ABSTRACT
Objectives
Diabetic nephropathy (DN) is one of the most serious microvascular complications of diabetes and is the most common cause of end-stage renal disease. Tripartite motif-containing (TRIM) proteins are a large family of E3 ubiquitin ligases that contribute to protein quality control by regulating the ubiquitin – proteasome system. However, the detailed mechanisms through which various TRIM proteins regulate downstream events have not yet been fully elucidated. The current research aimed to determine the function and mechanism of TRIM22 in DN.
Methods
DN models were established by inducing HK-2 cells using high glucose (HG) and diabetic mice (db/db mice). Cell viability, apoptosis, mitochondrial reactive oxygen species, and mitochondrial membrane potential were detected by Cell Counting Kit-8 and flow cytometry, respectively. Pathological changes were evaluated using hematoxylin and eosin, periodic acid schiff and Masson staining. The binding between TRIM22 and optic atrophy 1 (OPA1) was analyzed using co-immunoprecipitation. The m6A level of TRIM22 5′UTR was detected using RNA immunoprecipitation.
Results
TRIM22 was highly expressed in patients with DN. TRIM22 silencing inhibited HG-induced apoptosis and mitochondrial dysfunction in HK-2 cells. Promoting mitochondrial fusion alleviated TRIM22 overexpression-induced cell apoptosis, mitochondrial dysfunction in HK-2 cells, and kidney damage in mice. Mechanistically, TRIM22 interacted with OPA1 and induced its ubiquitination. Wilms tumor 1-associating protein (WTAP) promoted m6A modification of TRIM22 through the m6A reader insulin-like growth factor 2 mRNA-binding protein 1 (IGF2BP1).
Discussion
TRIM22 silencing inhibited the progression of DN by interacting with OPA1 and inducing its ubiquitination. Furthermore, WTAP promoted m6A modification of TRIM22 via IGF2BP1.
KEYWORDS: WTAP, m6A, TRIM22, diabetic nephropathy, mitochondrial dysfunction, OPA1
1. Introduction
Diabetic nephropathy (DN) is among the most critical microvascular complications of diabetes and is the most common cause of end-stage renal disease [1]. Microvascular lesions associated with diabetes mainly cause glomerular lesions. In recent years, evidence has shown that renal tubulointerstitial lesions caused by diabetes, such as renal interstitial fibrosis and tubular atrophy, play an important role in the progression of renal damage [2]. Tubular epithelial cells, which constitute a significant portion of the renal parenchyma, are susceptible to destruction during kidney injury [3]. However, the molecular mechanisms underlying tubular epithelial cell injury in DN remain unclear, warranting further studies on the optimal therapeutic approach.
Mitochondria are known as cellular powerhouses that produce adenosine triphosphate (ATP) and reactive oxygen species (ROS) and participate in cell apoptosis [4,5]. Mitochondrial dynamics, including fusion and fission, are vital for the metabolic regulation of cellular energy. Mitochondrial fusion is mediated by mitofusins (MFN1 and MFN2) and optic atrophy 1 (OPA1), whereas mitochondrial fission is mediated by dynamin-related protein 1 (DRP1) [6]. Several studies have demonstrated that disturbances in mitochondrial dynamics within the proximal tubules is a critical characteristic associated with DN [7,8]. Therefore, revealing the molecular mechanisms affecting mitochondrial dynamics in kidney tissues and cells may provide potential targets for the treatment of DN.
The tripartite motif-containing (TRIM) protein family is a subfamily of the Ring E3 ubiquitin ligase family, with over 70 TRIM proteins having been discovered so far [9]. Recent researches have shown that TRIM family proteins play important roles in transcriptional regulation, cell proliferation, cell metastasis, cell apoptosis, and tumor formation [10–12]. Several studies have demonstrated that the TRIM family proteins, which directly or indirectly act as regulatory proteins, are involved in the development of diabetic complications. For example, TRIM72, TRIM13, TRIM16, and TRIM18 may be potential therapeutic targets for the treatment of diabetic cardiomyopathy and DN [13,14]. Reports have shown that mitophagy is of great importance in maintaining mitochondrial dynamics. One study showed that TRIM21 mediated the ubiquitination of tyrosine aminotransferase to inhibit mitophagy in gallbladder cancer [15]. Moreover, another study found that the TRIM27–TBK1–SQSTM1/p62 pathway facilitated mitochondria clustering and mitophagy [16]. Therefore, TRIM family proteins may be an important regulator of mitochondrial dynamics. However, whether TRIM family proteins mediate DN progression by mediating mitochondrial dynamics remains unclear.
RNA methylation, one of the important aspects of epigenetic research, has been found to potentially mediate gene expression and splicing, RNA editing and stability, and mRNA lifespan and degradation [17]. N6-methyladenosine (m6A) is a widely present base modification behavior on mRNA that has become a research hotspot in recent years. m6A methylation modification, which is involved in methyltransferases, demethylases, and methylated reading proteins, is reversible and plays an important role in mitochondrial dynamics and the occurrence and development of DN. For example, Wilms’ tumor 1-associating protein (WTAP) is a methyltransferase that can promote m6A methylation of NLRP3 mRNA to induce cell pyroptosis and inflammation in DN [18] and mediate m6A modification of lncRNA Snhg1 to ameliorate myocardial injury via OPA1-dependent mitochondrial fusion [19]. Insulin-like growth factor 2 mRNA-binding protein 3 (IGF2BP3) is an m6A reader that can mediate CAMK1 mRNA stability through m6A modification to alleviate DN progression by inhibiting mitochondrial fission [20]. Several studies have confirmed the m6A modification of TRIM family members, such as TRIM59 [21], TRIM11 [22] and TRIM7 [23]. However, it remains unclear whether WTAP/IGF2BPs-mediated m6A modification regulates the stability of TRIM family members and thus mediates DN development.
The current study therefore aimed to explore the mechanism by which TRIM family members are involved in the process of DN development. Our study has been the first to demonstrate that TRIM22 expression is increased in patients with DN and high glucose (HG)-induced HK-2 human renal tubular epithelial cells. Moreover, we found that TRIM22 silencing inhibited HG-induced apoptosis and mitochondrial dysfunction in HK-2 cells, as well as alleviated kidney damage in mice. The mechanism underlying such findings is the interaction between TRIM22 and OPA1, inducing its ubiquitination. In addition, WTAP promoted m6A modification of TRIM22 through the m6A reader IGF2BP1. These findings provide insights into novel candidate targets and strategies for the clinical treatment of DN.
2. Materials and methods
2.1. Bioinformatics analysis
The Gene Expression Omnibus database was searched, and GSE30122 was obtained from the renal tubules of patients with diabetic nephropathy [24]. Gene expression differences between isolated control and DN tubular tissue were determined using statistical analysis (t-test unpaired, P < 0.05, fold change > 1.0). Gene Set Enrichment Analysis (GSEA) was used to identify enriched pathways in TRIM22-high versus TRIM22-low groups. A P value of < 0.05 and false discovery rate of < 0.25 indicated statistical significance.
2.2. Clinical sample collection
Human kidney biopsy tissues from patients with DN (n = 43) and normal kidney tissues from nephrectomies (n = 9) were obtained from Yueyang Hospital of Integrated Traditional Chinese and Western Medicine. Tissues were placed into RNALater and manually microdissected at 4°C for tubular compartment. Studies involving patients and specimens were approved by the Ethics Committee of Yueyang Hospital of Integrated Traditional Chinese and Western Medicine, Shanghai University of Traditional Chinese Medicine (approval number KYSKSB2020–091) and were conducted in accordance with the Declaration of Helsinki. Informed consent regarding the use of specimens was obtained from all patients.
2.3. Animals
Male C57BL/KsJ diabetic mice (db/db; 8 weeks old) were housed under a 12:12 h light – dark cycle, and their nutritional requirements were met through ad libitum feeding. Control mice were normal male C57BL/KsJ mice (db/m; 8 weeks old). db/db mice were randomly allocated into two groups (n = 6 per group), namely the db/db group and db/db + M1 group that received mitochondrial fusion promoter M1 (10 mg/kg; Sigma-Aldrich, SML0629) once a day via gavage. We then collected serum and urine samples 4 weeks after injection, dislocated their cervical vertebrae, and collected kidney tissues for hematoxylin and eosin (H&E) staining, periodic acid schiff (PAS) staining, and Masson staining as previously described [25]. The Creatinine Assay Kit, Urea Assay Kit, and Urine Protein Test Kit (all from Nanjing Jiancheng Bioengineering Institute) were used to measure serum creatinine, blood urea nitrogen, and urine protein levels, respectively. All animal studies were approved by the Ethics Committee of Yueyang Hospital of Integrated Traditional Chinese and Western Medicine, Shanghai University of Traditional Chinese Medicine (approval number YYLAC-2021–125).
2.4. Immunofluorescence microscopy
Fixed kidney tissues were stained with anti-OPA1 (Proteintech; 27733-1-AP) and corresponding second antibody. In this study, nucleic acid was visualized using 4′,6-diamidino-2-phenylindole solution and then observed under a confocal laser scanning microscope.
2.5. Cell culture
HK-2 cells were obtained from the ATCC and cultured in Dulbecco's Modified Eagle Medium with 10% (v/v) fetal bovine serum and 100 U/mL penicillin and streptomycin. HK-2 cells at 80% confluence were treated with 5.5 mM normal glucose (NG) with or without mitochondrial fusion promoter M1 for 48 h, 10 μM MG132 (proteasome inhibitor) for 4 h, or 30 mM HG for 6, 12, 24, or 48 h. Additionally, the NG group was treated with 24.5 mM mannitol.
2.6. Gene overexpression and knockdown
The TRIM22 or OPA1 gene was synthesized and inserted into a pLVX-Puro vector (Clontech, USA). To knockdown TRIM22, WTAP, or IGF2BP1 expression, three shRNAs targeting TRIM22 (shTRIM22-1, shTRIM22-2, and shTRIM22-3), two shRNAs targeting WTAP (shWTAP-1 and shWTAP-2) or IGF2BP1 (shIGF2BP1-1 and shIGF2BP1-2), as well as scramble shRNA (shNC) as negative control, were synthesized and inserted into a pLKO.1 vector. The shRNA sequences are listed in Table 1. Meanwhile, the recombinant plasmids and packaging vectors psPAX2 and pMD2G were co-transfected using Lipofectamine 2000 (Invitrogen, USA). The pcDNA3.1 vector was ligated using His-tagged mutant or full-length OPA1 cDNA, which was named His-OPA1 (K228R, K568R, and WT). In 293 T cells, His-OPA1 constructs, HA-Ub expression vector, and TRIM22 expression vector were co-transfected with Lipofectamine 2000.
Table 1.
shRNA sequences used in this study.
| Gene | Sequences (5′−3′) |
|---|---|
| Human TRIM22 shRNA-1 | GGAAGATGACATCAGACAA |
| Human TRIM22 shRNA-2 | GGATCAGAGACAAGTGAAA |
| Human TRIM22 shRNA-3 | GCATCACTGCAAAGATCAA |
| Human WTAP shRNA-1 | GCAAGAGTGTACTACTCAA |
| Human WTAP shRNA-2 | GGAACAGACTAAAGACAAA |
| Human IGF2BP1 shRNA-1 | GGCTCAGTATGGTACAGTA |
| Human IGF2BP1 shRNA-2 | AGCAAGATACCGAGACAAA |
| shNC | GGACGAGCTGTACAAGTAA |
2.7. Cell counting kit-8 (CCK-8)
HK-2 cells (3 × 103 cell/well) were seeded into 96-well plates and treated for 0, 12, 24, and 48 h. CCK-8 solution was incubated with each well for 4 h. Thereafter, cell viability was measured using a microplate reader.
2.8. Flow cytometry
For cell apoptosis analysis, propidium iodide (PI) staining and FITC-labeled annexin V were used. Following centrifugation, the cells were stained with Annexin V-FITC/PI for 15 min. CytoFLEX flow cytometry (BD Biosciences, USA) was used to evaluate apoptosis. Mitochondrial ROS was evaluated using the MitoSOX probe and analyzed using flow cytometry. Moreover, the mitochondrial membrane potential (MMP) ratio was calculated as red (JC-1 aggregates)/green (JC-1 monomers) fluorescence intensity using the JC-1 Assay Kit (C2006, Beyotime Institute of Biotechnology, Jiangsu, China) and analyzed via flow cytometry.
2.9. Measurement of ATP and ADP
ATP content was determined using the ATP Assay Kit (Abcam; ab83355), whereas ADP content was determined using the ADP Assay Kit (Abcam; ab83359). ATP and ADP concentrations were normalized to the corresponding total protein amounts from each sample.
2.10. Quantitative real-time PCR (RT-qPCR)
The TRIzol method was used to extract RNA from human renal tubules or HK-2 cells, and cDNA was reversed using cDNA reverse transcription reagent kit (Takara, Japan; RR047A). We performed RT-qPCR on an ABI 7500 fast machine (Applied Biosystems, USA) using the SYBR Premix EX Taq Kit. The primer sequences are listed in Table 2. Normalized gene expression was determined using the 2−ΔΔCT method. β-Actin was used as the control for RT-qPCR.
Table 2.
Primer sequences for real-time PCR assay.
| Gene | Primer pair sequences | Number of base pairs | Gene ID |
|---|---|---|---|
| TRIM22 | F: 5′-GAGAACCGCCTGGAAGAATTA-3′ | 205 bp | NM_006074.5 |
| R: 5′-ATCTGAGATGAGCGTGCTGG-3′ | |||
| OPA1 | F: 5′-CAGCGCATGCTTGCTATCAC-3′ | 218 bp | NM_015560.3 |
| R: 5′-AGAGCTTCAATGAAAGCATCAAGT-3′ | |||
| WTAP | F: 5′-GTAATGGTAGCTCCTCCCGC-3′ | 174 bp | NM_004906.5 |
| R: 5′-ACCCCGCACTGAGTTGATTT-3′ | |||
| IGF2BP1 | F: 5′-GCGATGAAGGCCATCGAAAC-3′ | 269 bp | NM_006546.4 |
| R: 5′-AGCTTCATGATGGCTTGCCT-3′ | |||
| ACTB | F: 5′-AGGATTCCTATGTGGGCGAC-3′ | 273 bp | NM_001101.5 |
| 5′-ATAGCACAGCCTGGATAGCAA-3′ |
2.11. Western blot
Protein samples were prepared in lysing buffer for Radio Immunoprecipitation Assay (RIPA). Sodium dodecyl sulfate-polyacrylamide gel electrophoresis was then conducted, followed by transfer onto membranes and blocking with 5% (v/v) skim milk. The membranes were incubated with primary antibodies against TRIM22 (Abcam; ab68071; 1:500, v/v), OPA1 (Biorbyt; orb337383; 1:1000, v/v), MFN1 (Abcam; ab221661; 1:1000, v/v), MFN2 (Abcam; ab205236; 1:2000, v/v), DRP1 (Abcam; ab184247; 1:1000, v/v), WTAP (Abcam; ab195380; 1:10000, v/v), and β-actin (Proteintech; 66009-1-Ig; 1:5000, v/v) at 4°C overnight. Incubation with goat anti-rabbit IgG and goat anti-mouse IgG (ZSGB-BIO, Beijing, China; ZB-2301, ZB-2305; 1:10000, v/v) was subsequently performed.
2.12. Protein stability assay
To examine OPA1 protein turnover, cycloheximide (CHX; 0.1 mg/mL) was added to the cell culture medium, after which the cells were harvested at the indicated time points. Following cell lysis, Western blot analysis was performed using anti-OPA1 and anti-β-actin antibodies. Subsequently, OPA1 protein levels were quantified relative to β-actin using ImageJ.
2.13. Co-immunoprecipitation (Co-IP) and ubiquitination analysis
A lysate of 293 T cells was prepared using RIPA buffer and reacted with anti-TRIM22 (USBiological, Salem, MA, USA; 134727) and anti-OPA1 (Abcam; ab42364) antibodies or control IgG (Santa Cruz Biotech.; sc-2027) and then with protein A/G Plus agarose. The immunoprecipitated complexes were analyzed using Western blot with anti-TRIM22, anti-OPA1, or anti-Ub.
2.14. Pull-down assay
After treatment, lysed cells were incubated with Ni2+-NTA agarose beads (Qiagen) followed by co-transfection with His-OPA1 constructs, HA-Ub, and TRIM22 expression vector. To separate the proteins, Western blotting was used to visualize the bands formed by the complexes.
2.15. m6A content analysis
Poly(A)+ RNA was purified using the GenElute™ mRNA Miniprep Kit (Sigma, Louis, MO, USA; MRN10) to measure m6A content. Briefly, binding solution and RNA were added to each well, which was then incubated for RNA binding. Thereafter, the diluted capture antibody was added into each well. Following incubation with detection antibody and enhancer solution, the wells were incubated for 1–10 min with developer solution. The reaction was stopped and determined on a microplate reader.
2.16. RNA immunoprecipitation assays
Following the manufacturer's protocol, RNA immunoprecipitation (RIP) assays were conducted using the Magna RIP RNA-Binding Protein Immunoprecipitation Kit. RNA – protein complexes were conjugated with anti-m6A, anti-IGF2BP1, or anti-IgG antibody. After incubation, agarose beads and protein A/G were incubated again. Finally, RNAs were purified using phenol:chloroform:isoamyl alcohol and subjected to RT-qPCR.
2.17. Luciferase reporter gene assays
The TRIM22 5′UTR sequence was cloned into the pGl3 vector. HK-2 cells were treated with 30 mM HG, transduced with shWTAP-1 and shWTAP-2, and transfected with the pGl3-TRIM22 5′UTR luciferase reporter plasmid and Renilla luciferase pRL-TK vector using Lipofectamine 2000 (Invitrogen). Firefly luciferase activity was normalized to Renilla luciferase activity using the manufacturer's protocol.
2.18. mRNA stability measurements
Samples were collected 0, 2, 4, and 6 h after treatment with actinomycin D (GlpBio, Montclair, CA, USA; GC16866). We then performed reverse transcriptase synthesis using oligo(dT) primers and measured mRNA levels via RT-qPCR.
2.19. Data analysis
All experiments were conducted at least three times independently. All data were processed using GraphPad Prism 8.4.2 and presented as mean ± standard deviation. The normality of variable distribution was assessed using the Shapiro–Wilk test, whereas the homogeneity of the variance was assessed using Levene’s test. Owing to the normal distribution of variables, the two-sided unpaired Student’s t-test (for comparison between two groups) or one-way analysis of variance followed by Dunnett’s post hoc test (for multi-group comparisons) were used for statistical analyses. A P value of <0.05 indicated statistical significance.
3. Results
3.1. TRIM22 was highly expressed in patients with DN
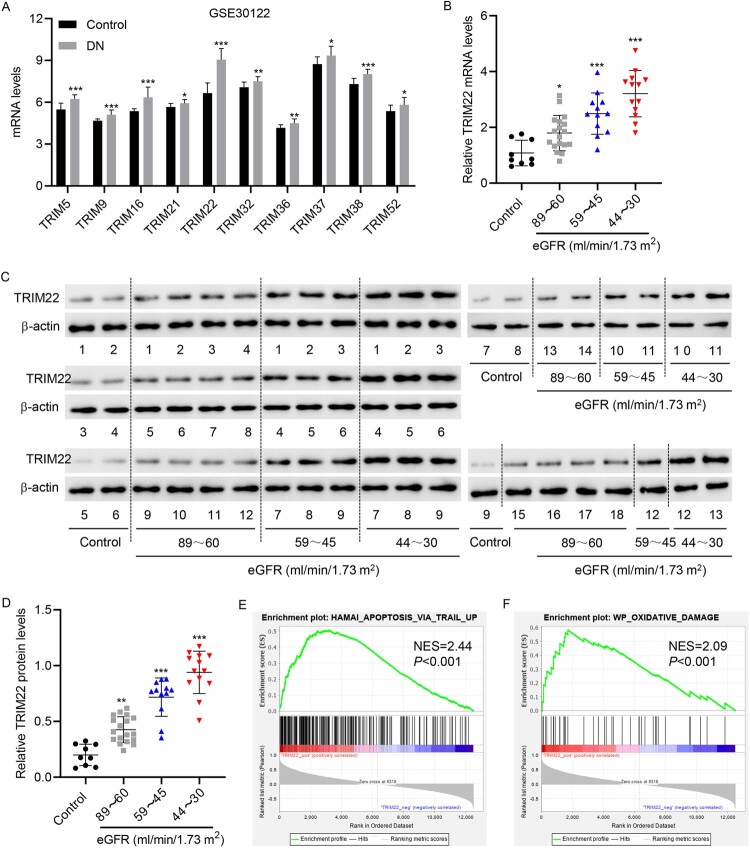
GSE30122 database analysis showed that the expressions of various TRIM family members, including TRIM5, 9, 16, 21, 22, 32, 36, 37, 38, and 52, were increased in the renal tubules of patients with DN compared with normal controls, with TRIM22 having the highest expression (P = 4.36E-07, fold change = 1.36) among the TRIM family members (Figure 1(A)). Therefore, TRIM22 was selected as the subject for study. The renal tubules of patients with DN admitted to our hospital were collected and divided into three groups based on eGFR (ml/min/1.73 m2) (the 89–60 group, 59–45 group, and 44–30 group). TRIM22 expression was significantly higher in the three groups than in the control group, with the 44–30 group having the highest mRNA and protein expression (Figure 1(B–D)). These findings suggest that TRIM22 may play an important role in the renal tubules of patients with DN. Moreover, TRIM22 expression was notably correlated with clinical characteristics, including hemoglobin A1c, hemoglobin, eGFR, BUN, serum creatinine, serum albumin, and albuminuria (Table 3). To further analyze the associated functions of TRIM22, GSEA was used to identify enriched pathways in patients with DN who had high and low expression of TRIM22. Accordingly, GSEA showed that the differentially expressed genes in the TRIM22-high and – low expression groups were enriched in HAMAI_APOPTOSIS_VIA_TRAIL_UP and WP_OXIDATIVE_DAMAGE pathways (Figure 1(E–F)), suggesting that TRIM22 may affect DN progression through the apoptosis and oxidative damage pathways.
Figure 1.
TRIM22 expression in patients with DN. (A) The GSE30122 database was used to analyze the expression of TRIM family members in the renal tubules of patients with DN. The renal tubules of patients with DN were collected and divided into three groups according to eGFR (ml/min/1.73 m2) (the 89–60 group: 18 cases; 59–45 group: 12 cases; and 44–30 group: 13 cases). Simultaneously, the renal tubules of nine normal controls undergoing renal puncture were collected. TRIM22 expression was detected using (B) RT-qPCR and (C, D) Western blotting. (E, F) GSEA analysis of the correlation between TRIM22 expression and HAMAI_ APOPTOSIS_ VIA_ TRAIL_ UP and WP_ OXIDATIVE_ DAMAGE signaling pathways. *P < 0.05, **P < 0.01, ***P < 0.001 versus control.
Table 3.
Clinical characteristics of patients with DN and control subjects.
| Variables | Group | p-value | ||
|---|---|---|---|---|
| Control (n = 10) | DN with low TRIM22 level (n = 18) | DN with high TRIM22 level (n = 25) | ||
| Age (years) | 53.1 ± 10.6 | 47.6 ± 10.2 | 53.1 ± 11.2 | 0.419a; 0.232b |
| Male (n, %) | 3 (30.0) | 10 (55.6) | 17 (68.0) | 0.191c; 0.833d |
| BMI (kg/m2) | 22.9 ± 1.96 | 31.7 ± 4.65 | 29.7 ± 4.36 | <0.001a; 0.182b |
| Hemoglobin A1c (%) | 5.26 ± 1.16 | 8.03 ± 1.24 | 9.19 ± 1.46 | <0.001a; 0.018b |
| Hemoglobin (g/dL) | 14.3 ± 0.42 | 13.3 ± 0.92 | 12.7 ± 0.71 | <0.001a; 0.013b |
| eGFR (mL/min/1.73 m2) | 95.8 ± 2.16 | 65.7 ± 11.1 | 49.3 ± 10.3 | <0.001a; <0.001b |
| BUN (mg/dL) | 11.9 ± 0.62 | 16.0 ± 1.96 | 18.4 ± 2.53 | <0.001a; 0.002b |
| Serum creatinine (mg/dL) | 0.79 ± 0.14 | 0.94 ± 0.21 | 1.14 ± 0.31 | <0.004a; 0.037b |
| Serum albumin (g/dL) | 4.39 ± 0.29 | 3.90 ± 0.46 | 3.66 ± 0.35 | <0.001a; 0.040b |
| Albuminuria (mg/day) | 2.61 ± 0.65 | 89.6 ± 23.9 | 121.9 ± 46.2 | <0.001a; 0.012b |
DN, diabetic nephropathy; BMI, body mass index; BUN, blood urea nitrogen; eGFR, estimated glomerular filtration rate. a, for differences among three groups using a Kruskal–Wallis test; b, for differences between TRIM22 low and high in DN groups using Mann–Whitney test. c, for differences among three groups and d, for differences between TRIM22 low and high in DN groups, using Chi square test.
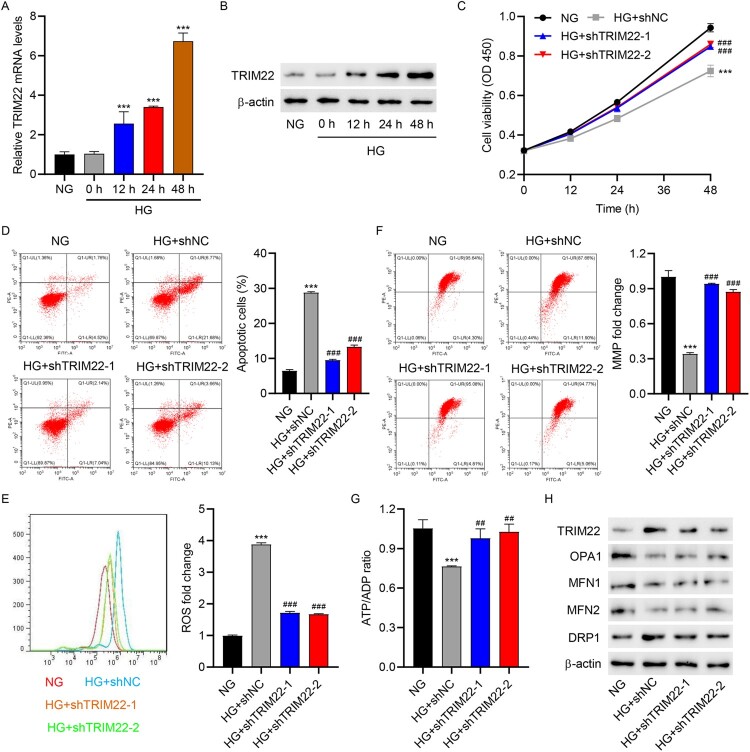
3.2. TRIM22 silencing inhibited HG-induced apoptosis and mitochondrial dysfunction in HK-2 cells
To test our hypothesis, cellular functional assays were performed. First, RT-qPCR and Western blotting were used to observe TRIM22 mRNA and protein expression under various HG exposure times. Our results showed that HG treatment significantly increased the expression of TRIM22 in HK-2 cells (Figure 2(A–B). Subsequently, TRIM22 was knocked down in HK-2 cells via TRIM22 shRNA lentivirus (shTRIM22-1, shTRIM22-2, and shTRIM22-3) transduction (Fig. S1A–S1B), after which cell viability and apoptosis were determined via the CCK-8 assay and flow cytometry, respectively. HG treatment significantly inhibited cell viability and promoted cell apoptosis, whereas TRIM22 shRNA lentivirus (shTRIM22-1 and shTRIM22-2) transduction significantly increased cell viability and inhibited cell apoptosis compared to shNC transduction in HG-induced HK-2 cells (Figure 2(C–D)). Researched had recently uncovered an interplay between ROS-induced oxidative stress and mitochondrial dynamics, indicating a correlation between oxidative damage and the control of mitochondrial shape [26]. Therefore, mitochondrial ROS, MMP, ATP/ADP levels, as well as expression of mitochondrial dynamics-related markers, was detected. Interestingly, HG significantly increased mitochondrial ROS levels and decreased MMP and ATP/ADP levels, whereas shTRIM22-1 and shTRIM22-2 transduction reversed such effects of HG (Figure 2(E–G)). Meanwhile, HG significantly promoted the protein expression of TRIM22 and DRP1 and inhibited the expression of OPA1, MFN1, and MFN2, whereas shTRIM22-1 and shTRIM22-2 transduction reversed the effects of HG on TRIM22 and OPA1 expression (Figure 2(H)). These results suggest that TRIM22 may regulate HG-induced HK-2 cell apoptosis and mitochondrial dysfunction via OPA1-dependent mitochondrial fusion.
Figure 2.
TRIM22 silencing inhibited HG-induced apoptosis and mitochondrial dysfunction in HK-2 cells. HK-2 cells were treated with high glucose (HG, 30 mM) (osmotic pressure was controlled with normal glucose concentration 5.5 and 24.5 mM mannitol, NG) to construct a renal tubular injury model of diabetes. TRIM22 expression was detected using (A) RT-qPCR and (B) Western blotting at 0, 6, 12, 24, and 48 h. HK-2 cells were transduced with TRIM22 shRNA lentivirus (shTRIM22-1 and shTRIM22-2) or scramble shRNA (shNC) and stimulated with HG for 48 h. (C) CCK-8 was used to determine cell viability. (D) Flow cytometry was used to detect cell apoptosis, (E) mitochondrial ROS, and (F) MMP. (G) Biochemical assay was used to determine the ATP/ADP ratio. (H) Western blotting was used to detect TRIM22, OPA1, MFN1, MFN2, and DRP1 expression. ***P < 0.001 versus NG; ##P < 0.01, ###P < 0.001 versus HG + shNC.
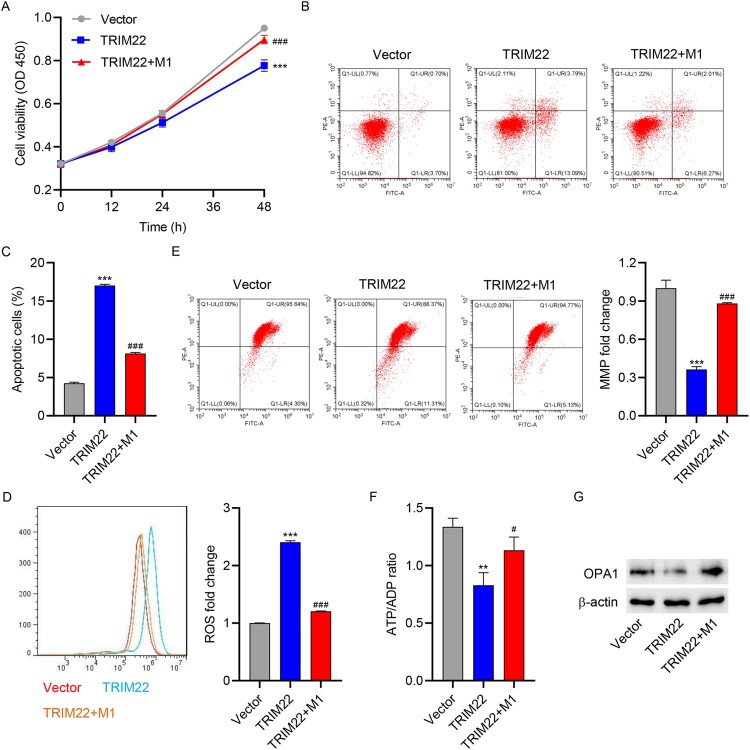
3.3. Promotion of mitochondrial fusion reversed TRIM22 overexpression-induced apoptosis and mitochondrial dysfunction in HK-2 cells
To further examine the role of mitochondrial fusion in TRIM22-induced apoptosis and mitochondrial dysfunction, TRIM22 was overexpressed in HK-2 cells (Fig. S1A–S1B) and then treatment with mitochondrial fusion promoter M1. Notably, our findings showed that TRIM22 overexpression significantly inhibited cell viability (Figure 3(A)) and promoted cell apoptosis (Figure 3(B–C)). Interestingly, TRIM22 overexpression increased mitochondrial ROS levels and decreased MMP and ATP/ADP levels (Figure 3(D–F)). Meanwhile, TRIM22 overexpression inhibited OPA1 expression (Figure 3(G)). However, treatment with mitochondrial fusion promoter M1 reversed the effects of TRIM22 overexpression on cell viability, apoptosis, mitochondrial dysfunction, and OPA expression (Figure 3(A–G)).
Figure 3.
Promotion of mitochondrial fusion relieved TRIM22 overexpression-induced apoptosis and mitochondrial dysfunction in HK-2 cells. HK-2 cells transduced with TRIM22 expression vector or blank vector were treated with 10 μM mitochondrial fusion inducer M1 alone or in combination for 48 h. (A) CCK-8 was used to determine cell viability. (B, C) Flow cytometry was used to detect cell apoptosis, (D) mitochondrial ROS, and (E) MMP. (F) Biochemical assay was used to determine the ATP/ADP ratio. (G) Western blotting was used to determine OPA1 expression. ***P < 0.001 versus vector; #P < 0.05, ###P < 0.001 versus TRIM22.
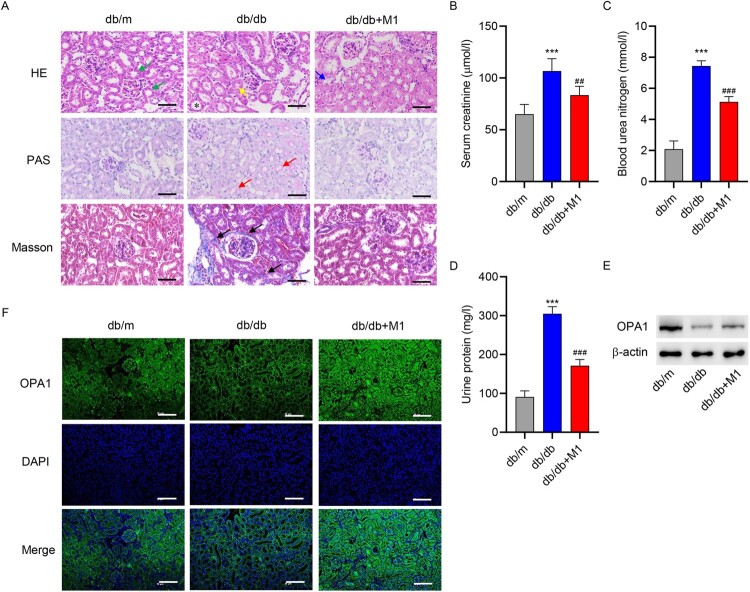
3.4. Promotion of mitochondrial fusion ameliorated kidney injury in mice
To further examine the role of mitochondrial fusion in DN in vivo, db/db mice were treated with mitochondrial fusion promoter M1, after which pathological changes, renal damage, and OPA1 expression were examined. H&E, PAS, and Masson staining indicated renal damage and fibrosis in diabetic mice. However, M1 treatment significantly ameliorated renal damage and fibrosis (Figure 4(A)). The levels of creatinine, urea nitrogen, and urinary protein were significantly increased in the db/db group, but M1 significantly reduced their levels (Figure 4(B–D)). After examining OPA1 expression in renal tissue using Western blot and immunofluorescence, we found a significantly lower OPA1 expression in the db/db group than in the control group, although M1 significantly increased its expression (Figure 4(E–F)).
Figure 4.
Promotion of mitochondrial fusion reversed kidney injury in mice. Eight-week-old diabetic mice were divided into a control group (db/m), model group (db/db), and model + mitochondrion fusion inducer M1 (db/db + M1) group that received intervention for 4 weeks. (A) HE, PAS, and Masson staining were used to analyze the pathological changes in renal tissues (scale bar, 50 μm). HE staining identified normal proximal tubules with narrow and irregular lumena, unclear cell boundaries, and the presence of brush border structures (green arrow) and abnormal proximal tubules with tubular dilatation (*), atrophy (yellow arrow), and loss of brush border integrity (blue arrow). PAS staining was used to identified renal tubular dilatation (red arrow). Masson staining identified extracellular matrix deposition (black arrow). Biochemical detection of (B) creatinine, (C) urea nitrogen and (D) urinary protein. (E) Western blotting and (F) immunofluorescence staining were used to determine OPA1 expression in renal tissues (scale bar, 100 μm). ***P < 0.001 versus db/m; ##P < 0.01, ###P < 0.001 versus db/db.
3.5. TRIM22 interacted with OPA1 and induced its ubiquitination
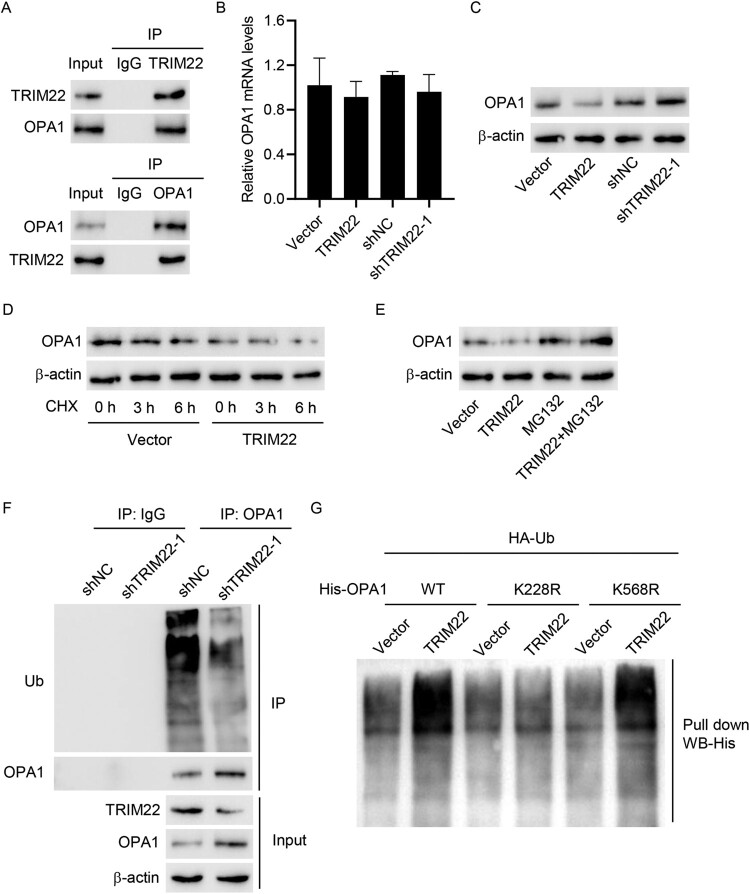
To further explore the mechanism underlying TRIM22, this study first used Co-IP to detect the binding activity of TRIM22 with OPA1 (Figure 5(A)). Accordingly, RT-qPCR revealed no significant difference in OPA1 mRNA expression after overexpression and interference with TRIM22, whereas Western blotting revealed decreased and increased OPA1 protein expression after overexpression and interference with TRIM22, respectively (Figure 5(B–C)). TRIM22 overexpression, along with CHX treatment, significantly reduced OPA1 expression in a time-dependent manner (Figure 5(D)). Additionally, TRIM22 overexpression combined with MG132 (a proteasome inhibitor) significantly increased OPA1 expression (Figure 5(E)). shTRIM22-1 transduction inhibited the ubiquitination of OPA1 and promoted its protein expression (Figure 5(F)). In the pull-down assay, cells were co-transfected with WT or mutant His-OPA1 constructs (K228R and K568R) along with TRIM22 expression vector and HA-Ub construct. Our results illustrated that K228R completely blunted TRIM22-induced OPA1 ubiquitination (Figure 5(G)), suggesting that the K228 site is essential for TRIM22-induced OPA1 ubiquitination.
Figure 5.
TRIM22 interacted with OPA1 and induced its ubiquitination. (A) Co-IP was used to detect the binding activity of TRIM22 and OPA1. TRIM22 shRNA lentivirus (shTRIM22-1), scramble shRNA (shNC), TRIM22 expression vector or blank vector was transfected into 293 T cells. (B) RT-qPCR and (C) Western blotting were used to detect OPA1 expression. (D) 293 T cells transfected with TRIM22 expression vector or blank vector were treated with protein synthesis inhibitor CHX, and Western blotting was used to determine OPA1 expression. (E) TRIM22 expression vector or blank vector were transfected into 293 T cells, which were subsequently treated with MG132 alone or in combination for 4 h and analyzed for OPA1 expression using Western blotting. (F) TRIM22 shRNA lentivirus (shTRIM22-1) or scramble shRNA (shNC) were transfected into 293 T cells, after which IP and Western blotting were used to detect the ubiquitination. (G) Cells were co-transfected with the His-OPA1 (WT) or mutant His-OPA1 constructs (K228R and K568R) along with TRIM22 expression vector or blank vector and HA-Ub construct and then analyzed using the pull-down assay.
3.6. OPA1 overexpression reversed TRIM22 overexpression-induced apoptosis and mitochondrial dysfunction
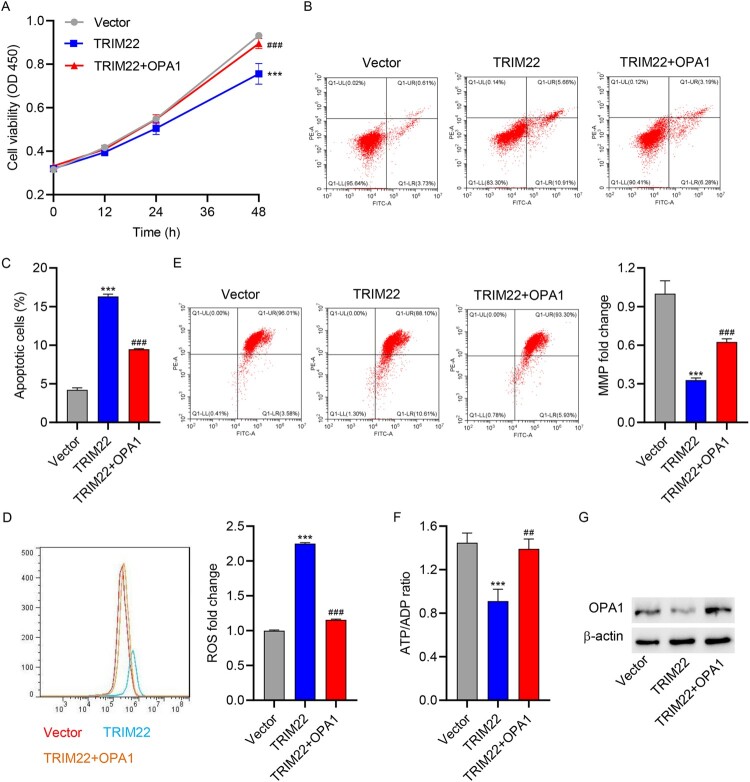
To investigate the role of OPA1 in TRIM22-induced apoptosis and mitochondrial dysfunction, HK-2 cells were co-transduced with TRIM22 and OPA1 expression vector for 48 h. TRIM22 overexpression significantly inhibited cell viability, whereas OPA1 overexpression significantly increased cell viability of HK-2 cells (Figure 6(A)). In addition, TRIM22 overexpression significantly promoted cell apoptosis, increased mitochondrial ROS levels, and decreased MMP and ATP/ADP levels, whereas OPA1 overexpression reversed the effects of TRIM22 (Figure 6(B–F)). Meanwhile, TRIM22 overexpression significantly inhibited the protein expression of OPA1, whereas OPA1 overexpression promoted the protein expression of OPA1 (Figure 6(G)).
Figure 6.
OPA1 overexpression relieved TRIM22 overexpression-induced apoptosis and mitochondrial dysfunction in HK-2 cells. Co-transduction of TRIM22 and OPA1 expression vector into HK-2 cells for 48 h. (A) CCK-8 was used to determine cell viability. (B, C) Flow cytometry was used to detect cell apoptosis, (D) mitochondrial ROS, and (E) MMP. (F) Biochemical assay was used to determine the ATP/ADP ratio. (G) Western blotting was used to detect OPA1 expression. ***P < 0.001 versus vector; ###P < 0.001 versus TRIM22.
3.7. WTAP promoted m6A modification of TRIM22 via IGF2BP1
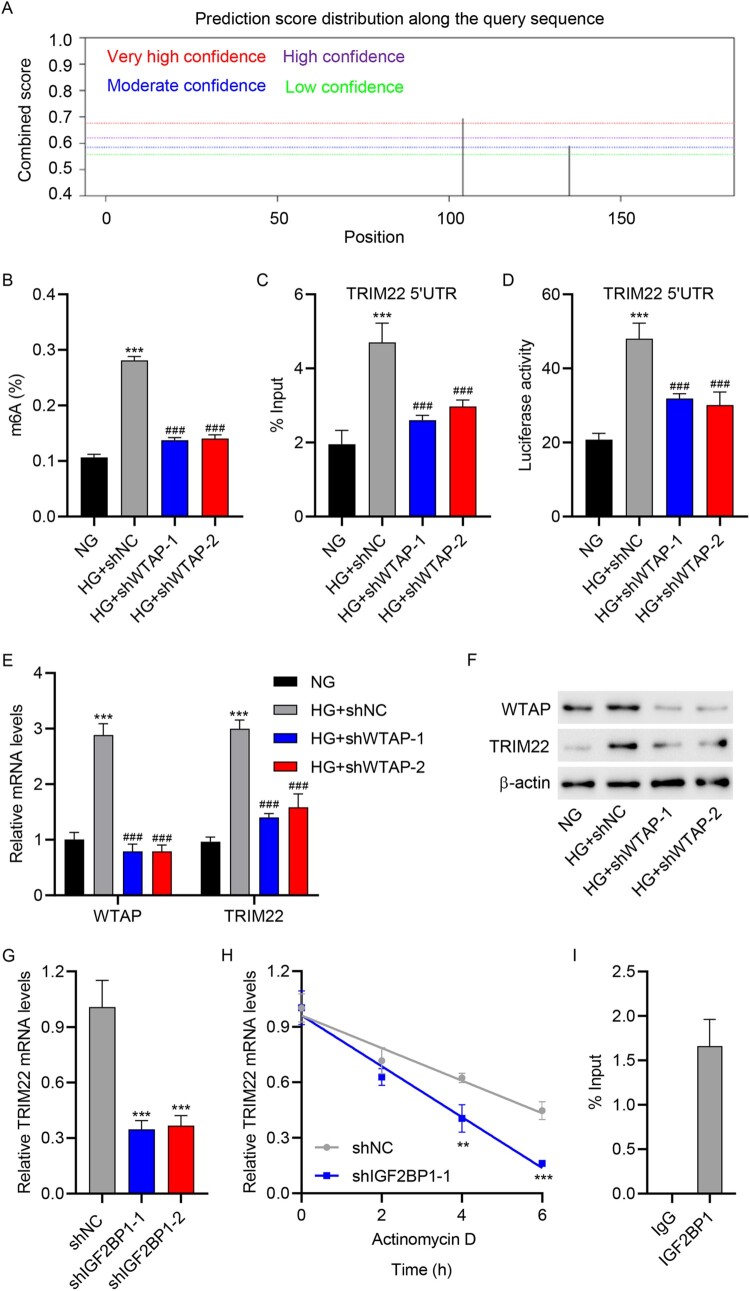
To investigate the regulation of TRIM22 in HG-induced HK-2 cells, we used the SRAMP website to predict the m6A modification of TRIM22 mRNA. SRAMP analysis revealed m6A modification sites in TRIM22 mRNA 5′UTR (Figure 7(A)). Subsequently, WTAP was knocked down in HK-2 cells via WTAP shRNA lentivirus (shWTAP2-1 and shWTAP-2) transduction, and the regulatory relationship between WTAP and TRIM22 was verified using ELISA, RIP, luciferase reporter, RT-qPCR, and Western blot assay. Notably, we found that shWTAP2-1 and shWTAP-2 transduction significantly inhibited global m6A levels and methylation levels of TRIM22 mRNA 5′UTR in HG-induced HK-2 cells (Figure 7(B–C)). In addition, the luciferase activity of TRIM22 mRNA 5′UTR was reduced following shWTAP2-1 and shWTAP-2 transduction in HG-induced HK-2 cells (Figure 7(D)). RT-qPCR and Western blotting revealed that shWTAP2-1 and shWTAP-2 transduction inhibited the expression of WTAP and TRIM22 in HG-induced HK-2 cells (Figure 7(E–F)). Next, IGF2BP1 was knocked down in HK-2 cells via IGF2BP1 shRNA lentivirus (shIGF2BP1-1 and shIGF2BP1-2) transduction (Fig. S1C–S1D), and the regulatory relationship between IGF2BP1 and TRIM22 was verified using RT-qPCR and RIP assays. As shown in Figure 7(G), shIGF2BP1-1 and shIGF2BP1-2 transduction significantly reduced the mRNA expression level of TRIM22. Moreover, following actinomycin D administration in HK-2 cells, we found that shIGF2BP1-1 transduction markedly reduced the stability of TRIM22 mRNA (Figure 7(H)). Furthermore, RIP assay found that TRIM22 mRNA 5′UTR was enriched in the anti-IGF2BP1 group but not in the anti-IgG group (Figure 7(I)). These data revealed that WTAP promoted m6A modification of TRIM22 via IGF2BP1.
Figure 7.
WTAP promoted the m6A modification of TRIM22 via the m6A reader IGF2BP1. (A) The SRAMP website was used to predict the m6A modification of TRIM22. HK-2 cells were transduced with WTAP shRNA lentivirus (shWTAP-1 and shWTAP-2) or scramble shRNA (shNC) and stimulated with HG for 48 h. (B) ELISA was used to determine m6A levels. (C) RIP was used to determine TRIM22 mRNA 5′UTR m6A levels. (D) Luciferase reporter gene assay was used to determine TRIM22 mRNA 5′UTR activity. (E) RT-qPCR and (F) Western blotting were used to determine the expression of WTAP and TRIM22. (G) HK-2 cells were transduced with IGF2BP1 shRNA lentivirus (shIGF2BP1-1 and shIGF2BP1-2) or scramble shRNA (shNC) and analyzed for TRIM22 expression using RT-qPCR. (H) HK-2 cells were transduced with IGF2BP1 shRNA lentivirus (shIGF2BP1-1) or scramble shRNA (shNC), followed by actinomycin D treatment for 0, 2, 4, and 6 h. The transcription level of TRIM22 was determined using RT-qPCR. (I) RIP-PCR was used to detect the binding of IGF2BP1 to TRIM22 mRNA 5′UTR. **P < 0.01, ***P < 0.001 versus NG or shNC; ###P < 0.001 versus HG + shNC.
3.8. OPA1 and WTAP expression in patients with DN
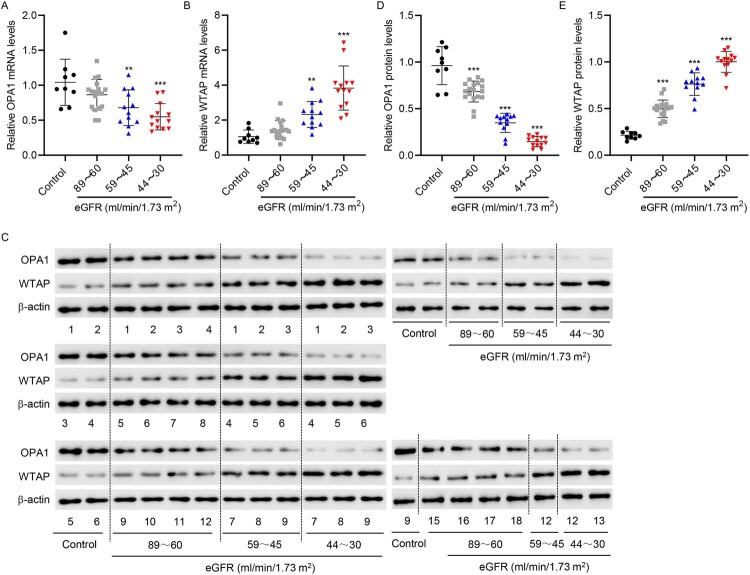
Renal tubules from patients with DN were collected and divided into three groups according to their eGFR (ml/min/1.73 m2) (the 89–60 group, 59–45 group, 44–30 group). OPA1 expression was significantly lower in the renal tubules of DN patients with low eGFR levels, whereas the opposite was observed for WTAP expression (Figure 8(A–E)).
Figure 8.
OPA1 and WTAP expression in patients with DN. The renal tubules of DN patients were collected and divided into three groups according to eGFR (ml/min/1.73 m2) (The 89–60 group: 18 cases; 59–45 group: 12 cases; and 44–30 group: 13 cases). Simultaneously, the renal tubules of nine normal controls undergoing renal puncture were collected. (A, B) RT-qPCR and (C – E) Western blotting were used to determine OPA1 and WTAP expression. **P < 0.01, ***P < 0.001 versus control.
4. Discussion
DN, one of the most pervasive microvascular complications, has become a vital concern globally [27–29]. The treatment of DN still remains challenging, with multiple mechanisms involved this process [30–32]. Here, we demonstrated that TRIM22 levels were upregulated in the renal tubules of patients with DN. TRIM22 knockdown promoted cell viability and inhibited apoptosis of HK-2 cells but upregulated mitochondrial fusion protein OPA1. Moreover, our findings showed that TRIM22 interacted with OPA1 and that its overexpression caused OPA1 downregulation via the ubiquitination of OPA1 at site K228. The presented findings improve our understanding of the role of TRIM22 in the progression of DN and provide a novel molecular target to prevent DN progression.
TRIM22 has been implicated in cell proliferation, differentiation, and death [33]. In line with this, evidence has shown that TRIM22 inhibits osteosarcoma development via the proteasome degradation pathways and autophagolysosomal degradation pathways [34]. Our data confirmed that TRIM22 expression was strongly upregulated in patients with DN. In addition, we found a positive relationship between TRIM22 expression and eGFR based on eGFR estimation in patients with DN and healthy people. Thus, our data suggests that abnormal TRIM22 expression is strongly associated with DN. GSEA database analysis focused on TRIM22-regulated genes identified the apoptosis pathway and the oxidative damage pathway as the most significantly enriched. Previous studies have shown that a caspase-dependent pathway mediated TRIM22 function through increased Bak expression [35]. Next, we found that high glucose induced the expression of TRIM22 and toxicity in HK-2 cells. TRIM22 overexpression has been shown to be anti-proliferative, a finding consistent with our results [36]. TRIM22 knockdown induced a high ratio of apoptotic cells. Moreover, mitochondrial damage promoted a significant decrease in MMP levels and ATP/ADP ratio and an increase in mitochondrial ROS production and TRIM22 expression. Excessive ROS production causes mitochondrial dysfunction, mainly characterized by loss of MMP and decreased ATP production [37]. Here, we showed that interference of TRIM22 attenuated HG-induced mitochondrial dysfunction by maintaining MMP levels, increasing the ATP/ADP ratio, and decreasing ROS generation. OPA1, MFN2, and MFN1, which are located in mitochondrial membrane, are crucial for mitochondrial fusion [38,39]. The present study revealed that HG affected mitochondrial dynamics and that TRIM22 knockdown reversed the HG-induced downregulation of OPA1. Collectively, TRIM22 could possibly regulate mitochondrial function and protect against mitochondrial injury in DN. Moreover, mitochondrial activator promoter M1 reduced TRIM22-induced HK2 cell damage. Evidence suggests that TRIM22 alters mitochondrial fusion-related proteins involved in respiration/ATP synthesis, affecting ROS production or other mitochondrial functions.
Mitochondrial membrane fusion protein OPA1 has also been associated with certain diseases [40,41]. Loss of OPA1 triggered ATP and MMP loss and induced mitochondrial fragmentation [42]. The OPA1-regulated process of mitochondrial fusion plays a critical role in cellular stress response [43–45]. Here, diabetic mice had lower levels of OPA1 protein than did non-diabetic mice, which was accompanied with histological changes during tubulointerstitial damage and prominent collagen deposition. Moreover, after evaluating the interaction between TRIM22 and OPA1, we found that TRIM22 overexpression promoted a reduction in OPA1 expression. one previous study reported that the ubiquitination-dependent degradation of OPA1 played a fundamental role in enhancing OPA1 [46]. Here, we demonstrated that the proteasome inhibitor MG132 prevented proteasomal degradation of OPA1, suggesting that OPA1 expression is regulated by posttranslational modification, particularly ubiquitination. TRIM22, a novel E3 ubiquitin ligase, involves proteasomal-mediated degradation of the protein [47,48]. Our data also suggests that TRIM22 induces OPA1 ubiquitination via the ubiquitination site K228. Moreover, OPA1 overexpression attenuated the inhibition of the cell viability of TRIM22-overexpressed cells. Meanwhile, OPA1 overexpression promoted mitochondrial balance, which manifested as a reduction in mitochondrial-derived ROS, increase in the ATP/ADP ratio, and stabilization of MMP. Recent studies have shown that m6A methylation modification plays an important role in the occurrence and development of DN [18, 49, 50]. The current research found that inhibition of WTAP significantly decreased m6A and methylation levels of TRIM22 and that WTAP promoted m6A modification of TRIM22 via the m6A reader IGF2BP1 in DN.
5. Conclusion
Taken together, our findings showed that WTAP/IGF2BP1-mediated m6A modification of TRIM22 promoted apoptosis and mitochondrial dysfunction in HG-induced HK-2 cells by suppressing mitochondrial fusion. By exploring the molecular mechanisms affecting mitochondrial dynamics, our data highlights the important role of TRIM22-mediated OPA1 ubiquitination in DN progression. Considering mitochondrial dynamics, the proposed WTAP/IGF2BP1–TRIM22–OPA1 axis opens up new avenues for exploring effective therapeutic strategies for DN.
Supplementary Material
Funding Statement
This study was supported by the National Natural Science Foundation of China (81973818) and Youth Project of National Natural Science Foundation of China (82004263).
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
All data presented in this study are included within the paper and its Supplementary files.
References
- 1.Pan S, Li Z, Wang Y, et al. A Comprehensive weighted gene co-expression network analysis uncovers potential targets in diabetic kidney disease. J Transl Int Med. 2023;10:359–368. doi: 10.2478/jtim-2022-0053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alicic RZ, Rooney MT, Tuttle KR.. Diabetic kidney disease: challenges, progress, and possibilities. Clin J Am Soc Nephrol. 2017;12:2032–2045. doi: 10.2215/CJN.11491116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li J, Shu L, Jiang Q, et al. Oridonin ameliorates renal fibrosis in diabetic nephropathy by inhibiting the Wnt/β-catenin signaling pathway. Ren Fail. 2024;46(1):2347462. doi: 10.1080/0886022X.2024.2347462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Amorim JA, Coppotelli G, Rolo AP, et al. Mitochondrial and metabolic dysfunction in ageing and age-related diseases. Nat Rev Endocrinol. 2022;18:243–58. doi: 10.1038/s41574-021-00626-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brand MD, Nicholls DG.. Assessing mitochondrial dysfunction in cells. Biochem J. 2011;435:297–312. doi: 10.1042/BJ20110162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moqbel SAA, Zeng R, Ma D, et al. The effect of mitochondrial fusion on chondrogenic differentiation of cartilage progenitor/stem cells via Notch2 signal pathway. Stem Cell Res Ther. 2022;13:127. doi: 10.1186/s13287-022-02758-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Han F, Wu S, Dong Y, et al. Aberrant expression of NEDD4L disrupts mitochondrial homeostasis by downregulating CaMKKβ in diabetic kidney disease. J Transl Med. 2024;22:465. doi: 10.1186/s12967-024-05207-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rizk FH, El Saadany AA, Atef MM, et al. Ulinastatin ameliorated streptozotocin-induced diabetic nephropathy: Potential effects via modulating the components of gut-kidney axis and restoring mitochondrial homeostasis. Pflugers Arch. 2023;475(10):1161–76. doi: 10.1007/s00424-023-02844-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hatakeyama S. TRIM family proteins: roles in autophagy, immunity, and carcinogenesis. Trends Biochem Sci. 2017;42(4):297–311. doi: 10.1016/j.tibs.2017.01.002 [DOI] [PubMed] [Google Scholar]
- 10.Hatakeyama S. TRIM proteins and cancer. Nat Rev Cancer. 2011;11(11):792–804. doi: 10.1038/nrc3139 [DOI] [PubMed] [Google Scholar]
- 11.Qian H, Chen L.. TRIM proteins in fibrosis. Biomed Pharmacother. 2021;144:112340. doi: 10.1016/j.biopha.2021.112340 [DOI] [PubMed] [Google Scholar]
- 12.Zhan W, Zhang S.. TRIM proteins in lung cancer: mechanisms, biomarkers and therapeutic targets. Life Sci. 2021;268:118985. doi: 10.1016/j.lfs.2020.118985 [DOI] [PubMed] [Google Scholar]
- 13.Wan T, Li X, Li Y.. The role of TRIM family proteins in autophagy, pyroptosis, and diabetes mellitus. Cell Biol Int. 2021;45(5):913–26. doi: 10.1002/cbin.11550 [DOI] [PubMed] [Google Scholar]
- 14.Chen Q, Gao C, Wang M, et al. TRIM18-regulated STAT3 signaling pathway via PTP1B promotes renal epithelial-mesenchymal transition, inflammation, and fibrosis in diabetic kidney disease. Front Physiol. 2021;12:709506. doi: 10.3389/fphys.2021.709506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu Z, Zhang J, Jia Z, et al. TRIM21-mediated ubiquitylation of TAT suppresses liver metastasis in gallbladder cancer. Cancer Lett. 2024;592:216923. doi: 10.1016/j.canlet.2024.216923 [DOI] [PubMed] [Google Scholar]
- 16.Garcia-Garcia J, Berge AKM, Overå KS, et al. TRIM27 is an autophagy substrate facilitating mitochondria clustering and mitophagy via phosphorylated TBK1. FEBS J. 2023;290:1096–116. doi: 10.1111/febs.16628 [DOI] [PubMed] [Google Scholar]
- 17.Li L, Xu N, Liu J, et al. m6A Methylation in cardiovascular diseases: from mechanisms to therapeutic potential. Front Genet. 2022;13:908976. doi: 10.3389/fgene.2022.908976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lan J, Xu B, Shi X, et al. WTAP-mediated N6-methyladenosine modification of NLRP3 mRNA in kidney injury of diabetic nephropathy. Cell Mol Biol Lett. 2022;27(1):51. doi: 10.1186/s11658-022-00350-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu L, Wu J, Lu C, et al. WTAP-mediated m6A modification of lncRNA Snhg1 improves myocardial ischemia-reperfusion injury via miR-361-5p/OPA1-dependent mitochondrial fusion. J Transl Med. 2024;22(1):499. doi: 10.1186/s12967-024-05330-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yuan D, Li H, Dai W, et al. IGF2BP3-stabilized CAMK1 regulates the mitochondrial dynamics of renal tubule to alleviate diabetic nephropathy. Biochim Biophys Acta Mol Basis Dis. 2024;1870:167022. doi: 10.1016/j.bbadis.2024.167022 [DOI] [PubMed] [Google Scholar]
- 21.Chen Y, Wu Y, Zhu L, et al. METTL3-Mediated N6-methyladenosine modification of Trim59 mRNA protects against sepsis-induced acute respiratory distress syndrome. Front Immunol. 2022;13:897487. doi: 10.3389/fimmu.2022.897487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang R, Li SW, Liu L, et al. TRIM11 facilitates chemoresistance in nasopharyngeal carcinoma by activating the β-catenin/ABCC9 axis via p62-selective autophagic degradation of Daple. Oncogenesis. 2020;9:45. doi: 10.1038/s41389-020-0229-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou C, Zhang Z, Zhu X, et al. N6-Methyladenosine modification of the TRIM7 positively regulates tumorigenesis and chemoresistance in osteosarcoma through ubiquitination of BRMS1. EBioMedicine. 2020;59:102955. doi: 10.1016/j.ebiom.2020.102955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Woroniecka KI, Park AS, Mohtat D, et al. Transcriptome analysis of human diabetic kidney disease. Diabetes. 2011;60(9):2354–69. doi: 10.2337/db10-1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang Z, Fu X, Zhou F, et al. Huaju Xiaoji formula regulates ERS-lncMGC/miRNA to enhance the renal function of hypertensive diabetic mice with nephropathy. J Diabetes Res. 2024;2024:6942156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Choi EH, Kim MH, Park SJ.. Targeting mitochondrial dysfunction and reactive oxygen species for neurodegenerative disease treatment. Int J Mol Sci. 2024;25(14):7952. doi: 10.3390/ijms25147952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhao X, Zhang X, Ran X, et al. Simple-to-use nomogram for evaluating the incident risk of moderate-to-severe LEAD in adults with type 2 diabetes: A cross-sectional study in a Chinese population. Sci Rep. 2020;10(1):3182. doi: 10.1038/s41598-019-55101-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jihua C, Cai C, Xubin B, et al. Effects of dexmedetomidine on the RhoA /ROCK/ Nox4 signaling pathway in renal fibrosis of diabetic rats. Open Med. 2019;14(1):890–8. doi: 10.1515/med-2019-0105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qin L, Qin W, Wang J, et al. Combined treatment of diabetic nephropathy with alprostadil and calcium dobesilate. Exp Ther Med. 2017;14:5012–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tang B, Li W, Ji TT, et al. Circ-AKT3 inhibits the accumulation of extracellular matrix of mesangial cells in diabetic nephropathy via modulating miR-296-3p/E-cadherin signals. J Cell Mol Med. 2020;24(15):8779–88. doi: 10.1111/jcmm.15513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim J, Moon E, Kwon S.. Effect of Astragalus membranaceus extract on diabetic nephropathy. Endocrinol Diabetes Metab Case Rep. 2014;2014:140063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lenoir O, Jasiek M, Hénique C, et al. Endothelial cell and podocyte autophagy synergistically protect from diabetes-induced glomerulosclerosis. Autophagy. 2015;11(7):1130–1145. doi: 10.1080/15548627.2015.1049799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fernandes AP, Águeda-Pinto A, Pinheiro A, et al. Evolution of TRIM5 and TRIM22 in bats reveals a complex duplication process. Viruses. 2022;14(2):345. doi: 10.3390/v14020345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Basu R, Lee J, Wang Z, et al. Loss of TIMP3 selectively exacerbates diabetic nephropathy. Am J Physiol Renal Physiol. 2012;303(9):F1341–F1352. doi: 10.1152/ajprenal.00349.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen C, Zhao D, Fang S, et al. TRIM22-mediated apoptosis is associated with bak oligomerization in monocytes. Sci Rep. 2017;7(1):39961. doi: 10.1038/srep39961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu W, Zhao Y, Wang G, et al. TRIM22 inhibits osteosarcoma progression through destabilizing NRF2 and thus activation of ROS/AMPK/mTOR/autophagy signaling. Redox Biol. 2022;53:102344. doi: 10.1016/j.redox.2022.102344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Su H, Hu C, Cao B, et al. A semisynthetic borrelidin analogue BN-3b exerts potent antifungal activity against Candida albicans through ROS-mediated oxidative damage. Sci Rep. 2020;10(1):5081. doi: 10.1038/s41598-020-61681-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guo XF, Gu SS, Wang J, et al. Protective effect of mesenchymal stem cell-derived exosomal treatment of hippocampal neurons against oxygen-glucose deprivation/reperfusion-induced injury. World J Emerg Med. 2022;13(1):46–53. doi: 10.5847/wjem.j.1920-8642.2022.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hsu JY, Jhang YL, Cheng PH, et al. The truncated c-terminal fragment of mutant ATXN3 disrupts mitochondria dynamics in spinocerebellar ataxia type 3 models. Front Mol Neurosci. 2017;10:196. doi: 10.3389/fnmol.2017.00196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Engmann O, Hortobágyi T, Pidsley R, et al. Schizophrenia is associated with dysregulation of a Cdk5 activator that regulates synaptic protein expression and cognition. Brain. 2011;134(8):2408–2421. doi: 10.1093/brain/awr155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Alam S, Abdullah CS, Aishwarya R, et al. Dysfunctional mitochondrial dynamic and oxidative phosphorylation precedes cardiac dysfunction in R120G-αB-crystallin-induced desmin-related cardiomyopathy. J Am Heart Assoc. 2020;9(23):e017195. doi: 10.1161/JAHA.120.017195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu W, Zhao D, Shah SZA, et al. OPA1 overexpression ameliorates mitochondrial cristae remodeling, mitochondrial dysfunction, and neuronal apoptosis in prion diseases. Cell Death Dis. 2019;10(10):710. doi: 10.1038/s41419-019-1953-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Caglayan S, Hashim A, Cieslar-Pobuda A, Jensen V, Behringer S, Talug B, et al. optic atrophy 1 controls human neuronal development by preventing aberrant nuclear DNA methylation. iScience. 2020;23:101154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cho C, Zeigler M, Mizuno S, et al. Reductions in hydrogen sulfide and changes in mitochondrial quality control proteins are evident in the early phases of the corneally kindled mouse model of epilepsy. Int J Mol Sci. 2022;23(3):1434. doi: 10.3390/ijms23031434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pang Y, Zhu Z, Wen Z, et al. HIGD-1B inhibits hypoxia-induced mitochondrial fragmentation by regulating OPA1 cleavage in cardiomyocytes. Mol Med Rep. 2021;24(2):549. doi: 10.3892/mmr.2021.12188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sanderson TH, Raghunayakula S, Kumar R.. Neuronal hypoxia disrupts mitochondrial fusion. Neuroscience. 2015;301:71–78. doi: 10.1016/j.neuroscience.2015.05.078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kirui J, Mondal A, Mehle A.. Ubiquitination upregulates influenza virus polymerase function. J Virol. 2016;90(23):10906–10914. doi: 10.1128/JVI.01829-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ji J, Ding K, Luo T, et al. TRIM22 activates NF-κB signaling in glioblastoma by accelerating the degradation of IκBα. Cell Death Differ. 2021;28(1):367–381. doi: 10.1038/s41418-020-00606-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sun Q, Geng H, Zhao M, et al. FTO-mediated m6 A modification of SOCS1 mRNA promotes the progression of diabetic kidney disease. Clin Transl Med. 2022;12(6):e942. doi: 10.1002/ctm2.942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lin Z, Lv D, Liao X, et al. CircUBXN7 promotes macrophage infiltration and renal fibrosis associated with the IGF2BP2-dependent SP1 mRNA stability in diabetic kidney disease. Front Immunol. 2023;14:1226962. doi: 10.3389/fimmu.2023.1226962 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data presented in this study are included within the paper and its Supplementary files.