Abstract
The human T-lymphotropic viruses (HTLVs) types 1 and 2 originated independently and are related to distinct lineages of simian T-lymphotropic viruses (STLV-1 and STLV-2, respectively). These facts, along with the finding that HTLV-1 diversity appears to have resulted from multiple cross-species transmissions of STLV-1, suggest that contact between humans and infected nonhuman primates (NHPs) may result in HTLV emergence. We investigated the diversity of HTLV among central Africans reporting contact with NHP blood and body fluids through hunting, butchering, and keeping primate pets. We show that this population is infected with a wide variety of HTLVs, including two previously unknown retroviruses: HTLV-4 is a member of a phylogenetic lineage that is distinct from all known HTLVs and STLVs; HTLV-3 falls within the phylogenetic diversity of STLV-3, a group not previously seen in humans. We also document human infection with multiple STLV-1-like viruses. These results demonstrate greater HTLV diversity than previously recognized and suggest that NHP exposure contributes to HTLV emergence. Our discovery of unique and divergent HTLVs has implications for HTLV diagnosis, blood screening, and potential disease development in infected persons. The findings also indicate that cross-species transmission is not the rate-limiting step in pandemic retrovirus emergence and suggest that it may be possible to predict and prevent disease emergence by surveillance of populations exposed to animal reservoirs and interventions to decrease risk factors, such as primate hunting.
Keywords: retrovirus, zoonosis, simian, exposures, diversity
Primate T-lymphotropic viruses (PTLVs) are deltaretroviruses composed of three distinct groups (PTLV-1, -2, and -3), which by conventional nomenclature are named simian T-lymphotropic viruses (STLVs) when found in nonhuman primates (NHPs) and human T-lymphotropic viruses (HTLVs) when found in humans, regardless of suspected zoonotic origin (1–9). PTLV-1 and PTLV-2 include both HTLV-1 and HTLV-2 and their simian analogs (STLV-1 and STLV-2, respectively) (1–5), whereas PTLV-3 comprises only simian viruses (6–9). The finding that HTLV-1 and HTLV-2 originated independently and are related to distinct lineages of STLVs (STLV-1 and STLV-2, respectively) (1, 2), combined with results showing that HTLV-1 diversity appears to have resulted from multiple cross-species transmissions of STLV-1 (1–5), suggests that contact between humans and infected NHPs may result in HTLV emergence. Like HIV, HTLV has spread globally to at least 22 million persons sexually, from mother to child, and by exposure to contaminated blood through transfusions and injection drug use (1, 10, 11). HTLV-1 causes adult T cell leukemia and HTLV-1-associated myelopathy/tropical spastic paraperesis and other inflammatory diseases in ≈2–5% of those infected (1, 10). HTLV-2 is less pathogenic than HTLV-1 and has been associated with a neurologic disease similar to HTLV-1-associated myelopathy/tropical spastic paraperesis (11).
There has been no evidence that STLVs cross into people occupationally exposed to NHPs in laboratories and primate centers, as has been documented for other primate retroviruses, including simian immunodeficiency virus, simian foamy virus (SFV), and simian type D retrovirus (12–15). Nevertheless, zoonotic transmission of STLV to human populations naturally exposed to NHPs through hunting or butchering, similar to that recently reported for SFV in African hunters (16), would be of particular public health significance because of the transmissible and pathogenic nature of this group of viruses among humans. Previous studies have not documented evidence of HTLV outside the PTLV-1 and PTLV-2 groups (1–9, 17, 18), although those studies were not focused on persons who reported direct and repeated exposure to the blood and body fluids of NHPs, which may increase the likelihood of human infection with zoonotic primate retroviruses (16, 19).
To determine whether previously undescribed HTLVs are present in persons exposed to the blood and body fluids of wild primate populations that are infected with STLV-1 and STLV-3 (7, 8), we examined persons from rural villages in southern Cameroon located near natural NHP habitats for evidence of PTLV infection. This population has been shown to have frequent primate exposure risks, including hunting, butchering, and keeping primate pets (16, 19).
Materials and Methods
Study Population and Ethical Approvals. Persons living in 11 rural villages in southern Cameroon proximal to forested and nonforested NHP habitats (Fig. 1) were invited to participate in a community-based HIV prevention campaign designed to provide information through the use of Cameroonian educators and counselors and therefore to decrease transmission. Participation in the study was voluntary. The study protocol was approved by the Johns Hopkins Committee for Human Research, the Cameroon National Ethical Review Board, and the HIV Tri-Services Secondary Review Board. Study participants completed questionnaires designed to determine behaviors that exposed participants to NHPs. Individuals were asked to identify and quantify their exposure to a range of NHPs, which were reliably classified into categories that could be easily distinguished by this population: chimpanzee, gorilla, and monkey. All personal identifiers were removed from questionnaires and matching specimens before testing to provide an anonymous, unlinked study population.
Fig. 1.
Map of study sites shown in relation to distribution of lowland forest (green) in central Africa. Sites with evidence of PTLV infection are represented with colored circles, which correspond to the PTLV(s) found at the site.
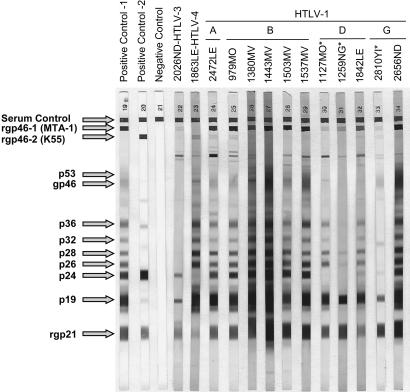
Specimen Preparation and Serology. Blood was collected from participants, transported to a central laboratory, processed into plasma and peripheral blood mononuclear cell aliquots, and stored at –80°C. Initial screening for HTLV antibodies in plasma specimens was performed by using the Vironostika HTLV-1/2 microelisa system (Organon-Teknika, Durham, NC) according to the manufacturer's instructions. This enzyme immunoassay contains purified HTLV-1 and HTLV-2 viral lysates and a recombinant HTLV-1 p21 envelope (Env) antigen. Reactive specimens were then tested by a Western blot (WB) test (HTLV Blot 2.4, Genelabs Diagnostics, Singapore) that contains disrupted HTLV-1 virions, a gp21 recombinant protein (GD21) common to both HTLV-1 and HTLV-2, and two HTLV type-specific recombinant Env peptides, MTA-1 and K55, which allow serologic differentiation of HTLV-1 and HTLV-2, respectively. Specimens that were reactive to the Gag (p24) and Env (GD21) proteins were considered seropositive. Seropositive specimens that were reactive to MTA-1 or K55 were considered HTLV-1-like or HTLV-2-like, respectively. Seropositive samples not reactive to either the MTA-1 or K55 peptides were considered HTLV positive, but untypeable. Specimens that were reactive to either p24 or GD21 alone or in combination with other HTLV proteins (p19, p26, p28, p32, p36, gp46, and p53) (Fig. 2) were considered indeterminate. The enzyme immunoassay and WB assay have been shown to be capable of detecting antibodies to a broad range of PTLVs (6, 8).
Fig. 2.
HTLV WB serologic pattern of infected African hunters. HTLV classification based on phylogenetic analyses is provided above specimen codes. Asterisks indicate specimens with indeterminate WB results. Reactivity to HTLV-specific proteins is indicated on the left.
PCR Detection and Sequence Analysis. DNA was prepared from uncultured peripheral blood mononuclear cells, and its integrity was confirmed by β-actin PCR as described (16). All DNA preparation and PCR assays were performed in a laboratory where only human specimens are processed and tested according to recommended precautions to prevent contamination. DNA specimens were first screened with a generic PTLV PCR assay capable of detecting 222-bp tax sequences from each of the three major PTLV groups (6, 17). Sequence analysis of this tax sequence provided broad genetic classification into each major PTLV group. Phylogenetic resolution within the PTLV-1 and PTLV-3 groups was done by using LTR sequences as described (6, 20). An overlapping portion of the 3′ HTLV-1 LTR from specimens 1259NG, 1127MO, 1842LE, and 2810YI was PCR amplified with the external primers 5VLTRext (20) and 1MNDR1 (5′-GTCGTGAATGAAAGGGAAAGGGGT-3′) followed by the internal primers Enh280 (20) and 1MNDR2 (5′-AGGGGTGGAACTTTCGATCTGTAA-3′). The tax (577-bp) and polymerase (pol) (709-bp) sequences of HTLV-3 and HTLV-4 were amplified by the use of nested PCR with primers designed from conserved PTLV regions. The external and internal tax primers are PTLVTPG [5′-T(C/T)ACCT(G/A)GGACCCCATCGATGGACG-3′] and PGTAXR1 [5′-GAIGA(T/C)TGIA(C/G)TAC(T/C)AAAGATGGCTG-3′] and PH2Rrev [5′-CCTTATCCCTCGICTCCCCTCCTT-3′] and PGTAXR2 [5′-TTIGGG(T/C)AIGGICCGGAAATCAT-3′], respectively. The external and internal pol primers are PGPOLF1 [5′-C(T/G)TTAAACCIGA(A/G)CGCCTCCAGGC-3′] and PGPOLR1 [5′-GG(T/C)(A/G)TGIA(A/G)CCA(A/G)(A/G)CIAG(T/G)GGCCA-3′] and PGPOLF2 [5′-AC(T/C)TGGT(C/T)(C/T)(G/C)(G/C)A(A/G)GGCCCTGGAGG-3′] and PGPOLR2 [5′-G(A/G)(T/C)(A/G)GGIGTICCTTTIGAGACCCA-3′], respectively. Additional diagnostic PCR with PTLV-specific primers was performed on specimens with negative results for the generic 222-bp tax fragments. Assays described previously were used for PTLV-1 env and STLV-3 LTR and HTLV-2 env PCR (6, 21). For HTLV-4 detection, a nested PCR assay was developed based on the HTLV-4 tax sequence by using the external primers 1863TF1 (5′-CTCCTTCTTTCAGTCCGTGCGGAG-3′) and 1863TR1 (5′-GGGGTAGTCAGGTTTGGCTGGTAT-3′) and the internal primers 1863TF2 (5′-CCTACCGCAACGGATGTCTTGAAA-3′) and 1863TR2 (5′-TATGGCGCCGGTGTGATGATAAAG-3′) and standard conditions to generate a 275-bp fragment. Percentage nucleotide divergence was calculated by using the gap program in the Wisconsin package (Genetics Computer Group, Madison, WI) (22). Sequences were aligned by using the clustal w program (23); gaps were removed, and distance-based trees were generated by using the Kimura two-parameter model in conjunction with the neighbor-joining method in the mega program (version 2.1) and maximum likelihood analysis in the paup* program as described in detail elsewhere (6, 14). The reliability of the final topology of the trees was tested with 1,000 bootstrap replicates.
Primate Taxonomic Nomenclature. The nomenclature used here is as defined elsewhere (24). NHPs were coded by using the first letter of the genus and the first two letters of the species names with their house names or codes within parentheses. Cercopithecus mona (Cmo; mona monkey), Cercopithecus neglectus (Cne; De Brazza's guenon), Cercopithecus mitis (Cmi; Sykes's monkey), Cercopithecus nictitans (Cni; greater spot-nosed guenon), Cercopithecus pogonias (Cpo; crested mona monkey), Chlorocebus aethiops (Cae; African green monkey), Cercocebus torquatus (Cto; red-capped mangabey), Cercocebus agilis (Cag; agile mangabey), Miopithecus ogouensis (Mog; talapoin monkey), Allenopithecus nigroviridis (Ani; Allen's swamp monkey), Mandrillus sphinx [Msp; mandrill (mnd)], Papio anubis [Pan; olive baboon (bab)], Papio cynocephalus (Pcy; yellow baboon), Papio hamadryas (Pha; sacred baboon), Papio ursinus (Ppu; chacma baboon), Papio papio (Ppa; Guinea baboon), Piliocolobus badius (Pba; red colobus monkey), Macaca tonkeana (Mto; Celebes macaque), Pan troglodytes (Ptr; chimpanzee), Pan paniscus (Ppn; bonobo), and Gorilla gorilla (Ggo; western lowland gorilla).
Results
Serologic Detection of PTLV-Like Infection in Persons Exposed to NHPs. Approximately 200 persons from each of 11 villages in southern Cameroon participated in the study (Fig. 1). Then, 930 persons (38.8%) who reported exposure to NHP blood and body fluids, mainly through hunting and butchering, were selected for testing for PTLV antibodies. A total of 97 (10.4%) specimens were enzyme immunoassay reactive; 90 (9.7%) of those were also reactive in the WB assay. A broad range of WB profiles were seen (Fig. 2), including HTLV-1-like (n = 10, 1.1%), HTLV-2-like (n = 5, 0.5%), HTLV-positive but untypeable (n = 13, 1.4%), and HTLV-indeterminate (n = 62, 6.7%).
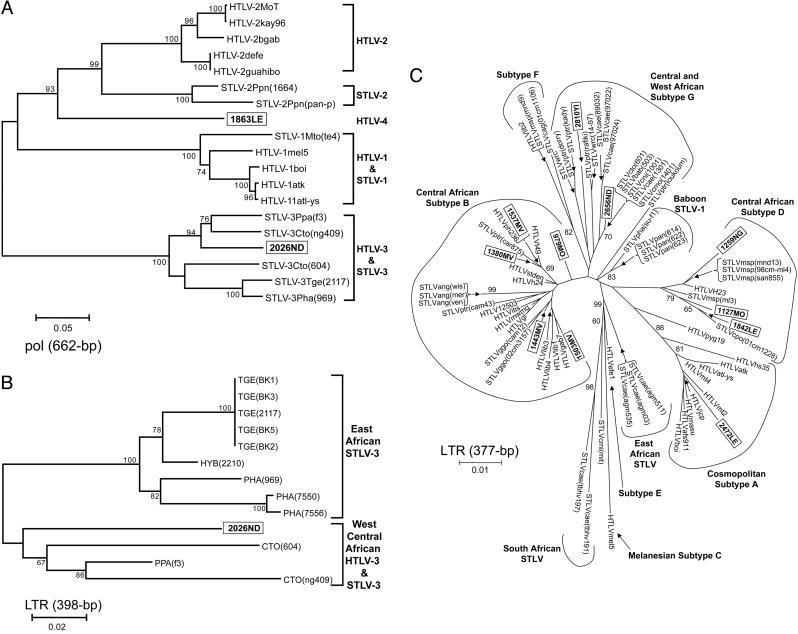
Identification of Human Infection with Novel PTLVs. DNA from peripheral blood mononuclear cells available from 86 of the 90 WB-reactive specimens was then subjected to PCR amplification of several viral regions. DNA was not available from 4 persons with HTLV-indeterminate WB results. Viral sequences from 13 persons were obtained by the use of this strategy and included sequences from 9 of 10 persons with HTLV-1-like WB reactivity, 1 of 5 persons with HTLV-2-like WB reactivity, and 3 of 58 persons with HTLV-indeterminate WB results (Table 1 and Fig. 2). PTLV sequences were not amplified from the peripheral blood mononuclear cells from any of the 13 persons with HTLV-positive but untypeable WB profiles. The 13 HTLV-infected persons were from six different lowland forest sites in southern Cameroon (Fig. 1) and included men and women who reported multiple opportunities for contact with the blood and body fluids of NHPs (Table 1). Viral sequences were analyzed phylogenetically (Fig. 3) along with African and global representatives of HTLV and STLV because PTLV diversity is influenced more by geography than by primate species (1–9).
Table 1. NHP exposures for HTLV-infected central African hunters.
| NHP exposure†
|
|||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HTLV WB profile*
|
HTLV group
|
Age, yr
|
Hunting technique
|
Hunt
|
Butcher
|
Pet
|
|||||||||||
| ID | Site | Nearest PTLV | Sex | m | c | g | m | c | g | m | c | g | Reported injuries | ||||
| 1842 | LE | 1 | 1 | Subtype D, mandrill STLV-1 | M | 32 | x | ||||||||||
| 1863 | LE | 2 | 4 | Distinct from all major PTLV groups | M | 48 | Snare | x | x | x | Bitten/scratched by wild animal | ||||||
| 2472 | LE | 1 | 1 | Subtype A, cosmopolitan | F | 27 | x | ||||||||||
| 979 | MO | 1 | 1 | Subtype B, central African | M | 30 | Gun | x | x | Bitten by monkey | |||||||
| 1127 | MO | IND | 1 | Subtype D, mandrill STLV-1 | M | 44 | Gun, snare | x | x | ||||||||
| 1380 | MV | 1 | 1 | Subtype B, central African | F | 55 | x | x | x | ||||||||
| 1443 | MV | 1 | 1 | Subtype B, central African | F | 71 | x | x | x | ||||||||
| 1503 | MV | 1 | 1 | Subtype B, central African | F | 75 | x | x | x | ||||||||
| 1537 | MV | 1 | 1 | Subtype B, central African | M | 39 | Gun, snare | x | x | ||||||||
| 2026 | ND | 1 | 3 | STLV-3 | M | 63 | Snare | x | x | ||||||||
| 2656 | ND | 1 | 1 | Subtype G, central West African STLV-1 | M | 65 | Gun | x | x | x | |||||||
| 1259 | NG | IND | 1 | Subtype D, mandrill STLV-1 | M | 71 | Gun, snare | x | x | Bitten/scratched by wild animal | |||||||
| 2810 | YI | IND | 1 | Subtype G, central west African STLV-1 | M | 55 | Snare | x | x | ||||||||
IND, indeterminate.
m, monkey; c, chimpanzee; g, gorilla.
Fig. 3.
Phylogenetic relationships of PTLV polymerase (pol; 662 bp) (A), STLV-3 LTR (398 bp) (B), and PTLV-1 LTR (377 bp) (C) sequences by neighbor-joining analysis. Sequences generated in the current study are shown in boxes. NHP taxon codes are provided in Materials and Methods. Support for the branching order was determined by 1,000 bootstrap replicates; only values of 60% or more are shown. Branch lengths are proportional to the evolutionary distance (scale bar) between the taxa.
The most notable finding was the discovery of a human virus that is distinct from all known PTLV lineages, with 26–34% and 18–25% nucleotide divergence in the conserved pol and tax genes, respectively, a range of divergence similar to that among PTLV-1, PTLV-2, and PTLV-3 (8). This virus formed a separate phylogenetic lineage with a long branch length and significant bootstrap support in both the pol (Fig. 3A) and tax (data not shown) trees. Identical topologies were obtained by using maximum likelihood analysis (data not shown). These results suggest a long independent evolution that is slightly closer to PTLV-2 than to PTLV-1 or PTLV-3. Phylogenetic analyses combined with GenBank blast searches show that this is the only known virus in this previously undescribed group. For these reasons, this virus, which we designate HTLV-4, qualifies as the original member of a novel species in the deltaretrovirus genus. Following the guidelines of the International Committee on Taxonomy of Viruses (25) and pending formal classification, we propose primate T-lymphotropic virus 4 (PTLV-4) as the name for this species and HTLV-4(1863LE) as the prototype strain. HTLV-4 was found in a 48-yr-old male hunter (1863LE) from the southern forests of Cameroon who had an HTLV-2-like WB result, but with weak reactivity to the p24 gag and K55 type-specific peptide (Fig. 1). He reported hunting monkeys, chimpanzees, and gorillas and being bitten and scratched by a wild animal, although he did not specify the animal.
We also document, with significant phylogenetic bootstrap support in both the pol (Fig. 3A) and tax (data not shown) regions, evidence of human infection within the PTLV-3 group. LTR sequences from this virus, which we designate HTLV-3, clusters with STLV-3 viruses present in west African NHPs (7, 8), as would be expected of a zoonotic infection acquired from infected primates in close geographic proximity (Fig. 3B). HTLV-3 was found in a 63-yr-old man (2026ND) from the southern forests of Cameroon who reported hunting and butchering monkeys (Table 1), and who had an HTLV-1-like WB result but with a weak MTA-1 type specific peptide reactivity (Fig. 1). The fact that this virus falls within the diversity of a group of STLVs originally identified >10 years ago (9) without evidence of a human counterpart to date (6–9, 17, 18) suggests that this infection was most likely acquired zoonotically through exposure to the blood or body fluids of a NHP hunted in this region (7, 8). However, the present data cannot exclude the possibility that this infection is not a recent cross-species transmission.
We also found a broad diversity of HTLV-1 viruses in this population. Of the 11 HTLV-1-like LTR sequences, 2 did not fall within any of the known HTLV-1 subtypes but clustered clearly within a clade that comprised only STLV-1 from central and west Africa (Fig. 3C). One of these viruses clustered with good bootstrap support with STLV-1 from monkeys in Cameroon (99% nucleotide identity) and was from a 65-yr-old man (2656ND) from the southern forests of Cameroon who reported that he hunted and butchered monkeys and kept a gorilla as a pet (Table 1). The second virus was phylogenetically related to STLV-1 recently identified in chimpanzees and red colobus monkeys (26) (97% nucleotide identity) and was from a 55-yr-old man (2810YI) who reported hunting and butchering monkeys (Table 1). Plasma from person 2810YI demonstrated an absence of p24 reactivity and thus showed an indeterminate WB profile. The presence of these viruses in NHPs and now hunters suggests that these persons were infected zoonotically. We refer to this distinct clade as HTLV-1 subtype G.
Three persons (1259NG, 1127MO, and 1842LE) from different villages had HTLV-1 subtype D viruses, known to infect geographically overlapping populations of humans, mandrills, and crested mona monkeys (Cercopithecus pogonias) in central Africa (Fig. 3C) (5). The HTLVs from these three individuals shared 97–98% nucleotide identity with STLVs found in mandrills and crested mona monkeys. Two of the three viruses were found in hunters, both with absent p24 reactivity and thus indeterminate WB profiles (Table 1), providing indirect evidence of cross-species transmission between humans and mandrills and humans and crested mona monkeys within subtype D, further supporting the claims of cross-species transmission of this subtype (5). These results are consistent with human infection with SFV from mandrills, which we documented previously (16), and suggest that the frequent hunting of mandrills may explain the widespread transmission of mandrill retroviruses.
Five persons (979MO, 1380MV, 1443MV, 1503MV, and 1537MV) were infected with HTLV-1 subtype B viruses, which are endemic among humans in central Africa and which are believed to have originated from STLV-1 in this region (1–4, 27) (Fig. 3C). Thus, these five subtype B viruses may have been acquired zoonotically from STLV-1-infected primates or from human-to-human transmission or both. For example, HTLVs found in two persons from the same village (1537MV and 1380MV) shared 99% nucleotide identity with a chimpanzee STLV-1 from Cameroon (28) and clustered with this STLV-1 with significant bootstrap support providing evidence for cross-species infections in this primate-exposed population (Fig. 3C). In addition, a 71-year-old woman (1443MV) from the same village, who reported butchering gorillas, was infected with a virus most closely related to STLV-1 found in gorillas from Cameroon (98% nucleotide identity) (7, 28), although without significant phylogenetic bootstrap support (Fig. 3C). The very close genetic relationships (>97% nucleotide identity) of these HTLV-1s with STLV-1s from monkeys and apes found in west central Africa strengthens the possibility of relatively recent cross-species infections occurring in this region. Interestingly, person 1503MV is also WB-positive for SFV (16), indicating that zoonotic transmission in an individual is not limited to a single retrovirus. Infection with more than one retrovirus may provide a biologic setting that could alter the pathogenicity and transmissibility of these viruses.
One person (2472LE) was infected with the HTLV-1 subtype A virus, a clade consisting of sequences from only globally disseminated HTLV-1 (1–4); thus, this infection was most likely acquired through human-to-human transmission.
DNA specimens from the remaining 73 WB reactive persons were all negative by the generic PCR assay for tax sequences and for four other sequences specific for each PTLV clade, including HTLV-4. These negative PCR results may be due to the presence of low proviral loads, seroconversion, infection with additional highly divergent viruses, abortive infections, or nonspecific cross-reactivity that commonly occurs in African samples and that has been reported previously to cause increased levels of HTLV seroindeterminate WB results (29).
Discussion
We have investigated the diversity of HTLV in persons exposed to NHPs in forested regions of southern Cameroon. Our results demonstrate that HTLV diversity is far greater than previously understood. Our discovery of HTLV-3 and HTLV-4, which are both distinct from HTLV-1 and HTLV-2, effectively doubles the number of known human viruses in the deltaretrovirus genus. Although a possible recent simian origin could be recognized for HTLV-3 and at least five other HTLV-1 viruses we identified, the origin of HTLV-4 is unclear because this virus does not have a known primate counterpart. However, our data, combined with a history of other STLVs jumping from primates to humans (1–5), suggest that HTLV-4 most likely represents an ancient or recent cross-species transmission from a NHP infected with a highly divergent STLV. The possibility that HTLV-4 represents a previously unrecognized virus that is being transmitted between humans suggests a need for more screening for this virus in central African populations. Additionally, because blood banks in central Africa do not typically screen for HTLV, further spread of these viruses among central Africans may be facilitated by blood donations from infected persons. The finding that HTLV-4 and HTLV-3 are serologically indistinguishable from HTLV-1 and HTLV-2 by current assays may explain why these viruses have not been previously identified and highlight the need for improved diagnostic assays to reliably and accurately detect these viruses. Although plasma samples from the HTLV-3- and HTLV-4-infected persons were detected by an ELISA containing purified HTLV-1 and HTLV-2 viral lysates, the overall sensitivity of this screening method for detecting these novel viruses is not known. Similarly, the sensitivity of detecting HTLV-3 and HTLV-4 by using screening assays employing only HTLV-1 antigens, or the use of recombinant proteins or synthetic peptides, is also not known.
Health exams of participants and collection of information regarding person-to-person contact were not included in the current study design, and thus we were unable to assess either disease associations or secondary transmission with these HTLV infections. Therefore, clinical evaluations and longitudinal epidemiological studies are needed for persons infected with HTLV-3, HTLV-4, and STLV-1-like viruses to determine whether these viruses cause disease and are transmissible among humans, as occurs with the other HTLVs (1, 10, 11). Although STLV-3 has been reported to transform or immortalize human CD4+ lymphocytes in vitro (9), further studies are needed to determine both the cellular tropism of HTLV-3 and HTLV-4 and the ability of these viruses to transform lymphocytes. In the current study peripheral blood mononuclear cells were only available for genetic characterization of these HTLVs.
Up to 28% of some populations in central Africa have been reported to have indeterminate HTLV serology results, and the majority of these persons are negative by PCR testing, which is consistent with our findings (29). However, our identification of divergent STLV-1-like viruses (subtypes D and G) in persons with indeterminate WB results is similar to the original report of HTLV-1 subtype E and F infection in Central Africans (30). These data suggest that PCR testing is required for further evaluation of HTLV infection in persons with indeterminate WB patterns and that molecular testing can lead to the identification of new HTLV-1 subtypes. We also observed a number of HTLV-positive but PCR-negative samples despite the use of an array of generic and specific PTLV primers. The reasons for the negative PCR results in some HTLV-positive samples and the majority of seroindeterminate specimens are not known and may be related to a number of factors, including infection with divergent PTLVs, low proviral loads, or low or nonspecific antibody reactivity (29).
Our results also suggest that contact with the blood and body fluids of NHPs is a major factor in the emergence of novel HTLVs, which are known to be transmissible among humans and have the potential to cause disease (1, 10, 11). Because the hunting and butchering of wild NHPs is widespread throughout central Africa (31, 32) and both STLV-1 and STLV-3 and other simian retroviruses are known to be highly prevalent among hunted NHPs (7, 8, 19, 33), it is suspected that the zoonotic transmission of STLV and other simian pathogens is not a restricted risk. The selection of persons naturally exposed to NHPs may have enriched for the identification of STLV-like viruses and novel PTLVs in our study population that were not seen in previous studies (17, 18). However, case-control and/or cohort studies that examine specimens from both persons in direct contact with NHPs and the NHPs they are exposed to may be necessary to confirm the relationship between primate exposure and HTLV emergence observed here. Nonetheless, to our knowledge this is the first study to use both behavioral data and testing of blood specimens from persons with primate exposures in a natural setting to address this important question of PTLV emergence.
Despite the global importance of the pandemic retroviruses HIV and HTLV, surprisingly little is known about the mechanisms of emergence for this class of pathogens. Although these viruses entered into humans through cross-species transmission from infected NHPs (1–3, 34), it is widely believed that the rarity of successful cross-species transmission is the primary constraint on retrovirus emergence. However, our findings suggest that cross-species transmission is not the rate-limiting step in pandemic retrovirus emergence and that factors which occur after cross-species transmission, such as viral adaptation and evolution (35), must play a role in disease emergence and global dissemination. The results of the current study and our previous report showing SFV infection in this population indicate active and frequent cross-species retrovirus infection of persons exposed to NHPs (16). Although primate hunting is an ancient behavior, our finding of HTLVs in Cameroon with high genetic identity to STLVs sympatric to this region supports the hypothesis that cross-species infections are ongoing. A combination of ecological and anthropological factors, including urbanization and the associated demand for wild game, the availability of firearms, and access to remote regions through expanded road networks, may have provided increased opportunities for these ongoing retroviral cross-species transmissions.
Our findings also suggest that it may be possible to predict and prevent viral emergence by longitudinal surveillance of exposed populations and by the establishment of interventions to decrease risk factors such as primate hunting. The increasing evidence that primate hunting is associated with the emergence of a range of simian retroviruses (16) calls for increased public health surveillance and clinical follow-up of persons exposed to the blood and body fluids of wild NHPs. We have shown that primate exposure risks are widespread among rural villages in central Africa (16, 19). Because these populations are increasingly connected to urban communities (36, 37), there is the potential for rapid and potentially undetected global spread of these simian pathogens. The current HIV pandemic is an example of how virulent retroviruses can rapidly spread after a cross-species transmission event (34). Effective strategies to reduce primate hunting are critically needed to prevent the transmission of additional primate pathogens and also to preserve this valuable but limited resource.
Acknowledgments
We thank the government of Cameroon for permission to undertake this study. We thank A. Boupda, L. Zekeng, S. Koulla-Shiro, H. Mbogos, L. Kaptue, K. Long, A. Weil, the Hopkins Cameroon Program staff, and the U.S. Embassy in Yaoundé for assistance in carrying out the work. This work was supported by an award from the U.S. Military HIV Research Program (to D.S.B.) and grants from the National Institutes of Health Fogarty International Center (International Research Scientist Development Award Grant 5 K01 TW000003-05 to N.D.W. and AIDS International Training and Research Program Grant2D43 TW000010-17-AITRP to Chris Beyrer, Johns Hopkins Bloomberg School of Public Health, Baltimore). Additional funding was provided by the National Geographic Society Committee for Research and Exploration (to N.D.W.) and an award from the Johns Hopkins Bloomberg School of Public Health, Center for AIDS Research (Grant P30 AI42855 to N.D.W.). This research was funded in part by an appointment to the Emerging Infectious Disease Fellowship Program (to A.D.G.) administered by the Association of Public Health Laboratories and funded by the Centers for Disease Control and Prevention.
Author contributions: N.D.W., W.H., E.M.-N., F.E.M., D.L.B., T.M.F., D.S.B., and W.M.S. designed research; N.D.W., J.K.C., A.D.G., V.S., U.T., J.N.T., A.T.P., M.L., E.M.-N., and W.M.S. performed research; N.D.W. and W.M.S. contributed new reagents/analytic tools; N.D.W., W.H., T.M.F., D.S.B., and W.M.S. analyzed data; and N.D.W., W.H., M.L., F.E.M., D.L.B., T.M.F., D.S.B., and W.M.S. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: HTLV, human T-lymphotropic virus; PTLV, primate T-lymphotropic virus; STLV, simian T-lymphotropic virus; NHPs, nonhuman primates; WB, Western blot; SFV, simian foamy virus.
Data deposition: The 28 HTLV sequences reported in this paper have been deposited in the GenBank database (accession nos. AY818406–AY818433).
References
- 1.Gessain, A. & Mahieux, R. (2000) Bull. Soc. Pathol. Exot. 93, 163–171. [PubMed] [Google Scholar]
- 2.Salemi, M., Desmyter, J. & Vandamme, A. M. (2000) Mol. Biol. Evol. 17, 374–386. [DOI] [PubMed] [Google Scholar]
- 3.Salemi, M., Van Dooren, S. & Vandamme, A. M. (1999) AIDS Rev. 1, 131–139. [Google Scholar]
- 4.Slattery, J. P., Franchini, G. & Gessain, A. (1999) Genome Res. 9, 525–540. [PubMed] [Google Scholar]
- 5.Mahieux, R., Chappey, C., Georges-Courbot, M. C., Dubreuil, G., Mauclere, P., Georges, A. & Gessain, A. (1998) J. Virol. 72, 10316–10322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Van Dooren, S., Shanmugam, V., Bhullar, V., Parekh, B., Vandamme, A. M., Heneine, W. & Switzer, W. M. (2004) J. Gen. Virol. 85, 507–519. [DOI] [PubMed] [Google Scholar]
- 7.Courgnaud, V., Van Dooren, S., Liegeois, F., Pourrut, X., Abela, B., Loul, S., Mpoudi-Ngole, E., Vandamme, A., Delaporte, E. & Peeters, M. (2004) J. Virol. 78, 4700–4709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meertens, L., Mahieux, R., Mauclere, P., Lewis, J. & Gessain, A. (2002) J. Virol. 76, 259–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goubau, P., Van Brussel, M., Vandamme, A. M., Liu, H. F. & Desmyter, J. (1994) Proc. Natl. Acad. Sci. USA 91, 2848–2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yamashita, M., Ido, E., Miura, T. & Hayami, M. (1996) J. Acquired Immune Defic. Syndr. Hum. Retrovirol. 13, S124–S131. [DOI] [PubMed] [Google Scholar]
- 11.Araujo, A. & Hall, W. W. (2004) Ann. Neurol. 56, 10–19. [DOI] [PubMed] [Google Scholar]
- 12.Khabbaz, R. F., Heneine, W., George, J. R., Parekh, B., Rowe, T., Woods, T., Switzer, W. M., McClure, H. M., Murphey-Corb, M. & Folks, T. M. (1994) N. Engl. J. Med. 330, 172–177. [DOI] [PubMed] [Google Scholar]
- 13.Heneine, W., Switzer, W. M., Sandstrom, P., Brown, J., Vedapuri, S., Schable, C. A., Khan, A. S., Lerche, N. W., Schweizer, M., Neumann-Haefelin, D., et al. (1998) Nat. Med. 4, 403–407. [DOI] [PubMed] [Google Scholar]
- 14.Switzer, W. M., Bhullar, V., Shanmugam, V., Cong, M. E., Parekh, B., Lerche, N. W., Yee, J. L., Ely, J. J., Boneva, R., Chapman, L. E., et al. (2004) J. Virol. 78, 2780–2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lerche, N. W., Switzer, W. M., Yee, J. L., Shanmugam, V., Rosenthal, A. N., Chapman, L. E., Folks, T. M. & Heneine, W. (2001) J. Virol. 75, 1783–1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wolfe, N. D., Switzer, W. M., Carr, J. K., Bhullar, V. B., Shanmugam, V., Tamoufe, U., Prosser, A. T., Torimiro, J. N., Wright, A., et al. (2004) Lancet 363, 932–937. [DOI] [PubMed] [Google Scholar]
- 17.Busch, M. P., Switzer, W. M., Murphy, E. L. & Heneine, W. (2000) Transfusion 40, 443–449. [DOI] [PubMed] [Google Scholar]
- 18.Vandamme, A. M., Van Laethem, K., Liu, H. F., Van Brussel, M., Delaporte, E., de Castro Costa, C. M., Fleischer, C., Taylor, G., Bertazzoni, U., Desmyter, J., et al. (1997) J. Med. Virol. 52, 1–7. [PubMed] [Google Scholar]
- 19.Wolfe, N. D., Prosser, A. T., Carr, J. K., Tamoufe, U., Mpoudi-Ngole, E., Torimiro, J. N., LeBreton, M., McCutchan, F. E., Birx, D. L. & Burke, D. S. (2004) Emerg. Infect. Dis. 10, 2094–2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meertens, L., Rigoulet, J., Mauclere, P., Van Beveren, M., Chen, G. M., Diop, O., Dubreuil, G., Georges-Goubot, M. C., Berthier, J. L., Lewis, J., et al. (2001) Virology 287, 275–285. [DOI] [PubMed] [Google Scholar]
- 21.Switzer, W. M., Pieniazek, D., Swanson, P., Samdal, H. H., Soriano, V., Khabbaz, R. F., Kaplan, J. E., Lal, R. B. & Heneine, W. (1995) J. Virol. 69, 621–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Womble, D. D. (2000) Methods Mol. Biol. 132, 3–22. [DOI] [PubMed] [Google Scholar]
- 23.Thompson, J. D., Higgins, D. G. & Gibson, T. J. (1994) Nucleic Acids Res. 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Groves, C. (2001) Primate Taxonomy (Smithsonian Inst. Press, Washington, DC).
- 25.Linial, M. L., Fan, H., Hahn, B., Löwer, R., Neil, J., Quackenbush, S., Rethwilm, A., Sonigo, P., Stoye, J. & Tristem, M. (2004) in Virus Taxonomy, Seventh Report of the International Committee on Taxonomy of Viruses, eds. Fauquet, C. M., Mayo, M. A., Maniloff, J., Desselberger, U. & Ball, L. A. (Elsevier, London), pp. 421–440.
- 26.Leendertz, F. H., Junglen, S., Boesch, C., Formenty, P., Couacy-Hymann, E., Courgnaud, V., Pauli, G. & Ellerbrok, H. (2004) J. Virol. 78, 4352–4356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mahieux, R., Ibrahim, F., Mauclere, P., Herve, V., Michel, P., Tekaia, F., Chappey, C., Garin, B., Van Der Ryst, E., Guillemain, B., et al. (1997) J. Virol. 71, 1317–1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nerrienet, E., Meertens, L., Kfutwah, A., Foupouapouognigni, Y., Ayouba, A. & Gessain, A. (2004) J. Gen. Virol. 85, 25–29. [DOI] [PubMed] [Google Scholar]
- 29.Mahieux, R., Horal, P., Mauclere, P., Mercereau-Puijalon, O., Guillotte, M., Meertens, L., Murphy, E. & Gessain, A. (2000) J. Clin. Microbiol. 38, 4049–4057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Salemi, M., Van Dooren, S., Audenaert, E., Delaporte, E., Goubau, P., Desmyter, J. & Vandamme, A. M. (1998) Virology 246, 277–287. [DOI] [PubMed] [Google Scholar]
- 31.Bowen-Jones, E. & Pendry, S. (1999) Oryx 33, 233–246. [Google Scholar]
- 32.Brashares, J. S., Arcese, P., Sam, M. K., Coppolillo, P. B., Sinclair, A. R. & Balmford, A. (2004) Science 306, 1180–1183. [DOI] [PubMed] [Google Scholar]
- 33.Peeters, M., Courgnaud, V., Abela, B., Auzel, P., Pourrut, X., Bibollet-Ruche, F., Loul, S., Liegeois, F., Butel, C., Koulagna, D., et al. (2002) Emerg. Infect. Dis. 8, 451–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hahn, B. H., Shaw, G. M., De Cock, K. M. & Sharp, P. M. (2000) Science 287, 607–614. [DOI] [PubMed] [Google Scholar]
- 35.Antia, R., Regoes, R. R., Koella, J. C. & Bergstrom, C. T. (2003) Nature 426, 658–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wolfe, N. D., Daszak, P., Kilpatrick, A. M. & Burke, D. S. (2005) Emerg. Infect. Dis., in press. [DOI] [PMC free article] [PubMed]
- 37.Schrag, S. J. & Wiener, P. (1995) Trends Ecol. Evol. 10, 319–324. [DOI] [PubMed] [Google Scholar]