Abstract
A fundamental, yet underexplored, materials system is the interface between biological molecules and inorganic surfaces. In an elemental approach to this problem, we have systematically examined the adhesion of amino acids to a series of inorganic surfaces including metals, insulators, and semiconductors. Significant differential adhesion is observed over the full complement of amino acids, determined largely by amino acid side-chain charge. Extensive mapping of the amino acid adhesion versus materials in multiple solutions is presented, with preliminary mechanisms derived from concentration and pH dependence. These results provide an empirical basis for building peptide to inorganic surface structures, and, using this adhesion data, we design inorganic nanostructures that are shown to selectively bind to prescribed primary peptide sequences.
Keywords: peptide adhesion, semiconductors
Interfaces have been a focus of intense research in condensed matter systems, not only naturally occurring boundaries between materials but also artificially produced junctions (1). Novel systems and phenomena (2) have been created and observed at interfaces formed by hard materials, such as insulators and semiconductors. Concurrent with development of inorganic structures, methods for artificial bulk synthesis of biological macromolecules have been developed, particularly peptide-based species (3). Given these accomplishments in producing inorganic and biological materials, it is a natural extension of materials development to inquire about the interfaces that may be formed between them. Can we understand inorganic to biological material interfaces by examining the interactions of the building blocks of biological systems, such as amino acids, nucleic acids, or energy storage macromolecules with the inorganic surfaces? If this interaction can be understood and controlled, a host of hybrid molecular structures and processes will be available for applications, fundamental condensed matter, and biological materials study.
Past work addressing biological to inorganic interfaces and, specifically, peptide adhesion to inorganic materials has generally involved use of complex biological components. Repeating polypeptides on the surface of Escherichia coli were found to selectively adhere to certain metallic surfaces (4). Biological population selection applied to the growth of bacteriophage (5) evolved surface peptides that specifically bind to semiconductor surfaces. Further use of this phage display method (6) allowed evolution of biological templates for nucleation of semiconductor nanowires or nanocrystals. Other efforts to specifically effect biological material adhesion to surfaces have used intermediary layering. Oligopeptides (7) containing a cell adhesion motif were adhered to gold surfaces. Various other methods (8–11) have been used to produce inorganic to biological interfaces, including self-assembled monolayers that bind to intermediate moieties containing cell binding ligands (12) of Arg-Gly-Asp. Rather than adhering biomaterials to inorganic surfaces through intermediate layers, if peptides can be adhered directly to inorganic surfaces, can rules for this process be derived from the constituent amino acid adhesive properties?
Our objective is to understand the interactions of amino acids with a distinct set of inorganic materials. We examine here the adhesion properties of the full complement of amino acids with inorganic materials used in present-day microfabrication and nanofabrication applied to electronic devices. Inorganic samples are exposed to solutions of fluorescently tagged peptides synthesized from a single amino acid, and persistent fluorescence is measured to quantify the adhered peptide density. Our fundamental finding is that amino acids show significant differential adhesion to different materials, with a strong correlation of adhesion to the side-chain charge. To deduce adhesion mechanisms, pH and concentration dependence are examined. These results provide an empirical basis for building peptide structures on inorganic surfaces. In this vein, the data are used to design an inorganic nanostructure that selectively binds to a prescribed primary peptide sequence: layered nanostructures fabricated on the length scale of an 11-residue peptide show specific adhesion to the peptide sequence. This result represents successful construction of a man-made inorganic molecular scale structure, selective to a simple biologically specific site.
Peptide to Surface Adhesion Tests
The adhesion tests involve fabricating a set of layered materials that are exposed to solutions of peptides, each comprised of a different amino acid, then measuring the amount of residual peptide on the surface through fluorescence signal output after washing. The peptides are all synthesized with between 8 and 10 residues of a single amino acid by using each of the 20 amino acids. Each peptide is tagged with a fluorescent marker on the N terminus with the C terminus free. The inorganic materials tested are representative of surfaces used in semiconductor processing: metals, semiconductors, and insulators. Layered structures are first formed (e.g., silicon nitride on gallium arsenide), and a pattern is produced on the top layer by using standard photolithographic pattern definition. Either the top layer is etched, resulting in a relief pattern, or metal is deposited in the pattern; both methods result in material contrast. Residual oxide is purposely not removed from any of the surfaces to simulate most common processing and resultant microstructures. The inorganic materials tested include five metals: Au, Pd, Pt, Ti, and Al. Gold and palladium form essentially no oxide layer, platinum and titanium form a thin oxide layer, and aluminum forms a robust oxide layer. Semiconductors examined are GaAs, with minor oxide formation, and AlGaAs, generating a thick oxide for 30% Al stoichiometry. The tested insulator layers, Si3N4 and SiO2, are amorphous. A more detailed review of materials and methods is available in Supporting Materials and Methods, which is published as supporting information on the PNAS web site.
Differential Adhesion of Peptides to Inorganic Surfaces
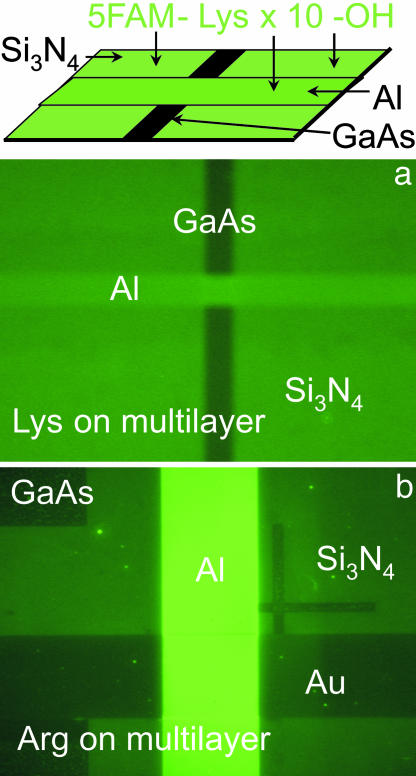
The fundamental result of this study, that amino acids adhere differentially to inorganic surfaces, is demonstrated by the examples in Figs. 1 and 2 (see Fig. 6, which is published as supporting information on the PNAS web site). Fig. 1b shows marked fluorescence output for polylysine adherent to patterned Si3N4 compared with the GaAs substrate. The fluorescence is uniform with minimal inhomogeneities. Other peptide/inorganic surfaces shown in Fig. 1 c–e likewise demonstrate homogeneous fluorescence intensity. The fluorescence output has been calibrated in comparison to known densities of fluorescence molecules: Fig. 1b results correspond to a peptide density of ≈22,000 peptides per μm2 for the lysine on Si3N4. At that surface coverage and with the longest dimension of the peptide (including fluorescence tag) ≈4 nm, the density corresponds to <1 peptide per square of the long dimension.
Fig. 1.
Adhesion of single amino acid constituted peptides to various inorganic surfaces as observed by using fluorescence microscopy. (a and b) Differential amino acid adhesion to inorganic surfaces as demonstrated in normal phase contrast (a) and fluorescence micrograph (b) of patterned structure used to test differential peptide adhesion. This structure is Si3N4 (deposited with plasma-enhanced chemical vapor deposition) on (100) GaAs with the pattern due to areas of the Si3N4 that have been removed by dry etching (reactive ion etching). This sample was then placed in a solution of peptide containing 10 Lys, N-terminated by a fluorescence molecule, then water washed. No evidence of fluorescence is apparent on the exposed GaAs, but the Si3N4 surface fluoresces homogeneously, corresponding to a peptide density of ≈2 × 104 peptides per μm2. (c–e) Fluorescence micrographs of amino acid (10-mer peptides) adhesion to multiple surfaces. The dark elements in each micrograph are the GaAs substrates to which minimal adhesion is observed.
Fig. 2.
Differential adhesion of single amino acid constituted peptides to multiple surfaces. Ten-oligomer peptides of Lys (a) and Arg (b) are exposed to multilayer materials. GaAs substrates are patterned with Si3N4, Al, and Au as shown. For each peptide, multiple intensities are present, demonstrating differential surface adhesion.
The peptide adhesion can vary with different inorganic surfaces: this finding is displayed in Fig. 2 a and b, where different fluorescent intensities are observed for different materials exposed to the same amino acid. As such, adhesion depends on both the material surface and the amino acid.
These results dictate that a full mapping is needed of the adhesion with respect to varied materials for each and all amino acids. Nine inorganic materials were tested for adhesion properties against the full complement (20) of single amino acid peptides. Additionally, a diverse set of solvents is necessary to begin to map these adhesion properties, with three chosen that include an organic solvent (DMSO), a biologically compatible buffered solution (Hepes), and water.
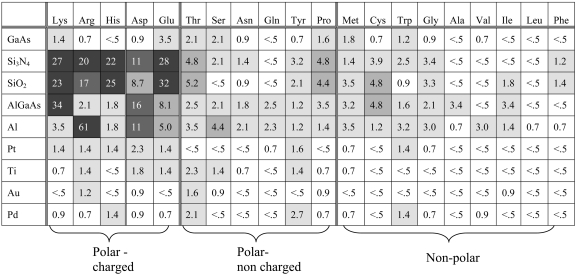
Table 1 displays the adhesion mapping in 1 mM H2O; specific inorganic material surfaces and amino acids are delineated there, with the respective adhered peptide densities. The other solvents tested are 0.25 mM Hepes and 0.25 mM DMSO. Similar adhesion patterns were observed with these solvents to those in Table 1, but with overall lower adhered densities, because the progression is made from H2O to Hepes to DMSO; these adhesion results are displayed in Tables 2 and 3, which are published as supporting information on the PNAS web site. Within the tables, the adhered density values have variability on repeat testing (repeat sample fabrication and repeat peptide solubilization and measurement) of ≈15%.
Table 1. Amino acid adhesion with solvent H2O.
Adhered peptide density (×103/μm2) as derived from fluorescence output for each of the 20 amino acids used to comprise 8- to 10-residue peptides, to each of 9 inorganic surfaces and in 1 mM H2O. HEPES and DMSO at concentrations of 0.25 mM were also tested with grossly similar results (see Supporting Materials and Methods). The variation in intensity preparation is ≈15%. The larger the value of adhesion, the darker shading used to aid in comparison between the matrices of amino acids and inorganic surfaces. Note the prevalence of the increased adhesion of the charged side groups to the insulators and the overall lack of interaction of the amino acids to the low-oxide forming metals.
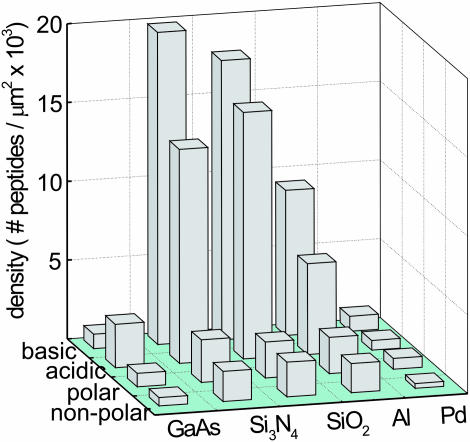
The overall mapping results can be reduced to a set of general trends. These data are summarized in Fig. 3. In this figure, the adhered peptide density is averaged for each peptide side-chain group (13) [polar (basic or acidic), polar but not charged, and nonpolar] on the respective inorganic surfaces and is averaged over the three different solvent results. The peptides with charged side groups, either basic or acidic, show marked adhesion to several, but not all, of the inorganic surfaces. This finding is in contrast to the reduced adhesion observed in the polar (not charged) and nonpolar peptides to most materials. However, the materials SiO2,Si3N4, and Al are apparently more adherent than the GaAs and Pd surfaces tested. From these results, it appears that the side-chain charge properties determine substantial differential adhesion to materials. These results also indicate that the use of multiple amino acids in the peptide chain overcomes the residual C terminus effects, because both significant adhesion and no apparent adhesion occur to identical surfaces by peptides containing the carboxylic acid terminus but possessing different amino acid species.
Fig. 3.
Summary of adhesion results for five representative inorganic surfaces and the four major groups of amino acids. The adhered peptide density is measured with fluorescence microscopy, and a comparison is made to a fluorescence standard; the raw data for this plot are contained in Tables 1, 2, 3. For each of the inorganic surfaces, data from all of the respective amino acids within each group (basic, acidic, etc.) are averaged over solvents H2O, Hepes, and DMSO to derive the adhered density. Note the general substantial adhesion of the charged amino acids (acidic and basic side chains) to the insulators Si3N4 and SiO2 in contrast to the metallic Pd interactions.
To explicitly examine the adhesion specificity and variability, we now review in more detail the results in the adhesion tables. For all three solvents, the peptides with charged side chains show pronounced adhesion to the Si-based insulators Si3N4 and SiO2. AlGaAs demonstrates a less consistent adhesion to the charged group, as is also shown by aluminum, which is coated by the native oxide. These results uniformly support the generalization that the charged side chains adhere to these insulators. At the other extreme, the nonpolar amino acids show minimal to no adhered peptide on metals that form little or no oxide (Pt, Pd, and Au). This finding holds for all three solvents. The metals are generally nonreactive, with some adhesion by the charged side group amino acids as more polar solvents are used.
Generally low reactivity is seen between the nonpolar peptides and Si-based insulators and the oxide surfaces of AlGaAs and aluminum. There is some variability in this finding with respect to both the solvents used and the different amino acids within this group. For example, the sulfur-containing side groups demonstrate more adhesion to the range of inorganics tested than is shown by the remainder of the nonpolar moieties. Surprisingly, some members of the nonpolar group show measurable (but not large) adhesion to the insulators and strong oxide surfaces.
As may be expected, the polar, but not charged, amino acids demonstrate large intragroup variability in adhesive properties with some solvent dependence. This group shows relatively strong adhesion to the insulators in water compared with the minimal reactivity in the organic solvent DMSO.
These tables both provide the basis to generalize the binding properties and, importantly, offer an empirical guide to these interactions. Although clear tendencies are present in these data, such as strong adhesion of the charged amino acids to the insulators or weak interaction of amino acids with nonoxidized metals, the remaining results provide empirical guidance for their respective interactions.
Mechanisms of Adhesion
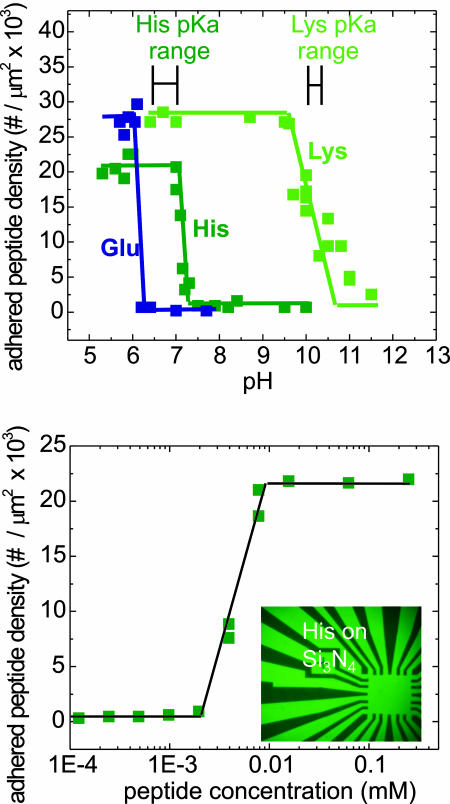
Given this empirical compilation of amino acid adhesion to surfaces, an understanding of the adhesion mechanisms is needed. The adhesion process was preliminarily examined by studying concentration and pH dependence. Several stronger adhesion sets were tested over a range of pH. The pH was progressively increased by the addition of NH4OH to the H2O solution, and the adhered peptide density was measured as before for each pH value. The results are shown in Fig. 4 Upper for the three charged side-group species of Lys, His, and Glu on Si3N4. All three amino acids show negligible adhesion at high pH, and, for each, the pH range is displayed over which adhesion drops from its value in nonbuffered 1 mM concentration to the value at high pH. Lys demonstrates this drop at approximately pH 10.2, which is near the intrinsic pKa of the side chain in that amino acid (13). Similarly, His shows an adhesion drop at approximately pH 7.4, close to the pKa value of the unassociated His side chain. These results offer a simple picture of the adhesion process for basic side group amino acids. The amorphous Si3N4 surface can present negatively charged sites (14) in solution. For pH values less than the pKa value of the amino acid applied to the surface, the peptide will be positively charged at the amino acid side group sites, providing an attractive interaction to the surface. Because the pH is increased through the respective amino acid pKa, the basic amino acid is neutralized and the attractive interaction is lost.
Fig. 4.
Examining mechanisms of amino acid adhesion. (Upper) Titration results examine adhered peptide density as a function of solution pH for three different peptides, each comprised of 10-mers of the respectively labeled amino acids on Si3N4. The predicted pKa of each free amino acid is shown; note the correspondence of adhesion change to each for Lys and His, but not for Glu. (Lower) Adhered peptide density dependence on peptide concentration by using His on Si3N4. Adhesion maintains the saturated value shown here beyond the graph's range up to 1 mM, and the drop to trace amounts occurs at the value roughly calculated to correspond to depletion of the peptide in the solvent, given the solvent volume and substrate area.
However, this simplistic picture is insufficient to describe the response of an acidic amino acid species, such as glutamic acid, as shown in Fig. 4 Upper. Here, a loss of adhesion is again observed with increasing pH, with the transition occurring at approximately pH 6.4. The pKa value for Glu's side chain is ≈4.2, so that above this pH value the peptide is negatively charged. The scenario of a predominantly negatively charged surface in the pH range of 4.2–6.4 is therefore inconsistent with the strong adhesion observed by this acidic moiety, suggesting that multiple adhesion sites of different polarizations may be present on these amorphous surfaces. This finding is the object of present investigation.
Concentration dependence for one adhesion reaction is shown in Fig. 4 Lower. In this study of His on Si3N4, a constant coverage is observed for an extremely large range of peptide concentrations, suggesting that a repulsive layer is achieved that does not allow further ordering on the material surface. The saturated surface coverage density corresponds to well less than 1 peptide per long peptide dimension squared, indicating that a substantial surface space charge effect may be at work. The adhered density drops to the observable limit at a concentration near 1 μM, corresponding to the value predicted for depletion of the peptide from the solution.
From these results, the mechanisms for surface adhesion appear to be more consistent with chemisorption than physisorption. Whereas there are clearly discernible strong charge attractions of the charged amino acids to the surfaces, the process of using a polar solution wash (H2O) establishes a relatively high threshold to observe adhesion. It is possible that a covalent attachment could be overwhelmed by this process. A weaker physisorption mechanism would, under this assumption, be unlikely to result in observable adhesion in these experiments.
Closer examination of the adhesion tables shows that, within the side-chain charge groups, systematic secondary differences in adhesion properties exist. Two examples include the comparison of aspartic to glutamic acids and serine to threonine. In both comparisons, the longer side chain shows stronger general adhesion, suggesting that steric structure may influence the adhesion mechanism. Both pairs demonstrate biological function differences in material crystallization processes (15, 16), again despite similar charge states, indicating the potential importance of geometry in addition to charge.
In considering the less pronounced interactions, a potential source of variability in the results is contaminants within the peptide preparations that could induce or block adhesion. The generally lower fluorescence outputs of the polar (not charged) and nonpolar groups would be influenced proportionally more by such contaminants.
These results present multiple mechanisms for modulating adhesion of different peptides to similar surfaces. The pH studies show that, for certain pH ranges, adhesion could be promoted for one amino acid species while being suppressed in another. In addition, strict control of peptide concentration can be used to vary the total adhered amounts, in conjunction with control of exposure time. A third method for modulation of adhesion may be through the use of different solvents.
Matching an Inorganic Structure to a Primary Peptide Sequence
The next level of complexity in the problem of peptide adhesion to inorganic surfaces asks whether an inorganic surface can be fabricated on the length scale of variations in a primary peptide sequence. This effort entails constructing an inorganic structure comprised of a sequence of components designed to spatially match a prescribed peptide sequence having amino acids that will adhere (or not adhere) to the inorganic component sequence by using the adhesion properties demonstrated above in this study.
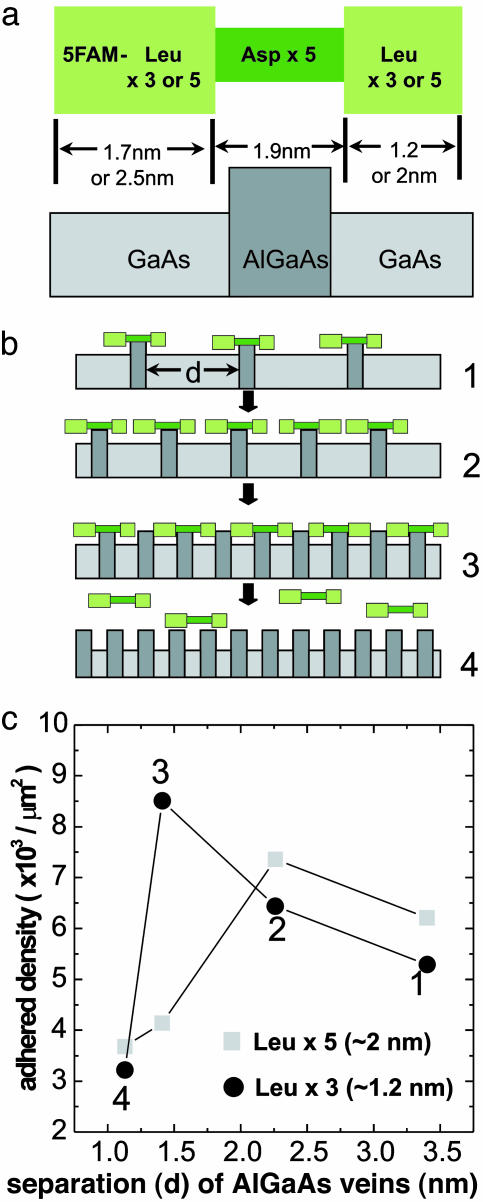
The experiment is shown schematically in Fig. 5 a and b. The inorganic surfaces are constructed by using molecular beam epitaxy (MBE) layering of GaAs and AlGaAs. Using the information gleaned in the above materials adhesion studies, we select a peptide sequence that includes a series of hydrophobic amino acids (three or five Leu), a center section of Asp that is known to adhere to AlGaAs, with the peptide terminated symmetrically with more Leu; this results in a peptide that is adhesive in the center to AlGaAs and nonadhesive at either end. Through MBE, a series of layered GaAs/AlGaAs structures are produced, with the layer thickness of the AlGaAs designed to be slightly less than the sequence length of the five central aspartic acids. A piece of this layered MBE sample is cleaved to expose the layering and etched to selectively remove only GaAs, leaving a profile of exposed AlGaAs veins. As designed, the peptide's aspartic acid section can adhere to these AlGaAs veins, with the leucines not adhering to the veins or the GaAs. In the experiment, a series of such MBE structures are produced with the AlGaAs layer thickness ≈0.8 nm but with different wafers constructed by using different distances d between veins of 1.1 nm up to 3.5 nm. At the larger vein separations, the peptide should adhere to the AlGaAs. However, as the AlGaAs layers are made close enough, the leucine sequences will abut the adjacent veins, inhibiting adhesion.
Fig. 5.
Matching an inorganic structure to a primary peptide sequence. (a) Schematic of peptides designed specifically to fit inorganic surface relief. (b) The relief surface is AlGaAs veins protruding from the GaAs background; four different samples were tested, each with different separation d of the AlGaAs veins. (c) The peptide of a with three leucines on each end shows adhesion to the AlGaAs veins for the three largest vein separations but shows abrupt adhesion loss for separation d less than the hydrophobic peptide end group length. The peptide with five leucines shows adhesion loss for the two smallest vein separations. Ten oligomer of aspartic acid applied to this series of samples does not show the dramatic drop in adhesion for the smallest vein separations.
Typical results of this test are shown in Fig. 5c. At the largest AlGaAs vein separation, the peptide demonstrates a strong adhesion to the growth layers containing the AlGaAs veins. As the vein separation is decreased, the fluorescence output increases due to the higher density of AlGaAs. For the peptide with three Leu on each end, the fluorescence output decreases dramatically as the vein-to-vein separation is made smaller than the peptide's hydrophobic tail dimension, in this case ≈1.3 nm. Similarly, the peptide symmetrically terminated with five Leu shows abrupt adhesion loss for a larger vein separation of nearly 2 nm. The results suggest that the steric interference of the leucines with the adjacent AlGaAs veins inhibits the adhesion of the peptide to the AlGaAs/GaAs profile. A 10-mer of aspartic acid applied to this series of samples does not show the dramatic drop in adhesion for the smallest vein separation. This result implies that inorganic materials can be fabricated to specifically adhere to prescribed peptide sequences by using the adhesive properties of the different amino acids.
Discussion
To summarize results, peptides comprised of single-species amino acids are shown to differentially adhere to a range of material surfaces. These adhesion properties can be correlated to the side groups defining the amino acids. The mechanisms for adhesion can in some instances be explained by the simple solution charge states of the peptides and surfaces. Using the adhesion maps derived here, primary peptide sequences were produced and inorganic structures were fabricated that show specific binding that can be controlled through the inorganic surface design.
Two directions of investigation are presented by these results: can the amino acid properties be used to affect surface properties of the inorganics, and can the inorganics be used to affect interaction of amino acid based adherents? The specificity of the amino acid adhesion to certain surfaces potentially offers mechanisms for selectively identifying, coating (as with a resist), or passivating prescribed material layers.
Broader topics are suggested by the question of how the inorganics can be used to affect interactions of amino acid based adherents, both by using simple single material surfaces and by using the high lateral spatial registry of MBE shown here. Among many possibilities, further efforts can address performing conjugation reactions on these surfaces, with the potential for producing a variety of adhered biological macromolecules. Add to this the ability to specifically position biomolecules, and synthetic control can be considered because of the possibilities for aligning biological macromolecules and affecting their interactions. The extreme spatial control achieved in MBE layered structures introduces the potential for biomaterial recognition. This spatial registry lends itself to incorporating electronic devices, as have already been achieved (17) to study macromolecular arrangements and properties.
Author contributions: R.L.W. and L.N.P. designed research; R.L.W. and K.W.B. performed research; K.W.W. and L.N.P. contributed new reagents/analytic tools; R.L.W. analyzed data; and R.L.W. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviation: MBE, molecular beam epitaxy.
References
- 1.Luth, H. (1995) Surfaces and Interfaces of Solid Materials (Springer-Verlag, Berlin).
- 2.Chakraborty, T. & Pietilainen, P. (1995) The Quantum Hall Effects (Springer-Verlag, Berlin).
- 3.Fields, G. B., ed. (1997) Solid-Phase Peptide Synthesis, Methods in Enzymology, (Academic, San Diego), Vol. 289.
- 4.Brown, S. (1997) Nat. Biotechnol. 15 269–272. [DOI] [PubMed] [Google Scholar]
- 5.Whaley, S., English, D. S., Hu, E. L., Barbara, P. F. & Belcher, A. M. (2000) Nature 405 665–668. [DOI] [PubMed] [Google Scholar]
- 6.Mao, C., Flynn, C. E., Hayhurst, A., Sweeney, R., Qi, J., Georgiou, G., Iversion, B. & Belcher, A. M. (2003) Proc. Natl. Acad. Sci. USA 100 6946–6951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang, S.G., Yan, L., Altman, M., Lassie, M., Nugent, H., Frankel, F., Lauffenburger, D. A., Whitesides, G. M. & Rich, A. (1999) Biomaterials 20 1213–1220. [DOI] [PubMed] [Google Scholar]
- 8.Wagner, P., Nock, S., Spudich, J. A., Volkmuth, W. D. & Chu, S. (1997) J. Struct. Biol. 119 189–201. [DOI] [PubMed] [Google Scholar]
- 9.Veiseh, M., Zareie, M. H. & Zhang, M. (2002) Langmuir 18 6671–6678. [Google Scholar]
- 10.Gleason, N. J., Nodes, C. J., Higham, E. M., Guchert, N., Aksay, I. A., Schwarzbauer, J. E. & Carbeck, J. D. (2003) Langmuir 19 513–518. [Google Scholar]
- 11.Alireza, R., Johnson, R., Lefkow, A. R. & Healy, K. E. (1999) Langmuir 15 6931–6939. [Google Scholar]
- 12.Yeo, W. S., Yousaf, M. N., Mrksich, M. (2003) J. Am. Chem. Soc. 125 14994–14995. [DOI] [PubMed] [Google Scholar]
- 13.Cantor, C. R. & Schimmel, P. R. (1997) Biophysical Chemistry: Part 1: The Conformation of Biological Macromolecules (Freeman, New York).
- 14.Quirk, M. & Serda, J. (2001) Semiconductor Manufactoring Technology (Prentice–Hall, Upper Saddle River, NJ).
- 15.Aizenberg, J., Lambert, G., Weiner, S. & Addadi, L. (2002) J. Am. Chem. Soc. 124 32–39. [DOI] [PubMed] [Google Scholar]
- 16.Lowenstam, H. A. & Weiner, S. (1989) On Biomineralization (Oxford Univ. Press, New York).
- 17.Natelson, D., Willett, R. L., West, K. W., Pfeiffer, L. N. (2000) Appl. Phys. Lett. 77 1991–1993. [Google Scholar]