Abstract
Mouse deafness mutations provide valuable models of human hearing disorders and entry points into molecular pathways important to the hearing process. A newly discovered mouse mutation named hurry-scurry (hscy) causes deafness and vestibular dysfunction. Scanning electron microscopy of cochleae from 8-day-old mutants revealed disorganized hair bundles, and by 50 days of age, many hair cells are missing. To positionally clone hscy, 1,160 F2 mice were produced from an intercross of (C57BL/6-hscy × CAST/EiJ) F1 hybrids, and the mutation was localized to a 182-kb region of chromosome 17. A missense mutation causing a critical cysteine to phenylalanine codon change was discovered in a previously undescribed gene within this candidate interval. The gene is predicted to encode an integral membrane protein with four transmembrane helices. A synthetic peptide designed from the predicted protein was used to produce specific polyclonal antibodies, and strong immunoreactivity was observed on hair bundles of both inner and outer hair cells in cochleae of newborn +/+ controls and +/hscy heterozygotes but was absent in hscy/hscy mutants. Accordingly, the gene was given the name “tetraspan membrane protein of hair cell stereocilia,” symbol Tmhs. Two related proteins (>60% amino acid identity) are encoded by genes on mouse chromosomes 5 and 6 and, together with the Tmhs-encoded protein (TMHS), comprise a distinct tetraspan subfamily. Our localization of TMHS to the apical membrane of inner ear hair cells during the period of stereocilia formation suggests a function in hair bundle morphogenesis.
Keywords: mouse, hair cell, stereocilia, tetraspan
Hearing loss is the most prevalent sensory disorder in human populations, occurring in ≈0.2–0.3% of all live births. More than half of these childhood cases are thought to be genetic (1). The development and maintenance of the intricate structures and complex mechanisms of the mammalian inner ear require the proper functioning and concerted interactions of hundreds of genes and their products. To date, >100 forms of human nonsyndromic deafness have been genetically mapped (Hereditary Hearing Loss Homepage (http://dnalab-www.uia.ac.be/dnalab/hhh), and many of the genes responsible have been identified and characterized (2). More than 200 mouse mutations are known to affect hearing and balance, and mouse models have been developed for >50 human hearing disorders (Hearing Impairment in Mice, www.jax.org/hmr/index.html). Because of the complex nature of the ear, it is likely that many more deafness-related genes remain to be discovered.
The inner ear is comprised of the vestibular region, which controls balance, and the cochlea, which is important in detecting, amplifying, and transmitting auditory information to the brain. In the organ of Corti of the cochlea, two distinct types of sensory cells, inner and outer hair cells, are essential for the transduction of sound into nerve impulses. Stereocilia are modified microvilli that project from the apical membranes of inner ear hair cells. The actin-filled stereocilia contain mechanically gated ion channels that open or close in response to sound-induced deflections and thus are crucial to the hearing process (3). Many mouse mutations have been valuable in identifying and characterizing genes that are important in the development and maintenance of hair cells and their stereocilia, including Myo7a, Myo6, Myo15, Cdh23, Pcdh15, Ush1c, Ush1g, Whrn, Actg1, and Espn (4).
Here, we describe a mouse mutation in a gene that encodes a protein we believe to be involved in the formation of hair cell stereocilia. We named the spontaneous mutation hurry-scurry (hscy) because of the characteristic rapid circling behavior of homozygous mutant mice. Mutant mice are also congenitally deaf. Using a positional cloning approach, we mapped hscy to chromosome (Chr) 17 and identified the underlying gene, which is predicted to encode an integral membrane protein with four transmembrane helices. Because the protein localized to hair cell stereocilia, we named it “tetraspan membrane protein of hair cell stereocilia,” gene symbol Tmhs. Its spatial and temporal expression pattern indicates a likely role in hair bundle morphogenesis.
Materials and Methods
Mice. The hscy mutation arose spontaneously at The Jackson Laboratory in a B6.MOR-Gusba line. Mutants were crossed to C57BL/6 mice for three generations followed by sibling matings to maintain the line. All mice were obtained from the Mouse Mutant Resource at The Jackson Laboratory, and all procedures involving their use were approved by the Institutional Animal Care and Use Committee.
Genetic Mapping. A pooled DNA strategy using microsatellite markers (5) was used to initially localize the mutation to Chr 17. DNAs from individual mice then were typed to refine the map position with the aid of the map manager computer program (6). PCR conditions for typing microsatellite markers were as described (7). Mutant mice (hscy/hscy) from the linkage cross were easily identified by their overt circling and head-shaking behavior. To distinguish +/+ and +/hscy genotypes of nonmutant recombinant mice, progeny tests with hscy/hscy mice were performed.
Auditory-Evoked Brainstem Response (ABR). Hearing in mice was assessed by ABR thresholds as described (8).
Histopathology and Scanning Electron Microscopy (SEM). Cross sections of the inner ear were obtained in the following manner. Mice were transcardially perfused in PBS followed by Bouin's fix. Inner ears were dissected out of the skull, decalcified in Bouin's for ≈2 weeks, and embedded in paraffin. Tissue sections were cut 4 μm thick and stained with hematoxylin/eosin. Tissues for SEM analysis were dissected and fixed in 2.5% glutaraldehyde in 0.1 M phosphate buffer (pH 7.2) for 3–4 h at 4°C followed by several washes in 0.1 M phosphate buffer. Bone and stria surrounding the cochlea were dissected away and the tectorial membrane removed to expose the organ of Corti. Tissues were processed in 2% osmium tetroxide, dehydrated, and dried. The organ of Corti was sputter-coated with gold and examined at 15 kV under a Hitachi (Tokyo) 3000N scanning electron microscope. For SEM analysis, the following numbers of mice of each genotype and developmental stage were examined: hscy/hscy [two postnatal day (P)0, one P8, three P15, one P50), +/hscy (two P0, one P8, one P15), and +/+ (two P15, one P50)].
Genomic DNA and RNA Isolation and cDNA Synthesis. Genomic DNA for PCR was prepared from mouse tail tips using the Hot Shot method (9). Total RNA from inner ear, whole brain, and kidney tissues was isolated with TRIzol reagent following the manufacturer's protocol (Invitrogen). Poly(A)+ mRNA for Northern blot analysis was isolated by using the PolyATract mRNA Isolation System (Promega). Mouse cDNA was synthesized by using SuperScript II reverse transcriptase according to the manufacturer's instructions (Invitrogen).
Northern Blot Hybridization. Northern blots were prepared and hybridized as described (10). Commercially prepared Northern blots from adult mouse tissues and mouse embryos (MTN blots, Clontech) also were used. The hybridization probe corresponded to nucleotides 22–875 of the XM_283418 cDNA sequence.
Production of Antibodies and Immunohistochemistry. A synthetic 16-aa peptide corresponding to the C-terminal end of the predicted mouse Tmhs-encoded protein (TMHS) was injected into rabbits, and high-titer antipeptide antiserum was collected and affinity purified by a commercial vendor (Alpha Diagnostics, San Antonio, TX). For immunofluorescence, tissues were dissected and fixed overnight in 4% paraformaldehyde, embedded in paraffin, and cut 4–6 μm. Embryonic stages were determined by checking vaginal plugs. Noon of the day the vaginal plugs were detected was considered embryonic day (E)0.5. Inner ears from mice older than P5 were decalcified in 7% EDTA/PBS for 1 week before embedding. Tissues were treated with 0.1% trypsin for 15 min at 37°C. After several PBS washes, the tissue sections were incubated overnight at 4°C with the anti-TMHS antibody (1:50) or anti-Myosin VIIa (1:500) (Affinity BioReagents, Golden, CO). Slides incubated without primary antibody were used as negative controls. Primary antibodies were detected with goat anti-rabbit Alexa Fluor 488 (1:500) (Molecular Probes). Images were visualized by using a Leitz (Wetzlar, Germany) DMRXE microscope and a Leica camera.
The following numbers of mice of each genotype and developmental stage were examined by immunohistochemistry with the anti-TMHS antibody: hscy/hscy (one E14.5, one E15.5, one E16.5, one E17.5, two P0, one P9, one P30, and one P60), +/hscy (one E14.5, one E17.5, one P0, one P9, one P30, and one P60), and +/+ (one E14.5, one E15.5, one E16.5, one E17.5, and two P0).
DNA Sequencing and Mutation Genotyping. Primers and sequencing methods are described in Supporting Text, which is published as supporting information on the PNAS web site.
Results
The hscy Phenotype. The overt phenotype of hscy homozygotes consists of circling behavior, frequent head shaking from side to side, and an inability to swim. The hearing ability of both mutant and control mice was measured by using ABR thresholds. We tested 15 mutant and 9 control mice between 26 and 131 days of age. None of the hscy/hscy mutant mice tested showed any response to auditory stimuli up to 110 dB sound pressure level. The ABR thresholds of control mice were in the range of normal hearing. The deafness and circling behavior are recessive and fully penetrant in mice on the C57BL/6 background and also in F2 hybrid linkage cross mice.
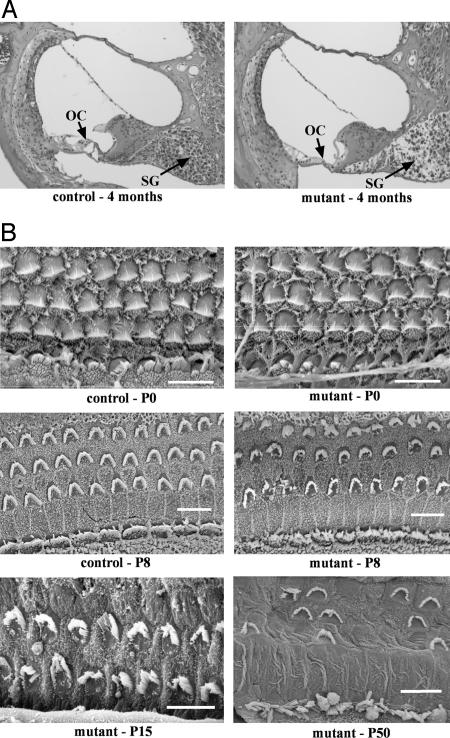
Examination of cochlear cross sections revealed severe degeneration of the organ of Corti in mutant but not control mice at 4 months of age, including loss of inner and outer hair cells and decreased spiral ganglia (Fig. 1A). Degeneration was more pronounced in the basal region of the cochlea than in the apex. Hair cell morphology was investigated further by SEM. At P0, mutant hair cells appear normal but begin to degenerate by P8 (Fig. 1B). At P8, stereocilia of both outer and inner hair cells in mutants are disorganized compared with those of controls. The outer hair cells have lost their rigid V-shaped pattern, and the inner hair cells have a more splayed appearance than those of controls. By P50, there are patches in the basal portion of the mutant cochlea in which the outer hair cells have completely degenerated, and inner hair cell stereocilia are severely splayed.
Fig. 1.
Cochlear pathology associated with the hscy mutation. (A) Cross sections through basal turns of cochleae from control and mutant mice at 4 months of age. Mutant cochleae exhibit severe degeneration of the organ of Corti (OC) and secondary degeneration of spiral ganglion cells (SG). (B) SEM examinations of OC surface preparations from cochleae of control and mutant mice at postnatal ages P0 and P8, and from mutant mice at P15 and P50. In P8 mutants, hair bundle disorganization and some hair cell loss is apparent; by P50, few stereocilia remain, and most outer hair cells have degenerated in basal portions of the cochlea.
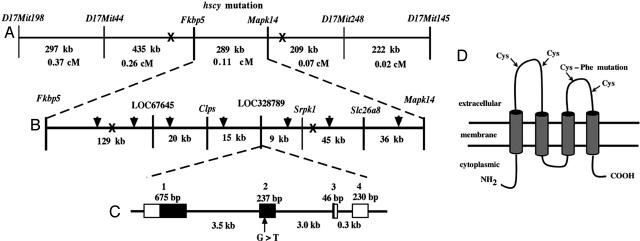
Positional Cloning and Mutant Gene Identification. To fine map the hscy mutation, we produced 1,160 F2 mice from an intercross between (C57BL/6-hscy × CAST/Ei) F1 hybrid mice. All F2 progeny produced from the linkage cross were genotyped at weaning for flanking markers, and only mice with informative recombinant chromosomes were further analyzed with additional markers. Crossover analysis with known markers limited the candidate interval to a 289-kb region between the genes Fkbp5 and Mapk14 (Fig. 2A). Using the available mouse genome sequence for the 289-kb candidate region, we designed custom primers to generate seven additional genomic CA repeat markers, which further narrowed the genetic interval to 182 kb (Fig. 2B). The narrowed region contained only two known genes, Clps and Srpk1, and two uncharacterized genes, LOC67645 and LOC328789. Of these four genes, we considered LOC328789 the most likely candidate because of its expression primarily in neural tissue, as represented by two full-length cDNAs, AK020389 (from diencephalon) and AK020670 (from neonate cerebellum), as well as 15 neurospecific ESTs (Unigene Mm.284760).
Fig. 2.
Positional cloning and characterization of the hscy gene. (A) Crossover exclusion refined the hscy mutation to the Fkbp5–Mapk14 interval. (B) Additional custom-designed markers (arrowheads) enabled further refinement of the candidate interval to an ≈182-kb region containing two known and two uncharacterized genes. (C) The uncharacterized gene (LOC328789) identified by the XM_283418 cDNA sequence is predicted to have four exons (shown as rectangles connected by lines representing introns); a G>T transversion was found in exon 2, within the protein coding sequence of this gene (black regions of exons). (D) Schematic representation of the predicted protein structure showing the four transmembrane domains and the two cysteines within each extracellular loop. The Cys>Phe change caused by the hscy mutation is indicated.
To evaluate the LOC328789 gene, we designed PCR primers to amplify overlapping regions of the 1,237-bp National Center for Biotechnology Information reference cDNA sequence XM_283418. We detected a substitution from G to T in DNA from hscy/hscy mutant mice at nucleotide 482 of the protein-coding portion of the cDNA (where nucleotide + 1 is the A of the ATG initiation codon). The mutation in cDNA is hence designated c.482G>T. No other sequence alterations were found in the cDNA of mutant mice. To verify the mutation detected in cDNA, we sequenced PCR fragments obtained from genomic DNA of three additional mutants as well as two additional C57BL/6J animals and a mouse of the genetically divergent CAST/Ei strain.
To confirm the correspondence of Tmhs genotypes with hscy phenotypes, we sequenced DNA from 99 mice produced from intercrosses of +/hscy heterozygotes and identified 22 with +/+ genotypes, 56 with +/hscy genotypes, and 21 with hscy/hscy genotypes. (DNA chromatographs are shown in Fig. 6, which is published as supporting information on the PNAS web site.) Mice with two copies of the c.482G>T mutation (hscy/hscy) always had a mutant phenotype, whereas mice with two copies of the G nucleotide (+/+) and mice with both G and T (+/hscy) always had normal phenotypes. The symbol Tmhs has been assigned to designate the previously unnamed and uncharacterized mouse gene (LOC328789) that underlies the hscy mutation.
Molecular Characterization of the Tmhs Gene and hscy Mutation. Comparison of the mouse cDNA sequence (XM_283418) with genomic DNA sequence (NT_039649) revealed that the Tmhs gene spans ≈7.9 kb and is composed of four exons (Fig. 2C). The protein coding sequence of XM_283418 (nucleotides 285–944) spans exons 1–3 and encodes a 219-aa protein. The hscy mutation occurs in exon 2 of Tmhs, which corresponds to nucleotide 766 of XM_283418. Exon 4 includes a B1 sine repeat (nucleotides 1058–1164 of XM_283418) that includes a potential polyA signal (ATTAAA). This signal appears to have been used to form most of the mRNAs and ESTs comprising UniGene Mm.284760 (such as AK020670, AW492878, BB125404, BB132044, BB258096, BB250980, and others), but a few transcripts apparently recognize a more 3′ signal (such as AK020389 and BB196812). The firstef computer program (11) analysis of genomic DNA strongly predicts a CpG-related promoter sequence and a first exon 5′ boundary near the 5′ end of the XM_283418 sequence, indicating that the actual transcription start site for Tmhs is likely to be within 100 bp of the 5′ end of this cDNA.
The G to T transversion causes a nonconservative amino acid change from cysteine to phenylalanine at amino acid position 161 (codon UGU to UUU). The amino acid mutation is thus designated C161F. The Tmhs gene is predicted by the tmhmm computer program (12) to encode a protein with four transmembrane helices and two extracellular loops, as shown schematically in Fig. 2D. The C161F mutation occurs in the second extracellular loop. Cysteine is a weakly polar hydrophilic amino acid with a thiol side chain, whereas phenylalanine is a nonpolar highly hydrophobic amino acid with an aromatic side chain. The large difference between these two amino acids would likely cause the mutant protein to be targeted for degradation early in development, consistent with the absence of protein expression that we observed in the mutant inner ear. Thiol side chains are essential for the formation of disulfide bonds between cysteine residues, which help to stabilize the tertiary structure of the protein. The cysteine mutated in hscy mice (C161F) is highly conserved in the orthologous proteins of other species, including rat, human, chicken, pufferfish, Drosophila, and Caenorhabditis elegans (Fig. 7, which is published as supporting information on the PNAS web site).
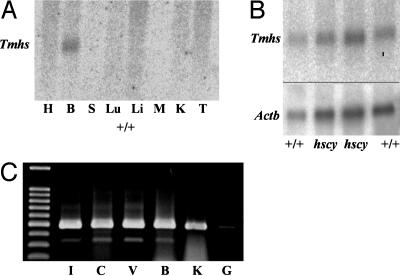
Tmhs Gene and Protein Expression. Gene expression was examined at the RNA level by performing Northern blot analysis and RT-PCR. A multiple tissue Northern blot from a commercial vendor (Clontech, BD Biosciences) consisting of poly(A)+ RNA from heart, brain, spleen, lung, liver, skeletal muscle, kidney, and testes of adult mice was hybridized with a Tmhs DNA probe. Of the eight tissues analyzed, a transcript was detected only in brain RNA (Fig. 3A), consistent with the neurospecific sources of database ESTs. The estimated size of the transcript (1.3 kb) agrees with the length of the XM_283418 cDNA sequence. A Northern blot of poly(A)+ RNA extracted from brains of +/+ control mice and hscy/hscy mutant mice and hybridized with the same probe revealed no differences in transcript levels between mutant and control mice (Fig. 3B). This result was anticipated, because the in-frame nature of the hscy missense mutation would not likely disrupt transcription.
Fig. 3.
Tmhs gene expression. (A) Commercial Northern blot of polyA+ RNA from multiple adult mouse tissues (H, heart; B, brain; S, spleen; Lu, lung; Li, liver; M, skeletal; muscle; K, kidney; and T, testes) hybridized with a Tmhs cDNA probe. (B) Northern blot of polyA+ RNA extracted from brains of adult C57BL/6J controls (+/+) and hscy/hscy mutants (hscy) hybridized with a Tmhs cDNA probe (Upper) and subsequently with a β actin control probe (Actb, Lower) to evaluate RNA loading concentrations. (C) RT-PCR of cDNA produced from total RNA extracted from adult C57BL/6J tissues (I, inner ear; C, cochlea; V, vestibule; B, brain; K, kidney; and G, genomic DNA). Tmhs PCR primers 730F (corresponding to exon 2 sequence) and 1212R (corresponding to exon 4 sequence) produced ≈480 bp of product from cDNA but failed to amplify a product from genomic DNA.
We used RT-PCR to examine Tmhs gene expression in the inner ear. Total RNA was extracted from the entire inner ear (cochlea + vestibule), cochlea, vestibule, brain, and kidney of an adult +/+ control mouse and used to make cDNA. Tmhs-specific PCR primers amplified the expected 480-bp product from each of the tissue-specific cDNAs, although a lower level of expression was seen in the kidney (Fig. 3C). Although not quantitative, these results verify inner ear expression of the gene.
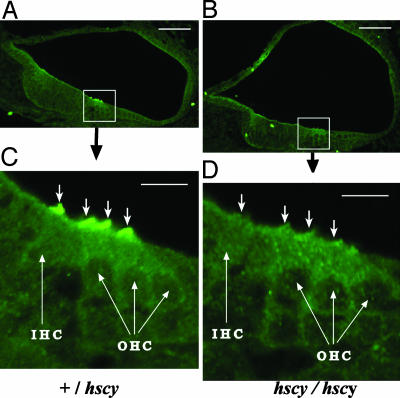
To examine the localization of the protein in the brain and inner ear, we used peptide antisera specific to the carboxyl terminal end of the predicted protein. We examined sagittal sections of brains from adult and newborn mice but did not detect any TMHS-specific immunofluorescence that was significantly above background (data not shown). In the inner ear, however, intense fluorescence clearly indicated a concentrated presence of TMHS on stereocilia of both inner and outer hair cells in newborn +/+ and +/hscy mice (Fig. 4 A and C). We saw no TMHS expression in cochlear hair cells of P0 hscy/hscy mutants (Fig. 4 B and D), confirming the specificity of the antibody and the severe consequence of the hscy missense mutation. The same TMHS expression pattern observed in P0 mice was seen in the inner ears of wild-type but not mutant mice at E16.5 and E17.5. We did not detect TMHS immunofluorescence in the inner ears of wild-type or mutant embryos or postnatal mice when examined at E14.5, E15.5, P9, P30, and P60.
Fig. 4.
Subcellular localization of TMHS protein. (A and B) Cross sections through the cochlear duct of a newborn +/hscy heterozygous control (A) and a hscy/hscy mutant (B) showing hair cell-specific localization. (Bar, 50 μm.) (C and D) Higher magnification of boxed regions from A and B. (Bar, 10 μm.) Intense immunofluorescence was observed on stereocilia (indicated by ↓) of both inner hair cells (IHC) and outer hair cells (OHC) in the +/hscy heterozygote (C) but not in the hscy/hscy mutant (D).
To verify that hair cells were intact in newborn hscy mutant mice, we examined the expression of myosin VIIa with a commercially available antibody. Myosin VIIa is expressed throughout the cell body in both inner and outer hair cells (13), and its expression appeared normal in cochleae of P0 hscy/hscy mice (Fig. 8, which is published as supporting information on the PNAS web site).
Because hscy/hscy mice exhibit circling and head-tossing behaviors characteristic of vestibular dysfunction, we also examined TMHS expression in the vestibular neuroepithelia of the inner ear. As in the cochlea, TMHS immunofluorescence was pronounced in the stereocilia of hair cells in the vestibular maculae and cristae of P0 wild-type mice (Fig. 9, which is published as supporting information on the PNAS web site).
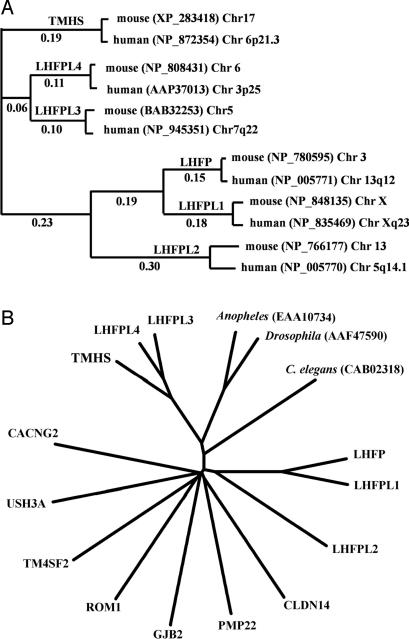
Related Sequences. A search of the Pfam, InterPro, Prosite, CDART, and Smart protein databases with the TMHS amino acid sequence resulted in no matches to any known protein domains. An Ensembl blast search of the public mouse genome database with the TMHS amino acid sequence revealed two closely related proteins (62–66% amino acid identity) that are encoded by genes located on mouse Chrs 5 and 6. These two related proteins were provisionally given the symbols LHFPL3 and LHFPL4 by database curators because of their ≈25% amino acid sequence similarity to the lipoma HMGIC fusion partner protein, LHFP (14). We used the clustalw computer program (15) to create a phylogram of amino acid sequence similarities to illustrate the orthologous relationships of the mouse and human genes encoding TMHS and LHFP-like proteins (Fig. 5A). The similarities of TMHS, LHFPL3, and LHFPL4 with one another (62–71% amino acid identities) are much higher than their similarities with LHFP, LHFPL1, or LHFPL2 (21–26% amino acid identities).
Fig. 5.
Protein relationships analyzed by clustalw multiple sequence alignments. (A) Phylogram of orthologous and paralogous relationships of mouse and human TMHS and LHFP-like proteins. GenBank reference nos. for these proteins and the chromosome positions of their respective genes are also shown. Numbers shown were calculated from pairwise distance scores. (B) Unrooted tree of tetraspan protein relationships. Representative members of all tetraspan subgroups are included. Only mouse sequences were used for these comparisons, except for Anopheles, Drosophila, and C. elegans.
The Tmhs gene is predicted to encode a four transmembrane domain protein, making it a member of the large tetraspan superfamily. This family includes the claudin tight junction proteins, the connexin gap junction proteins, clarins, proteolipid proteins, peripheral myelin, and epithelial membrane proteins, calcium channel γ-subunit-like proteins, and members of the tetraspanin family. We used clustalw multiple sequence alignments to compare the mouse TMHS protein sequence with representative members of these other tetraspan families in the mouse and with related sequences in other species (Fig. 6B). TMHS, LHFPL3, and LHFPL4 form a separate subfamily that is clearly distinct from all other tetraspan proteins.
Discussion
We present several lines of evidence that support Tmhs as the gene underlying the deafness and balance dysfunction of hscy mutant mice: (i) The hscy candidate gene region is genetically limited to only 182 kb, which includes the Tmhs gene. (ii) Tmhs genotypes (determined by DNA sequence analysis of the mutated 482G>T region) agree with hscy phenotypes in all >100 mice examined. (iii) The coisogenic nature of the hscy mutation (its spontaneous occurrence in a genetically homogeneous inbred strain) eliminates the possibility that the Tmhs 482G>T missense mutation is a population polymorphism. (iv) The cysteine residue of TMHS that is altered in hscy mutant mice is highly conserved in the orthologous proteins of other species from rats to C. elegans and is predicted to be essential for proper protein structure and function. (v) The absence of the TMHS protein in the inner ears of hscy/hscy mice demonstrates a causative connection between the hscy phenotype and the Tmhs gene product.
The human ortholog of the mouse Tmhs gene (GeneID 222662), which we designate TMHS, is located on chromosome 6p21.3. The hscy mutation of Tmhs thus may provide a model for human hearing disorders that map to this region, such as the dominant nonsyndromic disorders DFNA21 (16) or DFNA31 (17). According to the genetic and physical map positions of flanking markers (17), the TMHS gene is outside of the candidate regions for DFNA21 and DFNA31; however, because of the possibility of mapping errors, it may still be worthy of consideration as a candidate gene for these hearing disorders. Recently, a recessive nonsyndromic hearing disorder, DFNB53, has been mapped to the 6p21.3 region (Wenjie Chen, Hereditary Hearing Loss Homepage, http://webhost.ua.ac.be/hhh), and TMHS might also be considered a candidate gene for this newly mapped disorder.
In mammals, TMHS, LHFPL3, and LHFPL4 comprise a distinct subfamily within the large superfamily of tetraspan proteins (Fig. 5). In insects and worms, there appears to be a single orthologous protein that corresponds to the three-member mammalian subfamily. RNA sources for the cDNAs and ESTs found in the UniGene databases indicate that all three subfamily members are expressed primarily in neural tissues such as brain, spinal cord, and retina. Although the majority of expression database entries for Tmhs are from brain tissue, and our Northern blot results detected expression in the brain (Fig. 4A), the hscy mutant phenotype indicates that the inner ear is the primary site of TMHS function. By immunohistochemistry with specific antibodies, we detected a high level of protein expression specifically in stereocilia of inner ear hair cells but not in brain sections. One explanation for the seemingly contradictory results between gene and protein expression in the brain is that the TMHS protein may be distributed more diffusely among cells of the brain as compared with the much higher concentration found in hair cell stereocilia. Alternatively, Tmhs mRNA in brain may not be translated, or the protein may be unstable because of improper modification or localization.
Mutations of tetraspan proteins have been shown to underlie both human and mouse deafness disorders. Mutations of the gap junction proteins GJA1 (18), GJB2 (19), GJB3 (20), and GJB6 (21) are responsible for several human nonsyndromic deafness disorders, including DFNB1 and DFNA3. Mutations of the tight junction protein CLDN14 underlie DFNB29 (22), and mutations of clarin-1 underlie Usher syndrome type 3A (23). Targeted mutations of Cldn11 (24), Cldn14 (25), Gjb2 (26), and Gjb6 (27) cause hearing impairment in mice. The gap junction and tight junction tetraspan proteins that underlie hearing disorders are crucial for maintaining proper ion concentrations in the various subcompartments of the inner ear (2). Within the mammalian cochlea, the GJA1, GJB2, GJB3, and GJB6 gap junction proteins are found in supporting cells and in fibroblasts of the spiral ligament and limbus (18, 28, 29), the tight junction protein CLDN11 is expressed in the basal cell layer of the stria vascularis (24), and CLDN14 is expressed in hair cells and supporting cells (22). Clarin-1, the tetraspan protein underlying USH3A, is expressed only in hair cells and is thought to play a role in the formation or structure of the hair cell synapse (23). Although CLDN14 and clarin-1 are expressed in hair cells, they are localized to the basolateral membranes of these cells. TMHS is the only tetraspan protein known to localize to the apical membrane and stereocilia of hair cells, although other tetraspan proteins have been localized to the apical membranes of polarized epithelial cells in other tissues (30, 31).
The period of peak TMHS expression corresponds with the developmental period of stereocilia formation in inner ear hair cells of the mouse, between E15 and P8. These temporal and spatial patterns of Tmhs expression coupled with the early onset of stereocilia disorganization observed in hscy/hscy mutant mice indicate a likely role for this gene in hair bundle morphogenesis. Cdh23 (32, 33) and Pcdh15 (34, 35) encode cadherin proteins thought to be involved in hair bundle morphogenesis and whose dysfunction results in stereocilia phenotypes similar to Tmhs mutants. Cadherins are transmembrane proteins of adherens junctions that mediate cell–cell adhesion and connections to the cytoskeleton. Some tetraspan proteins recently have been shown to localize to adherens junctions and are thought to play a role in regulating cellular adhesion and epidermal morphology through F-actin attachments (36, 37). In a like manner, it is possible that TMHS may associate specifically with inner ear cadherins and help direct development and morphogenesis of the hair bundle.
Supplementary Material
Acknowledgments
We thank the following personnel from The Jackson Laboratory for their contributions: Amy Kiernan and Verity Letts for critical review of this manuscript, Heping Yu for ABR technical assistance, Peter Finger for EM imaging assistance, Priscilla Jewett for histology assistance, and Kenneth Bosom and Sandra Gray for mouse colony management. We also thank Dawn Young for the initial discovery of the mutant mice and Gordon Watson (Children's Hospital Research Center, Oakland, CA) for providing them to us. This work was supported by National Institutes of Health (NIH) Grants DC04301 and RR01183. The Jackson Laboratory institutional shared services are supported by NIH Grant CA34196.
Author contributions: K.R.J. designed research; C.M.L.-G., L.H.G., S.A.C., J.W., Q.Y.Z., and K.R.J. performed research; C.M.L.-G., L.H.G., S.A.C., and K.R.J. analyzed data; and C.M.L.-G. and K.R.J. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: ABR, auditory-evoked brainstem response; SEM, scanning electron microscopy; En, embryonic day n;Pn, postnatal day n.
References
- 1.Morton, N. E. (1991) Ann. N.Y. Acad. Sci. 630, 16–31. [DOI] [PubMed] [Google Scholar]
- 2.Friedman, T. B. & Griffith, A. J. (2003) Annu. Rev. Genomics Hum. Genet. 4, 341–402. [DOI] [PubMed] [Google Scholar]
- 3.Muller, U. & Littlewood-Evans, A. (2001) Trends Cell Biol. 11, 334–342. [DOI] [PubMed] [Google Scholar]
- 4.Frolenkov, G. I., Belyantseva, I. A., Friedman, T. B. & Griffith, A. J. (2004) Nat. Rev. Genet. 5, 489–498. [DOI] [PubMed] [Google Scholar]
- 5.Taylor, B. A., Navin, A. & Phillips, S. J. (1994) Genomics 21, 626–632. [DOI] [PubMed] [Google Scholar]
- 6.Manly, K. F., Cudmore, R. H., Jr., & Meer, J. M. (2001) Mamm. Genome 12, 930–932. [DOI] [PubMed] [Google Scholar]
- 7.Johnson, K. R., Gagnon, L. H., Webb, L. S., Peters, L. L., Hawes, N. L., Chang, B. & Zheng, Q. Y. (2003) Hum. Mol. Genet. 12, 3075–3086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zheng, Q. Y., Johnson, K. R. & Erway, L. C. (1999) Hear. Res. 130, 94–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Truett, G. E., Heeger, P., Mynatt, R. L., Truett, A. A., Walker, J. A. & Warman, M. L. (2000) BioTechniques 29, 52, 54. [DOI] [PubMed] [Google Scholar]
- 10.Johnson, K. R., Cook, S. A., Erway, L. C., Matthews, A. N., Sanford, L. P., Paradies, N. E. & Friedman, R. A. (1999) Hum. Mol. Genet. 8, 645–653. [DOI] [PubMed] [Google Scholar]
- 11.Davuluri, R. V., Grosse, I. & Zhang, M. Q. (2001) Nat. Genet. 29, 412–417. [DOI] [PubMed] [Google Scholar]
- 12.Sonnhammer, E. L., von Heijne, G. & Krogh, A. (1998) Proc. Int. Conf. Intell. Syst. Mol. Biol. 6, 175–182. [PubMed] [Google Scholar]
- 13.Self, T., Mahony, M., Fleming, J., Walsh, J., Brown, S. D. & Steel, K. P. (1998) Development (Cambridge, U.K.) 125, 557–566. [DOI] [PubMed] [Google Scholar]
- 14.Petit, M. M., Schoenmakers, E. F., Huysmans, C., Geurts, J. M., Mandahl, N. & Van de Ven, W. J. (1999) Genomics 57, 438–441. [DOI] [PubMed] [Google Scholar]
- 15.Chenna, R., Sugawara, H., Koike, T., Lopez, R., Gibson, T. J., Higgins, D. G. & Thompson, J. D. (2003) Nucleic Acids Res. 31, 3497–3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kunst, H., Marres, H., Huygen, P., van Duijnhoven, G., Krebsova, A., van der Velde, S., Reis, A., Cremers, F. & Cremers, C. (2000) Clin. Otolaryngol. 25, 45–54. [DOI] [PubMed] [Google Scholar]
- 17.Snoeckx, R. L., Kremer, H., Ensink, R. J., Flothmann, K., de Brouwer, A., Smith, R. J., Cremers, C. W. & Van Camp, G. (2004) J. Med. Genet. 41, 11–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu, X. Z., Xia, X. J., Adams, J., Chen, Z. Y., Welch, K. O., Tekin, M., Ouyang, X. M., Kristiansen, A., Pandya, A., Balkany, T., et al. (2001) Hum. Mol. Genet. 10, 2945–2951. [DOI] [PubMed] [Google Scholar]
- 19.Kelsell, D. P., Dunlop, J., Stevens, H. P., Lench, N. J., Liang, J. N., Parry, G., Mueller, R. F. & Leigh, I. M. (1997) Nature 387, 80–83. [DOI] [PubMed] [Google Scholar]
- 20.Xia, J. H., Liu, C. Y., Tang, B. S., Pan, Q., Huang, L., Dai, H. P., Zhang, B. R., Xie, W., Hu, D. X., Zheng, D., et al. (1998) Nat. Genet. 20, 370–373. [DOI] [PubMed] [Google Scholar]
- 21.Grifa, A., Wagner, C. A., D'Ambrosio, L., Melchionda, S., Bernardi, F., Lopez-Bigas, N., Rabionet, R., Arbones, M., Monica, M. D., Estivill, X., et al. (1999) Nat. Genet. 23, 16–18. [DOI] [PubMed] [Google Scholar]
- 22.Wilcox, E. R., Burton, Q. L., Naz, S., Riazuddin, S., Smith, T. N., Ploplis, B., Belyantseva, I., Ben-Yosef, T., Liburd, N. A., Morell, R. J., et al. (2001) Cell 104, 165–172. [DOI] [PubMed] [Google Scholar]
- 23.Adato, A., Vreugde, S., Joensuu, T., Avidan, N., Hamalainen, R., Belenkiy, O., Olender, T., Bonne-Tamir, B., Ben-Asher, E., Espinos, C., et al. (2002) Eur. J. Hum. Genet. 10, 339–350. [DOI] [PubMed] [Google Scholar]
- 24.Gow, A., Davies, C., Southwood, C. M., Frolenkov, G., Chrustowski, M., Ng, L., Yamauchi, D., Marcus, D. C. & Kachar, B. (2004) J. Neurosci. 24, 7051–7062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ben-Yosef, T., Belyantseva, I. A., Saunders, T. L., Hughes, E. D., Kawamoto, K., Van Itallie, C. M., Beyer, L. A., Halsey, K., Gardner, D. J., Wilcox, E. R., et al. (2003) Hum. Mol. Genet. 12, 2049–2061. [DOI] [PubMed] [Google Scholar]
- 26.Cohen-Salmon, M., Ott, T., Michel, V., Hardelin, J. P., Perfettini, I., Eybalin, M., Wu, T., Marcus, D. C., Wangemann, P., Willecke, K., et al. (2002) Curr. Biol. 12, 1106–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Teubner, B., Michel, V., Pesch, J., Lautermann, J., Cohen-Salmon, M., Sohl, G., Jahnke, K., Winterhager, E., Herberhold, C., Hardelin, J. P., et al. (2003) Hum. Mol. Genet. 12, 13–21. [DOI] [PubMed] [Google Scholar]
- 28.Lopez-Bigas, N., Olive, M., Rabionet, R., Ben-David, O., Martinez-Matos, J. A., Bravo, O., Banchs, I., Volpini, V., Gasparini, P., Avraham, K. B., et al. (2001) Hum. Mol. Genet. 10, 947–952. [DOI] [PubMed] [Google Scholar]
- 29.Lautermann, J., ten Cate, W. J., Altenhoff, P., Grummer, R., Traub, O., Frank, H., Jahnke, K. & Winterhager, E. (1998) Cell Tissue Res. 294, 415–420. [DOI] [PubMed] [Google Scholar]
- 30.Bosse, F., Hasse, B., Pippirs, U., Greiner-Petter, R. & Muller, H. W. (2003) J. Neurochem. 86, 508–518. [DOI] [PubMed] [Google Scholar]
- 31.Frank, M., van der Haar, M. E., Schaeren-Wiemers, N. & Schwab, M. E. (1998) J. Neurosci. 18, 4901–4913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Siemens, J., Lillo, C., Dumont, R. A., Reynolds, A., Williams, D. S., Gillespie, P. G. & Muller, U. (2004) Nature 428, 950–955. [DOI] [PubMed] [Google Scholar]
- 33.Di Palma, F., Holme, R. H., Bryda, E. C., Belyantseva, I. A., Pellegrino, R., Kachar, B., Steel, K. P. & Noben-Trauth, K. (2001) Nat. Genet. 27, 103–107. [DOI] [PubMed] [Google Scholar]
- 34.Alagramam, K. N., Murcia, C. L., Kwon, H. Y., Pawlowski, K. S., Wright, C. G. & Woychik, R. P. (2001) Nat. Genet. 27, 99–102. [DOI] [PubMed] [Google Scholar]
- 35.Ahmed, Z. M., Riazuddin, S., Ahmad, J., Bernstein, S. L., Guo, Y., Sabir, M. F., Sieving, P., Griffith, A. J., Friedman, T. B., Belyantseva, I. A., et al. (2003) Hum. Mol. Genet. 12, 3215–3223. [DOI] [PubMed] [Google Scholar]
- 36.Simske, J. S., Koppen, M., Sims, P., Hodgkin, J., Yonkof, A. & Hardin, J. (2003) Nat. Cell Biol. 5, 619–625. [DOI] [PubMed] [Google Scholar]
- 37.Kearsey, J., Petit, S., De Oliveira, C. & Schweighoffer, F. (2004) Eur. J. Biochem. 271, 2584–2592. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.