Abstract
Two distinct mechanisms can be envisioned for resistance of human immunodeficiency virus type 1 (HIV-1) reverse transcriptase (RT) to nucleoside analogs: one in which the mutations interfere with the ability of HIV-1 RT to incorporate the analog, and the other in which the mutations enhance the excision of the analog after it has been incorporated. It has been clear for some time that there are mutations that selectively interfere with the incorporation of nucleoside analogs; however, it has only recently been proposed that zidovudine (AZT) resistance can involve the excision of the nucleoside analog after it has been incorporated into viral DNA. Although this proposal resolves some important issues, it leaves some questions unanswered. In particular, how do the AZT resistance mutations enhance excision, and what mechanism(s) causes the excision reaction to be relatively specific for AZT? We have used both structural and biochemical data to develop a model. In this model, several of the mutations associated with AZT resistance act primarily to enhance the binding of ATP, which is the most likely pyrophosphate donor in the in vivo excision reaction. The AZT resistance mutations serve to increase the affinity of RT for ATP so that, at physiological ATP concentrations, excision is reasonably efficient. So far as we can determine, the specificity of the excision reaction for an AZT-terminated primer is not due to the mutations that confer resistance, but depends instead on the structure of the region around the HIV-1 RT polymerase active site and on its interactions with the azido group of AZT. Steric constraints involving the azido group cause the end of an AZT 5′-monophosphate-terminated primer to preferentially reside at the nucleotide binding site, which favors excision.
Although there are now combination therapies for human immunodeficiency virus type 1 (HIV-1) that are reasonably effective, the emergence of drug-resistant virus remains a serious problem. The approved HIV-1 therapies involve drugs that inhibit two viral enzymes, reverse transcriptase (RT) and protease (PR). RT inhibitors can be divided into two groups: nucleoside analogs and nonnucleoside inhibitors. All of the nucleoside analogs lack the 3′ OH on the ribose ring and, when incorporated into viral DNA by RT, act as chain terminators. Strains of HIV-1 that are resistant to nucleoside analogs, including zidovudine (AZT), have changes in RT. In most cases, it has been possible to prepare purified recombinant RTs that carry the mutations known to confer resistance to nucleoside analogs in vivo and to show that these same mutations also confer drug resistance in in vitro polymerization assays with the purified recombinant RT. However, despite the fact that AZT was the first nucleoside analog used to treat HIV-1 infections, AZT resistance has been difficult to study in vitro. A specific set of mutations (M41L, D67N, K70R, T215Y/F, and K219E/Q) was shown to confer resistance to AZT in vivo more than 10 years ago (10); however, it has not been possible to reliably detect resistance to AZT 5′-triphosphate (AZTTP) with recombinant RT carrying these AZT resistance mutations in simple in vitro polymerization assays.
To make matters even more confusing, there are mutations (for example, the multidrug resistance mutation Q151M) that do confer considerable resistance to AZTTP in simple in vitro polymerization reactions (19, 23). A number of possible solutions to this dilemma have been suggested, the most promising of which is this: although many of the mutations that confer resistance to nucleoside analogs do so by interfering with nucleoside incorporation into DNA, AZT resistance mutations lead to enhanced excision of AZT from the nascent DNA strand after it has been incorporated (1, 15). The simple polymerization assays originally used to study resistance in vitro failed to detect resistance to AZTTP because these simple assays did not contain all the chemical entities necessary for the excision reaction, and so this property of the enzyme was overlooked. In particular, the excision reaction, which is mechanistically the reverse of the normal polymerization reaction, requires a pyrophosphate donor which RT joins to the AZT at the 3′ primer terminus, excising it from the primer DNA.
From data already published (1, 15), it is clear that both wild-type and AZT-resistant RTs can carry out an excision reaction under carefully controlled conditions in vitro. However, several important questions remain. (i) What is the real pyrophosphate donor? Both pyrophosphate and ATP can serve as pyrophosphate donors in in vitro reactions. Which is the pyrophosphate donor in vivo?
(ii) If the mechanism of AZT resistance involves excision of the 3′ nucleoside, why is resistance specific for AZT? It is clear from careful studies done with AZT-resistant viruses in cell culture that the mutations associated with AZT resistance (M41L, D67N, K70R, T215Y/F, and K219E/Q) are selective for AZT and provide relatively little resistance to other nucleoside analogs. What causes the excision reaction to remove AZT from the 3′ end of a primer more efficiently than, for example, a dideoxy nucleoside?
(iii) What is the actual mechanism of AZT resistance? One of the puzzles is that several of the mutations known to be important for AZT resistance do not appear to be in positions to make close contact with either the DNA or with an incoming deoxynucleoside triphosphate (dNTP). What is the role of these mutations?
We have used structural analysis and biochemical assays to develop a model that provides reasonable answers to these questions and explains most of the available data. In this model, the pyrophosphate donor is ATP, and at least some of the mutations that confer resistance to AZT create or enhance an ATP binding site. The 3′ end of the primer strand can be in either of two positions in HIV-1 RT. Immediately after the incorporation of an incoming dNTP, the end of the primer strand is at the polymerase active site, where the incoming dNTP was bound. To simplify further discussion, this site hereafter will be called the N (nucleotide binding) site. However, after the dNTP is incorporated, nucleic acid translocates one base pair, by an undefined mechanism. This position (hereafter called the P, or priming, site) is the position that the primer occupies in the crystal structures of HIV-1 RT and template-primer in either the presence (8) or the absence (6, 9) of an incoming dNTP. The end of the primer must be in the N site for the excision reaction to be carried out. When the end of the primer is in the P site, there is room for the incoming dNTP to bind at the N site, but excision cannot occur. In the absence of a bound dNTP, the end of the primer can move to the N site; however, the presence of a bound dNTP would prevent the end of the primer from moving to the N site. If a dideoxy nucleotide is incorporated and there are normal dNTPs present, a stable closed ternary complex is formed (7, 8, 22). In this complex, the end of the primer is at the P site, and the N site is occupied by the dNTP. In contrast, when AZT is incorporated, the AZT interferes with the formation of the closed ternary complex (15). We believe that the problem is steric and that the large azido group interferes with either the ability of AZT to occupy the P site, or the ability of the incoming dNTP to enter the N site, or both. As a consequence, primers that have AZT at their 3′ ends have good access to the N site and are readily excised in the presence of an appropriate pyrophosphate donor. This explains the specificity of AZT resistance; nucleoside analogs that do not have a bulky 3′ substituent (for example, dideoxy and acyclic nucleosides) do not have good access to the N site after they have been incorporated into viral DNA, and they are not efficiently excised even in the presence of a pyrophosphate donor.
MATERIALS AND METHODS
Preparation of HIV-1 RT.
The open reading frames encoding wild-type HIV-1 RT and each of the M184 mutants were cloned into a plasmid similar to p6HRT-PROT (2, 3, 11). The plasmid is based on the expression vector pT5m and was introduced into the Escherichia coli strain BL21(DE3) pLysE (3, 11, 17, 20). After induction with isopropyl-β-d-thiogalactopyranoside, the plasmid expresses both the p66 form of HIV-1 RT (either wild type or a mutant) and HIV-1 PR. Approximately 50% of the overexpressed p66 RT is converted to the p51 form by HIV-1 PR, and p66-p51 heterodimers accumulate in E. coli. The p66-p51 heterodimers were purified by metal chelate chromatography (3, 11, 12).
Low dNTP extension assay.
The low dNTP extension assay has been described previously (7). Briefly, −47 sequencing primer (New England Biolabs) was 5′ end labeled with [γ-32P]ATP and T4 polynucleotide kinase. After purification, the labeled primer was annealed to single-stranded M13mp18 DNA (New England Biolabs) by heating and slow cooling. For each sample, 1.0 μg of wild-type RT or RT variant was added to the labeled template-primer in 25 mM Tris-Cl (pH 8.0), 75 mM KCl, 8.0 mM MgCl2, 2.0 mM dithiothreitol (DTT), 100 μg of bovine serum albumin (BSA)/ml, and 10.0 mM 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS). The reaction mixture was supplemented with 0.1, 0.5, or 2.0 μM concentrations of dATP, dCTP, dGTP, and dTTP. The reactions were allowed to proceed at 37°C for 15, 30, or 60 min and then halted by phenol-chloroform extraction. The samples were precipitated by the addition of one volume of isopropanol, fractionated by electrophoresis on a 6.0% polyacrylamide gel, and autoradiographed.
Strand displacement assay.
The construct PPT-PBS Litmus 28 (4) contains the polypurine tract (PPT), a long terminal repeat (U3, R, and U5), and the primer binding site (PBS) of HIV-1 RT cloned into the vector Litmus 28 (New England Biolabs). Single-stranded PPT-PBS sense DNA was generated from this clone using the M13 helper phage M13KO7 (4). As previously described (5), an oligonucleotide complementary to the PBS was 5′ end labeled with [γ-32P]ATP and T4 polynucleotide kinase. After purification, the labeled primer was annealed to the single-stranded PPT-PBS sense DNA described above, along with a 10-fold excess of four unlabeled DNA oligonucleotides, by heating and slow cooling. The four unlabeled oligonucleotides will hybridize to regions of the HIV-1 long terminal repeat 3′ of the labeled PBS oligonucleotide, and they are separated from each other by three nucleotide gaps. There is a 10-nucleotide gap between the 3′ end of the labeled PBS primer and the 5′ end of the first unlabeled oligonucleotide. For each sample, 1.0 μg of wild-type RT or RT variant was added to the labeled template-primer in 25 mM Tris-Cl (pH 8.0), 35 mM KCl, 8.0 mM MgCl2, 2.0 mM DTT, 100 μg of BSA/ml, 10.0 mM CHAPS, and 10.0 μM concentrations of dATP, dCTP, dGTP, and dTTP. The reactions were allowed to proceed at 37°C for 30 min and then halted by phenol-chloroform extraction. The samples were precipitated by the addition of one volume of isopropanol, fractionated by electrophoresis on a 6.0% polyacrylamide gel, and autoradiographed. T4 DNA polymerase, which does not have strand displacement activity, was included as a control.
Primer block excision and extension assay.
The primer used in these assays is complementary to the HIV-1 PBS sequence (5′ GTCCCTG TTCGGGCGCCA 3′). The primer was 5′ end labeled with [γ-32P]ATP and T4 polynucleotide kinase. After purification, the labeled primer was annealed to a fivefold excess of template oligonucleotide, which is based on sequence from the U5-PBS region of the HIV-1 genome (5′AGTCAGTGTGGACAATCTCTAGCAATGGCGCCCGAACAGGGACTTGAAAGCGAAAGTAAA 3′), by heating and slow cooling. The position in italics is normally an A in the pNL 4-3 sequence. It was changed to a C to alter a run of A residues. After the primer is annealed to the template, the underlined A residue will be the first base of the template strand after the double-stranded region. To block the primer, the template-primer was suspended in 25 mM Tris-Cl (pH 8.0), 35 mM KCl, 8.0 mM MgCl2, 2.0 mM DTT, 100 μg of BSA/ml, 10.0 mM CHAPS, and a 10.0 μM concentration of either AZTTP (Moravek Biochemicals), 2′,3′-dideoxythymidine 5′-triphosphate (ddTTP; Boehringer Mannheim), or 3′-deoxy-2′,3′-didehydrothymidine 5′-triphosphate (D4TTP; Moravek Biochemicals). A total of 1.0 μg of wild-type RT was added to the labeled template-primer, and the reactions were allowed to proceed at 37°C for 30 min and then halted by pheno-chloroform extraction. The samples were precipitated by the addition of one volume of isopropanol, followed by an ethanol precipitation. The blocked template-primer was then resuspended in 25 mM Tris-Cl (pH 8.0), 75 mM KCl, 8.0 mM MgCl2, 2.0 mM DTT, 100 μg of BSA/ml, and 10.0 mM CHAPS. The concentration of template-primer was 0.15 nM. Depending upon the experiment (see the figure legends for details), the reaction buffer was supplemented with varying amounts of dNTPs, nucleoside analogs (AZTTP, ddTTP, or D4TTP), and pyrophosphate donor (ATP or sodium pyrophosphate). A total of 1.0 μg of wild-type RT or variant RT was added to each reaction mixture with a final reaction volume of 50 μl; the approximate concentration of enzyme was 200 nM. The reactions were allowed to proceed for 10 min at 37°C and then halted by phenol-chloroform extraction. The samples were precipitated by the addition of one volume of isopropanol, fractionated by electrophoresis on a 15.0% polyacrylamide gel, and autoradiographed. The total amount of template-primer (blocked and unextended plus deblocked and extended) and the amount of full-length product were determined by using a PhosphorImager.
Modeling.
The programs SYBYL and O were used to prepare models of the excision complex with PPi and ATP. Starting from the structure described by Huang et al. (8), the dTTP was converted to AZTTP, and a few energy minimization steps were performed with SYBYL (if too many minimization steps are performed, a kink is introduced in the azido group that does not conform to the crystal structure of AZT). The beta and gamma phosphates of the AZTTP were removed and the alpha phosphate was connected to the 3′ OH of the primer strand. The resulting bond was too long, and O was used to manually adjust the torsion angles between the nucleotide originally at the primer terminus and the AZT 5′-monophosphate (AZTMP) to bring the P-O bond distance to an appropriate value (∼1.65 Å). Energy minimization was performed with SYBYL to optimize the position of the nucleotides and ensure that the conformations were favorable. All of the base-pairing contacts and the nucleic acid-protein contacts were checked manually. This complex represents the wild-type enzyme with an AZTMP-terminated primer at the N site. Using the program O, four AZT resistance mutations were introduced: M41L, K70R, L210W, and T215Y. The positions of the side chains were chosen so that the most preferred rotamers faced the presumptive ATP binding site. The position of W210 was adjusted to stack with Y215.
The position of the pyrophosphate was modeled based on the position of the beta and gamma phosphates of dTTP in the ternary complex (8). The starting configuration of the ATP was similar to the dTTP in the structure described by Huang et al. (8). The ATP was manually docked so that the purine moiety would stack on Y215. Both O and SYBYL were used to adjust the position and the torsion angles of the ATP so that its beta and gamma phosphates would be in positions similar to those of the beta and gamma phosphates of the dTTP in the ternary structure (8). Close contacts with the protein were relieved by manual adjustments of torsion angles and local energy minimizations.
The ternary complex (8) was used to develop a model to look for steric conflict when the 3′ end of the primer was AZTMP and the AZTMP occupied the P site. The AZTMP was built using a dTMP from the double-stranded DNA as a template. The 3′ OH of the dTMP was replaced with an azido group, and the conformational angles were adjusted by energy minimization. This AZTMP was introduced in place of the ddGMP at the primer terminus. The corresponding template nucleotide was changed from dC to dA. The C-1 and C-4 sugar ring positions of the AZTMP and the corresponding dA were the same as those for the sugars at the corresponding positions of the original structure.
RESULTS
Mutant characterizations.
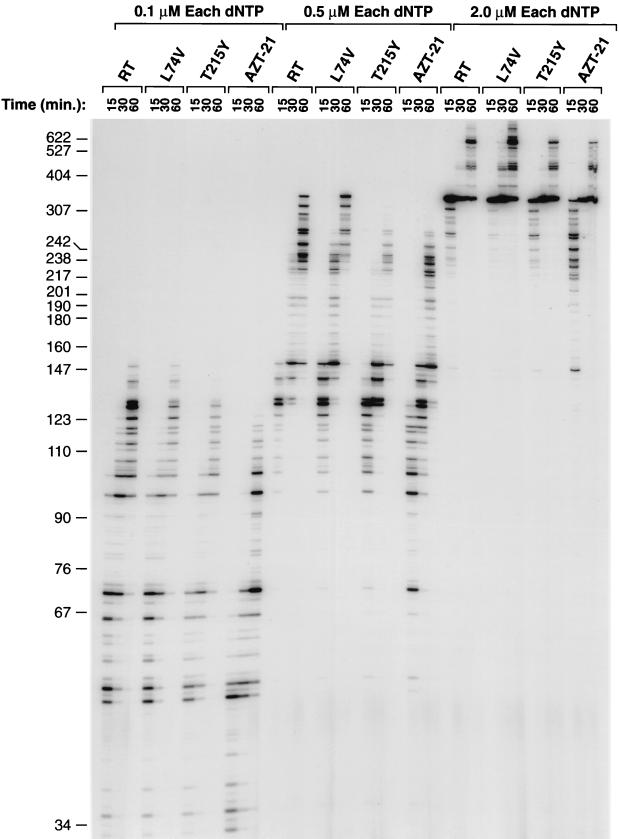
We tested RTs containing the simple AZT resistance mutation T215Y and the combination of AZT resistance mutations M41L, D67N, K70R, T215Y, and K219Q (designated AZT-21) against wild-type RT and a mutant RT resistant to the nucleoside analog dideoxyinosine (ddI) (L74V). Using poly(rC) · oligo(dG) as the template-primer, L74V had the same level of polymerase activity as wild-type RT. T215Y had a slightly decreased polymerase activity (approximately 90%), while AZT-21 had approximately 80% of the activity of wild-type RT (data not shown). Inhibition assays with AZTTP and poly(rA) · oligo(dT) showed that all of the RT variants were as sensitive to AZTTP (i.e., as likely to misincorporate AZTTP into the growing primer strand) as wild-type HIV-1 RT was, suggesting that the effects of the AZT resistance mutants are not at the level of AZTTP misincorporation (data not shown). In a low dNTP assay (Fig. 1), the RT mutants T215Y and AZT-21 were not able to polymerize as efficiently as wild-type RT or the drug-resistant variant L74V. In general, the RT with the five AZT resistance mutations in combination (AZT-21) was less efficient at polymerization than T215Y was (Fig. 1). In related assays, the mutants were tested for their processivity using both RNA and DNA templates. The results were similar to the results of the low dNTP assays described above (data not shown). The mutants were also assayed for their ability for strand displacement during polymerization. AZT-21 had the lowest strand displacement activity, followed by T215Y (data not shown). Both of the AZT-resistant variants, T215Y and AZT-21, were less efficient than wild-type RT or L74V. Again, these results match the results described above. All of these experiments showed that the AZT resistance mutations decrease the polymerization ability of HIV-1 RT but that the enzymes still retain high levels of activity. The level of activity of the enzymes in these assays was consistently RT ≈ L74V > T215Y > AZT-21.
FIG. 1.
The low dNTP extension assay tests the ability of wild-type and variant HIV-1 RTs to extend a primer using low concentrations of all four dNTPs. The strong pause site at approximately 350 nucleotides is probably due to a stem structure in the DNA which is used by the M13 bacteriophage for replication. When HIV-1 RT is polymerizing through this stem structure, the RT tends to pause.
Modeling experiments.
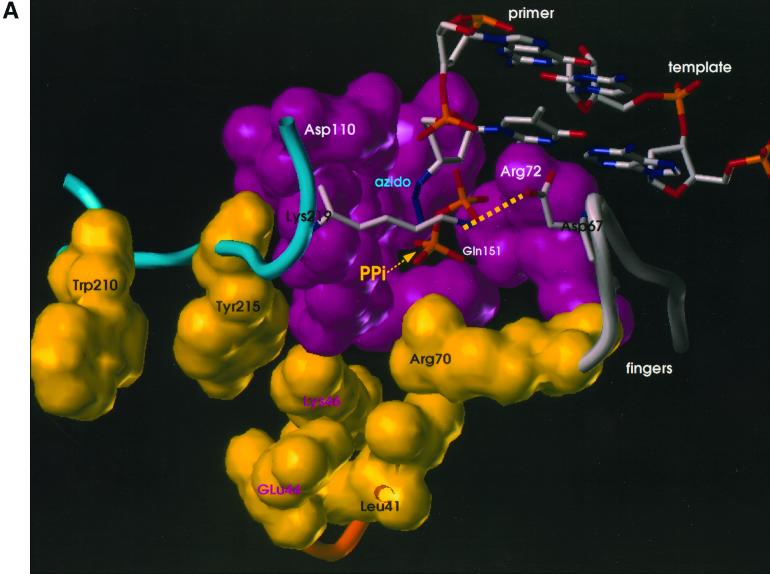
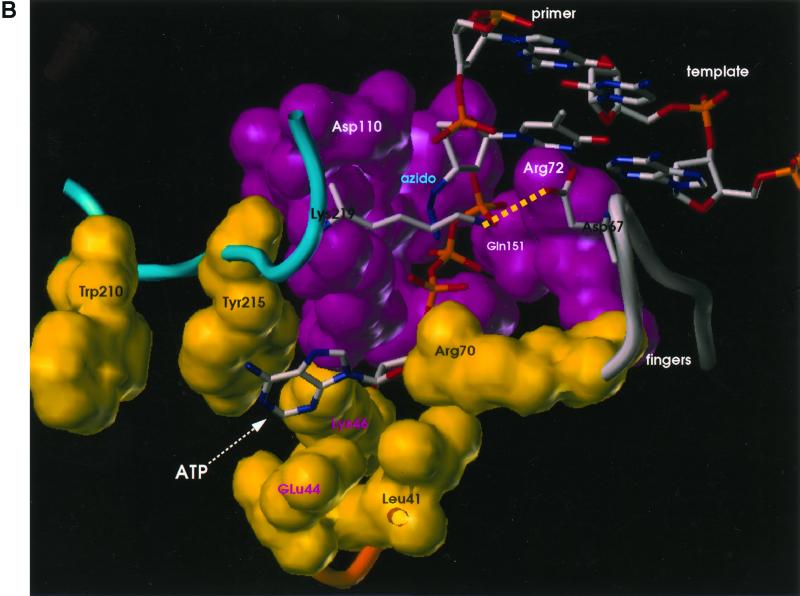
Since the mutations do not appear to affect the polymerase active site directly, how do they affect the excision of an AZTMP residue at the end of the primer strand? Two types of modeling experiments were performed. (i) Models were developed for the excision reaction in which the 3′ end of the primer strand was placed at the N site, and either PPi or ATP was bound at a position such that the phosphates of the bound ATP occupied positions corresponding to the positions of the beta and gamma phosphates of the incoming dNTP in the closed complex (8) (Fig. 2). These models should correspond to the structure of the enzyme-substrate complex just prior to excision. (ii) Starting with the structure of the binary (RT-DNA) complex with the end of the primer strand in the P site, a model was prepared with AZT at the 3′ end of the primer strand and an incoming dNTP at the N site (see Materials and Methods).
FIG. 2.
Models showing the binding of ATP and PPi to AZT-resistant HIV-1 RT. van der Waals surfaces are drawn for polymerase active site residues (magenta) and residues involved in ATP binding and AZT resistance (yellow). Mutated amino acids M41L, K70R, L210W, and T215Y are shown with black labels, and amino acids that could be involved with ATP binding but that are not mutated (E44, K46) are shown with magenta labels. The two terminal nucleotide base pairs of the template-primer are shown. The 3′ end of the primer is AZTMP; the azido group is labeled. AZT-21 has the amino acid substitutions M41L, D67N, K70R, T215Y, and K219Q. The amino acid substitution T215Y has been modeled here in order to show potential aromatic interactions with this residue. The wild-type amino acids at K219 and D67 were retained in the figure to show a potential salt bridge between the residues. As described in the text, the AZT resistance mutations at these residues will destroy this salt bridge and may increase the ability of the pyrophosphate donor to bind. K219 and D67 are shown as stick diagrams to avoid obscuring the pyrophosphate binding site. The presumptive salt bridge between Lys219 and Asp67 is shown as a dotted line (see text). Panel A shows the model with PPi bound, and panel B shows the model with ATP bound.
When the models with PPi and ATP bound in the position of the beta and gamma phosphates of the incoming dNTP were compared, it was obvious that several of the amino acids associated with AZT resistance could affect the binding of ATP but not of PPi. One of the mutant amino acids (T215Y/F) appeared to make direct contact with the adenine ring, potentially explaining the selection for a large hydrophobic amino acid on the surface of the AZT-resistant RT (Fig. 2).
The substitution of tryptophan at position 210 could help stabilize a tyrosine or phenylalanine at position 215 in a configuration that enhances the interaction with ATP. Figure 2 shows the wild-type amino acids at positions 219 (lysine) and 67 (aspartic acid). These amino acids could form a salt bridge that might interfere with access to the pyrophosphate binding site. Substitution of either amino acid (K219E/Q and/or D67N) would disrupt the salt bridge.
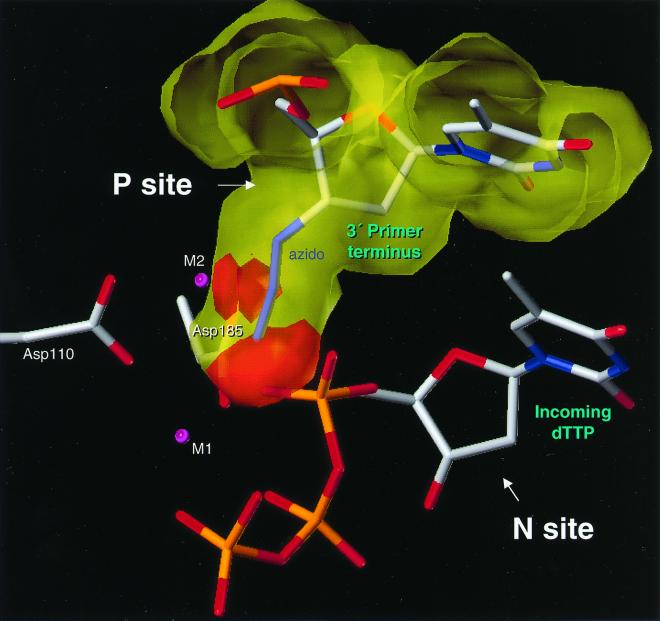
Placing AZTMP in the P site was also informative. In such a structure, if the 3′-most nucleotide in the primer is AZTMP and the end of the primer is placed in the P site, there is a steric clash between the azido group on the ribose ring and D185 (Fig. 3). The position of the azido group in the model suggests that it would also clash with an incoming dNTP, which could affect the ability of the enzyme to form the closed complex. Whether the effect is direct, on the positioning of the primer terminus, or indirect, on the ability of the incoming dNTP to bind and form a closed complex, the model suggests that AZT-terminated 3′ ends are more likely than other 3′ primer termini to be found at the N site in the position required for the excision reaction.
FIG. 3.
Steric hindrance when an AZT-terminated primer is bound to RT at the P site. The figure, based on the structure of the ternary RT-DNA-dNTP complex (8), shows that the distance between the azido of AZT and D185 would cause steric conflict; the distance between D185 and the first and second azido nitrogens is less than the sum of the van der Waals radii. The P and N sites are marked.
Excision reactions with ATP and PPi.
Although it is clear that both PPi and ATP can participate in excision reactions, the model shown in Fig. 2 predicts that, when wild-type RT and the AZT-resistant mutant are compared, there will be a difference in the relative ability of wild-type and AZT-resistant RT to excise the last nucleoside of the primer at moderate concentrations of ATP. Excision assays were performed using an AZTMP-terminated primer. In an attempt to make these in vitro assays mimic the in vivo reaction, the reactions were performed in the presence of AZTTP, all four dNTPs, either ATP or PPi, and a template that had several A's in it, to allow for the incorporation of AZTTP and the subsequent excision of the incorporated AZT during the reaction.
As shown in Fig. 4, there is a significant difference in the ability of the wild-type RT and the AZT-resistant enzymes to copy the entire template in the presence of ATP. Since there are only a few positions where AZT can be incorporated (or AZTMP can be excised), the difference is smaller than the level of resistance reported in vivo. This difference would be magnified if there were many sites to incorporate (and excise) AZTTP, as there is when the whole genome of HIV-1 is copied. However, this difference is not seen with PPi. Although PPi could support the excision of AZT, allowing RT to copy the entire template, the AZT-resistant enzymes were not more efficient than wild-type HIV-1 RT at synthesizing the full-length product if PPi was used in the assay. In fact, the AZT-resistant mutants were less efficient at excision with PPi than was wild-type RT. Therefore, the presence of PPi, either from release from dNTPs during the polymerization reaction or when present as contamination in the ATP or dNTP stock solutions will actually favor excision by the wild-type enzyme rather than by the AZT-resistant mutants. The ability of the various HIV-1 RTs to use PPi in the excision reaction was directly related to their rates of polymerization (compare Fig. 1 and 4). It should also be noted that any NTP, not just ATP, can serve as the pyrophosphate donor (data not shown). These results are what we predicted based on the model, and they agree with the data published by Meyer et al. (15) but not the data of Arion et al. (1).
FIG. 4.
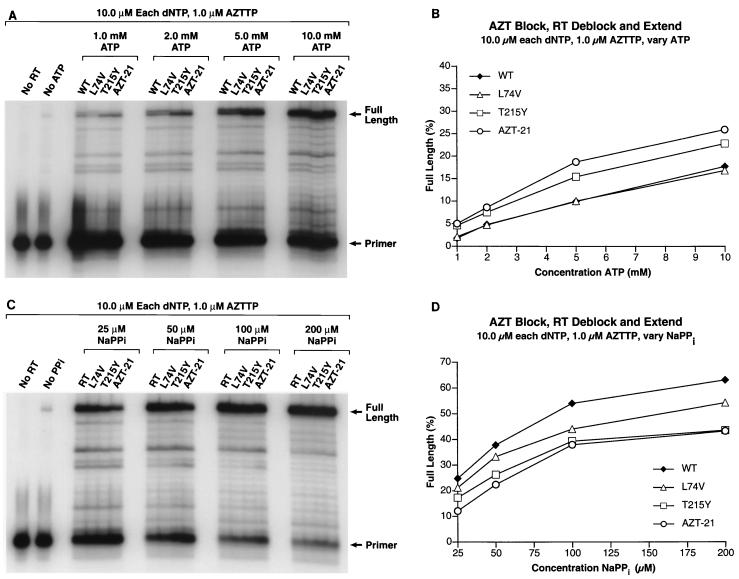
(A) The PBS primer was 5′ end labeled and annealed to the template as described in Materials and Methods. The 3′ end of the primer was blocked by the addition of an AZT residue. The ability of wild-type HIV-1 RT and the RT variants to remove the blocking AZT residue (deblocking) and to extend the freed end of the primer was tested in the presence of 10.0 μM concentrations of each dNTP, 1.0 μM AZTTP, and varying concentrations of ATP (1.0, 2.0, 5.0, and 10.0 mM) as the pyrophosphate donor. In the cell, nucleoside analogs will be present in the triphosphate form, and after a primer is deblocked there is a possibility that HIV-1 RT will add another nucleoside analog back on to the 3′ end of the primer rather than the normal dNTP, which in this case is dTTP. The addition of AZTTP to the reaction mixture is meant to reflect what can occur within the cell. A control lane with no added wild-type RT shows the pattern of the starting template-primer. A control lane to which has been added HIV-1 RT but no ATP indicates the amount of extendable primer. This could result from primer which did not get blocked by an AZT residue or from a low level of deblocking by the enzyme using the dNTPs as the pyrophosphate donor or a combination of both processes. The locations of the starting PBS primer and the fully extended primer are marked. (B) The gel in panel 4A was scanned by a PhosphorImager. In each lane, the amount of radioactivity in the full-length product was divided by the total amount of radioactivity to determine the percentage of full-length product. This value was plotted versus the level of ATP present in the reaction mixture. The percentage of full-length product in the No ATP control lane indicates that the background level is very low (<1.0%). (C) The 3′ end of the primer was blocked by the addition of an AZT residue, and the ability of wild-type HIV-1 RT and the RT variants to remove the blocking AZT residue and extend the freed end of the primer was tested in the presence of 10.0 μM concentrations of each dNTP, 1.0 μM AZTTP, and varying concentrations of NaPPi (25.0, 50.0, 100.0, and 200.0 μM) as the pyrophosphate donor. The locations of the starting PBS primer and the fully extended primer are marked. (D) The gel in panel C was scanned by a PhosphorImager. In each lane, the amount of radioactivity in the full-length product was divided by the total amount of radioactivity to determine the percentage of full-length product. This value was plotted versus the level of NaPPi present in the reaction mixture. The percentage of full-length product in the no NaPPi control lane indicates that the background level is very low compared to that for the reactions where NaPPi is present.
The data with ATP as the pyrophosphate donor show that the RT variant AZT-21 is more efficient than the RTs that contain the single amino acid substitution T215Y (Fig. 4A and B). This is consistent with a model where T215Y provides the main binding site for the ATP and some of the other amino acid substitutions help stabilize ATP binding.
Also of interest is the ability of the RTs, both wild-type and mutant, to add an untemplated base to the primer strand under low to moderate levels of ATP (Fig. 4A). The size of the full-length product is shown in the “No ATP” lane, and it is clear that wild-type RT and the L74V variant produced significant amounts of a product that is 1 nucleotide longer than the full-length product (both bands were considered full-length product in the PhosphorImager data). T215Y and AZT-21 also produced this untemplated product, but it accumulated to a lesser extent (Fig. 4A). This product was also produced if the primer was initially blocked with ddT (Fig. 5A). The extra band disappears when high levels of ATP (10.0 mM) are present and is not seen when PPi is the pyrophosphate donor. This extra band can also be seen in the control lane (No ATP lane) when high levels of dNTPs are present (Fig. 6A). Based on these observations, it appears that RT can use the excision reaction to remove an untemplated base (see Discussion).
FIG. 5.
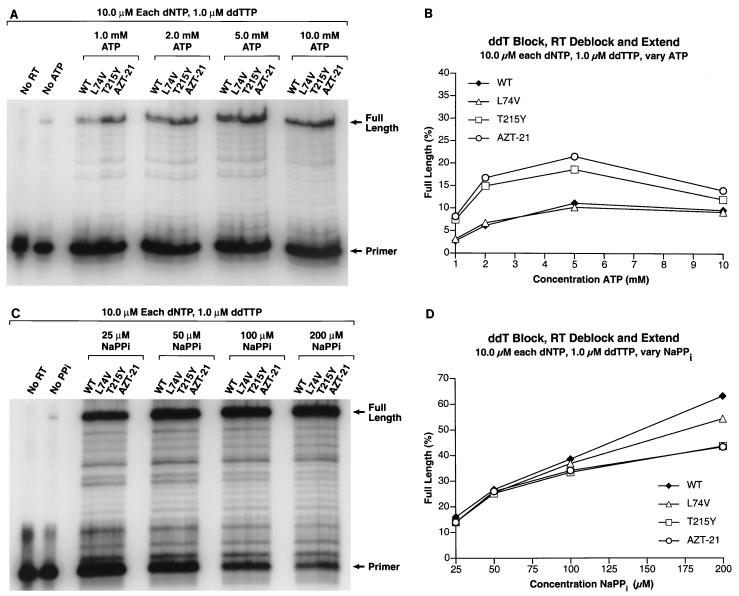
(A) The PBS primer was 5′ end labeled and annealed to the template as described in Materials and Methods. The 3′ end of the primer was blocked by the addition of a ddT residue. The ability of wild-type HIV-1 RT and the RT variants to remove the blocking ddT residue (deblocking) and extend the freed end of the primer was tested in the presence of 10.0 μM concentrations of each dNTP, 1.0 μM ddTTP, and varying concentrations of ATP (1.0, 2.0, 5.0, and 10.0 mM) as the pyrophosphate donor. Experiments using D4T as the blocking group gave similar results. (B) The gel in panel A was scanned by a PhosphorImager. In each lane, the amount of radioactivity in the full-length product was divided by the total amount of radioactivity to determine the percentage of full-length product. This value was plotted versus the level of ATP present in the reaction mixture. (C) The 3′ end of the primer was blocked by the addition of a ddT residue, and the ability of wild-type HIV-1 RT and the RT variants to remove the blocking ddT residue and extend the freed end of the primer was tested in the presence of 10.0 μM concentrations of each dNTP, 1.0 μM ddTTP, and varying concentrations of NaPPi (25.0, 50.0, 100.0, and 200.0 μM) as the pyrophosphate donor. The locations of the starting PBS primer and the fully extended primer are marked. (D) The gel in panel C was scanned by a PhosphorImager. In each lane, the amount of radioactivity in the full-length product was divided by the total amount of radioactivity to determine the percentage of full-length product. This value was plotted versus the level of NaPPi present in the reaction mixture.
FIG. 6.
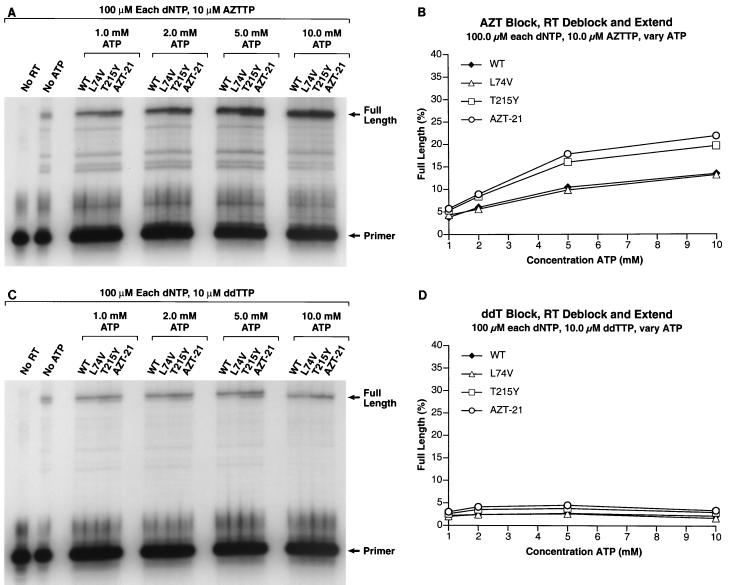
(A) The 3′ end of the primer was blocked by the addition of an AZT residue. The ability of wild-type HIV-1 RT and the RT variants to remove the blocking AZT residue (deblocking) and extend the freed end of the primer was tested in the presence of 100.0 μM concentrations of each dNTP, 10.0 μM AZTTP, and varying concentrations of ATP (1.0, 2.0, 5.0, and 10.0 mM) as the pyrophosphate donor. The ratio of dTTP:AZTTP remained at 10:1. The locations of the starting PBS primer and the fully extended primer are marked. (B) The gel in panel A was scanned by a PhosphorImager. In each lane, the amount of radioactivity in the full-length product was divided by the total amount of radioactivity to determine the percentage of full-length product. This value was plotted versus the level of ATP present in the reaction mixture. (C) The 3′ end of the primer was blocked by the addition of a ddT residue. The ability of wild-type HIV-1 RT and the RT variants to remove the blocking ddT residue (deblocking) and extend the freed end of the primer was tested in the presence of 100.0 μM concentrations of each dNTP, 10.0 μM ddTTP, and varying concentrations of ATP (1.0, 2.0, 5.0, and 10.0 mM) as the pyrophosphate donor. The ratio of dTTP:ddTTP remained at 10:1. The locations of the starting PBS primer and the fully extended primer are marked. (D) The gel in panel C was scanned by a PhosphorImager. In each lane, the amount of radioactivity in the full-length product was divided by the total amount of radioactivity to determine the percentage of full-length product. This value was plotted versus the level of ATP present in the reaction mixture. Experiments using D4TTP as the blocking group gave similar results.
Specificity of excision.
One possibility for selectivity would be that HIV-1 RT binds more tightly to a template-primer with AZTMP at the 3′ end of the primer, relative to a template-primer terminated with another nucleotide. In looking at the model in Fig. 3, it is possible that there is a difference in the overall ability of both wild-type and mutant RT to bind a double-stranded DNA with an AZT-terminated primer (relative to binding DNA duplexes whose primers are normally terminated or are terminated by a dideoxy nucleotide); however, there may be no difference in the overall binding affinity. The key issue is not the binding affinity but the position of the primer terminus relative to the polymerase active site (N site versus P site) and the ability to bind the incoming dNTP and form the closed complex. Excision can only occur when the primer terminus is in the N site; a dNTP bound at the N site will block excision.
When we tested the ability of wild-type and AZT-resistant HIV-1 RT to extend a dideoxy-terminated primer in the presence of the corresponding ddNTP, all four dNTPs, and ATP, the results obtained were those predicted by the model. Under appropriately chosen conditions, both wild-type and mutant HIV-1 RT can excise a dideoxy nucleotide from the primer terminus and can synthesize a full-length product in the presence of the relevant ddNTP, all four dNTPs, and either ATP or PPi. If the conditions are appropriately chosen (moderate concentrations of ATP), the mutant is more efficient than the wild-type enzyme at carrying out the excision reaction with dideoxy-terminated primers (Fig. 5).
If the AZT-resistant RT can efficiently excise a dideoxy from the end of a primer in vitro, why doesn't the AZT-resistant virus also show significant resistance to dideoxy nucleosides in vivo? The answer lies in the role played by the incoming dNTP. If the primer is terminated with a dideoxy nucleoside, then the incoming dNTP can bind normally, forming a closed (or dead-end) complex which is stable enough to be studied crystallographically (8). RT is unable to carry out the excision reaction when it is in a closed complex. Excision can only occur if the end of the primer is in the N site. In the closed complex, the end of the primer is in the P site and the bound dNTP prevents the primer excision. This model suggests that, if the end of the primer is AZTMP, the primer cannot be properly placed at the P site because of the steric hindrance between the azido group and D185 (Fig. 3). This idea can be tested biochemically. If the idea is correct, increasing the concentration of the dNTPs in the excision reaction mixture should cause the formation of the closed complex and prevent the excision of a dideoxy nucleoside from the end of the primer. However, even high levels of dNTPs should have little or no effect on the excision of AZT. Experiments were done to measure the ability of wild-type and mutant RTs to excise dideoxy nucleosides and AZT from the end of the primer. As expected, high concentrations of the cognate dNTP blocked the excision of the dideoxy nucleoside but had little effect on the excision of AZT (Fig. 6).
The higher concentrations of dNTPs in the reaction mixture may also be responsible for the higher level of full-length product in the control lane (Fig. 6). In our model, a dNTP could also act as a pyrophosphate donor. PhosphorImager analysis indicated that the level of full-length product in the AZTMP-blocked, No ATP control lane was two- to threefold less than the amount of full-length product with even the lowest level (0.1 mM) of ATP and wild-type RT (Fig. 6A). In the experiments with the ddTMP-blocked primer, there was less difference between the No ATP control lane and the other lanes, since excision was inhibited for all reactions (Fig. 6C).
Interestingly, with a dideoxy-terminated primer, the amount of full-length product decreased at the highest level of ATP (Fig. 5B). We have previously shown that ATP can bind at the RT active site, albeit in a configuration that appears unfavorable for incorporation (5). It appears that at a high enough concentration of ATP, ATP can bind at the N site and help to prevent the 3′ end of the primer from binding at the N site, where it could be excised. The AZT-blocked primer appears to be less sensitive than dideoxy-blocked primers to high concentrations of ATP, and it appears better able to compete for access to the N site at high concentrations of ATP.
DISCUSSION
Two distinct mechanisms can be envisioned for resistance of HIV-1 RT to nucleoside analogs: one in which the mutations interfere with the ability of HIV-1 RT to incorporate the analog, and the other in which the mutations enhance the excision of the analog after it has been incorporated. It has been clear for some time that there are mutations that selectively interfere with the incorporation of nucleoside analogs; however, it has only recently been proposed that AZT resistance can involve the excision of the nucleoside analog after it has been incorporated into viral DNA (1, 15). Although this proposal resolves some important issues, it leaves other questions unanswered. In particular, how do the AZT resistance mutations enhance excision, and what mechanism(s) causes the excision reaction to be relatively specific for AZT? We have used both structural and biochemical data to develop a model. In this model, several of the mutations associated with AZT resistance act primarily to enhance the binding of ATP, which is the most likely pyrophosphate donor in the in vivo excision reaction. These mutations serve to increase the affinity of RT for ATP so that, at physiological ATP concentrations, excision is reasonably efficient.
So far as we can determine, the specificity of the excision reaction for an AZT-terminated primer is not due to the mutations that confer resistance, but depends instead on the structure of the region around the HIV-1 RT polymerase active site and on its interactions with the azido group of AZT. The azido group of AZT appears to not interfere substantially with the binding of AZTTP at the N site or its incorporation into DNA. If the end of the primer is translocated to the P site, which would allow the incoming dNTP to bind at the N site, the azido group would have unfavorable interactions with D185. This steric clash could distort the end of the primer, which would also interfere with the binding of the incoming dNTP. There is biochemical support for this idea; a primer with an AZT-terminated end interferes with the ability of wild-type HIV-1 RT to form the closed complex with an incoming dNTP (15). This could be either an effect of the azido group (the AZT end could be in the P site, but the azido group could block the incoming dNTP from binding) or it could be the consequence of the AZT-terminated 3′ end preferentially occupying the N site, which would interfere with the binding of the incoming dNTP. Whether the effect is direct or indirect, both the model and the data support the idea that, in the presence of dNTPs, an AZT-terminated primer is much more likely to be bound at the N site than is a dideoxy-terminated primer for which there is no steric hindrance. We tested this idea directly by measuring the effects of dNTPs on the excision of either a dideoxy nucleoside or AZT. The presence of the appropriate incoming dNTP interferes with the excision of a dideoxy nucleoside, but it does not interfere with the excision of AZT. This is the basis of the selectivity of resistance for AZT; as has already been mentioned, this selectivity does not appear to be caused by the mutations that cause AZT resistance, but it appears to be an inherent property of the active site of HIV-1 RT. This effect occurs at normal levels of dNTPs (micromolar). However, ATP is present at much higher concentrations (millimolar). Although ATP does not bind to the active site of RT nearly as well as the incoming dNTP does, high concentrations of ATP can affect the excision of a dideoxy nucleoside from the 3′ end of the primer.
Under selective pressure from different nucleoside analogs, HIV-1 RT can develop resistance to multiple drugs. In some cases, it appears that a single RT can carry mutations that interfere with the incorporation of certain nucleoside analogs (the lamivudine [3TC] resistance mutation, M184V, for example), as well as the mutations (M41, D67N, K70R, L210W, and T215) that specifically facilitate the excision of AZT. When the M184V mutation is introduced into an RT in either the presence or the absence of the classical AZT resistance mutations, the enzyme becomes 3TC resistant. There is also a modest increase (ca. 5- to 10-fold) in the sensitivity of the enzyme to AZT (21); the explanation is that the M184V mutation modestly interferes with AZT excision. We propose that this is the result of an effect of the mutations at position 184 on the ability of the AZT-terminated primer to occupy the P site. As has already been discussed, if the primer has an AZT end, the incoming dNTP does not bind appropriately. Introducing an I or a V at position 184 not only changes the protein; the position of the template-primer is also altered by these mutations (18). As described above, we believe that there is steric hindrance for an AZTMP at the end of the primer with amino acid 185. Introducing either V or I at position 184 relaxes the constraints that prevent the formation of the closed complex with the incoming dNTP when the end of the primer is AZTMP, and it decreases the unfavorable interactions with the aspartic acid at position 185. This means that the 3TC resistance mutations reduce the amount of AZT excision. The fact that the introduction of the 3TC resistance mutations affects the AZT sensitivity of HIV-1 viruses that do, or do not, carry the classical AZT resistance mutations to approximately the same degree (about 5- to 10-fold) suggests that the wild-type HIV-1 RT carries out sufficient AZT excision to significantly affect the susceptibility of the wild-type virus to AZT. These data, taken in the context of the model, also suggest that, for the wild-type enzyme, the efficiency of the excision reaction is primarily limited by the relatively weak binding of ATP; the AZT resistance mutations resolve this deficiency by enhancing ATP binding.
Both wild-type HIV-1 RT and the AZT-resistant variants can excise AZT from the 3′ end of a primer both in vitro and in vivo, which raises a question: can these enzymes also excise misincorporated nucleotides? HIV-1 RT can add an untemplated base to the primer strand after it has completely copied a template (16). Although the addition of a nontemplated base is efficient in vitro, strand transfer points do not seem to be sites of increased mutation in vivo (24). In agreement with published data, we found that both wild-type HIV-1 RT and the AZT-resistant mutants efficiently add a nontemplated base. However, if there is sufficient ATP present, this nontemplated base can also be removed. This suggests that the excision reaction can correct certain DNA synthesis errors, and it raises the possibility that other types of incorporation errors could be corrected by an excision mechanism. However, the in vivo mutation rate of AZT-resistant HIV-1 is slightly higher than that of wild-type HIV-1 (13). It is possible, based on this information, that the RT excision reaction does not make a significant contribution to the fidelity of HIV-1 RT in vivo. However, it is also possible that the host DNA-dependent RNA polymerase plays a major role in determining the overall mutation rate in the HIV-1 life cycle.
There are RTs, typically from HIV-1 from patients who have been extensively treated with several nucleoside analogs, that have a large number of mutations in RT, and these viruses are broadly resistant to a variety of nucleoside analogs. In some patients, the RT has the suite of mutations associated with AZT resistance which we now believe cause excision and, in addition, it has a number of other mutations known to interfere with the incorporation of nucleoside analogs. Because some of these RTs have a large number of mutations, it is not a simple matter to predict the precise mechanism(s) of resistance associated with individual nucleoside analogs; however, it is possible, at least for some nucleoside analogs, that resistance is the result of the combined effects of a decreased ability to incorporate the analog and an enhanced ability to excise the analog after it has been incorporated.
ACKNOWLEDGMENTS
We are grateful to Hilda Marusiodis for help in preparation of the manuscript, to Pat Clark and Peter Frank for preparing purified HIV-1 RT, and to Lou Mansky for generously sharing unpublished information.
This research was sponsored by the National Cancer Institute, Department of Health and Human Services, under contract with ABL, and by the National Institute of General Medical Sciences.
REFERENCES
- 1.Arion D, Kaushik N, McCormick S, Borkow G, Parniak M A. Phenotypic mechanism of HIV-1 resistance to 3′-azido-3′-deoxythymidine (AZT): increased polymerization processivity and enhanced sensitivity to pyrophosphate of the mutant viral reverse transcriptase. Biochemistry. 1998;37:15908–15917. doi: 10.1021/bi981200e. [DOI] [PubMed] [Google Scholar]
- 2.Boyer P L, Ferris A L, Hughes S H. Cassette mutagenesis of the reverse transcriptase of human immunodeficiency virus type 1. J Virol. 1992;66:1031–1039. doi: 10.1128/jvi.66.2.1031-1039.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boyer P L, Ding J, Arnold E, Hughes S H. Subunit specificity of mutations that confer resistance to nonnucleoside inhibitors in human immunodeficiency virus type 1 reverse transcriptase. Antimicrob Agents Chemother. 1994;38:1909–1914. doi: 10.1128/aac.38.9.1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boyer P L, Lisziewicz J, Lori F, Hughes S H. Analysis of amino insertion mutations in the fingers subdomain of HIV-1 reverse transcriptase. J Mol Biol. 1999;286:995–1008. doi: 10.1006/jmbi.1998.2508. [DOI] [PubMed] [Google Scholar]
- 5.Boyer P L, Sarafianos S G, Arnold E, Hughes S H. Analysis of mutations at positions 115 and 116 in the dNTP binding site of HIV-1 reverse transcriptase. Proc Natl Acad Sci USA. 2000;97:3056–3061. doi: 10.1073/pnas.97.7.3056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ding J, Das K, Hsiou Y, Sarafianos S G, Clark A D, Jr, Jacobo-Molina A, Tantillo C, Hughes S H, Arnold E. Structure and functional implications of the polymerase active site region in a complex of HIV-1 RT with a double-stranded DNA template-primer and an antibody Fab fragment at 2.8 Å resolution. J Mol Biol. 1998;284:1095–1111. doi: 10.1006/jmbi.1998.2208. [DOI] [PubMed] [Google Scholar]
- 7.Gao H-Q, Boyer P L, Sarafianos S G, Arnold E, Hughes S H. The role of steric hindrance in 3TC resistance of human immunodeficiency virus type-1 reverse transcriptase. J Mol Biol. 2000;300:403–418. doi: 10.1006/jmbi.2000.3823. [DOI] [PubMed] [Google Scholar]
- 8.Huang H, Chopra R, Verdine G L, Harrison S C. Structure of a covalently trapped catalytic complex of HIV-1 reverse transcriptase: implications for drug resistance. Science. 1998;282:1669–1675. doi: 10.1126/science.282.5394.1669. [DOI] [PubMed] [Google Scholar]
- 9.Jacobo-Molina A, Ding J, Nanni R G, Clark A D, Jr, Lu X, Tantillo C, Williams R L, Kamer G, Ferris A L, Clark P, Hizi A, Hughes S H, Arnold E. Crystal structure of human immunodeficiency virus type 1 reverse transcriptase complexed with double-stranded DNA at 3.0 Å resolution shows bent DNA. Proc Natl Acad Sci USA. 1993;90:6320–6324. doi: 10.1073/pnas.90.13.6320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Larder B A, Kemp S D. Multiple mutations in HIV-1 reverse transcriptase confer high-level resistance to zidovudine (AZT) Science. 1989;246:1155–1158. doi: 10.1126/science.2479983. [DOI] [PubMed] [Google Scholar]
- 11.Le Grice S F, Grüninger-Leitch F. Rapid purification of homodimer and heterodimer HIV-1 reverse transcriptase by metal chelate affinity chromatography. Eur J Biochem. 1990;187:307–314. doi: 10.1111/j.1432-1033.1990.tb15306.x. [DOI] [PubMed] [Google Scholar]
- 12.Le Grice S F, Naas T, Wohlgensinger B, Schatz O. Subunit-selective mutagenesis indicates minimal polymerase activity in heterodimer-associated p51 HIV-1 reverse transcriptase. EMBO J. 1991;10:3905–3911. doi: 10.1002/j.1460-2075.1991.tb04960.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mansky L M, Bernard L C. 3′-azido-3′-deoxythymidine (AZT) and AZT-resistant reverse transcriptase can increase the in vivo mutation rate of human immunodeficiency virus type 1. J Virol. 2000;74:9532–9539. doi: 10.1128/jvi.74.20.9532-9539.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meyer P R, Matsuura S E, So A G, Scott W A. Unblocking of chain-terminated primer by HIV-1 reverse transcriptase through a nucleotide-dependent mechanism. Proc Natl Acad Sci USA. 1998;95:13471–13476. doi: 10.1073/pnas.95.23.13471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meyer P R, Matsuura S E, Mian A M, So A G, Scott W A. A mechanism of AZT resistance: an increase in nucleotide-dependent primer unblocking by mutant HIV-1 reverse transcriptase. Mol Cell. 1999;4:35–43. doi: 10.1016/s1097-2765(00)80185-9. [DOI] [PubMed] [Google Scholar]
- 16.Patel P H, Preston B D. Marked infidelity of human immunodeficiency virus type 1 reverse transcriptase at RNA and DNA template ends. Proc Natl Acad Sci USA. 1994;91:549–553. doi: 10.1073/pnas.91.2.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rosenberg A H, Lade B N, Chui D S, Lin S W, Dunn J J, Studier F W. Vectors for selective expression of cloned DNAs by T7 RNA polymerase. Gene. 1987;56:125–135. doi: 10.1016/0378-1119(87)90165-x. [DOI] [PubMed] [Google Scholar]
- 18.Sarafianos S G, Das K, Clark A D, Jr, Ding J, Boyer P L, Hughes S H, Arnold E. Lamivudine (3TC) resistance in HIV-1 reverse transcriptase involves steric hindrance with β-branched amino acids. Proc Natl Acad Sci USA. 1999;96:10027–10032. doi: 10.1073/pnas.96.18.10027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shirasaka T, Kavlick M F, Ueno T, Gao W Y, Kojima E, Alcaide M L, Chokekijchai S, Roy B M, Arnold E, Yarchoan R, et al. Emergence of human immunodeficiency virus type 1 variants with resistance to multiple dideoxynucleosides in patients receiving therapy with dideoxynucleosides. Proc Natl Acad Sci USA. 1995;92:2398–2402. doi: 10.1073/pnas.92.6.2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Studier F W, Moffatt B A. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol. 1986;189:113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- 21.Tian H, Whitcomb J M, Limoli K, Wrin T, Winslow G, Parkin N, Smith D, Lie Y S, Bakthiari M, Shugarts D, Schooley R T, Kuritzkes D, Petropoulos C J. Zidovudine/lamivudine co-resistance is preceded by a transient period of zidovudine hypersensitivity. Antivir Ther. 1998;3(Suppl. 1):22. [Google Scholar]
- 22.Tong W, Lu C D, Sharma S K, Matsuura S, So A G, Scott W A. Nucleotide-induced stable complex formation by HIV-1 reverse transcriptase. Biochemistry. 1997;36:5749–5757. doi: 10.1021/bi962410z. [DOI] [PubMed] [Google Scholar]
- 23.Ueno T, Shirasaka T, Mitsuya H. Enzymatic characterization of human immunodeficiency virus type 1 reverse transcriptase resistant to multiple 2′,3′-dideoxynucleoside 5′-triphosphates. J Biol Chem. 1995;270:23605–23611. doi: 10.1074/jbc.270.40.23605. [DOI] [PubMed] [Google Scholar]
- 24.Yu H, Jetzt A E, Ron Y, Preston B D, Dougherty J P. The nature of human immunodeficiency virus type 1 strand transfers. J Biol Chem. 1998;273:28384–28391. doi: 10.1074/jbc.273.43.28384. [DOI] [PubMed] [Google Scholar]