Abstract
Lizards and snakes putatively arose between the early Jurassic and late Triassic; they diversified worldwide and now occupy many different ecological niches, making them ideal for testing theories on the origin of ecological traits. We propose and test the “deep history hypothesis,” which claims that differences in ecological traits among species arose early in evolutionary history of major clades, and that present-day assemblages are structured largely because of ancient, preexisting differences. We combine phylogenetic data with ecological data collected over nearly 40 years to reconstruct the evolution of dietary shifts in squamate reptiles. Data on diets of 184 lizard species in 12 families from 4 continents reveal significant dietary shifts at 6 major divergence points, reducing variation by 79.8%. The most striking dietary divergence (27.6%) occurred in the late Triassic, when Iguania and Scleroglossa split. These two clades occupy different regions of dietary niche space. Acquisition of chemical prey discrimination, jaw prehension, and wide foraging provided scleroglossans access to sedentary and hidden prey that are unavailable to iguanians. This cladogenic event may have profoundly influenced subsequent evolutionary history and diversification. We suggest the hypothesis that ancient events in squamate cladogenesis, rather than present-day competition, caused dietary shifts in major clades such that some lizard clades gained access to new resources, which in turn led to much of the biodiversity observed today.
Keywords: cladogenesis, community ecology, historical ecology, squamate evolution
Squamate reptiles are ideal for testing theories on the origin of ecological traits. Their evolutionary history dates back to the early Jurassic or late Triassic (1–3), they have diversified on all major continents (4), and they occupy a remarkable diversity of ecological niches (4–7). One theory claims that ecological differences result from recent factors such as shifts in availability of different prey types or interspecific competition (competition hypothesis) in which species interactions for harvesting limited resources cause divergence in niche characteristics [food, time, or place (microhabitat)] (6, 8). This theory predicts that niche differences arose within a relatively recent ecological time frame. Evidence supporting this theory is of two types: (i) demonstrations that assemblages containing evolutionarily different species separate on at least one of three major niche axes (6), and (ii) demonstrations that species (e.g., Anolis lizards on islands) respond in predictable manners to introduction of ecologically and evolutionarily similar species, shifting microhabitats and/or diets (9, 10). Shifts in availability of different prey types for reasons other than competition could also stimulate diet shift under such a model (adaptation to current conditions). Such changes would be considered “shallow” in a phylogenetic sense because they occurred within an ecological time framework. Another theory claims that differences in ecological traits among species arose early in the evolutionary history of major clades (“deep history hypothesis”) such that present-day assemblages may coexist largely because of ancient preexisting differences (7, 11, 12). Evidence supporting this theory includes (i) demonstrations that dietary niche overlaps correlate with phylogenetic similarity in Amazonian lizards (7) and (ii) consistency of diets within South American xenodontine snakes independent of the assemblages in which they occur (11). Although most organismal biologists would expect trophic interactions to have a deep historical basis, our analysis ties one component of niche partitioning (diet) to cladogenic events in the Mesozoic.
In an earlier analysis, we and our colleagues suggested an association between ingestion of specific insects and a combination of feeding mechanisms and prey discrimination modes (7). Here, we use the best available data on lizard diets (184 species in 12 families from 4 continents) and alternate phylogenetic hypotheses of their relationships to quantitatively test two predictions of the deep history hypothesis: (i) a strong relationship between diet and evolutionary relationships of squamate reptiles should exist and (ii) nodes at which major dietary shifts occurred can be identified.
We chose lizard diets to test the deep history hypothesis because food is a major niche axis on which lizards separate in contemporary assemblages (5, 6) and because data we have collected over the past 35 years cut across both the evolutionary history of lizards and their global distribution.
Materials and Methods
Diets. During the past four decades, we have collected dietary data on 184 lizard species, including 91 Neotropical and 93 desert species from southwestern deserts of the United States; tropical rainforests of Nicaragua, Ecuador, and Brazil; semiarid regions of northeastern Brazil (Caatinga); Australian deserts; and the Kalahari Desert of Africa. Detailed methods for collection of lizards and identification and measurements of prey appear elsewhere (5, 6, 13, 14). All lizards were treated in accordance with federal, state, and university regulations (Animal Care Assurance 73-R-100, approved November 8, 1994, University of Oklahoma). Initial prey categories for desert and Neotropical lizards were nearly identical, which allowed us to reanalyze our data at various taxonomic levels. The original Neotropical lizard data set included 30 broad prey types (13), whereas the original desert lizard data set included 20 broad prey types (6). Most differences in prey categories resulted from prey specific to each area. Mollusks and earthworms, for example were not found in desert lizard stomachs. We used an expanded data set of volumetric dietary data (27 categories) for all of our combined species to construct an all-species dietary matrix. Although this procedure enters zero values for prey taxa missing in each data set, such taxa were generally rare in all other lizard diets. Consequently, effects of differences in prey availability among sites were minimal. Diets of pooled lizard samples at all sites were dominated by 7–12 abundant prey categories. The most common 12 prey categories accounted for 90% of all lizards' diets, and the first 7 categories accounted for 76% of all lizards' diets.
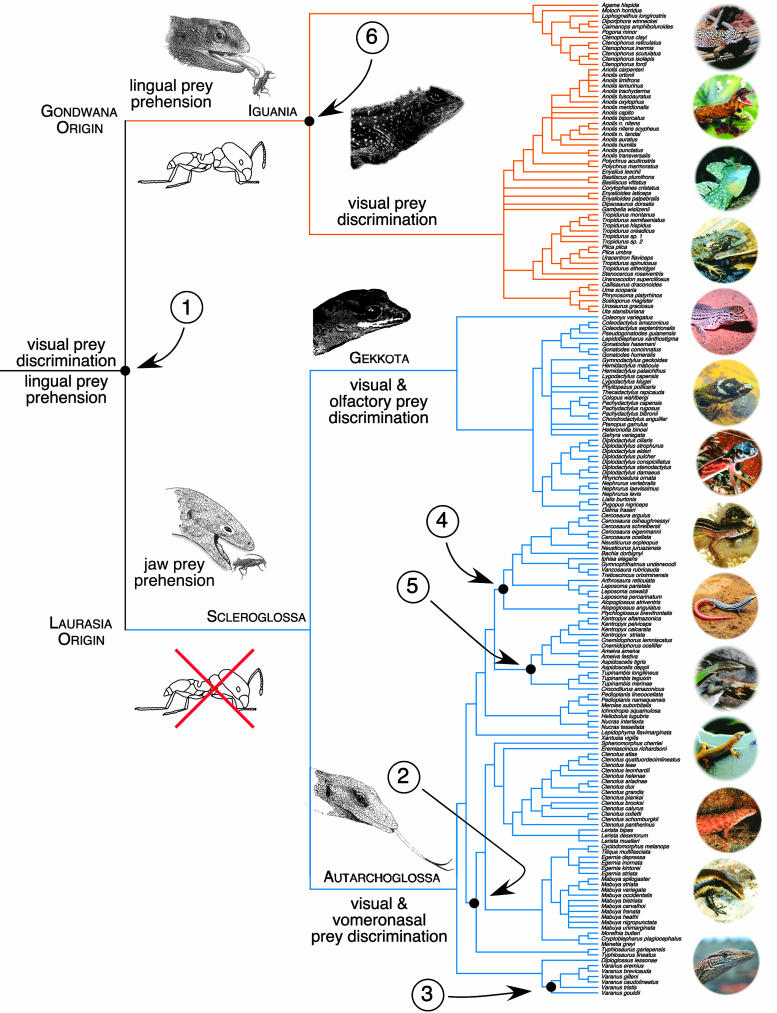
Phylogenetic Reconstruction. We constructed a composite phylogenetic hypothesis for the 184 lizard species based on 23 different studies (15–37) (Fig. 1), thus presenting a consensus view of lizard evolutionary history. For the present analysis, we consider species' membership in every clade at the family level or above. We also consider a recent competing phylogenetic hypothesis of squamate evolutionary history based on a combination of nuclear (RAG-1 and c-mos) and mitochondrial (ND2 region) genes, which suggests a nontraditional relationship among the three major clades, Iguania, Gekkota, and Autarchoglossa (38).
Fig. 1.
Phylogenetic hypothesis for 184 Neotropical and desert lizard species based on 23 different studies. Solid circles indicate six taxonomic groups that were significant in the stepwise canonical correspondence analysis (CCA); they are numerically labeled as follows: 1, Iguania/Scleroglossa; 2, Scincidae; 3, Varanidae; 4, Gymnophthalmidae; 5, Teiidae; 6, Agamidae/Iguanidae.
Ordination Analysis. To reconstruct the history of dietary change in lizards, we used CCA (39), a multivariate ordination procedure that directly associates variation in one matrix (lizard diets) with variation in another (lizard phylogeny). Relationships between ecological and phylogenetic characteristics of lizards have been previously examined by using such an analysis (40). Thus, here we ask whether an association exists between dietary composition and phylogeny, and we identify divergence points in the evolutionary history of lizards. CCA was performed with canoco 4.5 (41). The matrix consisting of 184 lizard species and proportional utilization coefficients of their combined 27 prey categories constituted the dependent variable. The independent variable was a matrix consisting of the 184 lizard species and their clade representations to the level of family. Because lizard size affects diet and covaries with clade, average snout-vent length of each lizard species was entered as a covariate. We used symmetric scaling and unimodal methods and downweighted rare prey categories. In the stepwise analysis, Monte Carlo permutation tests were performed on each variable by using 9,999 permutations. Each variable was tested manually one at a time to obtain F and P values. After each significant variable was included in the model, the subsequent variable that most reduced variance was tested and included if it was statistically significant (P < 0.05). This procedure was followed until subsequent variables were no longer significant.
Results and Discussion
Dietary variance was initially significantly reduced by 14 of the 19 clades, confirming a strong relationship between diet and evolutionary relationships of squamates (Table 1). We then tested individual clades one at a time, reduced residual variances, and repeated the process using the stepwise CCA analysis. This analysis revealed significant dietary shifts at six major divergence points (Table 2 and Fig. 1) reducing variation by a full 79.77% (Fig. 1). The six significant taxonomic groups in the CCA were Iguania/Scleroglossa, Scincidae, Varanidae, Gymnophthalmidae, Teiidae, and Agamidae/Iguanidae. The ordination faithfully represents the original data (only 20% of the variance is lost). Most striking is the diametrically opposed relationship between the two oldest squamate clades, Iguania and Scleroglossa (Figs. 2 and 3). Scleroglossans feed on prey types arranged at right angles to iguanian vectors. The most striking difference in diet between Iguania and Scleroglossa is that scleroglossans eat fewer ants, other hymenopterans, and beetles (4, 7). Reanalysis of our data using the alternative phylogenetic hypothesis (38) gave identical results with respect to the clades Iguania, Gekkota, and Autarchoglossa even though no “Scleroglossa” existed.
Table 1. Results of a phylogenetic CCA analysis on diets of 184 lizard species from New World tropics and both New and Old World deserts.
| Clade | Variation | Variation % | F value | P value |
|---|---|---|---|---|
| Iguania/Scleroglossa | 0.169 | 27.083 | 8.709 | 0.0001 |
| Iguanidae | 0.113 | 18.109 | 5.743 | 0.0001 |
| Varanidae | 0.105 | 16.827 | 5.306 | 0.0001 |
| Anguimorpha | 0.093 | 14.904 | 4.714 | 0.0001 |
| Scincidae | 0.089 | 14.263 | 4.463 | 0.0001 |
| Lacertoidea | 0.083 | 13.301 | 4.191 | 0.0001 |
| Gymnophthalmidae | 0.077 | 12.340 | 3.846 | 0.0002 |
| Scincomorpha | 0.075 | 12.019 | 3.753 | 0.0001 |
| Autarchoglossa | 0.074 | 11.859 | 3.704 | 0.0001 |
| Agamidae | 0.074 | 11.859 | 3.689 | 0.0006 |
| Teiidae | 0.052 | 8.333 | 2.570 | 0.0065 |
| Gekkota | 0.050 | 8.013 | 2.493 | 0.0027 |
| Lacertidae | 0.044 | 7.051 | 2.177 | 0.0465 |
| Gekkonidae | 0.040 | 6.410 | 1.996 | 0.0215 |
| Anguidae | 0.033 | 5.288 | 1.613 | 0.1520 |
| Pygopodidae | 0.033 | 5.288 | 1.617 | 0.1436 |
| Diplodactylidae | 0.020 | 3.205 | 0.972 | 0.4343 |
| Xantusiidae | 0.008 | 1.282 | 0.410 | 0.7941 |
| Eublepharidae | 0.004 | 0.641 | 0.213 | 0.9221 |
| Total variation | 0.630 |
Results are ranked by amount of residual variation reduced by each clade. Fourteen of the 19 clades significantly reduced variance.
Table 2. Summary of effects each clade had on reducing variation in diet.
| After inclusion of clades | Variation | Variation % | F value | P value |
|---|---|---|---|---|
| Iguania/Scleroglossa | 0.169 | 27.57 | 8.71 | 0.0001 |
| Scincidae | 0.097 | 15.82 | 5.11 | 0.0001 |
| Varanidae | 0.081 | 13.21 | 4.36 | 0.0003 |
| Gymnophthalmidae | 0.057 | 9.30 | 3.11 | 0.0006 |
| Teiidae | 0.046 | 7.50 | 2.54 | 0.0045 |
| Agamidae/Iguanidae | 0.039 | 6.36 | 2.14 | 0.0209 |
Results of stepwise CCA after inclusion of significant nested clades. Variation in diets is reduced by 79.8% by the six clades with P < 0.05.
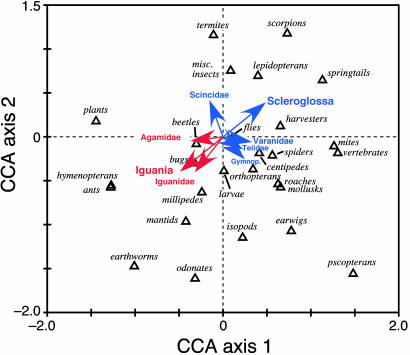
Fig. 2.
Biplot showing results of a canonical correspondence ordination analysis. This plot shows the position of each prey category on the first two axes of dietary niche space. Prey types that are eaten together are close to each other on this plot, whereas those that are seldom eaten by the same lizard species are far apart. The origin at 0.0, 0.0 represents the lowest common denominator or the overall lizard diet summed across all 184 species. Dietary generalists with broad food niches would lie in the interior of the plot near the origin, and specialists would be on peripheral areas of the diagram. Clades that significantly reduced variation are plotted with vectors radiating out from the origin. Lengths of vector arrows indicate significance strength, whereas tips represent the centroid of the prey types eaten by each clade. Scleroglossan clades are in blue, and iguanians clades are in red.
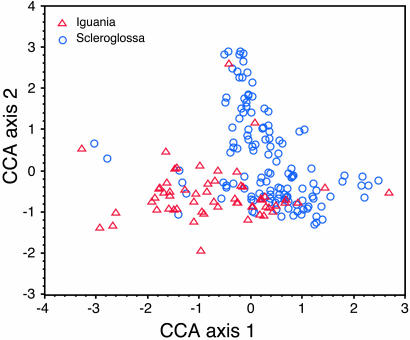
Fig. 3.
Plot showing positions of each species of iguanians (red triangles) and scleroglossans (blue circles) in the first two canonical correspondence axes of dietary niche space.
The first and most dramatic dietary divergence (27.57% of variation) occurred in late Triassic, when Iguania and Scleroglossa diverged from a pleurodont ancestor (3). The transition from the ancestral (iguanian-like) to the derived (scleroglossans) state (conventional phylogenetic hypothesis) includes transitions from (i) lingual to jaw prehension, (ii) dependence on visual cues to combined use of visual and chemical cues for prey detection and discrimination, (iii) ambush foraging mode to a more active foraging mode, and (iv) a plethora of behavioral, physiological, and morphological differences heretofore attributed to foraging mode (4, 7, 42, 43). Our results suggest that long ago acquisition of chemical prey discrimination, jaw prehension, and wide foraging opened up a new food resource base for scleroglossans, providing them access to sedentary and hidden prey that were unavailable to iguanians. Moreover, these dietary differences have persisted to the present day. This quantitative result is identical to the qualitative result reported earlier (7). Whether prey discrimination by chemical cues resulted in dropping many insects containing chemical defense mechanisms from lizard diets or simply resulted in selection of more profitable prey remains unexplored, although experimental evidence demonstrating that scleroglossans discriminate against foods containing alkaloids suggests the former (44, 45).
Iguanians occupy a different region in the CCA plot than do scleroglossans (Fig. 3). Iguanians feed on large numbers of ants, other hymenopterans, and beetles, prey that are detected visually and thus available to scleroglossans as well. Scleroglossans not only appear to avoid ants, other hymenopterans, and beetles, but they also feed on many prey types that are relatively unavailable to iguanians (termites, certain larvae, pupae, etc.). The great diversity of scleroglossan diets reflects the impact of chemical discrimination of prey, jaw prehension, and a more active lifestyle on ability to locate, discriminate, and handle cryptic or hidden prey.
Deep history of squamate reptiles appears to have played a profound role in determining lizard diets and accounts for a large portion of putative “niche partitioning” observed in phylogenetically diverse lizard assemblages throughout the world. Even though observed niche differences have ancient roots, partitioning of currently available niche space could still be strongly affected by the relative competitive abilities of current taxa that retain ancient differences. Current competition could still influence which taxa are present, although history has clearly determined many of the traits extant species possess. Species interactions must certainly drive niche segregation in lizard assemblages containing closely related species such as Ctenotus skinks in Australia and Anolis on islands. The impact of history on ecological characteristics can be visualized as a hierarchical network in which historical effects are greatest and species interactions are least among phylogenetically distant species, whereas, conversely, historical effects are minimal and species interactions are greatest among phylogenetically similar species occupying the same area.
Moreover, events that occurred in the remote past may have strongly influenced much of squamate biodiversity observed today. A biodiversity response is suggested when examining the impact of the Scleroglossa–Iguania divergence on the evolutionary history and diversification of squamates. These two sister clades are by definition the same age, but Iguania produced only about 1,230 extant species, whereas Scleroglossa produced about 6,000, half of which are “snakes.” As impressive as this difference is, it may have resulted from chance. Based on a Markov chain null model, and starting with the basal pair of branches (Iguania versus Scleroglossa), the three branch stage in the Markovian process sets in motion a cascade of increasing diversity along the original branch that first split, such that the disparity is maintained throughout the branching process. The expectation would be unequal clade representation. Thus an explanation may not be necessary. However, this expectation also does not rule out the possibility that greater diversification in one clade was driven by nonrandom factors. More impressive and more relevant to diversification are morphological and ecological shifts that have occurred repeatedly within Scleroglossa but never or rarely in Iguania. Limblessness has evolved repeatedly, with some scleroglossan species shifting to subterranean habits (46); nocturnality has evolved repeatedly (4, 47); and aquatic habits have evolved repeatedly, including diversification of one clade in warm oceans (sea snakes) (47). Limblessness, nocturnality, and subterranean lifestyles have never evolved in Iguania. Only a single iguanian species (Amblyrhynchus cristatus) enters an ocean, and it spends most of its time on land (48). The same characteristics that resulted in dietary shifts and divergences in scleroglossans resulted in major ecological shifts and diversification such that extant squamate assemblages are made up of species with varying degrees of deep-rooted differences that permit coexistence.
Our analysis implies that diet partitioning might be one of the first ecological responses to evolutionary change (e.g., shift from visual to chemical prey detection and discrimination) appearing among “species” during early diversification of a clade. It could be argued that shifting from one prey item to another requires no concomitant changes in morphology, body size, or a plethora of other traits, all of which should respond to other selective pressures as well. Morphology of both Anolis (49, 50) and Tropidurus (51), for example, responds relatively rapidly to extreme changes in habitat structure. Nevertheless, diets of most Anolis species are more similar to each other than to other lizard clades, and the same is true for Tropidurus. Lizards in seasonal environments often switch food based on availability (52–54), and some lizards change diets ontogenetically (55, 56), but shifts occurring at a population level are not nearly as drastic as shifts we have identified, particularly that at the Iguania–Scleroglossa divergence. The advent of chemosensory-based searching behavior by scleroglossan ancestors must have provided them access to an immense and diverse set of resources that were in place and relatively untapped by extant diurnal vertebrates. Lizards using visual cues for prey detection and discrimination would not have access to these resources. Impressive dietary divergence does occur within some smaller present-day lizard clades, suggesting that it has not only occurred in the past, but it is ongoing. For example, dietary diversity among liolaemid (iguanians) lizard species in southern South America is high, including multiple origins of herbivory (57). These lizards apparently diverged in the absence of scleroglossans (4, 7). Whether the high degree of dietary divergence among liolaemids is associated with shifts in mechanisms of prey detection and discrimination remains to be studied.
Our data, like data from almost all studies that attempt “global” hypothesis testing, suffer from grossly inadequate taxon sampling. One hundred eighty-four species may sound impressive, but it represents only 4.25% of “lizards” and 2.25% of squamates. Nevertheless, gathering these data has taken us most of our lives.
Acknowledgments
We thank G. R. Colli and A. Gainsbury, who generously gave us much needed assistance and guidance with CCA; our many colleagues, especially A. Bauer, W. E. Cooper, Jr., and K. Schwenk, who helped shape our thinking on global ecology of lizards; and R. May, W. E. Cooper, Jr., and J. B. Losos for commenting on the manuscript. We also thank Craig Guyer, Nick Gotelli, and Mike Kaspari for commenting on our comparison of numbers of extant iguanians versus scleroglossans and Tracy Heath for providing an important reference. E.R.P. also thanks the staffs of the Department of Zoology at the University of Western Australia and the Western Australian Museum plus the staff of the Department of Conservation and Land Management (CALM). Work in Brazil resulting in collection of lizard diet data was supported by National Science Foundation Grants DEB-9200779, DEB-9505518, and DEB-0415430 to L.J.V. and J. P. Caldwell. Brazilian agencies contributing to logistics include Instituto Nacional de Pesquisas da Amazonica (INPA), Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq, Portaria MCT no. 170, de 28/09/94), Instituto Brasileiro do Meio Ambiente e dos Recursos Naturais Renováveis (IBAMA, permit no. 073/94-DIFAS), and Museu Paraense E. Goeldi in Belém. A research agreement between the Sam Noble Oklahoma Museum of Natural History and the Museu Paraense E. Goeldi in collaboration with Dr. T. C. S. Avila-Pires made this possible. E.R.P.'s research has been supported by grants from the National Geographic Society, the John Simon Guggenheim Memorial Foundation, a senior Fulbright Research Scholarship, the Australian-American Educational Foundation, the University Research Institute of the Graduate School at the University of Texas at Austin, the Denton A. Cooley Centennial Professorship in Zoology at the University of Texas at Austin, the U.S. National Science Foundation, and the U.S. National Aeronautics and Space Administration. L.J.V. and E.R.P. thank their respective universities for Big 12 Faculty Fellowships.
Author contributions: L.J.V. and E.R.P. designed research, performed research, analyzed data, and wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviation: CCA, canonical correspondence analysis.
References
- 1.Estes, R. (1983) in Advances in Herpetology and Evolutionary Biology: Essays in Honor of Ernest E. Williams, eds. Rhodin, G. J. & Miyata, K. (Museum of Comparative Zoology, Harvard Univ., Cambridge, MA), pp. 365–398.
- 2.Evans, S. E. (1993) Rev. Paleobiol. 7, 55–65. [Google Scholar]
- 3.Evans, S. E. (2003) Biol. Rev. 78, 513–551. [DOI] [PubMed] [Google Scholar]
- 4.Pianka, E. R. & Vitt, L. J. (2003) Lizards: Windows to the Evolution of Diversity (Univ. of California Press, Berkeley).
- 5.Pianka, E. R. (1973) Annu. Rev. Ecol. Syst. 4, 53–74. [Google Scholar]
- 6.Pianka, E. R. (1986) Ecology and Natural History of Desert Lizards: Analyses of the Ecological Niche and Community Structure (Princeton Univ. Press, Princeton).
- 7.Vitt, L. J., Pianka, E. R., Cooper, W. E., Jr., & Schwenk, K. (2003) Am. Nat. 162, 44–60. [DOI] [PubMed] [Google Scholar]
- 8.Schoener, T. W. (1977) in Biology of the Reptilia, eds. Tinkle, D. W. & Gans, C. (Academic, New York), pp. 35–136.
- 9.Schoener, T. W. (1968) Ecology 49, 704–726. [Google Scholar]
- 10.Schoener, T. W. (1975) Ecol. Monogr. 45, 232–258. [Google Scholar]
- 11.Cadle, J. E. & Greene, H. W. (1993) in Species Diversity in Ecological Communities: Historical and Geographical Perspectives, eds. Ricklefs, R. E. & Schluter, D. (Univ. Chicago Press, Chicago), pp. 281–293.
- 12.Mayden, R. L. (1988) Syst. Zool. 37, 329–355. [Google Scholar]
- 13.Vitt, L. J. & Zani, P. A. (1996) Can. J. Zool. 74, 1313–1335. [Google Scholar]
- 14.Vitt, L. J., Zani, P. A. & Espósito, M. C. (1999) Oikos 87, 286–294. [Google Scholar]
- 15.Brehm, A., Jesus, J., Pinheiro, M. & Harris, D. J. (2001) Mol. Phylogenet. Evol. 19, 311–316. [DOI] [PubMed] [Google Scholar]
- 16.Castoe, T. A., Doan, T. M. & Parkinson, C. L. (2004) Syst. Biol. 53, 448–469. [DOI] [PubMed] [Google Scholar]
- 17.Doan, T. M. (2003) Zool. J. Linn. Soc. 137, 101–115. [Google Scholar]
- 18.Fitzgerald, L. A., Cook, J. A. & Aquino, A. L. (1999) Copeia 1999, 894–905. [Google Scholar]
- 19.Frost, D. R. (1992) Am. Mus. Novit. 3033, 1–68. [Google Scholar]
- 20.Frost, D. R. & Etheridge, R. (1989) Misc. Publ. Mus. Nat. Hist. Univ. Kansas 81, 1–65. [Google Scholar]
- 21.Frost, D. R., Etheridge, R., Janies, D. & Titus, T. A. (2001) Am. Mus. Novit. 3343, 1–38. [Google Scholar]
- 22.Glor, R. E., Vitt, L. J. & Larson, A. (2001) Mol. Ecol. 10, 2661–2668. [DOI] [PubMed] [Google Scholar]
- 23.Irschick, D. J., Vitt, L. J., Zani, P. A. & Losos, J. B. (1997) Ecology 78, 2191–2203. [Google Scholar]
- 24.Jackman, T. R., Larson, A., De Queiroz, K. & Losos, J. B. (1999) Syst. Biol. 48, 254–285. [Google Scholar]
- 25.Jennings, W. B., Pianka, E. R. & Donnellan, S. (2003) Syst. Biol. 52, 757–780. [PubMed] [Google Scholar]
- 26.Kluge, A. G. (1983) Copeia 1983, 465–475. [Google Scholar]
- 27.Kluge, A. G. (1987) Misc. Publ. Mus. Zool. Univ. Michigan 173, 1–54. [Google Scholar]
- 28.Kluge, A. G. (1995) Am. Mus. Novit. 3139, 1–23. [Google Scholar]
- 29.Macey, J. R., Schulte, J. A., Larson, A., Ananjeva, N. B., Wang, Y., Pethiyagoda, R., Rastegar-Pouyani, N. & Papenfuss, T. J. (2000) Syst. Biol. 49, 233–256. [DOI] [PubMed] [Google Scholar]
- 30.Mausfeld, P., Vences, M., Schmitz, A. & Veith, M. (2000) Mol. Phylogenet. Evol. 17, 11–14. [DOI] [PubMed] [Google Scholar]
- 31.Melville, J., Schulte, J. A. & Larson, A. (2001) J. Exp. Zool. 291, 339–353. [DOI] [PubMed] [Google Scholar]
- 32.Melville, J., Schulte, J. A. & Larson, A. (2004) Biol. J. Linn. Soc. 82, 123–138. [Google Scholar]
- 33.Reeder, T. W. (2003) Mol. Phylogenet. Evol. 27, 384–397. [DOI] [PubMed] [Google Scholar]
- 34.Reeder, T. W., Cole, C. J. & Dessauer, H. C. (2002) Am. Mus. Novit. 3365, 1–61. [Google Scholar]
- 35.Savage, J. M. & Guyer, C. (1989) Amph-Rept. 10, 105–116. [Google Scholar]
- 36.Schulte, J. A., II., Valladares, J. P. & Larson, A. (2003) Herpetologica 59, 399–419. [Google Scholar]
- 37.Whiting, A., Bauer, A. M. & Sites, J. W. (2003) Mol. Phylogenet. Evol. 29, 582–598. [DOI] [PubMed] [Google Scholar]
- 38.Townsend, T. M., Larson, A., Louis, E. & Macey, J. R. (2004) Syst. Biol. 53, 745–757. [DOI] [PubMed] [Google Scholar]
- 39.Ter Braak, C. J. F. (1986) Ecology 67, 1167–1179. [Google Scholar]
- 40.Giannini, N. P. (2003) Syst. Biol. 52, 684–695. [DOI] [PubMed] [Google Scholar]
- 41.Ter Braak, C. J. F. & Smilauer, P. (2002) canoco Reference Manual and User's Guide to canoco for Windows: Software for Canonical Community Ordination (Microcomputer Power, Ithaca, NY), Version 4.5.
- 42.Huey, R. B. & Pianka, E. R. (1981) Ecology 62, 991–999. [Google Scholar]
- 43.Schwenk, K. (2000) in Feeding, ed. Schwenk, K. (Academic, San Diego), pp. 175–291.
- 44.Cooper, W. E., Jr., Pérez-Mellado, V., Vitt, L. J. & Budzinsky, B. (2002) Physiol. Behav. 76, 297–303. [DOI] [PubMed] [Google Scholar]
- 45.Cooper, W. E., Jr., Caldwell, J. P., Vitt, L. J., Pérez-Mellado, V. & Baird, T. A. (2002) Can. J. Zool. 80, 655–663. [Google Scholar]
- 46.Wiens, J. J. & Slingluff, J. L. (2001) Evolution 55, 2303–2318. [DOI] [PubMed] [Google Scholar]
- 47.Greene, H. W. (1997) Snakes. The Evolution of Mystery in Nature (Univ. of California Press, Berkeley).
- 48.Carpenter, C. C. (1966) Proc. Calif. Acad. Sci. 34, 329–376. [Google Scholar]
- 49.Losos, J. B., Warheit, K. I. & Schoener, T. W. (1997) Nature 387, 70–73. [Google Scholar]
- 50.Losos, J. B., Jackman, T. R., Larson, A., deq Ueiroz, K. & Rodríguez-Schettino, L. (1998) Science 279, 2115–2118. [DOI] [PubMed] [Google Scholar]
- 51.Vitt, L. J., Caldwell, J. P., Zani, P. A. & Titus, T. A. (1997) Proc. Natl. Acad. Sci. USA 94, 3828–3832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dunham, A. E. (1981) Misc. Publ. Mus. Zool. Univ. Michigan 158, 1–62. [Google Scholar]
- 53.Durtsche, R. D. (1992) Oecologia 89, 85–89. [DOI] [PubMed] [Google Scholar]
- 54.Vitt, L. J. (1991) in The Ecology of Desert Communities, ed. Polis, G. A. (Univ. of Arizona Press, Tucson), pp. 250–276.
- 55.Durtsche, R. D. (2000) Oecologia 124, 185–195. [DOI] [PubMed] [Google Scholar]
- 56.Durtsche, R. D. (2004) Physiol. Biochem. Zool. 77, 459–470. [DOI] [PubMed] [Google Scholar]
- 57.Espinosa, R. E., Wiens, J. J. & Tracy, C. R. (2003) Proc. Natl. Acad. Sci. USA 101, 16819–16824. [DOI] [PMC free article] [PubMed] [Google Scholar]