Abstract
Genome hypermutation of different orthoretroviruses by cellular cytidine deaminases of the APOBEC3 family during reverse transcription has recently been observed. Lentiviruses like HIV-1 have acquired proteins preventing genome editing in the newly infected cell. Here we show that feline foamy virus (FFV), a typical member of the foamy retrovirus subfamily Spumaretrovirinae, is also refractory to genome deamination. APOBEC3-like FFV genome editing in APOBEC3-positive feline CRFK cells only occurs when the accessory FFV Bet protein is functionally inactivated. Editing of bet-deficient FFV genomes is paralleled by a strong decrease in FFV titer. In contrast to lentiviruses, cytidine deamination already takes place in APOBEC3-positive FFV-producing cells, because edited proviral DNA genomes are consistently present in released particles. By cloning the feline APOBEC3 orthologue, we found that its homology to the second domain of human APOBEC3F is 48%. Expression of feline APOBEC3 in APOBEC3-negative human 293T cells reproduced the effects seen in homologous CRFK cells: Bet-deficient FFV displayed severely reduced titers, high-level genome editing, reduced particle release, and suppressed Gag processing. Although WT Bet efficiently preserved FFV infectivity and genome integrity, it sustained particle release and Gag processing only when fe3 was moderately expressed. Similar to lentiviral Vif proteins, FFV Bet specifically bound feline APOBEC3. In particles from Bet-deficient FFV, feline APOBEC3 was clearly present, whereas its foamy viral antagonist Bet was undetectable in purified WT particles. This is the first report that, in addition to lentiviruses, the foamy viruses also developed APOBEC3-counter-acting proteins.
Keywords: cytidine deamination, virion infectivity factor, spumaretrovirus, restriction factor, zoonosis
The replication of viruses, their spread and disease potential in the infected individual, as well as their transmission to new hosts of the same or another species are tightly controlled by the interplay of several host- and virus-encoded functions. In the simplest case, the presence or absence of a single host function required for virus replication determines the permissiveness of the host. In many instances, however, resistance is actively mounted by the host through the expression of inactivating factor(s) directly interfering with the replication of the pathogen (1). As a consequence of the dynamic host/virus coevolution, virus-encoded defense strategies have been developed that are then also subjected to coevolutionary adaptation processes on either sides. An advanced understanding of the underlying molecular processes of antiviral resistance and virus counter-defense are essential for preventive and therapeutic purposes and also for basic research issues (2).
A case in point is the fact that the host-induced lethal editing of retroviral genomes by cellular APOBEC3 deaminases is specifically prevented by the HIV virion infectivity factor (Vif) (3–8). In this system, human APOBEC3F and -3G cytidine deaminases (hu3F and hu3G) specifically edit cytidine residues of the single-stranded DNA intermediate of retroviral reverse transcription leading to HIV genome degradation or hypermutation and subsequent error catastrophe. The protective effect of the HIV Vif accessory protein depends on strong and specific binding to cellular APOBEC3 proteins and their subsequent targeting to proteasome-mediated degradation (3, 4, 9–11). Both processes prevent packaging of APOBEC3 into HIV particles thus leading to the maintenance of genome integrity and replication competence. Although different APOBEC3 proteins can edit a variety of retroviral genomes and can even interfere by unknown means with the replication of the distantly related hepadnaviruses (12), the Vif-mediated defense strategy appears to be of high specificity. The Vif protein of the closely related simian immunodeficiency virus from African Green Monkey (SIVagm) does not protect against hu3G and, vice versa, nonhuman APOBEC3 proteins are resistant against HIV Vif-induced inactivation (3, 13), although a certain degree of cross-protection has been observed (14, 15). In selected cases, the species-specific APOBEC3–Vif interaction is determined by a single amino acid residue (16–19). Contrary to the APOBEC3-neutralizing activities of primate lentivirus Vif proteins, virtually nothing is known about APOBEC3-counteracting strategies used by simple retroviruses that do not have the capacity to encode a Vif-like protein.
Foamy viruses (FVs), the only members of the subfamily of Spumaretrovirinae, have complex genomes with the capacity to encode a Vif-like function. However, several features of FV replication are clearly different from the Orthoretrovirinae, e.g., the timing of reverse transcription and particle formation (reviewed in ref. 20); therefore, potential mechanisms to cope with the danger of genome editing are worth investigating for these viruses, too.
Here we show that the FFV-encoded accessory Bet protein inhibits cytidine deamination of the viral genome and, in particular, interferes with the antiretroviral activity of the feline APOBEC3 (fe3) protein described here. FFV Bet binds to fe3 but not to hu3G. Although fe3 is present in FFV particles derived from bet-deficient genomes, Bet is undetectable in WT FFV particles, as HIV Vif might be (21, 22). The importance of these findings to our understanding of virus-induced targeting of APOBEC3, as well as the effects on FV replication, will be discussed.
Materials and Methods
Cell Culture and cDNA Preparation. FFV-permissive feline CRFK cells, FeFAB cells, 293T cells, and FFV virions were propagated and used as described (23). Feline peripheral blood mononuclear cells (PBMCs) were isolated from EDTA-treated whole blood by Histopaque-1077 (Sigma) gradient centrifugation and cultured after activation with PHA (3 μg/ml) for 3 days in RPMI medium 1640 containing 15% FBS, 5 × 10–5 M 2-mercaptoethanol, 2 mM l-glutamine, and 100 units of human recombinant IL-2 per ml at 37°C and 5% CO2. For cDNA preparation, total RNA was isolated by using the RNeasy mini kit (Qiagen) according to the manufacturer. Total RNA (5 μg) was used to generate cDNA by using SuperScript III reverse transcriptase (Invitrogen).
Plasmids and DNA Transfection. FFV WT and Bet mutant plasmids pFeFV-BBtr and pFeFV-MCS and the eukaryotic FFV Bet expression plasmid have been described (23, 24). In pFeFV-BBtr, the 387-residue WT Bet is truncated after amino acid 116, whereas in pFeFV-MCS, few residues are exchanged and inserted at the same site. To increase gene expression, both Bet mutations were cloned into the CMV-IE promoter-driven FFV pCF-7 (25), resulting in mutants pCF-BBtr and pCF-MSC. The expression vector for hemagglutinin (HA)-tagged hu3G (phu3G-HA) (3) was a gift of Nathaniel R. Landau (The Salk Institute for Biological Studies, La Jolla, CA). Feline APOBEC3 (fe3) was identified by using 5′ and 3′ RACE reactions (5′/3′-RACE kit, Roche Diagnostics) employing mRNA from CRFK cells, the forward fAPO3F9 (5′-TGGAGGCAGCCTGGGAGGTG-3′) and reverse fAPO3F16 (5′-CTTGAGGGAGGAGGGAGGATG-3′) primers, and Pwo polymerase (Roche Diagnostics). Thirty cycles were run at 94°C for 30 s, 58°C for 1 min, and 72°C for 2 min. PCR products were cloned into pCR4Blunt-TOPO (Invitrogen), sequenced, and transferred into the EcoRI sites of pcDNA3.1(+) (Invitrogen) generating pfe3. Similarly, expression plasmid pfe3-HA encoding C-terminal HA-tagged fe3 was made by using forward fAPO3F18 (5′-TAGAAGCTTACCAAGGCTGGCGAGAGGAATGG-3′) and reverse fAPO3F19 (5′-AGCTCGAGTCAAGCGTAATCTGGAACATCGTATGGATACCTAAGGATTTCTTGAAGCTCTGC-3′) primers, sequenced, and cloned into the HindIII and XhoI sites of pcDNA3.1(+). DNA transfection into CRFK cells was done with Lipofectamine 2000 according to the manufacturer (Invitrogen), 293T cells were transfected by Ca-phosphate precipitation (26).
The fe3 cDNA PCR product was inserted into the BamHI and SalI sites of bacterial expression plasmid pGEX4T3, and the glutatione S-transferase-tagged fe3 fusion protein was purified by glutathione Sepharose chromatography as described (27) and used for antibody induction in rabbits (28).
Virological Methods. FFV particles were prepared from infected CRFK cells 3 or 5 days after infection. Particles were enriched from cell culture supernatant by sedimentation through 20% sucrose and resuspended in PBS as described (29). Particles were digested with the subtilisin protease to remove proteinaceous contaminants not incorporated into the virions (26).
Preparation of Particle-Derived Proviral DNA. To remove contaminating plasmid DNA, enriched FFV particles were treated for 2 h at 37°C with DNaseI according to the supplier (MBI Fermentas, St. Leon-Rot, Germany). The DNase was subsequently inactivated by adding EDTA to 2.5 mM, Proteinase K (Roche Diagnostics) to 0.2 mg/ml and incubation for 45 min at 72°C. Proteinase K was inactivated for 10 min at 98°C.
PCR Amplification, Cloning, and Analysis of Proviral FFV DNA. Virion-encorporated FFV DNA was amplified with sense primer 5′-CTTCTGGTTTGGACCTTACC-3′ and antisense primer 5′-GTTTTAGTAAGTGTAGCGGCGA-3′ using the proofreading Herculase DNA polymerase according to the manufacturer (Amersham Pharmacia). A total of 34 reaction cycles were run at 94°C for 30 s, 56°C for 40 s, and 75°C for 2 min. This PCR allowed amplification of unspliced FFV proviral DNA of ≈615 nt and spliced FFV proviral DNA of ≈330 nt and identification of the bet mutations. Reaction products were cloned by using the TOPO cloning kit as per the manufacturer's instructions (Invitrogen). Clones were identified by restriction enzyme digestion, and plasmid DNA was sequenced by using the DNA sequencer 377 (Applied Biosystems).
Immunoprecipitation and Western Blot Analysis. For coimmunoprecipitation of FFV-Bet and fe3 or hu3G, 293T cells were transfected with 2 μg of fe3-HA or human APOBEC3G-HA expression plasmid pfe3-HA or phu3G-HA and 2 μg of pFFV-Bet. After 2 days, cells were lysed in TLB (20 mM Tris, pH 7.4/137 mM NaCl/10% glycerol/2 mM EDTA, pH 8/1% Triton X-100/50 mM Na-β-glycerophosphate and protease inhibitors) and lysates cleared by centrifugation. For immunoprecipitation of fe3-HA or hu3G-HA, supernatants were incubated with anti-HA-beads (Roche Diagnostics) for 60 min at 4°C and washed five times with TLB. After boiling in electrophoresis sample buffer, samples were subjected to SDS/PAGE and immunoblotting (23). The FFV Bet, Env leader protein, and cat 8014 antisera have been described (23). Membranes were reacted with horseradish peroxidase-conjugated secondary antibodies (Amersham Pharmacia) and visualized by enhanced chemiluminescence (ECL, Amersham Pharmacia). For immunoblotting, identical amounts of cell extracts were used as determined by Roti-Quant protein quantification (Roth, Karlsruhe, Germany).
Results
Bet-Mutated FFV Genomes Are Edited in Feline CRFK Cells. We recently reported that CRFK cells display a nonpermissive phenotype when infected by bet-defective FFV (23). Similarly, Vif-minus feline immunodeficiency virus (FIV) is replication deficient in CRFK cells (30). In light of recent findings on the function of lentivirus Vif, we questioned whether expression of an APOBEC3-like cytidine-deaminase in CRFK cells might be involved in the restriction of bet-deficient FFV.
Therefore, we reexamined the replication of the previously described FFV bet mutants pFeFV-MCS and pFeFV-BBtr in CRFK cells (23). In clone pFeFV-MCS, only a few amino acids in the central part of Bet had been changed, and clone pFeFV-BBtr is characterized by a truncation of Bet at the same site (23). As described (23), the changes in bet resulted in a 102-to103-fold reduced titer of the mutants compared to WT FFV (data not shown). To identify the cause for the reduced titer, de novo synthesized FFV genomes were analyzed for the presence of APOBEC3-mediated C → U deamination of the DNA minus-strand resulting in G → A exchanges on the plus strand. For these studies, we took advantage of two specific features of FV reverse transcription: a substantial fraction of FV particles already contains full-length proviral DNA and part of this DNA specifically lacks the bet intron (see ref. 31). These intron-deficient, bet-spliced FFV DNAs are only generated after replication of the plasmid-encoded FFV genomes, and therefore cannot be derived from input DNA. CRFK cells were transfected with WT and bet-mutated FFV genomes. Released particles were purified 3 days later by sedimentation through sucrose and subjected to DNaseI digestion to remove plasmid DNA. The encapsidated, protected DNA was extracted and amplified by using PCR primers that allowed direct amplification of spliced and unspliced FFV DNA and confirmation of the introduced bet mutations. The sequencing data for spliced and unspliced FFV DNAs are summarized in Table 1. FFV WT genomes displayed a low mutation frequency with no preference for G → A exchanges. In contrast, G → A substitutions were highly enriched in DNAs from both bet mutants, independent of whether spliced or unspliced DNA was sequenced. The number of G → A exchanges varied between 1 and 11 per sequence (Table 1). Whereas all spliced cDNAs from both mutants contained at least one G → A exchange, some unspliced and thus even longer cDNAs of mutants pFeFV-MCS and pFeFV-BBtr did not. It is likely that these unmodified, full-length sequences were derived from input plasmid DNA and not from reverse-transcribed genomes despite the DNaseI digestion.
Table 1. Sequence characteristics of CRFK cell-derived, encapsidated FFV DNA genomes from WT and Bet mutant FFV proviruses.
| pFeFV-7 wt
|
pFeFV-MCS
|
pFeFV-BBtr
|
||||
|---|---|---|---|---|---|---|
| Spliced | Unspliced | Spliced | Unspliced | Spliced | Unspliced | |
| No. of sequences analyzed | 8 | 8 | 12 | 14 | 8 | 12 |
| Mutations G → A/other | 0/0 | 1/3 | 37/1 | 31/3 | 14/1 | 41/5 |
| Clones without G → A editing | 8 | 7 | 0 | 3 | 0 | 4 |
| Minimal no. of G → A per clone | 0 | 1 | 1 | 1 | 1 | 2 |
| Maximal no. of G → A per clone | 0 | 1 | 9 | 11 | 3 | 9 |
| Average no. of G → A per clone | 0 | 0.1 | 3.1 | 2.2 | 1.8 | 3.5 |
| G → A exchanges per 100 nucleotides | 0 | 0.02 | 1.01 | 0.38 | 0.60 | 0.59 |
When analyzing the minus strand for the sequence context in which the changes occurred, 68% were TTC to TTT changes, 14% were TCC to TCT exchanges, and in the remaining clones, at least one pyrimidine residue (NPyC or PyNC) preceded the altered C nucleotide (Table 2). PyPyC to PyPyT mutations are typical for APOBEC3-mediated editing of retroviral genomes (32, 33). In summary, these data indicate that CRFK cells express an APOBEC3-like deaminase (see below) and that FFV Bet counteracts this editing activity.
Table 2. Sequence context of minus strand C → T editing events in transfected Bet-mutant proviral DNA.
| Frequency, %
|
||
|---|---|---|
| Sequence exchange | In CRFK cells (n = 123) | In fe3-expressing 293T cells (n = 57) |
| CCC → CCT | 0 | 0 |
| CTC → CTT | 2 | 9 |
| TCC → TCT | 14 | 21 |
| TTC → TTT | 68 | 56 |
| XPyC → XPyT | 8 | 9 |
| PyXC → PyXT | 8 | 5 |
| PuPuC → PuPuT | 0 | 0 |
Pu, purin base; Py, pyrimidine base; X, any base.
To exclude the possibility that mutagenesis of bet interferes with the fidelity of FFV reverse transcription, we transfected WT pFeFV-7 and mutant pFeFV-BBtr into APOBEC-negative 293T cells and analyzed reverse-transcribed genomes from released particles for mutations. Under these conditions, the frequency and types of mutations were similar for WT and bet-mutated FFV genomes excluding a direct effect of Bet on the fidelity of the FFV RT (see below).
Characterization of Feline APOBEC3. To identify APOBEC3 expression in CRFK cells, degenerate primers derived from exons 3 and 6 of hu3G were used to amplify and clone the central part of the corresponding CRFK cell-derived feline APOBEC3 cDNA. The full-length feline APOBEC3 (fe3) cDNA was subsequently constructed by 5′- and 3′-RACE techniques. The 192-aa-long fe3 shows significant homology to the second (48.5%) and first (38.1%) domain of hu3F and to the single domain of hu3C (46.4%) cytidine deaminases (Fig. 1A). hu3F and fe3 consistently have a similar editing preference for the trinucleotide TTC (see Table 2), whereas hu3G prefers CCC (7, 34). When diagnostic PCR primers were used, substantial fe3 expression was detectable in CRFK cells and in PHA-activated feline PBMCs (Fig. 1B); the fe3 cDNA derived from PBMC was identical to that from CRFK cells.
Fig. 1.
The FFV Bet protein counteracts the antiviral activity of feline APOBEC. (A) Alignment of the amino acid sequence of feline APOBEC3 (fe3), human APOBEC3C (hu3C), and human APOBEC3F domain 1 (hu3F-A) and domain 2 (hu3F-B). Identical residues are marked. (B) Analysis of fe3 expression by RT-PCR of total RNA from feline CRFK cells and feline PBMCs. Neg, no cDNA added. (C) WT (pFeFV-7) and Bet mutant (pFeFV-BBtr) FFV genomes were cotransfected with pUC18 control DNA (vector) and pfe3 expression plasmid into 293T cells. The titer was determined by using FeFAB titration cells (23).
Fe3 Reduces the Titer of bet-Deficient FFV and Induces Genome Editing. The effect of fe3 coexpression with WT and bet-deficient FFV genomes was studied after transfection of 293T cells. For this purpose, the FFV titers were determined 2 days after transfection by using FeFAB reporter cells (23). Cotransfection of pfe3 reduced the WT FFV titer of pFeFV-7 up to 10-fold, whereas a 102-to103-fold reduction in titer was detected with the Bet-truncated pFeFV-BBtr mutant (Fig. 1C). This finding clearly demonstrates that FFV Bet efficiently counteracts the antiviral activity of feline APOBEC3.
As described for CRFK cell-mediated FFV genome editing, a total of 29 FFV DNA genomes released from WT and bet-mutant pFeFV-BBtr cotransfected with either pfe3-HA or pUC18 was analyzed. Fe3 overexpression in 293T cells resulted in 0.05 G → A exchanges per 100 nucleotides for the WT FFV genome compared with 0.13 G → A exchanges per 100 nucleotides when pUC control DNA was coexpressed. As expected, editing of the Bet mutant pFeFV-BBtr increased editing to 1.05 G → A exchanges per 100 nucleotides when fe3 was coexpressed, whereas few G → A exchanges (0.08 G → A exchanges per 100 nucleotides) occurred without fe3. The sequence context of the G → A exchanges by fe3 coexpression in 293T cells (Table 2, right column) is similar to that seen in CRFK cells expressing the endogenous fe3 deaminase activity. This finding indicates that the majority of FFV genome editing in CRFK cells can be attributed to the cloned fe3 or a closely related feline cytidine deaminase.
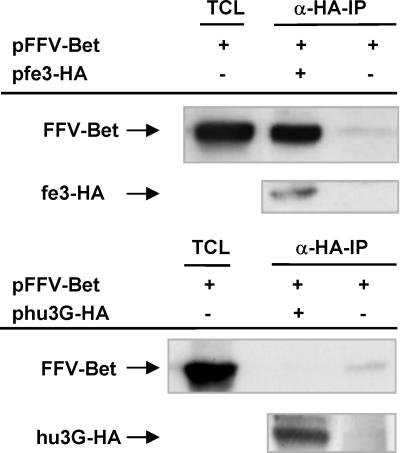
FFV Bet Specifically Binds to Feline APOBEC3. Because the fe3-encoded deaminase showed a Bet-dependent phenotype on FFV titer and genome editing, we analyzed by coimmunoprecipitation assays whether fe3 is specifically bound by FFV Bet. An FFV Bet expression plasmid was cotransfected into 293T together with plasmid pfe3-HA or control DNA cells, and lysates were subjected to coimmunoprecipitation using anti-HA beads, allowing detection of the HA-tagged fe3 protein (Fig. 2). Similar to HIV-1 Vif (3, 13), FFV Bet was coprecipitated by fe3-HA, whereas FFV Bet was not detected when the HA-tagged human APOBEC3G (hu3G-HA) protein was used, although it was clearly present in the lysate. These data demonstrate a species-specific binding of FFV Bet to the homologous fe3 but not the heterologous hu3G protein.
Fig. 2.
FFV Bet coimmunoprecipitates with feline APOBEC3, but not with human APOBEC3G. 293T cells were transfected with FFV Bet alone or with expression plasmids encoding HA-tagged fe3 or hu3G. As positive controls, total cell lysates (TCL) were prepared 2 days after transfection and FFV Bet was detected by immunoblotting using α-FFV-Bet antiserum (lane 1, upper line in each panel). For detection of APOBEC-associated FFV-Bet, α-HA-immunoprecipitations (α-HA-IP) were performed and FFV Bet was detected by immunoblot analysis (lanes 2 and 3, upper line). α-HA-immunoprecipitation was controlled by a α-HA immunoblot (lower line in each panel).
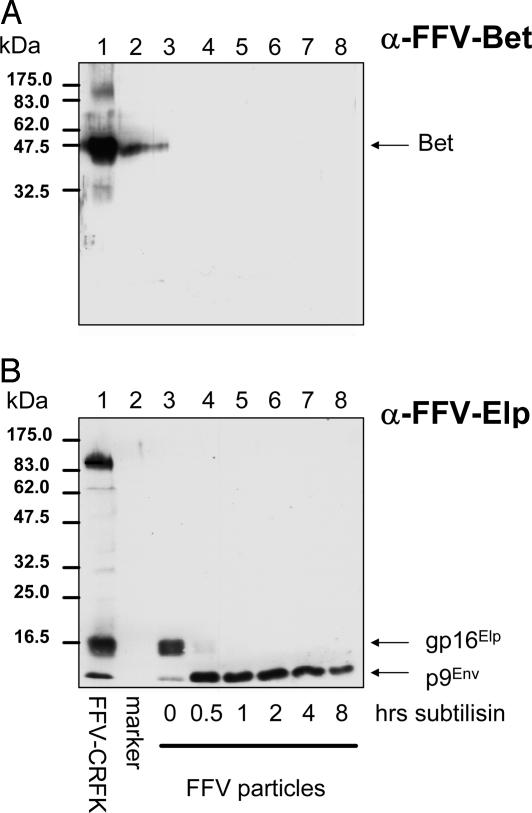
FFV Bet Is Not Incorporated in Virions. To determine whether the abundantly expressed cytoplasmic Bet that efficiently interacts with host cell-encoded fe3 is a component of viral particles, immunoblotting studies were performed. Particles were harvested from the supernatant of CRFK cells 5 days after WT FFV infection. The virions were subjected to subtilisin digestion to remove any Bet that was merely attached to the surface but not incorporated into FFV particles (26). Whereas low amounts of Bet were detectable in undigested FFV particles, subtilisin treatment completely eliminated Bet-specific signals, indicating that Bet was only copurified with virus particles (Fig. 3A). The conditions of subtilisin treatment were controlled by following digestion of the 16-kDa ectodomain of the FFV Env leader protein (Elp) to the 9-kDa membrane-protected product (Fig. 3B) (26). In lentiviruses, the APOBEC3-protecting Vif protein is found in released virions in most laboratories, but other groups have failed to detect Vif in virions (21, 22).
Fig. 3.
FFV Bet is undetectable in purified FFV particle preparations. FFV particles enriched from supernatants of FFV-infected CRFK cells by sedimentation through a sucrose cushion were digested with subtilisin for different time points as indicated below the blot and analyzed by immunoblotting. In parallel, antigen from FFV-infected CRFK cells and molecular mass markers were applied (lanes 1 and 2). The blot in A was reacted with a serum against FFV Bet, the blot in B was probed with an Elp antiserum. The positions of FFV Bet, the Env leader protein gp16Elp, and the Env signal peptide p9Env (26) are indicated at right, those of marker proteins are given at left.
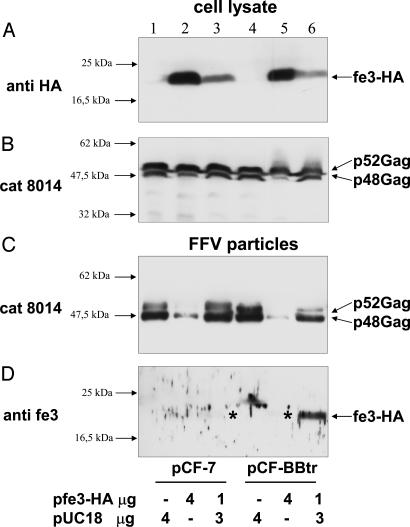
Fe3 Interferes with FFV Particle Release and Accumulates in Particles from bet-Deficient Genomes. We analyzed whether fe3 expression affects FFV gene expression and release or composition of particle. To this end, WT pCF-7 and Bet mutant pCF-BBtr proviruses were cotransfected with decreasing amounts of plasmid pfe3-HA into 293T cells (Fig. 4). In cellular extracts, HA-tagged fe3 was clearly detectable as a discrete band of ≈22 kDa (Fig. 4A). The overall expression level of fe3-HA was not altered in WT versus mutant Bet-expressing cells. The expression level of FFV Gag was also not affected by fe3-HA coexpression; however, the processing of the FFV p52 Gag precursor to the p48 Gag cleavage product was consistently reduced on overexpression of fe3-HA (Fig. 4B), whereas Pol processing appeared normal (data not shown). The observations that fe3 stability is not affected by Bet and that increasing amounts of fe3 interfere with Gag but not with Pol processing were confirmed in independent experiments (data not shown).
Fig. 4.
Effect of fe3 on FFV Gag expression and particle release. 293T cells were cotransfected with 4 μg of WT pCF-7 and mutant pCF-BBtr plus either pUC18 control DNA or 4 or 1 μg of pfe3-HA expression plasmids as indicated below the blots. Two days after transfection, cellular extracts (A and B) and released FFV particles (C and D) were harvested and analyzed by immunoblotting using an HA-specific antibody (A), cat serum 8014 (B and C), and the fe3-specific antiserum (D). The positions of marker proteins are given at the left margin. The positions of fe3-HA and p48 and p52 Gag are indicated. The asterisks mark faint fe3-HA bands in WT (lane 3) and mutant particles (lane 5) only visible after overexposure (data not shown). For virions from bet-deficient genomes, twice the amount was loaded (C and D, lanes 4–6).
We then analyzed the cell culture supernatants for WT and mutant particle release. When the Gag-reactive cat serum 8014 was used, cotransfection of high amounts (4 μg) of pfe3-HA strongly reduced release of particles derived from WT and bet-deficient proviruses, whereas lower amounts of fe3 only affected release from bet-deficient FFV genomes (Fig. 4C). A parallel blot reacted with the fe3-specific serum clearly revealed low amount of fe3-HA in particles from bet-deficient FFV genomes (Fig. 4D). In WT particles, miniscule amounts of fe3-HA were detectable only after overexposure of the blot (marked by asterisks in lane 3). For pCF-BBtr-derived virus, the amount of fe3-HA detected paralleled the release of particles: the low-level release with high fe3 concentrations resulted in only trace amounts of fe3 in the particle fraction, whereas moderate particle budding (at 1 μg of pfe3-HA DNA) was paralleled by an increased fe3 release. These data show that WT Bet inhibits fe3 packaging into FFV particles.
Discussion
We report here that bet-deficient genomes of FFV, a representative member of the retroviral Spumaretrovirinae subfamiliy, are edited in a cell type-dependent manner. We show that bet-deficient FFV genomes are susceptible to APOBEC3-mediated genome editing in feline CRFK cells. The cytidine deamination of FFV genomes already takes place in the virus-producing cell and not exclusively in the newly infected cell as shown for different orthoretroviruses including lentiviruses (3, 5–8). Coimmunoprecipitation assays and cotransfection of fe3 expression plasmids with WT and bet-mutated FFV genomes indicate that Bet, in addition to its other known functions (23, 35–37), counteracts cellular APOBEC3 activities. Infectivity of Bet-deficient FFV is reduced not only by genome deamination but also by an APOBEC3-induced reduction of particle release.
Studying APOBEC3-mediated FV genome editing and viral countermeasures, we identified a previously undescribed feline orthologue of the APOBEC3 deaminase family. The fe3 gene consists of a single APOBEC3 domain that displays significant homology (48.5%) to the second domain of hu3F. This genetic relatedness is consistent with the hu3F-like editing context described here for fe3. Fe3 mRNA is substantially expressed in feline PBMCs and CRFK cells.
Expression of fe3 is consistent with the nonpermissive phenotype of CRFK cells toward replication of bet-mutated FFV, and fe3 or a related APOBEC3 deaminase might also restrict replication of Vif-deleted FIV in CRFK cells (30). We assume that the APOBEC3-inactivating function is also required for FFV replication in its presently unknown target cells in cats explaining why Bet is maintained in vivo and in vitro (20, 25, 31).
APOBEC3-mediated genome deamination is considered to be an efficient barrier against lentiviral interspecies transmission events because the Vif proteins tend to counteract only APOBEC3 proteins of the cognate host species (3). The binding characteristics presented here for FFV Bet parallel these findings. Thus, genome deamination of FFV in humans after a zoonotic transmission is not likely to be prevented by FFV Bet. In preliminary experiments, we even found strong editing of the WT FFV genome by human and African green monkey APOBEC3G with a concomitant reduction in viral titers and similar effects of fe3 on the primate (human) FV (data not shown). This may explain why zoonotic transmission of FFV has not been detected (38).
The data presented indicate that FFV Bet binds to fe3. Together with the high-level cytoplasmic expression of Bet in all cell culture systems studied (31), this may point to an active sequestration of APOBEC3 away from the sites of FV particle assembly. This active sequestration of fe3 is in line with the observation that functional inactivation of Bet correlated with accumulation of fe3 in released virus particles. The alternative mechanism, that Bet may direct APOBEC3 proteins to proteasome-mediated degradation as is well documented for Vif (4, 9–11), appears unlikely because intracellular fe3 levels were not affected by FFV Bet. The fact that subtle mutations of Bet in clone pFeFV-MCS destroyed its protective potential as severely as truncating Bet at the same site indicates that this central part of Bet either directly affects its function, e.g., during APOBEC3 binding, or that this sequence is absolutely required for proper protein folding. The high concentrations of Bet may be not only required for APOBEC3 sequestration, but also to the other Bet functions, e.g., in establishing and maintaining persistence (35), reactivation from latency (36), intercellular trafficking (37), or particle release (23).
We cannot provide an explanation for why overexpression of fe3 reduced only intracellular Gag processing and particle release, two processes that may be functionally linked. However, WT Bet can partially suppress this reduction in Gag processing provided that fe3 is expressed at only moderate levels. Because the replication pathway of FVs is in part reminiscent of that of hepatitis B virus, the APOBEC3-mediated effect on FV particle release may be related to the presently unknown inhibitory mechanism of hu3G directed against hepatitis B virus pregenome packaging (12, 39).
The most distinguishing feature in the APOBEC3G-mediated editing of FV genomes in contrast to orthoretroviruses is the timing of deamination: in orthoretroviruses, editing only occurs in the newly infected cell. In contrast, deamination of FFV genomes by fe3 is already clearly detectable in genomes packaged into released particles. We obtained similar data for the primate FV (data not shown) that can be edited by different APOBEC3 deaminases (40). However, we cannot exclude the possibility that editing also takes place after virus release or further increases during postentry reverse transcription (41). The early onset of FV genome editing is most probably related to the fact that FV reverse transcription already starts before or during particle formation and release in the virus-producing cell. This finding may explain our observation that only low amounts of fe3 are present in particles from bet-deficient FFV genomes, because, in FVs, the virus-producing cell, and not the newly infected cell, is the major site of APOBEC3 action.
Acknowledgments
We thank Andreas Hunziker for expert DNA sequencing, Astrid Schwantes and Björn-Philipp Kloke for valuable suggestions, Hannelore Constabel for technical help, Nathaniel R. Landau for reagents, Jennifer Reed for critically reading the manuscript, and Lutz Gissmann for continuous support. Part of this study was supported by The Stanley Medical Research Institute Grant 03R-411 (to M.L.).
Author contributions: M.L. and C.M. designed research; M.L., F.R., P.B., H.M., N.K., Y.-B.K., U.R., M.B., and C.M. performed research; A.S. contributed new reagents/analytic tools; M.L., F.R., P.B., H.M., U.T., E.F., and C.M. analyzed data; and M.L., U.T., E.F., K.C., and C.M. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: Vif, virion infectivity factor; FV, foamy virus; PBMC, peripheral blood mononuclear cell; HA, hemagglutinin.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AY971954).
References
- 1.Bieniasz, P. D. (2004) Nat. Immunol. 5, 1109–1115. [DOI] [PubMed] [Google Scholar]
- 2.Harris, R. S. & Liddament, M. T. (2004) Nat. Rev. Immunol. 4, 868–877. [DOI] [PubMed] [Google Scholar]
- 3.Mariani, R., Chen, D., Schröfelbauer, B., Navarro, F., König, R., Bollman, B., Münk, C., Nymark-McMahon, H. & Landau, N. R. (2003) Cell 114, 21–31. [DOI] [PubMed] [Google Scholar]
- 4.Sheehy, A. M., Gaddis, N. C. & Malim, M. H. (2003) Nat. Med. 9, 1404–1407. [DOI] [PubMed] [Google Scholar]
- 5.Mangeat, B., Turelli, P., Caron, G., Friedli, M., Perrin, L. & Trono, D. (2003) Nature 424, 99–103. [DOI] [PubMed] [Google Scholar]
- 6.Zheng, Y. H., Irwin, D., Kurosu, T., Tokunaga, K., Sata, T. & Peterlin, B. M. (2004) J. Virol. 78, 6073–6076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wiegand, H. L., Doehle, B. P., Bogerd, H. P. & Cullen, B. R. (2004) EMBO J. 23, 2451–2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang, H., Yang, B., Pomerantz, R. J., Zhang, C., Arunachalam, S. C. & Gao, L. (2003) Nature 424, 94–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stopak, K., de Noronha, C., Yonemoto, W. & Greene, W. C. (2003) Mol. Cell 12, 591–601. [DOI] [PubMed] [Google Scholar]
- 10.Mehle, A., Strack, B., Ancuta, P., Zhang, C., McPike, M. & Gabuzda, D. (2004) J. Biol. Chem. 279, 7792–7798. [DOI] [PubMed] [Google Scholar]
- 11.Yu, X., Yu, Y., Liu, B., Luo, K., Kong, W., Mao, P. & Yu, X. F. (2003) Science 302, 1056–1060. [DOI] [PubMed] [Google Scholar]
- 12.Turelli, P., Mangeat, B., Jost, S., Vianin, S. & Trono, D. (2004) Science 303, 1829. [DOI] [PubMed] [Google Scholar]
- 13.Yu, Q., Chen, D., Konig, R., Mariani, R., Unutmaz, D. & Landau, N. R. (2004) J. Biol. Chem. 275, 53379–53386. [DOI] [PubMed] [Google Scholar]
- 14.Takeuchi, H., Kao, S., Miyagi, E., Khan, M. A., Buckler-White, A., Plishka, R. & Strebel, K. (2004) J. Biol. Chem. 280, 375–382. [DOI] [PubMed] [Google Scholar]
- 15.Gaddis, N. C., Sheehy, A. M., Ahmad, K. M., Swanson, C. M., Bishop, K. N., Beer, B. E., Marx, P. A., Gao, F., Bibollet-Ruche, F., Hahn, B. H. & Malim, M. H. (2004) J. Virol. 78, 12041–12046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schröfelbauer, B., Chen, D. & Landau, N. R. (2004) Proc. Natl. Acad. Sci. USA 101, 3927–3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu, H., Svarovskaia, E. S., Barr, R., Zhang, Y., Khan, M. A., Strebel, K. & Pathak, V. K. (2004) Proc. Natl. Acad. Sci. USA 101, 5652–5657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bogerd, H. P., Doehle, B. P., Wiegand, H. L. & Cullen, B. R. (2004) Proc. Natl. Acad. Sci. USA 101, 3770–3774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mangeat, B., Turelli, P., Liao, S. & Trono, D. (2004) J. Biol. Chem. 279, 14481–14483. [DOI] [PubMed] [Google Scholar]
- 20.Rethwilm, A. (2003) Curr. Top. Microbiol. Immunol. 277, 1–26. [DOI] [PubMed] [Google Scholar]
- 21.Kao, S., Akari, H., Khan, M. A., Dettenhofer, M., Yu, X. F. & Strebel, K. (2003) J. Virol. 77, 1131–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dettenhofer, M. & Yu, X. F. (1999) J. Virol. 73, 1460–1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Alke, A., Schwantes, A., Kido, K., Flötenmeyer, M., Flügel, R. M. & Löchelt, M. (2001) Virology 287, 310–320. [DOI] [PubMed] [Google Scholar]
- 24.Zemba, M., Alke, A., Bodem, J., Winkler, I. G., Flower, R. L., Pfrepper, K., Delius, H., Flügel, R. M. & Löchelt, M. (2000) Virology 266, 150–156. [DOI] [PubMed] [Google Scholar]
- 25.Schwantes, A., Ortlepp, I. & Löchelt, M. (2002) Virology 301, 53–63. [DOI] [PubMed] [Google Scholar]
- 26.Geiselhart, V., Schwantes, A., Bastone, P., Frech, M. & Löchelt, M. (2003) Virology 310, 235–244. [DOI] [PubMed] [Google Scholar]
- 27.Sehr, P., Muller, M., Hopfl, R., Widschwendter, A. & Pawlita, M. (2002) J. Virol. Methods 106, 61–70. [DOI] [PubMed] [Google Scholar]
- 28.Roesler, U., Scholz, H. & Hensel, A. (2003) Int. J. Syst. Evol. Microbiol. 53, 1195–1199. [DOI] [PubMed] [Google Scholar]
- 29.Wilk, T., de Haas, F., Wagner, A., Rutten, T., Fuller, S., Flügel, R. M. & Löchelt, M. (2000) J. Virol. 74, 2885–2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shacklett, B. L. & Luciw, P. A. (1994) Virology 204, 860–867. [DOI] [PubMed] [Google Scholar]
- 31.Löchelt, M. (2003) Curr. Top. Microbiol. Immunol. 277, 27–61. [DOI] [PubMed] [Google Scholar]
- 32.Yu, Q., König, R., Pillai, S., Chiles, K., Kearney, M., Palmer, S., Richman, D., Coffin, J. M. & Landau, N. R. (2004) Nat. Struct. Mol. Biol. 11, 435–442. [DOI] [PubMed] [Google Scholar]
- 33.Harris, R. S., Bishop, K. N., Sheehy, A. M., Craig, H. M., Petersen-Mahrt, S. K., Watt, I. N., Neuberger, M. S. & Malim, M. H. (2003) Cell 113, 803–809. [DOI] [PubMed] [Google Scholar]
- 34.Liddament, M. T., Brown, W. L., Schumacher, A. J. & Harris, R. S. (2004) Curr. Biol. 14, 1385–1391. [DOI] [PubMed] [Google Scholar]
- 35.Saib, A., Koken, M. H., van der Spek, P., Peries, J. & de The, H. (1995) J. Virol. 69, 5261–5268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Meiering, C. D. & Linial, M. L. (2002) Proc. Natl. Acad. Sci. USA 99, 15130–15135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lecellier, C. H., Vermeulen, W., Bachelerie, F., Giron, M. L. & Saib, A. (2002) J. Virol. 76, 3388–3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Butera, S. T., Brown, J., Callahan, M. E., Owen, S. M., Matthews, A. L., Weigner, D. D., Chapman, L. E. & Sandstrom, P. A. (2000) J. Am. Vet. Med. Assoc. 217, 1475–1479. [DOI] [PubMed] [Google Scholar]
- 39.Rösler, C., Köck, J., Malim, M. H., Blum, H. E. & von Weizsäcker, F. (2004) Science 305, 1403. [DOI] [PubMed] [Google Scholar]
- 40.Russell R. A., Wiegand, H. L., Moore M. D., Schäfer, A., McClure, M. O. & Cullen, B. R. (2005) J. Virol., in press. [DOI] [PMC free article] [PubMed]
- 41.Delelis, O., Saib, A. & Sonigo, P. (2003) J. Virol. 77, 8141–8146. [DOI] [PMC free article] [PubMed] [Google Scholar]