onset | 3.7 [0.8–3.2] | 0.001 | 2.0[1.7–11.3] | 0.01 | |
| ICU admission history | No | 1.00 | - | 1.00 | - |
| Yes | 10.3 [3.7–17.1] | 0.001* | 4.3 [2.4–11.2] | 0.01 | |
| Length of admission | ≤ 1 WK | 1.00 | 1.00 | 1.00 | - |
| 1–2 WKs | 0.8 [0.3–1.7] | 0.9 | 0.7 [0.1–1.2] | 0.06 | |
| 2–3 Wks | 3.8 [0.2–2.0] | 0.01 | 2.1 [2.3–8.5] | 0.01* | |
| 3–4 WKs | 7.7 [1.2–16.3] | 0.01 | 4.4 [3.0–15.9] | 0.001** | |
| ≥ 4 WKs | 9.1 [2.3–21.4] | 0.001* | 3.7 [1.6–19.4] | 0.0001** | |
| Pregnancy status at the time of infections suspected | Pregnant, not in labor | 1.00 | - | 1.00 | - |
| Pregnant, in labor | 0.2[0.1–0.6] | 0.32 | 0.2 [0.1–1.1] | 0.7 | |
| Postpartum | 7.3 [2.5–14.4] | 0.001* | 5.4 [2.1–10.2] | 0.001** | |
| Postabortion | 12.3 [7.7–20.4] | 0.001* | 9.8 [5.7–16.3] | 0.001*** | |
| Gravidity | Primigravida | 1.00 | - | 1.00 | - |
| Multigravida | 3.3 [0.1–4.6] | 0.03 | 2.2 [1.1–2.6] | 0.01 | |
| Parity | Nulliparous | 1.00 | - | 1.00 | - |
| Multiparous | 2.4[1.1–3.7] | 0.01 | 2.0 [1.3–4.7] | 0.009 | |
| Mode of birth | Vaginal birth | 1.00 | - | 1.00 | - |
| Cesarean | 4.3 [2.4–10.2] | 0.01 | 4.1 [2.0–9.2] | 0.001* | |
| Prolonged rupture of membranes | No | 1.00 | - | 1.00 | - |
| Yes | 12.3 [6.2–22.4] | 0.0001* | 8.1 [5.6–11.3] | 0.001** | |
| Septic abortion | No | 1.00 | - | 1.00 | - |
| Yes | 14.3 [3.7–19.4] | 0.001* | 5.3 [2.2–14.4] | 0.0001** | |
| Dilatation and curettage | No | 1.00 | - | 1.00 | - |
| Yes | 4.2 [2.4–11.5] | 0.001* | 2.2 [1.3–13.4] | 0.001** | |
| Chorioamnionitis | No | 1.00 | - | 1.00 | - |
| Yes | 12.3 [1.6–25.6] | 0.002* | 8.3 [2.2–19.1] | 0.001* | |
| Recurrent UTI | No | 1.00 | - | 1.00 | - |
| Yes | 5.4[2.9–13.2] | 0.02 | 3.3[1.6–13.2] | 0.01 | |
| Pulmonary infections Pneumonia | No | 1.00 | - | 1.00 | - |
| Yes | 2.9 [2.1–5.2] | 0.01* | 2.0 [1.5–3.8] | 0.01* | |
| Surgery/indwelling device/catheter | No | 1.00 | - | 1.00 | - |
| Yes | 3.7 [1.8–5.8] | 0.001* | 2.1 [1.4–6.2] | 0.01* | |
| Cervical/breast cancer | No | 1.00 | - | 1.00 | - |
| Yes | 6.1[2.2–17.3] | 0.01* | 5.2[1.2–11.4] | 0.001*** | |
| Hypertension | No | 1.00 | - | - | - |
| Yes | 0.4[0.2–1.4] | 0.39 | - | - | |
| Diabetes mellitus | No | 1.00 | - | - | - |
| Yes | 0.8[0.2–2.1] | 0.15 | - | - | |
| Within the past 3 months history of comorbidities | No | 1.00 | - | 1.00 | - |
| Yes | 7.8 [4.3–12.7] | 0.001* | 5.1. [3.8–11.1] | 0.001** | |
| Within 8wks antibiotic treatment | Yes | 1.00 | - | 1.00 | - |
| No | 16.3[9.3–19.7] | 0.01* | 15.8 [9.3–32.7] | 0.001*** | |
| History of alcohol intake | Never | 1.00 | 1.00 | 1.00 | - |
| Yes, some time alcohol and tela | 4[1.2–7.4] | 0.03 | 1.1 [0.4–1.9] | 0.2 | |
| Yes, frequently alcohol beer, tela | 8[2.9–11.3] | 0.001 | 3.2 [1.6–4.9] | 0.01* | |
Antimicrobial resistance profile of K. pneumoniae
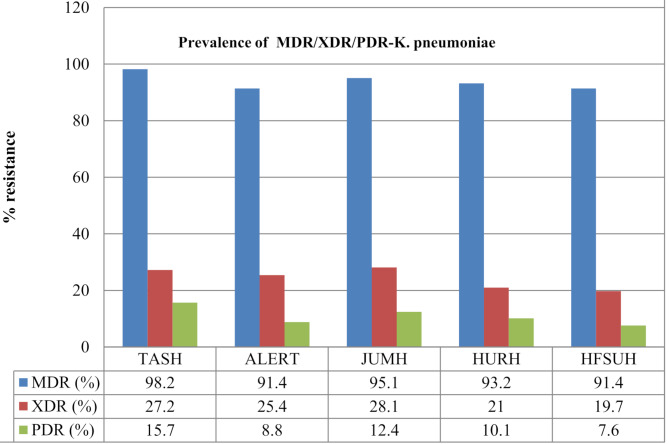
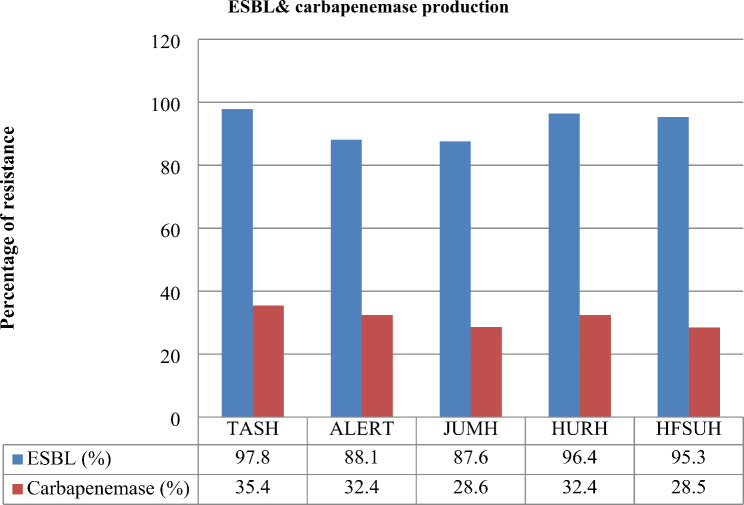
Six hundred nine non-repetitive K.pneumoniae isolates were found in various clinical samples, including urine, blood, wounds, CSF, and others. About 42.5% of the CRKP isolates were isolated from urine, followed by 35.5% from blood, surgical wound exudates (12.2%), CSF (6.4%), and other sources (3.4%) (Table 1). In terms of the antimicrobial resistance profile, 93.9% of the K. pneumoniae isolates were multidrug-resistant (MDR). Among these strains, 24.3% were XDR strains, followed by PDR strains (10.9%). 41% of the K. pneumoniae strains were resistant to carbapenems. The prevalence rates of ESBL- and carbapenemase-producing K. pneumoniae were 93.0% and 40.2%, respectively. All the ESBL-producing K.pneumoniae isolates were resistant to ceftriaxone, cefazolin, cefotaxime, ampicillin-sulbactam, ceftazidime, aztreonam, and cefepime. However, the resistance rates to other antibiotics, such as trimethoprim-sulfamethoxazole, tobramycin, ciprofloxacin, gentamicin, and piperacillin-tazobactam varied from 39.0 to 93.3% (Figs. 1 and 2). About 86.3% of the K. pneumoniae isolates in the urine samples and 75.6% in the blood samples were ESBL-producing strains (Table 1).
Fig. 1.
Prevalence of MDR/XDR/PDR-K. pneumoniae strains isolated from maternal with sepsis in five tertiary hospitals in Ethiopia, 2024. ALERT = ALERT Hospital; HFSH = Hiwot Fana Specialized Hospital; JUMC = Jimma University Medical Center; HUSH = Hawassa University Specialized Hospital and TASH = Tikur Anbessa Specialized Hospital
Fig. 2.
Prevalence of ESBL-/carbapenemase-producing K.pneumoniae strains isolated from maternal sepsis in five tertiary hospitals in Ethiopia, 2024. ALERT = ALERT Hospital; HFSH = Hiwot Fana Compressive Specialized Hospital; JUMC = Jimma University Medical Center; HUSH = Hawassa University Specialized Hospital and TASH = TikurAnbessa Specialized Hospital
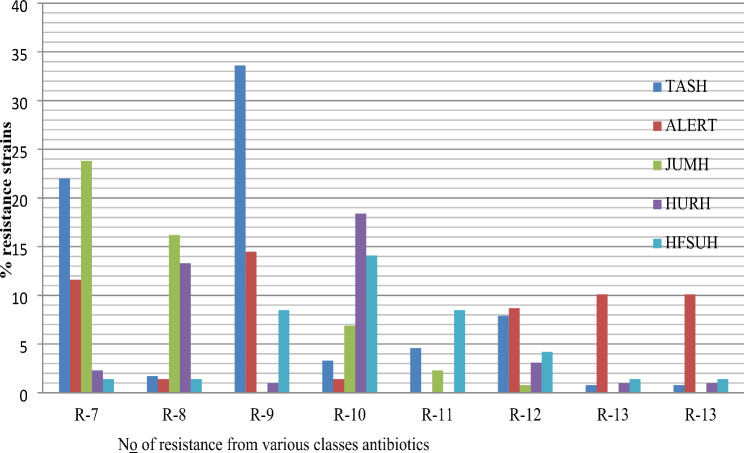
Among the K. pneumoniae isolates, the lowest resistance rates were to amikacin, tigecycline, meropenem, and ertapenem, with resistance rates of 16.4%, 29.1%, 27.6%, and 35.3%, respectively. In contrast to ESBL-producing strains, carbapenemase-producing K. pneumoniae was more common in blood samples (78.4%). Furthermore, the percentages of K. pneumoniae resistant to the R-8, R-9, R-10, R-11, and R-12 antibiotics were 34.0%, 57.7%, 44.1%, 15.4%, 24.7% and 13.3%, respectively (Fig. 3). The prevalence of carbapenem-resistant strains varies among hospitals (TASH, ALERT, JUMH, HURH, and HFSUH), ranging from 28.5 to 35.4%. Similarly, the prevalence of ESBL-producing K. pneumoniae ranged from 83.1 to 97.8% (Fig. 3).
Fig. 3.
Degree of resistance K. pneumoniae isolated from maternal with sepsis in five tertiary hospitals in Ethiopia, 2024. ALERT = ALERT Hospital; HFCSH = Hiwot Fana Specialized Hospital; JUMC = Jimma University Medical Center; HUSH = Hawassa University Specialized Hospital and TASH = TikurAnbessa Specialized Hospital
Risk factors associated with maternal sepsis
This study revealed that multiple factors increased the likelihood of contracting maternal sepsis. Recurrent UTIs (OR = 5.4; 95% CI: 2.9–13.2, P = 0.02); ICU admission (OR = 10.3; 95%CI: 3.7–17.1, P = 0.001); repeated abortions (OR = 14.3; 95%CI: 3.7–19.4, P = 0.001); caesarean delivery (OR = 4.3; 95%CI: 2.4–10.2, P = 0.01); history of not using antimicrobial agents within eight weeks (OR = 16.3; 95%CI: 9.3–19.7, P = 0.01); indwelling catheters (OR = 3.7; 95%CI: 1.8–5.8, P = 0.001); gravidity (OR = 3.3; 95%CI: 1.1–4.6, P = 0.03); post abortion (OR = 12.3; 95%CI: 7.7–20.4, P = 0.001); and alcoholism (OR = 5.2; 95% CI: 2.9–11.3, P = 0.001) were associated with having maternal sepsis of multidrug resistance K. pneumoniae (Table 2).
Table 2.
Biavariate and multivariate regression analysis of risk factors associated with maternal sepsis in five tertiary hospitals in Ethiopia, 2024
| Variable | categories | All women who had enrolled in the study | All women who had culture-positive for K.pneumoniae | ||
|---|---|---|---|---|---|
| N = 5613 | % | N = 609 | % | ||
|
Hospital clusters Central-Ethiopia |
Tikur Anbessa Specialized Hospita | 2317 | 41.3 | 241 | 39.6 |
| ALERT Hospital | 609 | 10.8 | 69 | 11.3 | |
| Western-Ethiopia | Jimma University Medical Center | 1013 | 18.1 | 130 | 21.3 |
| Southern-Ethiopia | Hawassa University Specialized Hospital | 903 | 16.1 | 98 | 16.1 |
| Eastern-Ethiopia | Hiwot Fana Specialized Hospital | 771 | 13.7 | 71 | 11.7 |
| Age (yrs) Mean +/- SD | < 20 | 590 | 10.5 | 81 | 13.3 |
| 21–30 | 1,927 | 34.3 | 143 | 23.5 | |
| 31–40 | 2,237 | 39.9 | 259 | 42.5 | |
| 41–49 | 859 | 15.3 | 126 | 20.7 | |
| Onset of infection | Present on admissions | 3186 | 56.8 | 171 | 28.1 |
| Hospital onset | 2427 | 43.2 | 438 | 71.9 | |
| Length of admission | ≤ 1 WK | 868 | 15.5 | 29 | 4.7 |
| 1–2 WKs | 1106 | 19.7 | 50 | 8.2 | |
| 2–3 Wks | 1983 | 35.3 | 79 | 13.0 | |
| 3–4 WKs | 954 | 17.0 | 188 | 30.9 | |
| ≥ 4 WKs | 702 | 12.5 | 263 | 43.2 | |
| Educational level | Cannot read and write | 807 | 14.4 | 250 | 41.0 |
| Elementary | 1549 | 27.6 | 219 | 36.0 | |
| High school/preparatory | 2284 | 40.7 | 79 | 13.0 | |
| College and above | 973 | 17.3 | 61 | 10.0 | |
| Residence | Urban | 1443 | 25.7 | 128 | 21.0 |
| Rural | 4170 | 74.3 | 481 | 79.0 | |
| Ward | ICU | 2459 | 43.8 | 298 | 49.0 |
| Gyn/Ob ward | 1358 | 24.2 | 136 | 22.3 | |
| Surgical | 921 | 16.4 | 77 | 12.7 | |
| Medical ward | 617. | 11.0 | 67 | 11.0 | |
| EOPD | 258 | 4.6 | 31 | 5.0 | |
| Antibiotics treatment history | No | 679 | 12.1 | 79 | 13.0 |
| yes | 4934 | 87.9 | 530 | 87.0 | |
| History of alcohol intake | Never | 1740 | 31.0 | 109 | 17.9 |
| Yes, some time(local tela) | 2975 | 53.0 | 239 | 39.2 | |
| Yes, common alcohol beer, tela | 898 | 16.0 | 261 | 42.9 | |
| Have ever heard about maternal sepsis | Mass media(TV/Radio/others) | - | 0.0 | - | - |
| Health profession/spouses | 130 | 2.3 | 24 | 3.9 | |
| Never heard about sepsis | 5483 | 97.7 | 585 | 96.1 | |
| Clinical specimens | Urine | 2073 | 36.9 | 259 | 42.5 |
| Blood | 2284 | 40.7 | 216 | 35.5 | |
| CSF | 881 | 15.7 | 39 | 6.4 | |
| Wound/surgical | 304 | 5.4 | 74 | 12.2 | |
| Others | 71 | 1.3 | 21 | 3.4 | |
On the basis of these findings, women who experienced septic abortion (two or more times more common) developed sepsis fivefold more likely to be infected with MDR strains than women who did not [AOR = 5.3; 95% CI = 2.2–14.4]; P = 0.0001]. Women who had a history of extended hospitalization (more than four weeks) developed sepsis three times more likely to be infected with MDR strains than did those who had never been hospitalized [(AOR = 3.7; 95% CI = 1.6–19.4); P = 0.0001]. Those women who had a history of dilatation and curettage developed sepsis twice [(AOR = 2.2; 95% CI = 1.3–13.4; P = 0.001)] as likely to be infected with MDR strains compared with those who did not. Women who were admitted to the ICU developed sepsis at a fourfold greater risk of being infected with MDR strains than did those who had never been admitted to the ICU did [AOR = 4.3; 95% CI = 2.4–11.2; P = 0.01]. Similarly, women who had indwelling catheters or invasive medical care developed sepsis threefold [(AOR = 2.1; 95% CI = 1.4–6.2; P < 0.01)] more likely to be infected with MDR strains than did those who did not. Women who had recurrent UTIs after childbirth were three times more likely to develop sepsis from MDR strains than were those who did not have a UTI [AOR = 3.3; 95% CI = 1.6–13.2]; P = 0.01]. Additionally, women who experienced a cesarean section wound infection after delivery were three times more likely to develop maternal sepsis than were those who did not have an infection [AOR = 4.1; 95% CI = 2.0–9.2]; P = 0.001]. Overall, these findings revealed that antimicrobial-resistant MDR strains are a significant challenge in maternal sepsis treatment and a major cause of morbidity and mortality in pregnant women and newborns (Table 2).
Treatment and management of maternal sepsis
This study revealed that among women who developed sepsis, 71.9% of maternal sepsis cases caused by K. pneumoniae were diagnosed at the hospital (Table 1). Upon arrival at tertiary hospitals, most women are initially administered antibiotics such as ampicillin, ciprofloxacin, trimethoprim-sulfamethoxazole, cefepime, aztreonam, ceftriaxone, amoxicillin, gentamicin, clindamycin, vancomycin, and piperacillin-tazobactam before arriving at the hospital. Furthermore, 44% of the sepsis patients received one or more cephalosporins, carbapenems, or tigecycline. These factors contribute to a negative bacterial culture, causing a delay in corrective measures being taken and leading to confusion with complicated pregnancy cases (Table 1). A total of 87.0% of women were treated for sepsis with high doses of broad-spectrum antibiotics, including vancomycin and carbapenems. However, 21–42% of these cases can lead to poor maternal outcomes and contribute to the development of long-term maternal outcomes (Table 2).
In the case of severe maternal sepsis, physicians often use a combination of two or three cephalosporins, carbapenems, aminoglycosides, and polymyxins. For instance, they prescribed tigecycline combination therapy, avibactam/ceftazidime, and imipenem/meropenem to treat maternal sepsis caused by MDR/XDR/PDR K. pneumoniae (Figs. 2 and 4). For those resistant to last-resort antibiotics, treatment with a combination of carbapenem with amikacin, tigeycline, or levofloxacin has shown positive outcomes (Fig. 4). After treatment with two or more antibiotics in combination, patients treated with meropenem and amikacine recovered with less hospitalization than did the other patients (Fig. 3).
Fig. 4.
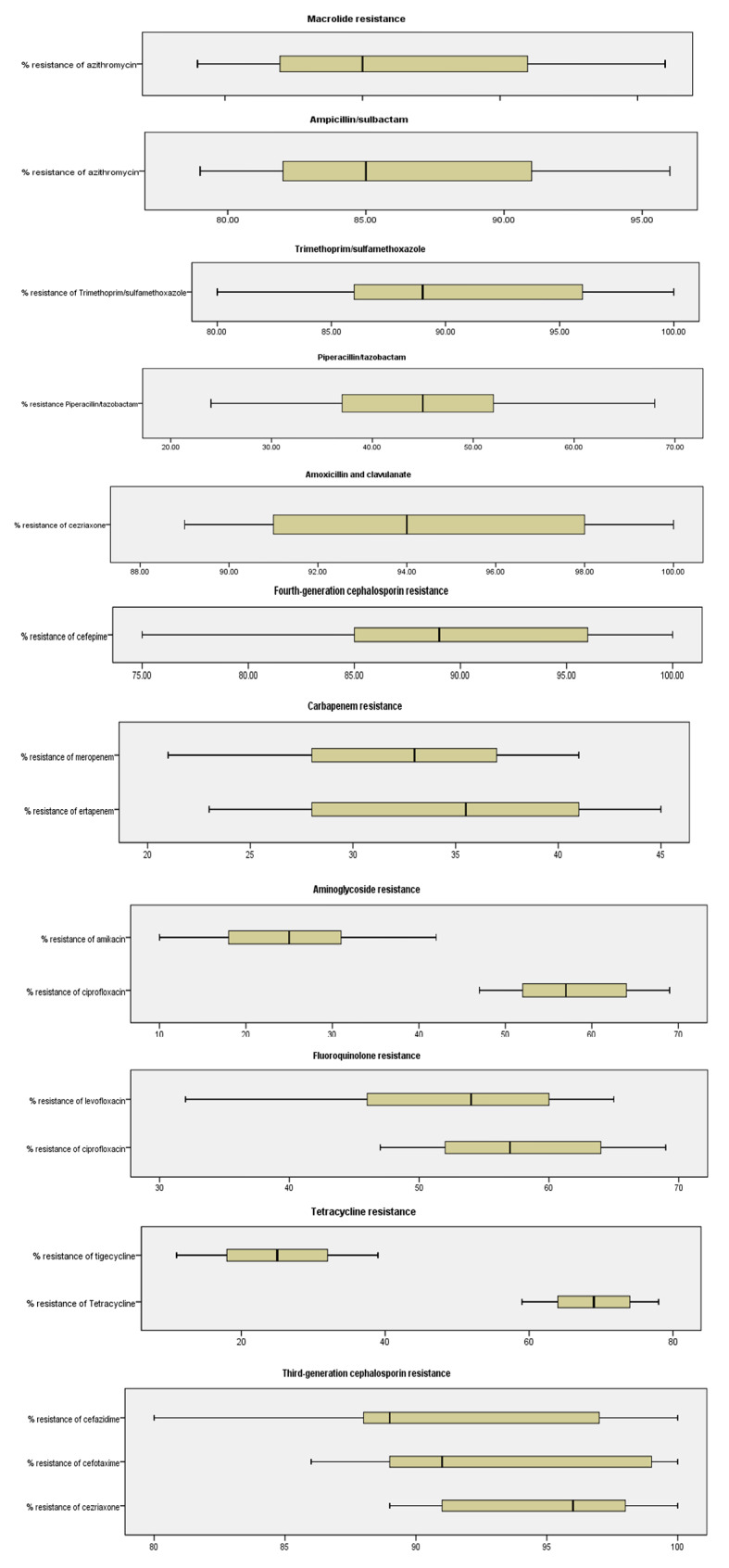
Percentage resistance to antimicrobials in K. pneumoniaeisolated from maternal sepsis cases in five tertiary hospitals in Ethiopia, 2024
This study revealed that women who developed maternal sepsis from CRKP were hospitalized for an average of 44 days. Overall, maternal sepsis caused by carbapenem-/cephalosporin-resistant K.pneumoniae is a neglected and deadly condition that has not received sufficient attention because it has short-term and long-term maternal health complications. Therefore, to reduce the impact of maternal sepsis, we need to focus on patient-centered care and promote IPC and AMS strategies. These measures can be implemented by facilitating early diagnosis and sepsis surveillance, creating public awareness, and strengthening IPC and AMS measures without delay.
Discussion
This study revealed that women who were suspected of having sepsis were included at five tertiary hospitals in three-fourths of the country during the study period. These hospitals serve as referral centers for three-fourths of the population of 125 million people. The study revealed that 97% of women who had experienced sepsis were unaware of the harmful symptoms and potential risks of this illness to the developing fetus. The awareness of sepsis is low. Studies have shown that 2.6% of the population in Japan, and 4.2% of the population in Singapore [8, 31, 42]. In contrast, studies in Sweden and Germany revealed that the prevalence of population awareness of sepsis was 92% and 88.6%, respectively [2, 9, 41].
In terms of residence, 74.3% of the maternal sepsis patients were from rural areas. A total of 56.8% of maternal sepsis patients had delayed treatment, making it difficult to determine whether symptoms were from infection or pregnancy complications upon arrival at the hospital. Although these suspected cases were treated with carbapenem and vancomycin, the number of patients infected with antibiotics resistant to those last-resort antibiotics has increased, which is unacceptable in the studied hospitals [10, 20, 24]. Clinicians face significant challenges in regard to the early recognition and diagnosis of sepsis in pregnant women. It mimics signs and symptoms of complicated pregnancy and needs urgent work to identify infection before its prognosis is improved to sepsis [4, 7–9, 31, 32, 43]. Additionally, alcoholism was associated with sepsis in mothers in this study. Those who frequently consumed alcohol, such as beer and ”Tela”, and were infected with K. pneumoniae had a greater likelihood of developing severe symptoms, accounting for 42.9% of the patients. These findings are consistent with many related studies [1, 3, 6, 16, 17, 34, 36, 39]. The maternal sepsis incidence and treatment of K. pneumoniae vary by region. This could be due to different factors such as sample type, geographic location, patient demographics, immune status, and co-infections, which can affect the magnitude and strain type of this bacterium, which is consistent with our findings in five tertiary hospitals [10, 15, 24, 35, 37].
This study revealed that K. pneumoniae was resistant to a high percentage (39–100%) of the tested antibiotics, including last-resort options such as carbapenems and tigecycline. These findings suggest that carbapenem and cephalosporin antibiotics have become less effective in treating superbugs. Without addressing the rapid spread of CRKP strains, alternative treatments may not be available in the near future. These findings are consistent with many studies carried out in other counties [34–37]. On the other hand, this bacterium is one of the most commonly acquired bacteria in both nosocomial and community infections and is highly spreading and developing antimicrobial resistance worldwide [10–14, 23, 24].
The prevalence of MDR K. pneumoniae was high at 93.9% (ranging from 91.4 to 98.2%). This finding is consistent with those of studies carried out by Tsegaye et al. 86.5% [34], Aberaet al. 45.2% [35], Belay et al. 84% [36], Beyene et al. 94.5% [46], and Awoke et al. 98.5% [37]. Overall, those studies demonstrated that the number of isolates evaluated for each study, the geographic variation, the studied population, and the test method used all affect the prevalence of MDR-K. pneumoniae. Another study by Cassini et al. reported various prevalences of antimicrobial-resistant infections across Europe [19]. Similarly, Lee et al. reported that there was variation in the strains, prevalence, and burden of carbapenemase-producing K. pneumoniae that disseminated worldwide [18]. Forero-Hurtado et al. reported antimicrobial resistance via blaKPC genes and other genes, such as blaNDM-1, and plasmid-mediated quinolone resistance [13, 22] cross-transmitted from K. pneumoniae to different species [15]. Similarly, this could involve vertical or horizontal genome transfer [17, 20, 21, 28]. We are actively working toward addressing the challenge of multidrug-resistant strains, including K. pneumoniae, which pose a significant obstacle to achieving our goal of reducing maternal and newborn morbidity and mortality rates in a sustainable manner [7, 18, 19, 27]. In the present study, the prevalence of XDR K.pneumoniae was 28.1%, which is higher than the rates reported in studies by Abera et al. 7.7% [35], and Beyene et al. 8.8% [46]. The percentage obtained in this study is slightly lower than that reported by Tsegayeet al. 43.3% [34]; similarly, the prevalence of PDR K. pneumoniae was 28.1%, which is higher than that reported in studies performed by Beyene et al.. 0.8%[ 46], Awoke et al.. 1.5% [37] and Tsegayeet al. 1.8% [34].
Our study of K. pneumoniae revealed that various strains that produce enzymes, namely carbapenemase and ESBL, which are responsible for causing antimicrobial resistance, have become resistant to antimicrobial drugs. A total of 41.7% of the K. pneumoniae strains were non-susceptible to carbapenems, and 40.2% produced carbapenemase. The prevalence of carbapenemase-producing strains reported in China ranges from 20.4 to 21.9% [11]. However, this figure is higher than that in the studies conducted by Abera et al., 15.4% [35], and Tsegaye et al., 9% [34]. The percentage obtained in this study is slightly lower than that reported by Beyene et al.., which was 30% [46]. On the other hand, 87.6–93.04% of K. pneumoniae strains were ESBL-producing strains. The highest frequency of this profile was found in hospitals in the central part of the country. Similarly, the western, southern, and eastern regions account for 87.6%, 96.4%, and 95.3%, respectively, of the profile. This study is similar to studies that reported a high percentage of ESBL-producing K. pneumoniae nationwide [3, 34–37] and even on a global scale [12, 14, 16, 17, 38].
Although sepsis is a significant healthcare problem, comprehensive data on sepsis caused by multidrug-resistant bacteria and the consequences of sepsis leading to exacerbated maternal morbidity and mortality is lacking. In this investigation, we employed binary and multivariate logistic regression models to pinpoint putative risk factors associated with K. pneumoniae-induced maternal sepsis. The findings revealed that women with specific clinical outcomes, such as septic abortion [AOR = 5.3; 95% CI = 2.2–14.4]; P = 0.0001], indwelling catheters[(AOR = 2.1; 95% CI = 1.4–6.2; P < 0.01)], alcoholism alcoholism [(OR = 5.2; 95% CI: 2.9–11.3, P = 0.001], ICU admission[AOR = 4.3; 95% CI = 2.4–11.2; P = 0.01], cesarean delivery[AOR = 4.1; 95% CI = 2.0–9.2]; P = 0.001], a history of not using antimicrobial agents within eight weeks[(AOR = 3.7; 95% CI = 1.6–19.4); P = 0.0001], and recurrent UTIs[AOR = 3.3; 95% CI = 1.6–13.2]; P = 0.01], history of dilatation and curettage [(AOR = 2.2; 95% CI = 1.3–13.4; P = 0.001)] were at greater risk of developing sepsis. These findings are similar to those of studies on the risk factors associated with sepsis, which exacerbates severe morbidity and mortality at community and health facilities (Table 2); these findings have also been reported by many researchers [7, 8, 18, 19, 25, 28, 32, 33, 42]. The occurrence of risk factors associated with maternal factors (host factors) [1, 27, 34, 43] and bacterial factors [8, 29] has led to the widespread spread of highly virulent strains [18, 20, 21, 28] and antimicrobial resistance, which includes strains that are pandrug resistant [8, 11, 26].
The management of sepsis encompasses fluid resuscitation for hypotension, empirical antimicrobial treatment, and vasoactive agents (norepinephrine) to reduce the risk of maternal complications. Although treatments have been used to reduce maternal mortality and morbidity caused by sepsis, an increase in antimicrobial-resistant bacterial infections has had adverse consequences. In this context, sepsis caused by CRKP and ESBL-producing K. pneumoniae has resulted in severe maternal sepsis, leading to millions of deaths, socioeconomic crises, and security issues. Figure 3 shows a high incidence of CRKP and ESBL-producing K. pneumoniae, which compromises the effectiveness of last-resort antibiotics such as carbapenem and tigecycline. These findings are similar to those of previous studies on pulmonary infections (pneumonia), surgical site infections and UTIs associated with CRKP [5, 9, 12, 19].
During the study period, it was observed that there were difficulties in promptly recognizing and treating CRKP because of the delayed reporting of culture results. This delay poses a challenge for gynecologists in reducing the risk of adverse maternal sepsis outcomes. These findings are similar to those of previous studies [3, 5, 11, 22, 30, 34, 37, 38, 40]. Treatment of maternal sepsis with antimicrobial agents involves a general gram-negative bacteremia algorithm, which uses broad-spectrum β-lactam antibiotics such as ciprofloxacin, trimethoprim-sulfamethoxazole, cefepime, aztreonam, ceftriaxone, amoxicillin, gentamicin, clindamycin, vancomycin, and piperacillin-tazobactam as part of empiric antimicrobial therapy. These findings are consistent with those of other studies [1, 21, 44, 47].
Conclusions
This study revealed that the overall prevalence of maternal sepsis caused by carbapenem- or cephalosporin-resistant K. pneumoniae is high. This highly contagious and ubiquitous nature of superbugs poses a seriously threat to public health and serious attention needs to be given to combat transmission. Hence, improving awareness, early diagnosis, person-centered care, IPC measures, integrated surveillance, efficient antimicrobial stewardship, improved sanitation and monitoring interventions are crucial to tackle preventable maternal sepsis caused by bacteria.
Acknowledgements
We would like to express our sincere gratitude to EPHI, AAU and Ghana University for providing financial support for our research. We are thankful to the Clinical Bacteriology and Mycology Unit of EPHI, TASH, ALERT, JUMH, HURH, and HFSUH for their expert assistance and for allowing us to use their laboratory facilities. Additionally, we would like to extend our thanks to Ebissa Fekede and Degafu Beyene for their unwavering support and encouragement throughout the project.
Abbreviations
- ARGs
Antimicrobial resistance genes
- CLSI
Clinical Laboratory Standard Institute
- CRKP
Carbapenem-resistant Klebsiella pneumoniae
- ESBL
Extended Spectrum Beta Lactamase
- IPC
Infection prevention and control
- MDR
Multidrug Resistant
- MGEs
Mobile genetic elements
- MHA
Muller Hinton agar
- MHB
Muller Hinton Broth
- MIC
Minimal inhibitory concentration
- PDR
Resistant to all antimicrobial classes
- UTI
Urinary tract infection
- XDR
Extreme drug resistance
- WHO
World Health Organization
Author contributions
EG designed and performed laboratory work and analyzed the data. BE, BA, HA, GD and TST were supervising overall project. All authors reviewed and approved the final manuscript.
Funding
This research was funded by AAU (4592/001/22/IOB), EPHI, and Ghana University for data collection, laboratory reagents and other costs. All institutes had no role in the study design, in the writing of the report, and in the decision to submit the article for publication.
Data availability
No datasets were generated or analysed during the current study.
Declarations
Ethics approval and consent to participate
This study was conducted after it is ethically reviewed and approved by the Research and Ethical Review Committee of Institute of Biotechnology, Addis Ababa University, Addis Ababa, Ethiopia.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Amir M, Brown JA, Rager SL, Sanidad KZ, Ananthanarayanan A, Zeng MY. Maternal microbiome and infections in pregnancy. Microorganisms. 2020;8(12):1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.World Health Organization. Global report on the epidemiology and burden of sepsis: current evidence, identifying gaps and future directions. WHO. 2020;56. https://iris.who.int/handle/10665/334216.
- 3.Okafor O, Roos N, Abdosh AA, Adesina O, Alaoui Z, Romero WA, et al. International virtual confidential reviews of infection-related maternal deaths and near-miss in 11 low- and middle-income countries - case report series and suggested actions. BMC Pregnancy Childbirth. 2022;22(1):431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rudd KE, Johnson SC, Agesa KM, Shackelford KA, Tsoi D, Kievlan DR, et al. Global, regional, and national sepsis incidence and mortality, 1990–2017: analysis for the global burden of disease study. Lancet. 2020;395(10219):200–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shields A, De Assis V, Halscott T. Top 10 pearls for the recognition, evaluation, and management of maternal sepsis. Obstet Gynecol. 2021;138(2):289–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jash S, Sharma S. Pathogenic infections during pregnancy and the consequences for fetal brain development. Pathogens. 2022;11(2):193. [DOI] [PMC free article] [PubMed]
- 7.Ornaghi S, Maraschini A, Buoncristiano M, Corsi Decenti E, Colciago E, Cetin I, et al. Maternal sepsis in Italy: a prospective, population-based cohort and nested case-control study. Microorganisms. 2023;11(1):1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fleischmann-Struzek C, Rudd K. Challenges of assessing the burden of sepsis. Med Klin Intensivmed Notfmed. 2023;118(Suppl 2):68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Randolph JL, Chan K, Albright A, Chen A. Delays in administration of the second antibiotic dose in patients with severe sepsis and septic shock. Hosp Pharm. 2021;56(4):247–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bassetti M, Garau J. Current and future perspectives in the treatment of multidrug-resistant Gram-negative infections. J Antimicrob Chemother. 2021;76:IV23–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baguiya A, Bonet M, Cecatti JG, Brizuela V, Curteanu A, Minkauskiene M, et al. Perinatal outcomes among births to women with infection during pregnancy. Arch Dis Child. 2021;106(10):946–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lan P, Jiang Y, Zhou J, Yu Y. A global perspective on the convergence of hypervirulence and carbapenem resistance in Klebsiella pneumoniae. J Glob Antimicrob Resist. 2021;25:26–34. [DOI] [PubMed] [Google Scholar]
- 13.Hammad HA, Mohamed IS, El-Badawy O, Zakaria AM, Shabaan L, Aly SA. pKpQIL-like plasmid contributes to the dissemination of blaNDM-1 and plasmid mediated quinolone resistance determinants among multi drug resistant Klebsiella pneumoniae in Assiut University Hospital, Egypt. Iran J Microbiol. 2023;15(2):208–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tang M, Li J, Liu Z, Xia F, Min C, Hu Y, et al. Clonal transmission of polymyxin B-resistant hypervirulent Klebsiella pneumoniae isolates coharboring bla NDM-1 and bla KPC-2 in a tertiary hospital in China. BMC Microbiol. 2023;23(1):1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Forero-Hurtado D, Corredor-Rozo ZL, Ruiz-Castellanos JS, Márquez-Ortiz RA, Abril D, Vanegas N, et al. Worldwide dissemination of blaKPC gene by novel mobilization platforms in Pseudomonas aeruginosa: a systematic review. Antibiotics (Basel). 2023;12(4):658. [DOI] [PMC free article] [PubMed]
- 16.Argimón S, David S, Underwood A, Abrudan M, Wheeler NE, Kekre M, et al. Rapid genomic characterization and global surveillance of klebsiella using pathogenwatch. Clin Infect Dis. 2021;73:S325–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang J, Xu Y, Wang M, Li X, Liu Z, Kuang D, et al. Mobilizable plasmids drive the spread of antimicrobial resistance genes and virulence genes in Klebsiella pneumoniae. Genome Med. 2023;15(1):1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zaman T, uz, Alrodayyan M, Albladi M, Aldrees M, Siddique MI, Aljohani S, et al. Clonal diversity and genetic profiling of antibiotic resistance among multidrug/carbapenem-resistant Klebsiella pneumoniae isolates from a tertiary care hospital in Saudi Arabia. BMC Infect Dis. 2018;18(1):1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sahle SG, Weldemariam S, Mehari M, ab, Abraha TA. Determinants of puerperal sepsis among post-partum mothers in Mekelle city public hospitals, Tigray, Ethiopia, 2021: a case control study. BMC Womens Health. 2023;23(1):1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang F, Yan X, Bai J, Xiang L, Ding M, Li Q, et al. Identification of the BolA protein reveals a novel virulence factor in K. pneumoniae that contributes to survival in host. Microbiol Spectr. 2022;10(5):1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou C, Zhang H, Xu M, Liu Y, Yuan B, Lin Y, et al. Within-host resistance and virulence evolution of a hypervirulent carbapenem-resistant Klebsiella pneumoniae ST11 under antibiotic pressure. Infect Drug Resist. 2023;16:7255–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huynh B, Passet V, Rakotondrasoa A, Diallo T, Kerleguer A. Klebsiella pneumoniae carriage in low-income countries: antimicrobial resistance, genomic diversity and risk factors. Gut Microbes. 2020;11(5):1287–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rocha J, Henriques I, Gomila M, Manaia CM. Common and distinctive genomic features of Klebsiella pneumoniae thriving in the natural environment or in clinical settings. Sci Rep. 2022;12(1):1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee CR, Lee JH, Park KS, Kim YB, Jeong BC, Lee SH. Global dissemination of carbapenemase-producing Klebsiella pneumoniae: Epidemiology, genetic context, treatment options, and detection methods. Front Microbiol. 2016;7:1–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen F, Wang Y, Zhang W, Cai Y, Cai Y, Zhao T, et al. A functional polymorphism-mediated disruption of EGR1/ADAM10 pathway confers the risk of sepsis progression. mBio. 2019;10(4):e01663–19. [DOI] [PMC free article] [PubMed]
- 26.Han M, Chen Z, He P, Li Z, Chen Q, Tong Z, et al. YgiM may act as a trigger in the sepsis caused by Klebsiella pneumoniae through the membrane-associated ceRNA network. Front Genet. 2022;13:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Melkie A, Dagnew E. Burden of puerperal sepsis and its associated factors in Ethiopia: a systematic review and meta-analysis. Arch Public Heal. 2021;79(1):1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dai G, Xu Y, Kong H, Xie W, Wang H. Risk factors for carbapenem-resistant Klebsiella pneumoniae infection and associated clinical outcomes. Am J Transl Res. 2021;13(6):7276–81. [PMC free article] [PubMed] [Google Scholar]
- 29.Brizuela V, Cuesta C, Bartolelli G, Abdosh AA, Abou Malham S, Assarag B, et al. Availability of facility resources and services and infection-related maternal outcomes in the WHO global maternal sepsis study: a cross-sectional study. Lancet Glob Heal. 2021;9(9):e1252–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang P, Liu C, Wu Z, Zheng J, Yi J, Wu N, et al. Clinical outcomes, microbiological characteristics and risk factors for difficult-to-treat resistance to Klebsiella pneumoniae infection. Infect Drug Resist. 2022;15(August):5959–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Parsons Leigh J, Brundin-Mather R, Moss SJ, Nickel A, Parolini A, Walsh D, et al. Public awareness and knowledge of sepsis: a cross-sectional survey of adults in Canada. Crit Care. 2022;26(1):1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu L, Sun X, Ma X. Systematic review and meta-analysis of mortality of patients infected with carbapenem-resistant Klebsiella pneumoniae. Ann Clin Microbiol Antimicrob. 2017;16(1):1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li Z, Liu X, Lei Z, Li C, Zhang F, Wu Y, et al. Genetic diversity of polymyxin-resistance mechanisms in clinical isolates of carbapenem-resistant Klebsiella pneumoniae : a multicenter study in China. Microbiol Spectr. 2023;27(2):e0523122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Id AA, Bitew A, Fentaw S, Tsige E, Assefa D, Lejisa T, et al. Phenotypic characterization of carbapenem non-susceptible gram-negative bacilli isolated from clinical specimens. PLoS ONE. 2021;2(12):e0256556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Alemayehu T. Prevalence of multidrug-resistant bacteria in Ethiopia: a systematic review and meta-analysis. J Glob Antimicrob Resist. 2021;26:133–9. [DOI] [PubMed] [Google Scholar]
- 36.Regassa BT, Tosisa W, Eshetu D, Beyene D, Abdeta A, Assefa A, et al. Antimicrobial resistance profiles of bacterial isolates from clinical specimens referred to Ethiopian Public Health Institute: analysis of 5-year data. BMC Infect Dis. 2023;23(1):1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Awoke T, Teka B, Seman A, Sebre S, Yeshitela B, Aseffa A, et al. High prevalence of multidrug-resistant Klebsiella pneumoniae in a tertiary care hospital in Ethiopia. Antibiotics. 2021;10(8):1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Arena F, Pilato V, Di, Vannetti F, Fabbri L, Antonelli A, Coppi M, et al. Population structure of KPC carbapenemase- producing Klebsiella pneumoniae in a long- term acute- care rehabilitation facility: Identification of a new lineage of clonal group 101, associated with local hyperendemicity. Microb Genom. 2020;6(1):e000308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhou Y, Ai W, Cao Y, Guo Y, Wu X, Wang B, et al. The co-occurrence of NDM-5, MCR-1, and FosA3-encoding plasmids contributed to the generation of extensively drug-resistant Klebsiella pneumoniae. Front Microbiol. 2022;12:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.GLASS whole-genome sequencing for surveillance of antimicrobial resistance. Geneva: World Health Organization. 2020.
- 41.Lee CR, Lee JH, Park KS, Jeon JH, Kim YB, Cha CJ, et al. Antimicrobial resistance of hypervirulent Klebsiella pneumoniae: Epidemiology, hypervirulence-associated determinants, and resistance mechanisms. Front Cell Infect Microbiol. 2017;21:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Argimón S, David S, Underwood A, Abrudan M, Wheeler NE, Kekre M, et al. Rapid genomic characterization and global surveillance of klebsiella using pathogenwatch. Clin Infect Dis. 2021;73(Suppl_4):S325–335. [DOI] [PMC free article] [PubMed]
- 43.Chen KJ, Chen YP, Chen YH, Liu L, Wang NK, Chao AN, et al. Infection sources and Klebsiella pneumoniae antibiotic susceptibilities in endogenous klebsiella endophthalmitis. Antibiotics. 2022;11(7):1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhu J, Wang T, Chen L, Du H. Virulence factors in hypervirulent. Klebsiella pneumoniae Front Microbiol. 2021;12:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huang Y, Li J, Wang Q, Tang K, Cai X, Li C. Detection of carbapenem-resistant hypervirulent Klebsiella pneumoniae ST11-K64 co-producing NDM-1 and KPC-2 in a tertiary hospital in Wuhan. J Hosp Infect. 2023;131:70–80. [DOI] [PubMed] [Google Scholar]
- 46.Beyene D, Bitew A, Fantew S, Mihret A, Evans M. Multidrug-resistant profile and prevalence of extended spectrum β-lactamase and carbapenemase production in fermentative gram-negative bacilli recovered from patients and specimens referred to NRL, Addis Ababa, Ethiopia. PLoS ONE. 2019;14(9):1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Clinical and Laboratory Standards Institute (CLSI). Performance Standards for Antimicrobial Susceptibility Testing. 32nd ed. CLSI supplement M100. CLSI, USA, 2022.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No datasets were generated or analysed during the current study.