Abstract
Recent evidence indicates that potential interactions within metabolic, protein–protein interaction, and transcriptional regulatory networks are used differentially according to the environmental conditions in which a cell exists. However, the topological units underlying such differential utilization are not understood. Here we use the transcriptional regulatory network of Escherichia coli to identify such units, called origons, representing regulatory subnetworks that originate at a distinct class of sensor transcription factors. Using microarray data, we find that specific environmental signals affect mRNA expression levels significantly only within the origons responsible for their detection and processing. We also show that small regulatory interaction patterns, called subgraphs and motifs, occupy distinct positions in and between origons, offering insights into their dynamical role in information processing. The identified features are likely to represent a general framework for environmental signal processing in prokaryotes.
Keywords: cellular networks, regulation, transcription
Transcriptional regulatory (TR) networks govern cellular life by initiating and mediating gene expression in response to environmental and intracellular cues that result in the execution of cellular programs such as metabolic adjustments, sporulation, or cell division. The nodes of this network are transcription units (genes or operons) together with their protein products, whereas the links connecting them correspond to TR interactions mediated by transcription factor (TF) proteins. The genome-scale identification of TF-binding sites, their binding specificities, and their condition-dependent utilization (1–7) results in increasingly comprehensive data sets amenable for analysis of TR-network topology and function.
Previous studies analyzing the TR-network topology of the prokaryote Escherichia coli and the eukaryote Saccharomyces cerevisiae have demonstrated that their TR networks share several characteristics such as the exponential distribution of in-degree connectivity, the scale-free distribution of out-degree connectivity, and the very low number of feedback circuits except for self-regulation (8–11). In addition, the same small-scale connectivity patterns [e.g., the feed-forward loop (FFL) and bifan motif] are overrepresented in both TR networks (12–15), suggesting that their topology has evolved to accomplish similar tasks in various organisms (14). Recent studies of motif dynamics (16–17) generated the first insights into their information-processing capabilities, although the position of motifs within TR networks and their aggregation into larger topological structures (10) may modify their dynamic behavior.
Despite these advances, there is a clear need to decipher the system-level organization of dynamic TR-network utilization (5) triggered by a various environmental and intracellular cues. Here, based on the inherent directionality of TR interactions in E. coli (11, 12), we identify topological units of environmental signal processing (called origons) as TR subnetworks originating at a distinct class of TFs. Using microarray data, we demonstrate that environmental signals affect significantly only the origons rooted at sensor TFs specialized for their detection. We also show that small-scale regulatory interaction patterns (subgraphs and motifs) occupy distinct positions in and between origons that, together with their filtering properties, are suggestive of their role in information processing. Taken together these results suggest that E. coli uses specific topological units of its TR network to detect the elementary components (modes) of complex environmental signals and subsequently develop a response by reassembling these elementary modes near the output layer of the network.
Methods
Databases and Software. The publicly available data on the TR network of E. coli MG1655 (www.weizmann.ac.il/mcb/UriAlon/Network_motifs_in_coli/ColiNet-1.1) (12), based predominantly on the RegulonDB database (18), was used for our work. The network originally contained 423 operons and 578 regulatory interactions, reduced to 418 operons and 519 links by the removal of all operons that have only autoregulatory links.
We downloaded microarray data from two public sources: A Systematic Annotation Package for Community Analysis of Genomes (ASAP) at the University of Wisconsin (Madison) (https://asap.ahabs.wisc.edu/annotation/php/ASAP1.htm) and the Oklahoma University Microarray Core Facility database (http://chase.ou.edu/macro/runs.php). The ASAP database contained 91 experiments on E. coli MG1655 transposon insertion mutants (as of June 2004), 50 using cDNA microarrays and 41 using Affymetrix (Santa Clara, CA) oligoarray platforms and 41 aerobic-shift experiments on E. coli K-12 MG1655 using Affymetrix oligoarray platforms; the Oklahoma University database contained 104 experiments performed on E. coli cells in various conditions.
For network representation we used the program pajek (http://vlado.fmf.uni-lj.si/pub/networks/pajek). For the rest of our programming (as described below), we used matlab (Mathworks, Natick, MA), c, and perl.
Microarray Data Assembly. Within the three microarray experiment sets, we considered the following as control: in the 50 cDNA microarray experiments, 11 experiments designated as “wild-type, standard-growth conditions”; in the 41 Affymetrix experiments, five experiments described as “wild-type, standard-growth conditions”; and in the 41 aerobic shift experiments, three designated as “wild-type aerobic.” To construct log ratios, we first calculated the average estimated transcript copy number (ETCN) within the control experiments for each of the three experimental data sets. Next, we divided the ETCNs from all other experiments by the control ETCNs and calculated the base-10 logarithm of the resulting values. Finally, we renormalized the resulting log ratios such that the average of all log ratios for each experiment became zero. The Oklahoma University data set already contained log ratios, and thus these data were only renormalized to yield average log ratios equal to 0.
We assembled expression tables from the four different data sets after renormalizing the data again such that the mean of all gene-expression profiles became 0, and the standard deviation of all expression values within each set of experiments became 1. The rows in the obtained tables corresponded to E. coli genes, and the columns corresponded to various experiments. Because the E. coli TR network used here consists of 855 genes distributed among 418 operons, we reduced the expression tables to rows corresponding only to these 855 genes. The resulting tables contained normalized expression values (log ratios) LRr,c in Nr = 855 rows and Nc columns, corresponding to Nc microarray experiments.
Node–Node Correlation. Because the rows were normalized to zero mean, we calculated the node–node cross-correlation coefficients cori,j as
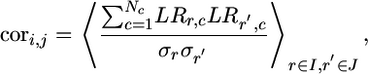 |
[1] |
where the averages are taken over all genes belonging to operons i and j, and the standard deviation of the gene from row r is
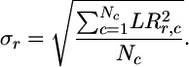 |
[2] |
Node–Signal Covariance. We calculated the node–signal covariance between node i and signal Sc with Nc binary values as
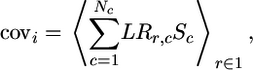 |
[3] |
where the averages are taken over all genes belonging to operon I.
Double z Scores. We consider an origon to be significantly affected by the external signal if it is significantly enriched in signal-affected nodes compared to the same number of nodes chosen randomly.
First, to identify the nodes significantly affected by a certain signal, we calculate covNS(n) for all n = 1, 2,..., N nodes within the network. Then we calculate a z score z(n) for each individual node, defined as
 |
[4] |
where μNS and σNS are the mean and standard deviation of covNS(n), respectively.
To determine origons significantly affected by the external signal, we calculate the average z score μO = 〈zNS〉O over all NO nodes within an origon. Then we repeatedly select NO nodes at random within the E. coli TR network to estimate the mean, μR and standard deviation, σR of the quantity 〈zNS〉R. Finally, we calculate the double z score, ZNS, for the origon as
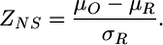 |
[5] |
Modeling the Dynamics of Small Subgraphs. To simulate subgraph dynamics, we used the built-in delay differential equation solver dde23 from matlab using mass-action kinetic modeling and quasiequilibrium approximation for reactions occurring at much faster time scales than the others. First, we modeled the alteration of sensor TF-binding activity by a small metabolite. Next, we modeled RNA polymerase binding and transcription initiation, elongation, and termination. Finally, we modeled translation initiation, elongation, and termination. In the simulations, we used generic constants found in the literature, and we varied them over reasonable ranges to test the robustness of our conclusions. Because of space limitations, we describe the details of the modeling in the supporting information.
Supporting Information. For more information, see Supporting Appendices 1–3 and Tables 2–5, which are published as supporting information on the PNAS web site.
Results
Directionality of Regulatory Interactions Defines TR Subnetworks. The E. coli TR network is defined by its nodes (operons and their protein products) and the links connecting them (TR interactions mediated by binding of TFs to the promoter regions of operons) (8–12). Because all links are unidirectional (12), and the network contains no cycles other than autoregulatory loops (8), the 418 nodes can be arranged hierarchically into five regulatory layers as described previously (11) (Fig. 1A). We define layers as the set of nodes for which the longest path connecting them to the input layer has one, two, three, or at most four links. The layers reflect the flow of information from 76 input nodes (representing sensor TFs that are not regulated transcriptionally by any other TF) to 312 output nodes (representing non-TF proteins) (12, 19).
Fig. 1.
Definition and characteristics of transcriptional subnetworks. (A) Schematic representation of the E. coli TR network. Nodes are genes/operons and their protein products, and links are TR interactions between them. White nodes represent TF-encoding operons in the input layer (layer 0); they are not regulated transcriptionally by any other TFs. Nodes located farther from the input layer are increasingly darker. The color of the links indicates activation (green), repression (red), or both activation and repression (yellow). Nodes marked by purple circles of larger size within the light-purple shaded area form a signal-affected transcriptional subnetwork, or origon, rooted at the node in layer 0. The numbers on the right indicate the number of nodes in the corresponding layers of the E. coli TR network. (B) The rob origon is shown as an example from the E. coli TR network. (C) The origon network. Circles represent origons labeled by their root node, which directly or indirectly regulates all other nodes in the origon. The radius of circles is proportional to the base-2 logarithm of the number of operons (nodes) in that origon. Black circles represent origons with tree topology, and red circles represent origons with FFL-tree topology, an example of the latter being shown in B. Two origons are connected if they share at least one node, with the thickness of links being proportional with the number of nodes in common.
Besides TFs in the input layer, most TFs from intermediate regulatory layers (such as AraC, ArcA, etc.) also mediate environmental signaling into the TR network, because their conformation and activity are affected by specific changes in the environment (e.g., metabolite availability, oxygen pressure, etc.). However, the expression of sensor TFs from lower layers can also be transcriptionally regulated by other TFs, in contrast to the operons in the input layer that can only be regulated by their own protein products. Therefore, we differentiate topological inputs (operons from layer 0) from sensory inputs (TFs with environment-dependent activity).
Because of link directionality and the sparseness of the E. coli TR network (8, 12, 19) (see the supporting information for a description of the network's topological characteristics), TFs regulate only a limited number of operons (nodes). Thus, the set of operons regulated directly or indirectly by a given TF form a transcriptional subnetwork, rooted at the given TF. All such subnetworks are parts of other subnetworks except for the ones originating at the 76 topological input nodes, to which we refer as origons (see Discussion) and label them according to their input node (Fig. 1A). Because the input layer contains 76 operons, there are a total of 76 origons in the E. coli TR network, each affected by different environmental signal(s) (see the supporting information for their detailed description). An actual example of an E. coli origon is shown in Fig. 1B.
If an environmental signal affects the activity or expression level of a single sensor TF, only the expression of the genes within its origon should be affected transcriptionally, the perturbation gradually percolating toward the output layer (Fig. 1A). In turn, when several sensors are affected simultaneously by complex external signals, signal processing may involve propagation through isolated origons or signal combination by connected origons. To examine their connectedness, we redrew the TR network with origons as nodes. Two nodes in this origon network are connected if they share at least one node (operon). As shown in Fig. 1C, 45 of the 76 origons form a single connected component, indicating that a perturbation applied to their input node will affect nodes in the lower layers of some other origons, as well. In contrast, 25 origons are isolated from the rest of the network, indicating that they carry out transcriptional responses to specific stimuli independently, and six origons form three additional two-node clusters.
Distinct Environmental Signals Affect Origons Selectively. The role of the E. coli TR network is to sense various environmental changes, process the obtained information, and develop a response, allowing the bacterium to dynamically adapt to continuously changing conditions. Let us assume that the environment is constant except for a single stimulus (factor). If the input nodes to each of the 76 origons constituting the network were responsive to different types of environmental changes, then only the origon rooted at the sensor for this stimulus should be affected. To test this hypothesis we focused on the crp (Fig. 12) and fnr origons (Fig. 14), rooted at the genes encoding the proteins FNR and CRP, respectively. FNR is a sensor TF that rapidly changes its conformation in the absence of oxygen, with a concomitant increase in its promoter-binding affinity (20, 21). In contrast, the DNA-binding affinity of CRP is not oxygen-dependent. Taking this into account, we used publicly available microarray data to decipher the transcriptional response to a stimulus known to affect the promoter-binding activity of the protein FNR. We grouped the available microarray data (22, 23) into two classes of experiments. The first class, in which wild-type and mutant strains of E. coli MG1655 were repeatedly sampled in aerobic and anaerobic conditions, is the best (and only) available data set for a repeated FNR-specific perturbation. The second class of experiments, in which aerobically grown wild-type and mutant E. coli strains were exposed to various nutrients, acid, heat shock, etc., represents a non-FNR-specific perturbation. Both data sets represent non-CRP-specific perturbations.
First, we reconstructed the FNR-specific “external signal” as a binary series of 39 values (0 for anaerobic and 1 for aerobic growth), based on oxygen availability in the growth media in the 39 aerobic-shift microarray experiments (Fig. 2A). Second, to characterize how repeated aerobic–anaerobic shifts affect the mRNA expression of individual nodes, we calculated the node–signal covariance (covNS; Fig. 2A) between their expression profile and the external signal. Third, we calculated z scores (13) (zNS; Fig. 2A) for every node to measure its affectedness (deviation from the covNS of other nodes within the network). Finally, we compared zNS histograms (Fig. 2B) for nodes located within specific origons or chosen randomly from the TR network (see Methods).
Fig. 2.
Validation of the origon concept by microarray data. (A) Definition of node–signal covariance (covNS) and node–node cross-correlation (corNN). The quantity covNS characterizes the impact of the changes in environmental oxygen concentration (39 binary values of [O2] in the top graph) on the intracellular mRNA expression [log ratios (LRs): vertical axes in the bottom three graphs] of three representative single-gene nodes (sodA, adhE, and cpxP). The z score, zNS, measures how different the covNS value of a node is from other nodes in the network. The quantity corNN characterizes the similarity between the mRNA expression profiles of the different genes. (B) Histograms of covNS (Left) within the crp and fnr origons (red bars) versus equal number of nodes chosen randomly (black bars) for FNR-specific (aerobic-shift) perturbation. The corresponding zNS histograms are shown (Center), as are the double z scores, ZNS, for all origons (Right) (the ZNS values for the fnr and crp origons are circled). The dashed black line corresponds to a cutoff of ZNS = 2. (C) Histograms of zNN within the crp and fnr origons (red bars) versus equal number of nodes chosen randomly (black bars) for FNR-specific (aerobic-shift) and nonspecific perturbation. The ZNN values for all origons are shown (Right)(fnr and crp origons are circled, and the dashed black line corresponds to a cutoff of ZNN = 2). (D) The predicted binary node–node correlation (within the narL origon) between two nodes is equal to the product of link types along any path connecting the two nodes in a nondirectional version of the origon. Link types are considered as follows: +1, activating; –1, repressing; 0, dual. (E) Predictions are only indicative of measured node–node cross-correlations in the fnr origon for FNR-specific stimulus. Along the horizontal and vertical axes are operons within the crp or fnr origon so that the diagonal contains values calculated for an operon with itself. The agreement between predicted binary (Left) and measured (Center) cross-correlations is calculated as the product of the predictions and measurements (Right) and the corresponding averages 〈P × M〉. All values in the right column are between –1 and 1, and correct predictions result in positive values, as indicated by the color scale shown at the bottom.
The histogram of zNS of all nodes within the crp origon (Fig. 2B Lower) is similar to the averaged histogram of zNS for the same number of nodes chosen randomly. In contrast, the zNS histogram for nodes within the fnr origon (Fig. 2B Upper) is substantially flatter than the same histogram for randomly selected nodes, indicating the increased number of strongly positive or negative covariances. To statistically compare the flatness of these distributions, we defined double z scores (ZNS; see Methods) based on zNS. The double z scores (Fig. 2B) for the fnr and crp origons were 6.04 and 0.85, respectively, indicating that nodes within the fnr origon are significantly affected by the FNR-specific signal, whereas nodes within the crp origon are not. In fact, the only five origons affected significantly (Z > 2) by the FNR-specific stimulus are rooted at the anoxic sensor FNR (Z = 6.04), the redox sensor SoxR (Z = 3.66), the regulator of nitrite- and nitrate-based anaerobic respiration and fermentation NarL (Z = 2.92), the σ factor involved in the utilization of various nitrogen sources RpoN (Z = 2.39), and the degradative (anoxic) arginine decarboxylase system AdiA–AdiY (Z = 2.18). Because the zNS distributions are non-Gaussian (Fig. 2B), we also used rank scores (24) to test the validity of our findings, determining the percentage of cases when the mean zNS of randomly chosen nodes exceeded the mean zNS within an origon. After this procedure, the list of significantly affected origons (P < 0.05) was identical with the one found by using double z scores, indicating that our findings are robust to the statistical method applied. The small number of significantly affected origons (also observed for other stimuli, such as H2O2 treatment, diauxic shift, and UV exposure; see the supporting information) confirms our hypothesis that external perturbations influence only a limited set of origons, suggesting that origons are meaningful topological units of the TR network, dynamically used for environmental signal processing.
Consistency Between Known TR Interactions and Microarray Data. After investigating how mRNA expression levels in the E. coli TR network reflect external changes, we studied how internal variations in the mRNA expression of various nodes relate to each other. To this end, we calculated node–node correlations, i.e., the cross-correlation between the expression profiles of two nodes (corNN, Fig. 2A). Given two nodes, we measured the deviation of their correlation from the corNN of randomly chosen node pairs by z scores (zNN) again. We compared the zNN values of node pairs located within the crp and fnr origons to zNN values of node pairs chosen randomly from the network. For an FNR-specific stimulus, the number of outlying zNN values is much higher for nodes within the fnr origon than for nodes chosen randomly or within the crp origon (Fig. 2C). We again used double z scores to statistically characterize the observed differences and obtained for FNR-specific perturbation Z = 21.5 for nodes within the fnr origon, compared to Z = –9.2 for nodes within the crp origon. For non-FNR-specific perturbations (Fig. 2C), both double z scores were <2 (Z = 0.56 for the fnr origon and Z = 0.16 for the crp origon), indicating that node–node correlations were not significantly affected in either of the two origons. Extending the analysis to all origons reveals that 17 origons (fnr, yhdG_fis, purR, fruR, argR, lrp, cysB, uidR, nlpD_rpoS, narL, rhaSR, mlc, adiA_adiY, nhaR, soxR, glcC, birA_murA) are characterized by significant (Z > 2) node–node correlations. Besides the known anoxic regulators FNR and NarL, this list contains origons rooted at redox sensors (SoxR), σ factors (RpoS), or TFs initiating reduced amino acid and nucleotide synthesis and transport in conditions of stress (PurR, ArgR, CysB, Lrp) (25).
We also examined whether the measured corNN values agree with the ones predicted by the type of interaction (activation or repression) between a TF and its target. We associated a binary value (1 or –1) with each node within an origon depending on how its expression level is expected to change when the expression of the root node is altered. We defined the predicted binary cross-correlation between two nodes as the product of the values associated with them. Dual interactions, in which TFs can be both activating or repressing, were not considered. Fig. 2D illustrates such predictions for the narL origon. Fig. 2E illustrates the predicted binary cross-correlations between every pair of nodes in the crp and fnr origons (P, left column of plots), the measured covNN values between the same pair of nodes (M, center column of plots), and the quality of predictions estimated as the product of the predicted and measured gene–gene cross-correlations (P × M, right column of plots). Increasingly lighter colors in the plots correspond to increasingly higher values, clearly illustrating that for FNR-specific stimulus, the predicted and measured corNN values agree substantially more within the fnr origon than within the crp origon. To quantitatively characterize this difference, we calculated averages over the plots in Fig. 2E Right, obtaining the values of (〈P × M〉, from top to bottom) 0.3049, 0.0454, 0.0693, and, 0.0377, respectively. These values indicate again that cross-correlations are very noisy and have little predictive value of actual transcriptional regulation for nonspecific perturbations applied to both the fnr and crp origon. In contrast, FNR-specific perturbation applied to the fnr origon results in a 10-fold increase in the quality of the predictions.
Motifs and Subgraphs Occupy Distinct Positions Within Origons and Filter Environmental Signals Differently. The effect of many environmental signals is initiated through altered sensor TF activity, followed by the dynamical propagation of the perturbation to lower layers, eventually altering the expression of all genes within an origon. This propagation takes place through small motifs (12), or subgraphs (15), that connect subsequent regulatory layers to each other. To elucidate the type and information-processing function of subgraphs encountered by the propagating signal, we next investigated the position and abundance of three-node subgraphs in the whole TR network as well as with respect to individual origons. Fig. 3A shows all three-node subgraphs found within the E. coli TR network: divergence (DIV), convergence (CNV), cascade (CAS), and FFL, all of which [composed of single regulatory interactions (SRIs)], connect two or three consecutive layers of the TR network. To examine the relative abundance of all three-node subgraphs present in the E. coli TR network, we used a randomization protocol (13) preserving all five layers and keeping the network acyclic (see the supporting information). Our analysis confirms that FFL subgraphs are significantly more abundant than expected (13). However, the abundance of DIV and CAS also deviates from the random expectation, with DIV occurring more frequently, whereas CAS is less frequent than expected (Table 1 and supporting information).
Fig. 3.
Filtering properties and dynamics of small subgraphs. (A) Directed small subgraphs: SRI, DIV, CNV, CAS, and FFL are shown. The nodes are labeled by X, Y, and Z progressively, depending on their distance from the input layer of the subgraph (layer 0). (B) Schematic illustration of SRIs, the basic building unit of all three-node subgraphs, reflecting the mechanism considered in our modeling. The signal SX activates the input TF, PX. Once activated, 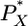 binds to the operator region of gene Y and allows binding of RNA polymerase PR to its promoter region. The polymerase–operator complex initiates transcription, resulting in the synthesis of an mRNA molecule RY after a delay τR, which, bound by a ribosome PP, is translated into the output protein PY after a delay τP. (C) Time courses of output protein levels after a periodic perturbation for SRIs and DIVs (black line), CASs (blue line), and FFLs (red line). The amplitude of fluctuations at the output of the FFL is nearly the same as at the output of the SRI, although it is substantially reduced because of the stronger filtering properties of CAS. (D) Frequency response of SRI and DIV (black circles), CAS (blue squares), and FFL (red triangles) showing the amplitude of fluctuations at the output of subgraphs (vertical axis) versus the frequency of the signal applied to their input (horizontal axis). All subgraphs are low-pass filters, but CAS has a much stronger filtering effect than either the SRI (DIV) or FFL.
binds to the operator region of gene Y and allows binding of RNA polymerase PR to its promoter region. The polymerase–operator complex initiates transcription, resulting in the synthesis of an mRNA molecule RY after a delay τR, which, bound by a ribosome PP, is translated into the output protein PY after a delay τP. (C) Time courses of output protein levels after a periodic perturbation for SRIs and DIVs (black line), CASs (blue line), and FFLs (red line). The amplitude of fluctuations at the output of the FFL is nearly the same as at the output of the SRI, although it is substantially reduced because of the stronger filtering properties of CAS. (D) Frequency response of SRI and DIV (black circles), CAS (blue squares), and FFL (red triangles) showing the amplitude of fluctuations at the output of subgraphs (vertical axis) versus the frequency of the signal applied to their input (horizontal axis). All subgraphs are low-pass filters, but CAS has a much stronger filtering effect than either the SRI (DIV) or FFL.
Table 1. Abundance of three-node subgraphs in the TR network of E. coli and the randomized version of this network.
| Subgraph | CNV | DIV | CAS | FFL |
|---|---|---|---|---|
| Abundance in real network | 227 | 4777 | 160 | 42 |
| Abundance in randomized network | 231.92 ± 8.05 | 4339.3 ± 132 | 186.69 ± 7.08 | 9.50 ± 4.17 |
| z score | 0.61 | 3.31 | 3.77 | 7.79 |
The corresponding z scores (13) (listed in the bottom row) indicate the deviation of the values for the real network from those for the randomized network. The corresponding distributions are shown in Fig. 6.
Based on the randomization protocol described above, we expected to find 5.07 ± 4.44 CNV subgraphs within individual origons, whereas the probability of finding no CNV subgraph in any origons is 0.11. It is surprising that CNV is completely absent from any of the individual E. coli origons. Therefore, the apparent role of CNV subgraphs is to combine perturbations propagating in different origons. We also found that the majority of origons are trees (Fig. 1C), containing only DIV and CAS subgraphs (Fig. 9). However, seven origons also contain FFL subgraphs in addition to their backbone tree structure (Figs. 12–18). Thus, origons can be classified into those with tree structure and those with FFL-tree structure (Fig. 1C) (see the supporting information for details). An example of an origon with tree structure is shown in Fig. 2C, and an origon with FFL-tree structure [containing aggregated FFL subgraphs (10)] is shown in Fig. 1B.
For free-living bacteria such as E. coli, environmental signals (e.g., available carbon sources, pH, or oxygen) change frequently. To understand how the effect of fluctuating environmental changes propagate within individual origons, we developed dynamical models of the elementary topological units (subgraphs) connecting subsequent layers (Fig. 3A) by using a generic mass-action kinetic model (26) and taking into account the time delay required for mRNA and protein synthesis (27, 28). We applied perturbation to each of the subgraphs by altering the activity of their input node through the periodically changing concentration of a signaling molecule (see the supporting information for details) and investigated their response amplitude through the concentration of the protein product of their output node (Fig. 3B). As shown in Fig. 3 C and D, the response amplitude of SRIs (Fig. 3A) decreases with increasing frequency of the input signal. Therefore, SRIs are low-pass filters, as described previously (29), i.e., they allow slow fluctuations to pass through but filter out fast signal fluctuations. Because SRIs are the basic building blocks of all three-node subgraphs, they all possess filtering properties to some degree. When keeping all parameters used in the simulation of the various subgraphs identical, we find that DIV (composed of two parallel SRIs) has an identical filtering effect to SRI. In contrast, the effect of the two subsequent SRIs comprising a CAS is combined, resulting in the strongest signal filtering. Finally, in FFLs with an AND-type promoter logic (30, 31), the strong filtering effect is again substantially reduced (Fig. 3 C and D) because of the extra link directly connecting their input and output nodes. Thus, in contrast to what was suggested previously (12), FFLs might have the role to diminish the strong filtering effects of CASs and/or combine two related signals from two sensory inputs (see Discussion).
Discussion
Our analysis indicates that in E. coli, distinct transcriptional subnetworks (called origons) are responsible for environmental perturbation processing. In relation to existing concepts of multigene regulatory structures, origons are more complex entities than modulons but less complex than stimulons (32). Specifically, all nodes within a modulon must be controlled directly by a common, “pleiotropic” regulator, whereas in the origon they can be controlled indirectly by percolation of altered transcriptional levels through the network. On the other hand, origons are subnetworks originating at a single TF, whereas stimulons include all nodes affected by an environmental signal and are composed of all the origons rooted at TFs sensitive to the signal.
Based on their complexity, environmental perturbations can be classified as elementary (change of a single factor on a constant environmental background) or complex (simultaneous changes of two or more environmental factors).
The transcriptional response of the cell will depend on the type of perturbation, the structure of the affected origon, and its interconnectivity with other origons. If an elementary perturbation is highly specific to one (or few) origon(s), it will affect only operons within them. On the other hand, complex perturbations involving related signals (e.g., two different sugars) typically affect two or more sensory proteins within an origon, resulting in a combined response within individual origons. In this case, processing of the incoming signals is mediated by CAS and/or FFL subgraphs that combine related signals within the same origon. Finally, complex perturbations involving unrelated stimuli (e.g., oxygen and one sugar) are often processed independently and then combined by overlapping origons through CNVs. In addition to signal combination by sensor TFs, the target promoters of FFLs and CNVs use combinatorial logic (30) (with a dependence on signal intensity, promoter strength, etc.) that needs to be determined experimentally (16, 33).
The origon concept suggests that at the transcriptional regulatory level cells perceive their environment by first dissecting complex external signals into elementary perturbations (or modes) processed by individual origons and then developing a response by reassembling the elementary modes near the output of the TR network. Decomposition into such elementary modes is common in signal analysis (Fourier components). Thus, the origon concept may also explain why dimensionality-reduction techniques such as singular-value decomposition (34–36) or network-component analysis (37) have been successful in uncovering biologically significant information. Because the origon concept is intimately related to the directed nature of the TR network, the identification of similar topological units in undirected (protein–protein interaction) and reversibly directed (metabolic) networks will require alternative approaches. Yet, to fully understand the response to external stimuli in these networks, one cannot disregard the origon-specific directed flow of environmental signals characterizing transcriptional regulation.
Supplementary Material
Acknowledgments
We thank Q. K. Beg for compiling Table 4 for supporting information; R. Dobrin, E. Ravasz, and A. Vázquez for discussion; and J. J. Collins for comments on the manuscript. Research at the University of Notre Dame, Northwestern University, and University of Pittsburgh was supported by the U.S. Department of Energy, National Institutes of Health, and National Science Foundation.
Author contributions: G.B., A.-L.B., and Z.N.O. designed research; G.B. performed research; G.B. contributed new reagents/analytic tools; G.B. analyzed data; and G.B., A.-L.B., and Z.N.O. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: TR, transcriptional regulatory; TF, transcription factor; FFL, feed-forward loop; DIV, divergence; CNV, convergence; CAS, cascade; SRI, single regulatory interaction.
References
- 1.Ren, B., Robert, F., Wyrick, J. J., Aparicio, O., Jennings, E. G., Simon, I., Zeitlinger, J., Schreiber, J., Hannett, N., Kanin, E., et al. (2000) Science 290, 2306–2309. [DOI] [PubMed] [Google Scholar]
- 2.Lieb, J. D., Liu, X., Botstein, D. & Brown, P. O. (2001) Nat. Genet. 28, 327–334. [DOI] [PubMed] [Google Scholar]
- 3.Zeitlinger, J., Simon, I., Harbison, C. T., Hannett, N. M., Volkert, T. L., Fink, G. R. & Young, R. A. (2003) Cell 113, 395–404. [DOI] [PubMed] [Google Scholar]
- 4.Harbison, C. T., Gordon, D. B., Lee, T. I., Rinaldi, N. J., Macisaac, K. D., Danford, T. W., Hannett, N. M., Tagne, J. B., Reynolds, D. B., Yoo, J., et al. (2004) Nature 431, 99–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Luscombe, N. M., Babu, M. M., Yu, H., Snyder, M., Teichmann, S. A. & Gerstein, M. (2004) Nature 431, 308–312. [DOI] [PubMed] [Google Scholar]
- 6.Pritsker, M., Liu, Y. C., Beer, M. A. & Tavazoie, S. (2004) Genome Res. 14, 99–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mukherjee, S., Berger, M. F., Jona, G., Wang, X. S., Muzzey, D., Snyder, M., Young, R. A. & Bulyk, M. L. (2004) Nat. Genet. 36, 1331–1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thieffry, D., Huerta, A. M., Perez-Rueda, E. & Collado-Vides, J. (1998) BioEssays 20, 433–440. [DOI] [PubMed] [Google Scholar]
- 9.Guelzim, N., Bottani, S., Bourgine, P. & Kepes, F. (2002) Nat. Genet. 31, 60–63. [DOI] [PubMed] [Google Scholar]
- 10.Dobrin, R., Beg, Q. K., Barabási, A.-L. & Oltvai, Z. N. (2004) BMC Bioinformatics 5, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ma, H. W., Buer, J. & Zeng, A. P. (2004) BMC Bioinformatics 5, 199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shen-Orr, S., Milo, R., Mangan, S. & Alon, U. (2002) Nat. Genet. 31, 64–68. [DOI] [PubMed] [Google Scholar]
- 13.Milo, R., Shen-Orr, S. S., Itzkovitz, S., Kashtan, N. & Alon, U. (2002) Science 298, 824–827. [DOI] [PubMed] [Google Scholar]
- 14.Conant, G. C. & Wagner, A. (2004) Nat. Genet. 34, 264–242. [DOI] [PubMed] [Google Scholar]
- 15.Vázquez, A., Dobrin, R., Sergi, D., Eckmann, J.-P., Oltvai, Z. N. & Barabási, A.-L. (2004) Proc. Natl. Acad. Sci. USA 101, 17940–17945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mangan, S., Zaslaver, A. & Alon, U. (2003) J. Mol. Biol. 334, 197–204. [DOI] [PubMed] [Google Scholar]
- 17.Setty, Y., Mayo, A. E., Surette, M. G. & Alon, U. (2003) Proc. Natl. Acad. Sci. USA 100, 7702–7707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Salgado, H., Gama-Castro, S., Martinez-Antonio, A., Diaz-Peredo, E., Sanchez-Solano, F., Peralta-Gil, M., Garcia-Alonso, D., Jimenez-Jacinto, V., Santos-Zavaleta, A., Bonavides-Martinez, C., et al. (2004) Nucleic Acids Res. 32, D303–D306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martinez-Antonio, A. & Collado-Vides, J. (2003) Curr. Opin. Microbiol. 6, 482–489. [DOI] [PubMed] [Google Scholar]
- 20.Kiley, P. J. & Beinert, H. (1998) FEMS Microbiol. Rev. 22, 341–352. [DOI] [PubMed] [Google Scholar]
- 21.Crack, J., Green, J. & Thomson, A. J. (2004) J. Biol. Chem. 279, 9278–9286. [DOI] [PubMed] [Google Scholar]
- 22.Allen, T. E., Herrgard, M. J., Liu, M., Qiu, Y., Glasner, J. D., Blattner, F. R. & Palsson, B. O. (2003) J. Bacteriol. 185, 6392–6399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chang, D. E., Smalley, D. J. & Conway, T. (2002) Mol. Microbiol. 45, 289–306. [DOI] [PubMed] [Google Scholar]
- 24.Middendorf, M., Ziv, E. & Wiggins, C. H. (2005) Proc. Natl. Acad. Sci. USA 102, 3192–3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Neidhardt, F. C., Curtiss, F., Ingraham, J. L., Lin, E. C. C., Low, K. B., Magasanik, B., Reznikoff, W. S., Riley, M., Schaechter, M. & Umbarger, H. E. (1996) Escherichia coli and Salmonella typhimurium Cellular and Molecular Biology (Am. Soc. Microbiol., Washington, DC), 2nd Ed.
- 26.Bolouri, H. & Davidson, E. H. (2002) BioEssays 24, 1118–1129. [DOI] [PubMed] [Google Scholar]
- 27.Santillan, M. & Mackey, M. C. (2001) Proc. Natl. Acad. Sci. USA 98, 1364–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Santillan, M. & Mackey, M. C. (2004) Biophys. J. 86, 1282–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Simpson, M. L., Cox, C. D. & Sayler, G. S. (2004) J. Theor. Biol. 229, 383–394. [DOI] [PubMed] [Google Scholar]
- 30.Buchler, N. E., Gerland, U. & Hwa, T. (2003) Proc. Natl. Acad. Sci. USA 100, 5136–5141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mangan, S. & Alon, U. (2003) Proc. Natl. Acad. Sci. USA 100, 11980–11985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Neidhardt, F. C., Ingraham, J. & Schaechter, M. (1990) Physiology of the Bacterial Cell: A Molecular Approach (Sinauer, Sunderland, MA).
- 33.Kalir, S. & Alon, U. (2004) Cell 117, 713–720. [DOI] [PubMed] [Google Scholar]
- 34.Alter, O., Brown, P. O. & Botstein, D. (2000) Proc. Natl. Acad. Sci. USA 97, 10101–10106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Holter, N. S., Mitra, M., Maritan, A., Cieplak, M., Banavar, J. R. & Fedoroff, N. V. (2000) Proc. Natl. Acad. Sci. USA 97, 8409–8414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yeung, M. K., Tegner, J. & Collins, J. J. (2002) Proc. Natl. Acad. Sci. USA 99, 6163–6168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liao, J. C., Boscolo, R., Yang, Y. L., Tran, L. M., Sabatti, C. & Roychowdhury, V. (2003) Proc. Natl. Acad. Sci. USA 100, 15522–15527. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.





