Abstract
Interferon (IFN)-induced 2′-5′ oligoadenylate synthetase (2-5A synthetase)/RNase L, PKR, and Mx proteins are considered to be the principal antiviral protein pathways through which IFN induces an antiviral state. It was previously reported that human parainfluenza virus type 3 (HPIV3) multiplication was inhibited by IFN-α in human lung epithelial cells A549 and that MxA was found to contribute to the inhibition process (Zhao et al., Virology 220:330–338, 1996). Viral primary transcription was dramatically inhibited in A549 cells after IFN-α treatment, but a step following primary transcription was inhibited in U87-MxA cells constitutively expressing MxA. Here we have investigated the role of MxA, believed to be cell type specific, and other antiviral pathways in the inhibition of viral primary transcription. Our data indicate that a novel IFN-induced pathway(s) is involved in the inhibition of primary transcription. This is based on the following findings: (i) IFN-α inhibited viral primary transcription in U87-MxA and other cell types including cells lacking MxA; (ii) cells constitutively expressing 2-5A synthetase had no antiviral effect against HPIV3; and (iii) primary transcription occurred in the absence of protein synthesis, a step of PKR target. The novel antiviral pathway(s) was induced by both IFN-α and IFN-γ to establish an effective antiviral state against HPIV3. By using IFN-α-signaling mutant cells, we found that IFN-γ could elicit antiviral effect against HPIV3 without cross talk with the IFN-α-signaling pathway. These data provide the first evidence that a novel antiviral pathway(s) contributes to the antiviral action of IFN against a nonsegmented negative-strand RNA virus by targeting the primary transcription.
Human parainfluenza virus type 3 (HPIV3), a paramyxovirus, is a significant cause of serious respiratory tract disease such as bronchiolitis, pneumonia, and croup in newborns and infants (2, 4, 21). The viral infection begins, following entry into host cells, with transcription of the genome RNA by a virion-associated RNA-dependent RNA polymerase, a process known as primary transcription (2, 4, 21). Genome replication by the same RNA polymerase is initiated after translation of the viral mRNAs. The newly synthesized viral genome RNAs are eventually packaged, and the mature virions bud out from the plasma membrane of the host cell (2, 4, 21). Interferon (IFN) can induce an antiviral state against HPIV3, but the exact mechanism by which IFN exerts its antiviral effect has not been elucidated (38).
The signal transduction pathways of alpha/beta IFN (IFN-α/β) and gamma IFN (IFN-γ) have been extensively studied (5, 33, 36). They transmit signals to the cell interior through distinct receptor complexes, IFNAR for IFN-α/β and IFNGRa for IFN-γ. Ligand-induced stimulation of the receptor complex results in the activation of receptor-associated Janus kinases (JAKs), specifically JAK1 and TYK2 for IFN-α/β and JAK1 and JAK2 for IFN-γ. After activation of JAKs, signal transducers and activators of transcription (STATs) are activated by phosphorylation leading to the formation of IFN-stimulated gene factor 3 (ISGF3) comprised of STAT1, STAT2, and p48 for IFN-α/β and IFN-γ-activated factor (GAF), a STAT1 homodimer, for IFN-γ. These complexes translocate to the nucleus and induce a large number of proteins, some of which possess antiviral activities. A concerted action of the antiviral proteins leads to the establishment of an antiviral state.
At present, three identifiable antiviral pathways have been implicated in the IFN-mediated inhibition of viruses (19, 24, 27, 31–33, 36): (i) the 2–5A synthetase/RNase L pathway, which degrades viral RNAs following activation by double-stranded RNA (dsRNA); (ii) the dsRNA-activated protein kinase (PKR), which inhibits mRNA translation in infected cells by phosphorylating the translation initiation factor eIF-2α; and (iii) the Mx proteins (Mx1 in mice and MxA in humans), whose precise mode of action is yet to be elucidated. In a previous study, it has been shown that HPIV3 multiplication is strongly inhibited by IFN-α (38). By using human neurogliomal U87-MxA cells constitutively expressing MxA, we showed that MxA contributes to the antiviral action of the IFN-α. The target of MxA was found to be a step following primary transcription in U87-MxA cells, although primary transcription was inhibited by IFN-α in human lung epithelial A549 cells.
Human MxA (76 kDa) is a cytoplasmic protein (1) which is rapidly induced in response to acute viral infections (28). Its role against RNA viruses of the families Orthomyxoviridae (13, 14, 20, 25, 26), Bunyaviridae (12, 18), Rhabdoviridae (26, 30), Paramyxoviridae (28, 29, 38), and Togaviridae (22) has been demonstrated. These studies indicate that the viral target recognition by MxA is virus and cell type specific, inhibiting transcription (28, 30) or mRNA translation (29) or transportation of nucleocapsids (20). However, the mechanism by which MxA is able to inhibit such a diverse array of viruses with varied viral target recognition is not clearly understood.
Inhibition of viral primary transcription is one of the strategies by which IFN elicits an antiviral effect against some nonsegmented negative-strand RNA viruses (28, 30). Of the three antiviral pathways, this process is the one in which MxA was found to be involved (28, 30). However, in the case of some members of this class of viruses, either the IFN's effect on primary transcription has not been investigated or the antiviral protein responsible for inhibiting the primary transcription remains uncharacterized (37). In the case of HPIV3, MxA was apparently not involved in the inhibition of primary transcription in U87-MxA cells (38). This raised the question of whether MxA can inhibit viral primary transcription in a cell-type-specific manner. Alternatively, an antiviral protein other than MxA may directly target the viral primary transcription. In this context, it is noteworthy that recently Zhou et al. (39) demonstrated the existence of alternative antiviral pathways against some viruses. Therefore, it is of interest to identify the antiviral pathway involved in the inhibition of HPIV3 primary transcription.
In this study, we have investigated the contribution of individual antiviral pathways to the IFN-dependent inhibition of HPIV3 primary transcription. Our data indicate that a novel antiviral protein(s) is involved in the inhibition of viral primary transcription and that MxA targets a step which follows the primary transcription. The novel antiviral protein appears to play a major role in the IFN action against HPIV3, while MxA plays an additional role.
MATERIALS AND METHODS
Virus, cells, antibodies, and IFNs.
HPIV3 (HA-1, NIH 47885) was propagated in CV-1 cells (ATCC CCL 70) as described previously (6, 7). Human lung epithelial cells (A549 [ATCC CCL 185]) were maintained in minimal essential medium (Gibco-BRL, Gaithersburg, Md.), and CV-1 cells were maintained in Dulbecco's modified Eagle's medium (DMEM) (Gibco-BRL), each supplemented with 10% fetal bovine serum (FBS), penicillin, streptomycin, and glutamine. Stably MxA-transfected human glioblastomal cell line U87-MxA and vector-transfected cell line U87-CL4 were kindly provided by Sibylle Schneider-Schaulies, Institute for Virology and Immunology, Wurzburg, Germany. These cells were maintained in minimal essential medium containing 10% fetal calf serum and G418 (500 μg/ml). IFN-α-signaling mutant cells (15) U1A, U1A(KD), and the wild-type cells 2fTGH were maintained in DMEM. Cells WtP69#9, constitutively expressing a 69-kDa isoform of 2-5A synthetase, and PDR2-hyg, containing an empty vector (16), were maintained in DMEM. Polyclonal antibodies against MxA and HPIV3 RNP were raised in rabbits. The IFN-α was purchased from Sigma Biochemicals, St. Louis, Mo., and IFN-γ was purchased from Roche Biochemicals, Indianapolis, Ind.
Plaque assay.
Effects of IFNs on the production of infectious HPIV3 virions in different cell types were studied using confluent monolayers of cells in 6-well plates. The cells were treated with IFN at the concentrations indicated in individual experiments for 12 h and then infected with HPIV3 at multiplicities of infection (MOI) indicated in individual experiments. Culture supernatants were collected at 40 h postinfection, unless otherwise stated, and the infectious virus yield was measured by plaque assay on CV-1 cells (8).
Similarly, the effect of MxA on the production of infectious HPIV3 virions was measured by using U87-MxA cells in 6-well plates. U87-CL4 cells served as the control. Both cell lines were infected with HPIV3 at the MOI indicated for individual experiments. At 40 h postinfection, culture supernatants were collected and infectious virus yields were quantitated by plaque assay.
Western blot.
Protein concentration was determined by using a Bio-Rad protein assay kit (Bio-Rad Laboratories, Richmond, Calif.) according to the manufacturer's protocol. Soluble proteins (20 μg) from infected and uninfected cells were resolved in a 10% polyacrylamide–sodium dodecyl sulfate gel followed by Western blotting onto a nitrocellulose membrane (35). Polyclonal antibodies against MxA, raised in rabbits, were used for the detection of MxA. Protein bands were visualized by staining with horseradish peroxidase-conjugated goat anti-rabbit antibody followed by enhanced chemiluminescence according to the manufacturer's protocol (Amersham).
Northern blot.
Total cellular RNA was isolated using RNA-STAT following the manufacturer's protocol (Tel-Test, Friendswood, Tex.). About 10 μg of RNA was analyzed in 1% agarose-formaldehyde gel and transferred onto nitrocellulose membrane. The blot was treated with 32P-labeled N cDNA probe for hybridization followed by washing and autoradiography.
Immunofluorescence.
Immunofluorescent staining and microscopy were carried out essentially as described previously (17). Briefly, cells were washed with phosphate buffered saline (PBS) and fixed in 3.7% formaldehyde in PBS for 15 min and then quenched with 50 mM NH4Cl-PBS for 15 min. The cells were then permeabilized with a permeabilization buffer containing 0.1% Triton X-100, 5% glycine, and 5% heat-inactivated FBS in PBS. Cells were incubated with anti-RNP antibody, raised in rabbit, for 1 h at room temperature. After the specified time, the cells were washed with PBS and incubated with fluorescein isothiocyanate (FITC)-conjugated goat anti-rabbit immunoglobulin antibody. The primary and secondary antibodies were diluted in permeabilization buffer. After being mounted, the cells were viewed using a Zeiss microscope.
RNA isolation and primer extension.
Cells (106) were treated with IFN-α or IFN-γ at 1,000 U/ml for 12 h or left untreated. The cells were then treated with cycloheximide (CHX) at 10 μg/ml for 2 h. After this incubation time, the cells were infected with HPIV3 at an MOI of 5 and incubated further in the presence of IFN and CHX. At 6 h postinfection, total cellular RNAs were extracted by using RNA-STAT according to the manufacturer's protocol (Tel-Test). The presence of equal amounts of 28S RNA in these samples was confirmed by analyzing them in agarose gel followed by ethidium bromide staining. Primer extension analysis using 1 μg of RNA was carried out following the procedure as described previously (9). Briefly, a negative-sense oligo which primes on the N mRNA was 5′ end labeled using [γ-32P]ATP and polynucleotide kinase using the manufacturer's protocol (Roche Biochemicals). The radiolabeled primer and the RNA were incubated in a reverse transcription reaction using Moloney murine leukemia virus reverse transcriptase at 42°C according to the manufacturer's protocol (Roche Biochemicals). The 93-nucleotide-long extension products were separated on a 6% polyacrylamide–7 M urea gel followed by autoradiography. The radiolabeled bands were quantitated by PhosphorImager (Molecular Dynamics, Sunnyvale, Calif.).
RESULTS
MxA is not required for IFN-α-mediated inhibition of HPIV3 primary transcription.
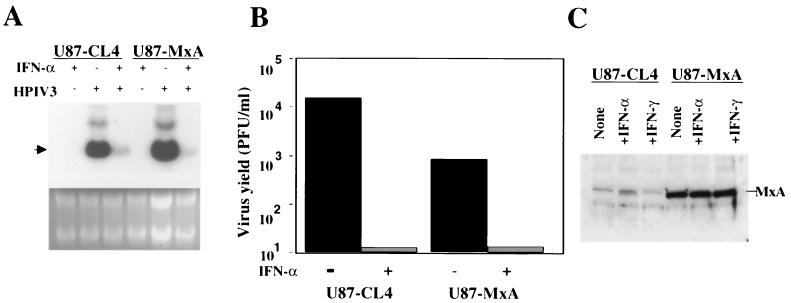
It was previously reported that IFN-α conferred a high degree of resistance to HPIV3 in A549 cells (38). The inhibitory effect of IFN-α occurred at the level of viral primary transcription but not at the level of virus entry (38). Development of an IFN-α-induced antiviral state correlated with the induction of MxA, suggesting its role in the inhibition process. By using U87-MxA cells, constitutively expressing MxA, it was found that MxA contributed to the antiviral action of IFN-α but that a step other than primary transcription was targeted (38). To further investigate the role of MxA in the inhibition of primary transcription, believed to be cell type specific (28–30), we first studied whether IFN-α could inhibit HPIV3 primary transcription in U87-MxA cells. The U87-MxA and empty-vector-transfected U87-CL4 cells were pretreated with IFN-α (1,000 U/ml) for 12 h followed by CHX (10 μg/ml) for 2 h. Cells were then infected with HPIV3 at an MOI of 5 and incubated in the presence of IFN-α and CHX. At 6 h postinfection, the level of viral major primary transcript, N mRNA, was determined by Northern hybridization. As shown in Fig. 1A, the accumulation of N mRNA was decreased in U87-MxA cells by about 30% compared to that in U87-CL4 cells (38). IFN-α treatment, on the other hand, resulted in the reduction of N mRNA accumulation in both U87-CL4 and U87-MxA cells by about 80%. The input genome RNA, however, could not be detected under these conditions. Therefore, we confirmed that the effect was on transcription but not on the virus entry by infecting cells with radiolabeled virions followed by immunoprecipitation (data not shown). These findings clearly indicate that MxA has no effect on the IFN-α- mediated inhibition of viral primary transcription. Next, to determine the effect of IFN-α on the production of infectious virions, we carried out plaque assay. As shown in Fig. 1B, constitutively expressed MxA in U87-MxA cells inhibited infectious virus production by only 1 log compared to that in U87-CL4 cells. In contrast, production of infectious virions was virtually abolished in both U87-CL4 and U87-MxA cells after IFN-α treatment. To confirm that the observed inhibition of virus production in these cells following IFN-α treatment was not due to induction of MxA, we determined the level of MxA by Western blotting using anti-MxA antibody. As shown in Fig. 1C, MxA was constitutively overexpressed in U87-MxA cells and was not significantly induced in U87-CL4 and U87-MxA cells after IFN-α treatment. These data indicate that an antiviral pathway(s) other than MxA plays a major role in the development of IFN-α-mediated antiviral state against HPIV3.
FIG. 1.
Inhibition of HPIV3 primary transcription in U87-MxA cells after IFN-α treatment. (A) Determination of viral primary transcription by primer extension using N mRNA-specific primer. Cells were pretreated with IFN-α (1,000 U/ml) for 12 h followed by treatment with CHX (10 μg/ml) for 2 h. The cells were then infected with HPIV3 at an MOI of 5 and incubated further in the presence of 10 μg/ml of CHX. At 6 h postinfection, cells were harvested and mRNA synthesis was measured by Northern blot hybridization using 32P-labeled N cDNA probe as described in Materials and Methods. The arrowhead indicates the migration position of N mRNA, and the upper band is bicistronic mRNA. (B) Effect of IFN-α on the production of infectious HPIV3 virions in U87-MxA cells. The production of infectious HPIV3 virions in the culture medium was determined by plaque assay at 24 h postinfection. (C) Western blot analysis of MxA in U87-CL4 and U87-MxA cells after treatment with 1,000 U/ml of IFN-α or IFN-γ, as indicated, for 12 h. Results are representative of three independent experiments.
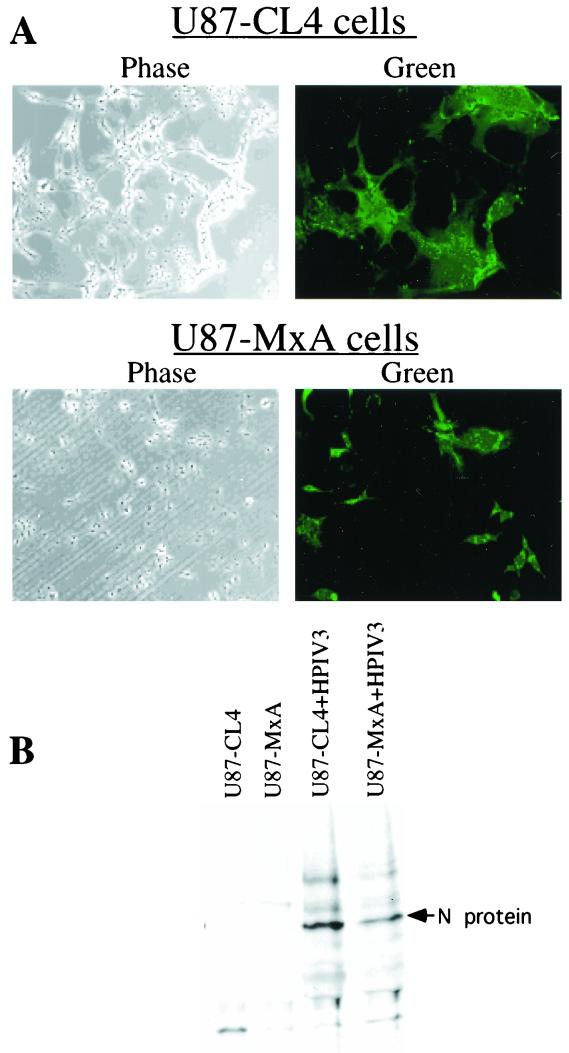
To gain insight into the antiviral action of MxA against HPIV3, we investigated whether the decreased production of infectious virions in U87-MxA cells correlated with decreased accumulation of intracellular viral RNP. The U87-CL4 and U87-MxA cells were infected with HPIV3 at an MOI of 0.1. By plaque assay, we found that virus production was decreased by more than 1 log in U87-MxA cells compared to U87-CL4 cells (data not shown). Under these conditions, intracellular RNP was detected by immunofluorescent labeling using anti-RNP antibody. As shown in Fig. 2A, intracellular RNP was significantly decreased in U87-MxA cells compared to that in U87-CL4 cells. To determine quantitatively the inhibition of intracellular viral RNP accumulation, we carried out metabolic labeling of infected cells with [35S]methionine followed by immunoprecipitation using anti-RNP antibody. As shown in Fig. 2B, accumulation of the viral RNP was decreased more than twofold in U87-MxA cells. It is important to note that the accumulation of intracellular RNP is significantly inhibited in U87-MxA cells, but it does not fully account for the dramatic inhibition of infectious virus production (more than 1 log). These data indicate that MxA most likely targets both replication and budding steps.
FIG. 2.
Accumulation of intracellular HPIV3 RNP in U87-CL4 and U87-MxA cells. (A) Cells, grown on coverslips, were infected with HPIV3 at an MOI of 0.1. At 12 h postinfection, the cells were fixed, permeabilized, and stained with anti-RNP antibody followed by FITC-conjugated secondary antibody as described in Materials and Methods. (B) Cells were infected with HPIV3 at an MOI of 0.1. At 12 h postinfection, cells were labeled with [35S]methionine (50 μCi/ml) for an additional 12 h. Cell lysate was then prepared and immunoprecipitated with anti-RNP antibody.
IFN-α inhibits viral primary transcription in both MxA-expressing and -nonexpressing cells.
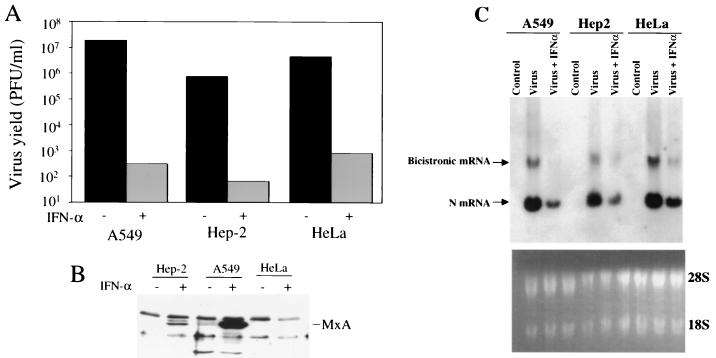
To investigate whether MxA could inhibit viral primary transcription by interacting with an IFN-α-induced protein, we took advantage of a cell line lacking MxA such as HeLa (32), a low producer such as HEp-2, and a high producer such as A549 (38). These cells were pretreated with IFN-α (1,000 U/ml) and were infected with HPIV3 at an MOI of 0.1. At 24 h postinfection, the production of infectious virions was quantitated by plaque assay. As shown in Fig. 3A, IFN-α inhibited the production of infectious virions by about 3 log in all these cell lines. Induction of MxA in these cells was determined by Western blotting using anti-MxA antibody. As shown in Fig. 3B, expression of MxA was extremely high in A549, moderate in HEp-2, and undetectable in HeLa cells, suggesting that IFN-α is able to effectively inhibit HPIV3 multiplication in the absence of MxA. By Northern blot analysis, we determined whether viral primary transcription was similarly inhibited after IFN-α treatment. As shown in Fig. 3C, N mRNA was inhibited by about 75% in A549, 65% in HEp-2, and 60% in HeLa cells. Bicistronic mRNA synthesis was similarly inhibited. These findings clearly indicate that an IFN-α-induced antiviral pathway other than MxA plays a major role in the antiviral action of IFN-α against HPIV3 by targeting the viral primary transcription.
FIG. 3.
Inhibition of HPIV3 multiplication, and induction of MxA in different cell lines by IFN-α. (A) Infectious virions released in the culture medium. Inhibition of the release of infectious virions in different cells after IFN-α (1,000 U/ml) treatment was determined by plaque assay. (B) Induction of MxA by IFN-α. The expression of MxA under different conditions as indicated was determined by Western blot analysis using anti-MxA antibody followed by enhanced chemiluminescence (Amersham). The migration position of MxA is shown. Two other protein bands present in all cell types are nonspecifically interacting proteins. (C) Effect of IFN-α on viral primary transcription. Cells were infected with HPIV3 at an MOI of 5, and at 6 h postinfection RNA was isolated. The RNA was analyzed in 1% agarose-formaldehyde gel and hybridized with 32P-labeled N cDNA probe followed by autoradiography. Results are representative of three independent experiments.
Viral primary transcription is inhibited by a novel antiviral pathway(s) induced by both IFN-α and IFN-γ.
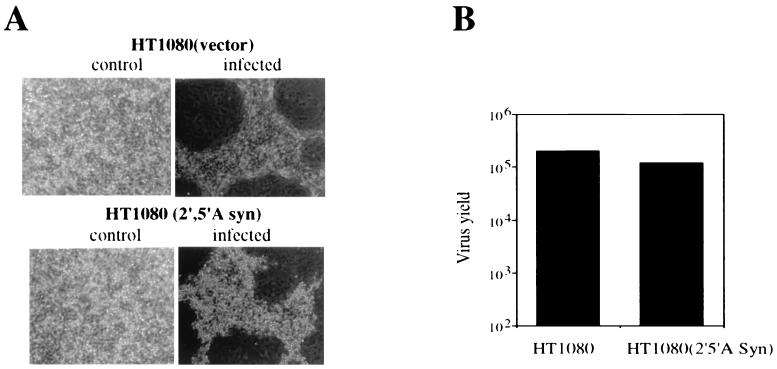
To begin identification of the antiviral pathway(s) that inhibit HPIV3 transcription, we first focused on the other well-characterized antiviral pathways. We determined the contribution of the 2-5A synthetase/RNase L pathway in the inhibition of primary transcription. Virus replication was examined in cells constitutively expressing 2-5A synthetase (16). Cells constitutively expressing a 69-kDa form of 2-5A synthetase were infected with HPIV3 at 0.1 MOI, and virus production was determined at 24 h postinfection. Empty-vector-transfected cells were used as the control. As shown in Fig. 4A, robust syncytium formation was seen in both 2-5A synthetase-expressing and control vector-transfected cells. By plaque assay, we found that the levels of production of progeny virions in these cells were virtually similar, indicating that the 2-5A synthetase/RNase L pathway has no role in the antiviral action of IFN-α against HPIV3 (Fig. 4B). Involvement of PKR in the IFN-α-mediated inhibition of viral primary transcription can be ruled out by the fact that primary transcription was studied in the presence of protein synthesis inhibitor CHX. These data indicate that an antiviral pathway(s) other than the three established antiviral pathways, hereafter referred to as a novel antiviral pathway(s), contributes to the IFN-α action against HPIV3 by inhibiting the viral primary transcription.
FIG. 4.
Replication of HPIV3 in cells constitutively expressing 2–5A synthetase. Cells constitutively expressing a 69-kDa form of 2–5A synthetase (HT1080 2–5A syn) and control vector-transfected cells (HT1080-Vector or HT1080) were infected with HPIV3 at an MOI of 0.1. (A) syncytium formation at 24 h postinfection. (B) Production of progeny virions. Infectious virus release was measured by plaque assay in the culture medium at 24 h postinfection. The results are representative of three independent assays.
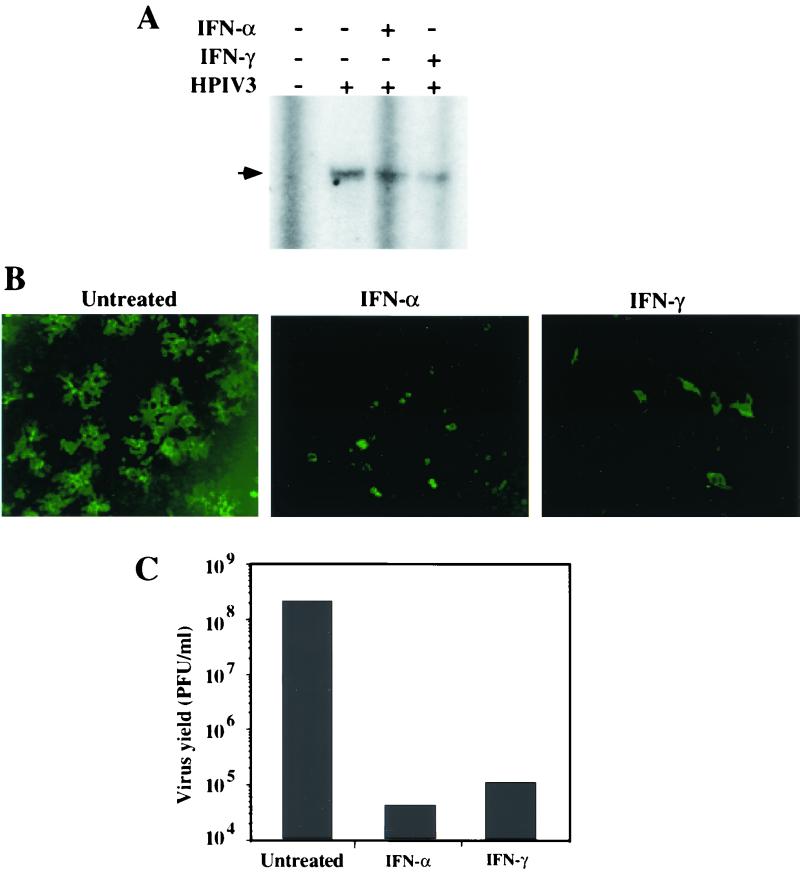
In light of differential induction of some of the antiviral proteins by IFN-α and IFN-γ (11, 32), we investigated the inhibition of HPIV3 primary transcription in IFN-α- and IFN-γ-treated cells. A549 cells were separately pretreated with IFN-α and IFN-γ and infected with HPIV3 at an MOI of 5. Cells were then incubated in the presence of CHX. At 6 h postinfection, cells were harvested and the accumulation of viral N mRNA was determined by primer extension analysis. As shown in Fig. 5A, both IFN-α and IFN-γ inhibited the N mRNA accumulation. PhosphorImager quantitation revealed that inhibitions of N mRNA by IFN-α and IFN-γ were each about 55%. The similarity in the levels of inhibition of N mRNA by IFN-α and IFN-γ suggests that the novel antiviral pathway(s) is induced by both IFN-α and IFN-γ. Moreover, MxA had no influence on the antiviral activity of the novel pathway(s) because its induction was strictly mediated by IFN-α (data not shown).
FIG. 5.
Antiviral effects of IFN-α and IFN-γ against HPIV3 in A549 cells. (A) Effects of IFNs on the primary transcription of HPIV3. A549 cells were pretreated with IFN-α or IFN-γ at 1,000 U/ml for 12 h followed by CHX (10 μg/ml) for 2 h. The cells were infected with HPIV3 at a MOI of 5 and incubated in the presence of IFN and CHX. At 6 h postinfection, cells were harvested and accumulation of N mRNA was determined by primer extension analysis as described in Materials and Methods. The arrowhead indicates the 93-nucleotide extension product representing N mRNA synthesis. (B) Effects of IFNs on the accumulation of intracellular viral RNP. The cells, grown on coverslips, were treated with IFNs for 12 h followed by infection with HPIV3 at an MOI of 1.0. At 12 h postinfection, the cells were fixed, permeabilized, and stained with anti-RNP antibody followed by FITC-conjugated secondary antibody as described in Materials and Methods. (C) Effects of IFNs on the production of infectious HPIV3 virions. The cells were treated with IFNs (1,000 U/ml) for 12 h followed by infection with HPIV3 at an MOI of 0.1. At 40 h postinfection, the release of infectious virions was measured by plaque assay. Results are representative of three independent experiments.
To determine whether intracellular viral RNP was similarly decreased, A549 cells were separately treated with IFN-α and IFN-γ for 12 h and subsequently infected with HPIV3 at an MOI of 0.1. At 12 h postinfection, intracellular viral RNP was detected by immunofluorescent labeling using anti-RNP antibody. As shown in Fig. 5B, intracellular viral RNP was significantly decreased in both IFN-α- and IFN-γ-treated cells.
To determine whether the effects of IFN-α and IFN-γ on primary transcription were reflected at the level of production of progeny virions, we carried out plaque assay. The A549 cells were pretreated separately with IFN-α and IFN-γ and subsequently infected with HPIV3 at an MOI of 0.1. At 40 h postinfection, release of infectious virions in the culture medium was determined by plaque assay. As shown in Fig. 5C, both IFN-α and IFN-γ inhibited the virus yield by more than 3 log. Together, these data indicate that both IFN-α and IFN-γ induce a novel antiviral pathway(s) to inhibit HPIV3 primary transcription.
IFN-γ can develop antiviral state against HPIV3 in IFN-α-signaling mutant cells.
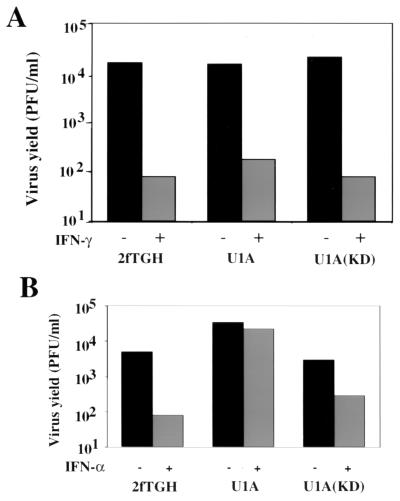
Previous reports indicated that the antiviral effect of IFN-γ against some viruses is influenced by the IFN-α/β signaling pathway (3, 10, 34). To determine whether the antiviral effect of IFN-γ against HPIV3 is similarly influenced by the IFN-α/β signaling pathway, IFN-α-signaling mutant cells U1A and U1A(KD) were used; they are defective in IFN-α signaling fully and partially, respectively (15). Empty-vector-transfected cells 2fTGH were used as the control. The cells were pretreated with IFN-γ and then infected with HPIV3 at an MOI of 0.1. To confirm the defects in IFN-α signaling, these cells were similarly treated with IFN-α and infected with HPIV3. At 24 h postinfection, production of infectious virions was quantitated by plaque assay. As shown in Fig. 6A, IFN-γ inhibited infectious virus production in the 2fTGH cells by more than 2 log, and similar inhibition was seen in the mutant cells U1A and U1A(KD). In contrast, IFN-α inhibited the virus production by about 2 log in 2fTGH cells but failed to do so in U1A cells (Fig. 6B). Consistent with the previous report that IFN-α signaling is partially restored in U1A(KD) cells (15), the antiviral effect of IFN-α against HPIV3 was also less pronounced (inhibited by 1 log) in these cells (Fig. 6B). These results indicate that IFN-γ-mediated inhibition of HPIV3 multiplication can occur efficiently without a requirement of synergism or cross talk with the IFN-α-signaling pathway as reported in some other viral systems (3, 10, 34).
FIG. 6.
Effects of IFN-α and IFN-γ on HPIV3 replication in IFN-α-signaling mutant cells. (A) Effect of IFN-α on the production of infectious HPIV3 virions. The cells were treated with IFN-α (1,000 U/ml) for 12 h followed by infection with HPIV3 at an MOI of 0.1. At 24 h postinfection, the release of progeny virions was measured by plaque assay. (B) Effect of IFN-γ on the production of HPIV3 virions. The cells were pretreated with IFN-α and infected with HPIV3 as above. At 24 h postinfection, the release of progeny virions was measured. U1A and U1A(KD) represent the Tyk2-null and Tyk2-kinase-defective cells, respectively. 2fTGH represents the empty-vector-transfected cells. Results are representative of three independent experiments.
DISCUSSION
In this communication we have shown that IFN-α-induced MxA and a pathway(s) besides the three well-characterized antiviral pathways, referred to as a novel antiviral pathway(s), contribute to the antiviral action of IFN-α against HPIV3. The novel antiviral pathway(s) targets the viral primary transcription, while MxA targets the steps following primary transcription of the virus multiplication cycle. Moreover, our data suggest that both IFN-α and IFN-γ induced the novel antiviral pathway(s), and consequently both cytokines developed effective antiviral states against HPIV3. By using IFN-α-signaling mutant cells, we found that the IFN-γ-mediated antiviral effect against HPIV3 does not require a synergism or cross talk with the IFN-α signaling pathway (3, 10, 34).
The role of MxA against HPIV3 is similar to the findings with vesicular stomatitis virus (VSV) and measles virus, belonging to the group of nonsegmented negative-strand RNA viruses of families Rhabdoviridae and Paramyxoviridae, respectively (28–30). MxA has also been shown to inhibit multiplication of other viruses such as influenza virus, Thogoto virus, La Crosse virus, and Semliki Forest virus, representing different virus families (12, 18, 20, 22). Despite this inhibitory potential of MxA against a broad range of viruses, the precise mechanism of the inhibition remains unclear. In the case of HPIV3, the viral multiplication is inhibited in U87-MxA cells at the steps following primary transcription, perhaps replication and budding (38). Likewise, MxA was shown to inhibit measles virus (29) and Semliki Forest virus (22) multiplication by targeting a step following primary transcription. In the case of measles virus, the viral envelope glycoprotein mRNA translation was affected, whereas for the Semliki Forest virus the replication step was targeted. Thus, it remains to be seen whether HPIV3 envelope glycoprotein mRNA translation is similarly affected by MxA, resulting in an impairment of virus budding. Our immunofluorescent and immunoprecipitation analyses of viral RNP in U87-MxA cells (Fig. 2) indeed suggest such a possibility because the intracellular RNP level, although significantly reduced in U87-MxA cells, was not sufficient to account for the dramatic reduction of infectious virus production. This suggested a role for MxA in the inhibition of HPIV3 glycoprotein mRNA translation, thereby affecting virus budding. In addition, the inhibition of RNP accumulation in U87-MxA cells, compared to U87-CL4 cells, indicates that the HPIV3 replication step, as in the Semliki Forest virus (22), may be affected. In that case, as observed with the Semliki Forest virus, the HPIV3 RNA polymerase could be a target for MxA. Further studies are needed to investigate these possibilities.
The HPIV3 primary transcription was dramatically inhibited in U87-MxA cells following IFN-α treatment but not by the constitutively expressed MxA (Fig. 1A). This indicated that there was no defect in the cell type per se but rather that MxA targeted a step other than primary transcription. However, our experiments cannot rule out the possibility that MxA is able to inhibit HPIV3 primary transcription in other cell types in a manner similar to what is observed with measles virus (28). Importantly, these findings indicated a role for a novel antiviral pathway(s) in the inhibition of HPIV3 primary transcription. Moreover, our data clearly indicate that the novel antiviral pathway is able to inhibit HPIV3 primary transcription to establish an effective antiviral state in the absence of MxA (Fig. 3). Thus, it is apparent that different antiviral proteins target at least two different steps of the HPIV3 multiplication cycle. This is not surprising, because a large number of studies indicated that the concerted actions of several antiviral proteins are involved in the IFN-induced antiviral state against any given virus, and some of these proteins may perform partially overlapping functions (5, 19, 24, 27, 31–33, 36). In the case of nonsegmented negative-strand RNA viruses, MxA was found to inhibit the VSV primary transcription (30), while PKR targeted the mRNA translation step (23). Newcastle disease virus (NDV) multiplication was similarly inhibited by IFN by targeting the viral primary transcription and envelope glycoprotein mRNA translation, although the antiviral proteins have not been characterized (37). In agreement with these findings, in the case of HPIV3, MxA was found to inhibit a posttranscriptional step, while a novel antiviral protein inhibited the primary transcription. Moreover, constitutively expressed MxA in U87-MxA cells showed less-pronounced inhibition of HPIV3 at a higher MOI (38). But IFN-α treatment of the same cell type induced the novel antiviral protein to target primary transcription, and as a result, establishment of an effective antiviral state was seen. This clearly indicated that the novel antiviral protein plays a major role in the IFN action against HPIV3.
Most of the IFN antiviral studies in the past were focused on determining the role of the three antiviral protein pathways; however, the relative contributions of individual IFN-induced proteins against a particular virus were not assessed. Recently, a study generating single-, double-, and triple-knockout mice (39) has assessed the contributions of the three antiviral pathways to IFN action. These studies indicated that although the three antiviral pathways contributed significantly, alternative pathways of IFN action were found to play a role against VSV and encephalomyocarditis virus. Our data clearly indicate that such a novel antiviral pathway is operative against HPIV3, targeting the viral primary transcription. We noted that primary transcription is inhibited more strongly in U87 cells (Fig. 1) than in A549 cells (Fig. 3 and 5). The reason for this difference is presently unclear. Nonetheless, the novel antiviral pathway(s) plays a major role in the antiviral action of both IFN-α and IFN-γ, unlike MxA and many other antiviral proteins which are exclusively induced by IFN-α (5, 11, 19, 24, 27, 31–33, 36, 35). Our data, however, cannot rule out the possibility that different proteins are involved in these two cases, although similar inhibition levels of primary transcription and infectious virus production by IFN-α and IFN-γ argue against this possibility. Thus, it seems that the novel antiviral protein plays an important role in the IFN-γ action against HPIV3. Moreover, this antiviral action of IFN-γ does not require a synergism or cross talk with the IFN-α/β signaling pathway (3, 10, 34) because the antiviral effect is seen also in U1A cells.
In conclusion, our results provide evidence that a novel antiviral pathway(s) is involved in the inhibition of an RNA virus transcription. The novel antiviral pathway(s) appears to play a major role in the antiviral action of IFNs against HPIV3. Thus, our findings open up a new area of research looking into the interaction of novel antiviral proteins with the viral minimal transcription and replication unit to mediate IFN-induced antiviral effect. In the case of HPIV3, the recently developed in vitro transcription and in vivo minigenome replication as well as protein-protein interaction systems (9) that involve RNP containing three viral proteins, N, P, and L, can be used as a tool. Further studies towards identification and characterization of the novel antiviral protein(s) are under way.
ACKNOWLEDGMENTS
We thank Amiya K. Banerjee for constructive criticisms and valuable suggestions during this work. We thank George R. Stark for providing the IFN-signaling mutant cell lines. We thank Ganes C. Sen and Arundhuti Ghosh for providing cell lines constitutively expressing 2-5A synthetase. We thank Mairead Commane for help in maintaining the IFN-signaling mutant cells.
This work was supported by U.S. Public Health Service grant AI3207.
REFERENCES
- 1.Aebi M, Fah J, Hurt N, Samuel C E, Thomis D, Bazzigher L, Pavlovic J, Haller O, Staeheli P. cDNA structures and regulation of two interferon-induced human Mx proteins. Mol Cell Biol. 1989;9:5062–5072. doi: 10.1128/mcb.9.11.5062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Banerjee A K, Barik S, De B P. Gene expression of negative strand RNA viruses. Pharmacol Ther. 1991;51:47–70. doi: 10.1016/0163-7258(91)90041-j. [DOI] [PubMed] [Google Scholar]
- 3.Cantell K, Pirhonen J. IFN-gamma enhances production of IFN-alpha in human macrophages but not in monocytes. J Interferon Cytokine Res. 1996;16:461–463. doi: 10.1089/jir.1996.16.461. [DOI] [PubMed] [Google Scholar]
- 4.Collins P L, Chanock R M, McIntosh K. Parainfluenza viruses. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 1205–1241. [Google Scholar]
- 5.Darnell J E, Jr, Kerr I M, Stark G R. Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science. 1994;264:1415–1421. doi: 10.1126/science.8197455. [DOI] [PubMed] [Google Scholar]
- 6.De B P, Burdsall A L, Banerjee A K. Role of cellular actin in human parainfluenza virus type 3 genome transcription. J Biol Chem. 1993;268:5703–5710. [PubMed] [Google Scholar]
- 7.De B P, Lesoon A, Banerjee A K. Human parainfluenza virus type 3 transcription in vitro: role of cellular actin in mRNA synthesis. J Virol. 1991;65:3268–3275. doi: 10.1128/jvi.65.6.3268-3275.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.De B P, Gupta S, Gupta S, Banerjee A K. Cellular protein kinase C ζ regulates human parainfluenza virus type 3 replication. Proc Natl Acad Sci USA. 1995;92:5204–5208. doi: 10.1073/pnas.92.11.5204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De B P, Hoffman M A, Choudhary S, Huntley C C, Banerjee A K. Role of NH2- and COOH-terminal domains of the P protein of human parainfluenza virus type 3 in transcription and replication. J Virol. 2000;74:5886–5895. doi: 10.1128/jvi.74.13.5886-5895.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DeMaeyer E, DeMaeyer-Guignard J. Interferons and other regulatory cytokines. New York, N.Y: Wiley; 1988. [Google Scholar]
- 11.Der S D, Zhou A, Williams B R G, Silverman R H. Identification of genes differentially regulated by interferon α, β, or γ using oligonucleotide arrays. Proc Natl Acad Sci USA. 1998;95:15623–15628. doi: 10.1073/pnas.95.26.15623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frese M, Kochs G, Feldmann H, Hertkorn C, Haller O. Inhibition of bunyaviruses, phleboviruses, and hantaviruses by human MxA protein. J Virol. 1996;70:915–923. doi: 10.1128/jvi.70.2.915-923.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frese M, Kochs G, Meier-Dieter U, Siebler J, Haller O. Human MxA protein inhibits tick-borne Thogoto virus but not Dhori virus. J Virol. 1995;69:3904–3909. doi: 10.1128/jvi.69.6.3904-3909.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frese M, Weeber M, Weber F, Speth V, Haller O. Mx1 sensitivity: Batken virus is an orthomyxovirus closely related to Dhori virus. J Gen Virol. 1997;78:2453–2458. doi: 10.1099/0022-1317-78-10-2453. [DOI] [PubMed] [Google Scholar]
- 15.Gauzzi M C, Velazquez L, McKendry R, Mogensen K E, Fellous M, Pellegrini S. Interferon-α-dependent activation of Tyk2 requires phosphorylation of positive regulatory tyrosines by another kinase. J Biol Chem. 1996;271:20494–20500. doi: 10.1074/jbc.271.34.20494. [DOI] [PubMed] [Google Scholar]
- 16.Ghosh A, Sarkar S N, Sen G C. Cell growth regulatory and antiviral effects of the P69 isozyme of 2–5 (A) synthetase. Virology. 2000;266:319–328. doi: 10.1006/viro.1999.0085. [DOI] [PubMed] [Google Scholar]
- 17.Gupta S, De B P, Banerjee A K. Involvement of actin microfilaments in the replication of human parainfluenza virus type 3. J Virol. 1998;72:2655–2662. doi: 10.1128/jvi.72.4.2655-2662.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kanerva M, Melen K, Vaheri A, Julkunen I. Inhibition of puumala and tulu hantaviruses in vero cells by MxA protein. Virology. 1996;224:55–62. doi: 10.1006/viro.1996.0506. [DOI] [PubMed] [Google Scholar]
- 19.Kerr I M, Stark G R. The antiviral effects of the interferons and their inhibition. J Interferon Res. 1992;12:237–240. doi: 10.1089/jir.1992.12.237. [DOI] [PubMed] [Google Scholar]
- 20.Kochs G, Haller O. Interferon-induced human MxA GTPase blocks nuclear import of Thogoto virus nucleocapsids. Proc Natl Acad Sci USA. 1999;96:2082–2086. doi: 10.1073/pnas.96.5.2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lamb R A, Kolakofsky D. Paramyxoviridae: the viruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 1177–1204. [Google Scholar]
- 22.Landis H, Simon-Jodicke A, Kloti A, Di Paolo C, Schnorr J J, Schneider-Schaulies S, Hefti H P, Pavlovic J. Human MxA protein confers resistance to Semliki Forest virus and inhibits the amplification of a Semliki Forest virus-based replicon in the absence of viral structural proteins. J Virol. 1998;72:1516–1522. doi: 10.1128/jvi.72.2.1516-1522.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee S B, Bablanian R, Esteban M. Regulated expression of the interferon-induced protein kinase p68 (PKR) by vaccinia virus recombinants inhibits the replication of vesicular stomatitis virus but not that of poliovirus. J Interferon Cytokine Res. 1996;16:1073–1078. doi: 10.1089/jir.1996.16.1073. [DOI] [PubMed] [Google Scholar]
- 24.McNair A N B, Kerr I M. Viral inhibition of the interferon system. Pharmacol Ther. 1992;56:79–95. doi: 10.1016/0163-7258(92)90038-2. [DOI] [PubMed] [Google Scholar]
- 25.Pavlovic J, Haller O, Staeheli P. Human and mouse Mx proteins inhibit different steps of the influenza virus multiplication cycle. J Virol. 1992;66:2564–2569. doi: 10.1128/jvi.66.4.2564-2569.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pavlovic J, Zurcher T, Haller O, Staeheli P. Resistance to influenza virus and vesicular stomatitis virus conferred by expression of human MxA protein. J Virol. 1990;64:3370–3375. doi: 10.1128/jvi.64.7.3370-3375.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Samuel C E. Antiviral actions of interferon: interferon-regulated cellular proteins and their surprisingly selective antiviral activities. Virology. 1991;183:1–11. doi: 10.1016/0042-6822(91)90112-o. [DOI] [PubMed] [Google Scholar]
- 28.Schneider-Schaulies S, Schneider-Schaulies J, Schuster A, Bayer M, Pavlovic J, ter Meulen V. Cell type-specific MxA-mediated inhibition of measles virus transcription in human brain cells. J Virol. 1994;68:6910–6917. doi: 10.1128/jvi.68.11.6910-6917.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schnorr J J, Schneider-Schaulies S, Simon-Jodicke A, Pavlovic J, Horisberger M A, ter Meulen V. MxA-dependent inhibition of measles virus glycoprotein synthesis in a stably transfected human monocytic cell line. J Virol. 1993;67:4760–4768. doi: 10.1128/jvi.67.8.4760-4768.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schwemmle M, Weining K C, Richter M F, Schumacher B, Staeheli P. Vesicular stomatitis virus transcription is inhibited by purified MxA protein. Virology. 1995;206:545–554. doi: 10.1016/s0042-6822(95)80071-9. [DOI] [PubMed] [Google Scholar]
- 31.Sen G C, Lengyel P. The interferon system: a bird's eye view of its biochemistry. J Biol Chem. 1992;267:5017–5020. [PubMed] [Google Scholar]
- 32.Staeheli P. Interferon-induced proteins and the antiviral state. Adv Virus Res. 1990;38:147–200. doi: 10.1016/s0065-3527(08)60862-3. [DOI] [PubMed] [Google Scholar]
- 33.Stark G R, Kerr I M, Williams B R G, Silverman R H, Schreiber R D. How cells respond to interferons. Annu Rev Biochem. 1998;67:227–264. doi: 10.1146/annurev.biochem.67.1.227. [DOI] [PubMed] [Google Scholar]
- 34.Takaoka A, Mitani Y, Suemori H, Sato M, Yokochi T, Noguchi S, Tanaka N, Taniguchi T. Cross talk between interferon-γ and -α/β signaling components in caveolar membrane domains. Science. 2000;288:2357–2360. doi: 10.1126/science.288.5475.2357. [DOI] [PubMed] [Google Scholar]
- 35.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vilcek J, Sen G C. Interferons and other cytokines. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 375–399. [Google Scholar]
- 37.Yu Z, Gotoh B, Hamaguchi M, Nagai Y. Antiviral action of interferon-beta on Newcastle disease virus: selectivity to the hemagglutinin-neuraminidase gene expression. Med Microbiol Immunol. 1995;184:45–52. doi: 10.1007/BF00216789. [DOI] [PubMed] [Google Scholar]
- 38.Zhao H, De B P, Das T, Banerjee A K. Inhibition of human parainfluenza virus-3 replication by interferon and human MxA. Virology. 1996;220:330–338. doi: 10.1006/viro.1996.0321. [DOI] [PubMed] [Google Scholar]
- 39.Zhou A, Jayashree M, Paranjape J, Der S D, Williams B R G, Silverman R H. Interferon action in triply deficient mice reveals the existence of alternative pathways. Virology. 1999;258:435–440. doi: 10.1006/viro.1999.9738. [DOI] [PubMed] [Google Scholar]