Abstract
Because CD30 is highly expressed on Hodgkin's lymphoma and anaplastic large cell lymphoma, it is a promising target for immunotherapy. Soluble CD30, the extracellular domain of CD30 that is shed from the cells, can reduce the effects of CD30-targeting agents by competitive binding. In this study, we identified two epitopes on membrane-associated CD30 that are missing on soluble CD30 probably because of a conformational change upon shedding. These epitopes are potentially superior targets for immunotherapy because targeting them should be free from the competitive effects of soluble CD30. We studied 27 anti-native CD30 mAbs that were assigned to 8 different topographical epitopes. Soluble CD30 was prepared from culture supernatants of L540 cells or Karpas 299 cells. In an ELISA, the mAbs to two epitopes, Ep2 (amino acids 107–153) and Ep7 (amino acids 282–338), showed less than a 2% average cross-reactivity to soluble CD30 compared with a CD30-Fc fusion protein. In addition, these mAbs bound to CD30 on cells in the presence of an excess of soluble CD30. These epitopes (Ep2 and Ep7) are, therefore, more efficiently presented on cell-associated CD30 than on soluble CD30 (membrane-specific epitopes). Also, soluble CD30 in the sera of mice bearing L540 tumors did not form immune complexes with the membrane-specific mAbs analyzed by size-exclusion chromatography. In contrast, mAbs to the other epitopes reacted with both soluble CD30 and membrane CD30. Our results suggest that it may be possible to find membrane-specific epitopes on other immunotherapy target molecules.
Keywords: antibody therapy, Hodgkin's lymphoma, soluble CD30, monoclonal antibody, conformation
Human CD30 is a promising target for cancer immunotherapy, because CD30 is highly expressed in Hodgkin's disease and anaplastic large-cell lymphoma but is expressed on a small subset of normal lymphocytes (1–3). Several different immunotherapy strategies have been tried by using CD30 as the target (4–12). We have been developing immunotoxins targeting CD30 (13, 14), encouraged by our recent clinical trials with immunotoxins targeting other lymphoma antigens CD22 and CD25 (15, 16) that show that immunotoxins can be useful for treating some types of hematological malignancies (17).
CD30 is a 105- to 120-kDa type I transmembrane glycoprotein (18) containing six cysteine-rich domains (CRDs). The first three and the last three each make up a ligand-binding site (unpublished data). The extracellular domain of CD30 is released from the cell as a soluble 85- to 90-kDa protein, upon cleavage by cell membrane-anchored metalloproteinases (19–22). This type of cleavage is common in type I membrane proteins including other immunotherapy targets such as CD25 (23) and the erb-B2 protein (24). The level of soluble CD30 is elevated according to the progression status of the lymphoma (3, 19, 25, 26). The cleavage site(s) of CD30 has not been precisely identified, but it is assumed to be close to the plasma membrane, by analogy with other soluble receptors (20) and by studies using various deletion mutants of CD30 (22). The shedding is an important consideration for immunotherapy because the soluble form of membrane proteins can neutralize antibody-based therapeutic reagents before they reach their target on the cell membrane and/or alter the biodistribution of these agents. The binding of soluble CD30 to anti-CD30 Fv was reported in clinical studies using an immunotoxin or a bispecific recombinant antibody both derived from an anti-CD30 mAb, Ki-4 (11, 12). Also, in a mouse model, coadministration of metalloproteinase inhibitors enhanced the efficacy of a CD30 immunotoxin probably by preventing the generation of soluble CD30 (9). Despite the possible inhibitory effects on antibody-based therapy, the biochemical and antigenic characteristics of soluble CD30 need further investigation.
Many previous studies using therapeutic antibodies suggested the importance of selecting an appropriate epitope (Ep) (27, 28); the exact mechanism of the advantageous effect is still largely uncharacterized. A possible mechanism is the difference among epitopes in the susceptibility of competition by soluble forms of target antigens. To develop a useful anti-CD30 immunotoxin, we have focused our attention on the affinity and epitope of the immunotoxin (13). Our previous studies with immunotoxins demonstrated that high affinity is a key in increasing the efficacy of immunotoxins (13, 29–31), but the advantage of targeting particular epitopes remains unclear.
In this study, we examine the antigenic structure of soluble CD30 using a large panel of mAbs (total of 27). These include 21 mAbs that we produced against the extracellular domain of CD30 and 6 isolated by others (2, 13, 32–37). We have used these to identify eight different epitopes on CD30 (13, 37, 38). We also located these epitopes in the amino acid sequence of CD30 and found that three of the eight epitopes overlap with two duplicated CD30-ligand (CD30-L) binding sites on CD30 (unpublished data). We have examined the reactivity of these mAbs with soluble CD30. Surprisingly, we found that two of the eight epitopes on CD30 are unique for the membrane-type CD30 molecule and absent on soluble CD30. We call these membrane-specific epitopes. They are located in the middle of the CD30 molecule, suggesting that a dynamic conformational change in CD30 occurs upon shedding to destroy or bury the structures of the two membrane-specific epitopes. We conclude that mAbs against membrane-specific epitopes are potentially useful for CD30-targeted immunotherapy because there should be no competition by soluble CD30.
Materials and Methods
Cells. A431/CD30 (14) is a stable transformant of A431 cells that express CD30 on the cell surface. CD30-positive HD lines, L540, KM-H2, and L591; CD30-positive anaplastic large cell lymphoma lines, Karpas 299 and SU-DHL-1; and a CD30-negative (very weak) line, HL60, were cultured in Iscove's modified Dulbecco's medium (Invitrogen) supplemented with 10% FBS (HyClone).
mAbs. A total of 27 anti-CD30 mAbs were used in this study. The mAbs to the extracellular domain of human CD30 include 21 mAbs established by us (13, 37) and 6 mAbs previously produced by others (2, 32–36). Their characteristics are summarized in Table 1.
Table 1. Properties of anti-CD30 mAbs used in this study.
| Name | CL* | Kd,† nM | Western blot‡ | T-Ep§ | Ep location¶ | LBI∥ | Ref. |
|---|---|---|---|---|---|---|---|
| Ber-H2 | 1 | 3.0 | + | 1 | 66-107, 224-282 | + | 36 |
| T21 | 1 | 3.5 | + | 1 | 66-107, 224-282 | + | 13 |
| HRS-4 | 1 | 5.3 | + | 1 | 66-107, 224-282 | + | 34 |
| T13 | 1 | 3.1 | + | 1 | 66-107, 224-282 | + | 13 |
| Ki-4 | 1 | 3.7 | + | 1 | 66-107, 224-282 | + | 32 |
| T6 | 1 | 13.0 | + | 1 | 66-107, 224-282 | + | 13 |
| T103 | 1 | 2.7 | - | 1 | 66-107, 224-282 | + | 37 |
| T420 | 2b | 1.9 | - | 1 | 66-107, 224-282 | + | 37 |
| T105 | 1 | 4.2 | + | 2 | 107-153 | - | 37 |
| T215 | 1 | 5.9 | + | 2 | 107-153 | - | 37 |
| T411 | 2b | 7.5 | - | 2 | 19-153 | - | 37 |
| T426 | 2b | 6.4 | - | 2 | 19-153 | - | 37 |
| T214 | 1 | 7.7 | - | 2 | 107-153 | - | 37 |
| HeFi-1 | 1 | 2.4 | - | 3 | 19-153 | - | 33 |
| T107 | 1 | 4.5 | - | 4 | 19-153, 224-383 | + | 37 |
| T406 | 2a | ND** | - | 4 | 19-153, 224-383 | - | 37 |
| T24 | 1 | 2.2 | - | 5 | 19-153, 224-383 | + | 13 |
| T427 | 2a | 0.9 | - | 5 | 19-153, 224-383 | + | 37 |
| T25 | 1 | 6.4 | - | 6 | 282-338 | - | 13 |
| T112 | 1 | 5.8 | - | 6 | 282-338 | - | 37 |
| T405 | 2b | 5.7 | + | 7 | 282-338 | - | 37 |
| T408 | 2a | 6.4 | + | 7 | 282-338 | - | 37 |
| Ki-1 | 3 | ND | ND | 8†† | 19-68 | - | 2 |
| M67 | 1 | 6.6 | - | 8 | 19-68 | - | 35 |
| T104 | 1 | 7.4 | - | 8 | 19-68 | - | 37 |
| T302 | 1 | 12.4 | - | 8 | 19-68 | - | 37 |
| T201 | 1 | 8.3 | - | 8 | 19-153 | - | 37 |
All mAbs react to native CD30 on various cells in a FACS analysis (37).
1, IgG1; 2a, IgG2a; 2b, IgG2b; 3, IgG3. All mAbs possess κ light chain.
Affinity to recombinant CD30-HFc fusion proteins determined by an ELISA (37).
Reactivity to recombinant CD30-HFc fusion protein in Western blot (37).
Determined by mutual competition of all pair of the mAbs for the binding to CD30-HFc (38).
Minimum fragments of CD30 showing reactivity with the mAbs (unpublished data). The numbers indicate the amino acid residues of human CD30 sequence (GenBank accession no. NM_001243.2). The predicted extracellular domain is amino acids 19-383. CD30 contains two duplicate regions; thus, some mAbs reacted with two different fragments.
Ligand-binding inhibition. Inhibitory effects on interaction between CD30-ligand and CD30 (unpublished data).
Not determined
Unpublished data.
Preparation of Recombinant CD30-Human IgG1-Fc Fusion Proteins. The extracellular domain of CD30 was expressed as a fusion protein with the Fc portion of human IgG (13, 37). CD30-Fc maintains the same conformation as membrane-associated CD30 as verified by its reactivity with conformational dependent mAbs (unpublished data). Ig superfamily receptor translocation associated 2 (IRTA2)-Fc fusion protein (39) was used as the control in some assays.
Preparation and Characterization of Soluble CD30. L540 or Karpas 299 cells were grown at high concentrations (up to 2 × 107 per ml) in two-compartment cell-culture flasks (INTEGRA CL 1000 devises, INTEGRA Biosciences, Ijamsville, MD) to harvest soluble CD30 in the culture supernatants. For some experiments, the soluble CD30 was purified by using an affinity column (NHS-activated Sepharose 4 Fast Flow, Amersham Pharmacia Biosciences) on which anti-CD30 mAbs T420 and T427 (to different epitopes) were immobilized according to the manufacturer's instructions. The acid eluates (pH 2.7) containing soluble CD30 were immediately neutralized with 1 M Tris (pH 8.0) and extensively dialyzed against PBS. The purified soluble CD30s and the CD30-producing cell lysates were analyzed by SDS/PAGE or Western blotting, using a mixture of the anti-CD30 mAbs Ber-H2, T105, and T405 as described in ref. 37. To analyze the aggregation formation of the soluble CD30, the soluble CD30s in the culture supernatants were separated on a size-exclusion column (TSK SW4000, 7.5 mm × 300 mm, Tosoh Bioscience, Montgomeryville, PA) with PBS as the mobile phase, and each fraction was analyzed by CD30 ELISA (see below). A part of the purified soluble CD30 sample was also in-gel-digested with chymotrypsin or trypsin and analyzed by liquid chromatography/tandem MS to determine the peptide sequences for the protein identification (Laboratory of Proteomics and Analytical Technologies, National Institutes of Health).
CD30 ELISA. The level of soluble CD30 was measured by a sandwich ELISA with purified CD30-Fc as the standard. Microtiter plates were coated with 200 ng per 50 μl per well of rat anti-mouse IgG1 mAb (90-6551, Zymed) in PBS and added with 200 ng per 100 μl per well of anti-CD30 mAb T107 (mouse IgG1). After washing, 100 μl of samples (soluble CD30) and CD30-Fc standards with another anti-CD30 mAb (2 μg/ml) T420 (mouse IgG2b) in blocking buffer were added. For detection, incubation with horseradish peroxidase rat anti-mouse IgG2b mAb (04-6320, Zymed; 1/500 in blocking buffer with 0.05% Tween 20) and a tetramethylbenzidine substrate kit (Pierce) were used.
Effects of Soluble CD30 on the Binding of Anti-CD30 mAbs to CD30. In the first experiment, the competitive effects of soluble CD30 on the binding of the mAbs to CD30-Fc were evaluated in a competitive ELISA. Microtiter plates were coated with 20 ng of CD30-Fc per well, added with an appropriate dilution of anti-CD30 mAbs (5–20 ng/ml) with various dilutions of CD30-Fc (1–100 ng/ml), soluble CD30 (1–100 ng/ml soluble CD30s obtained in L540 or Karpas 299 cell culture supernatants), or IRTA2-Fc (1–100 ng/ml) as a negative control, and incubated for 2 h at room temperature. The bound mAbs were detected with horseradish peroxidase-labeled rat anti-mouse κ mAb (04-6620, Zymed; 1/500 dilution).
In the second experiment, the binding of each mAb to membrane CD30 on L540 cells was examined by FACS in the presence of soluble CD30, of CD30-Fc, or of IRTA2-Fc. Each mAb (100 ng/ml) was mixed with a 5-fold excess of soluble CD30 from L540 cells, CD30-Fc, or IRTA2-Fc, and then reacted with L540 cells (4 × 106 cells per ml) in PBS containing 5% FBS and 0.1% sodium azide. FACS analysis was carried out as described in ref. 37.
In the third experiment, the interaction between soluble CD30 and the selected anti-CD30 mAbs was analyzed by separation of the immune complex on a sizing column. Purified mAb (0.1 mg) was radiolabeled with 1 mCi of Na131I (PerkinElmer) (1 Ci = 37 GBq) in 0.1 M phosphate buffer at pH 7.5, using 1.5-ml polypropylene tubes coated with 10 μg of Iodogen (Pierce). After 5 min of incubation at room temperature, the sample was purified by HPLC on a TSK column (SW4000, Tosoh). Peak fractions were selected, incubated with CD30-human Fc (HFc), soluble CD30 from L540 cells, or serum of the mouse inoculated with L540 cells, and then injected to Superose 6 (Amersham Pharmacia Biosciences). The radioactivity of eluate was monitored.
Results
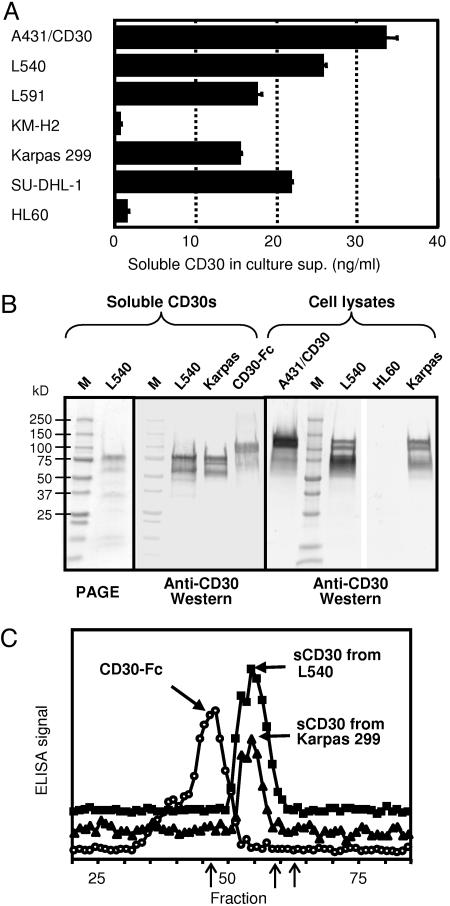
Preparation and Characterization of Soluble CD30. Soluble CD30 is produced in patients with Hodgkin's lymphoma or anaplastic large-cell lymphoma as well as by many cell lines derived from them (19, 22, 40, 41). Fig. 1A showed the levels of soluble CD30 in the culture supernatants from various cells lines. All CD30-positive cells except KM-H2 produced significant amounts of soluble CD30 (4 × 105 cells produced 15–35 ng of soluble CD30 in 36 h), whereas CD30-negative HL60 cells produced no soluble CD30. Soluble CD30 from L540 and Karpas 299 cells accumulated in the culture medium in a time-dependent manner (data not shown).
Fig. 1.
Characterization of soluble CD30s produced by cell lines. (A) Soluble CD30 levels measured by a sandwich ELISA in the culture supernatants of various cell lines. Cells were inoculated at 4 × 105 per ml and cultured for 36 h. Standard deviations of triplicate measurement are shown as bars. (B) SDS/PAGE and Western blot analysis of the soluble CD30 and cell lysates. Two micrograms of affinity-purified soluble CD30 protein were separated in 4–20% gradient SDS/PAGE gel under reducing conditions (Left). Twenty nanograms of purified proteins or 40 μg of total cell lysates were used for the Western blotting (Center and Right). (C) Size-exclusion chromatography analysis of soluble CD30s and CD30-Fc. One hundred nanograms of each protein in 0.5% BSA was separated on a TSK SW4000 column, and 0.2-ml fractions were measured by an anti-CD30 sandwich ELISA. From the standard proteins (three arrows in the fraction line: Fr.46.5 = 670,000, Fr.9 = 158,000, and Fr.63 = 44,000), the molecular masses of the major peaks of CD30-Fc and sCD30s were estimated as 645 and 317 kDa, respectively.
Soluble CD30 produced by L540 and Karpas 299 cells was purified on an affinity column on which anti-CD30 mAbs, T420 (Ep1) and T427 (Ep5), were immobilized. These purified soluble CD30s were analyzed by SDS/PAGE (Fig. 1B Left) and by Western blotting with a pool of anti-CD30 mAbs (Fig. 1B Center). The major top band in the purified soluble CD30 from L540 cells migrated ≈80 kDa in size (Fig. 1B Left), consistent with the reported size of soluble CD30. The same size bands were predominantly stained in the immunoblots of both the purified CD30 preparations and a CD30-Fc recombinant protein, although a few bands with smaller sizes were seen. liquid chromatography/tandem MS analysis of the upper band of soluble CD30 from L540 cells gave six fragments, which correspond to amino acids 51–63, 105–124, 114–126, 125–145, 265–270, and 300–320 of CD30. These results indicate that the upper band of the soluble CD30 preparations comprise the extracellular domain of CD30. The smaller minor bands in the purified soluble CD30 in SDS/PAGE were different in size from the bands stained in the Western blot, indicating that these purified samples contain smaller proteins that are not related to CD30 and that smaller bands in the immunoblot were probably small amounts of the degradation products of soluble CD30. Consistent with this, seven peptides derived from the second and third bands of the PAGE were identified as derived from unrelated human proteins (data not shown). We conclude that the purified CD30 preparation contained ≈66% CD30 (from the intensity analysis of the bands) and that the rest were unrelated proteins. We also analyzed CD30 proteins in cell lysates by the same Western blotting methods (Fig. 1B Right). Three bands (120, 105, and 80 kDa in size) were detected in all of the CD30-producing cells (A431/CD30, L540, and Karpas 299) although the intensity of these bands varied, but not in CD30-negative cells (HL60). These three bands correspond to two membrane-associated CD30 antigens (120 and 105 kDa) and a precursor molecule without glycosylation (90 kDa) described in ref. 42. These results show that the soluble CD30 protein is smaller than the membrane-type CD30s as expected, although it was not clearly distinguishable in size from the precursor protein.
The soluble CD30 from L540 and Karpas 299 cells was also analyzed by size-exclusion chromatography. As shown in Fig. 1C, soluble CD30 from both cells types eluted in the same fractions with similar shaped peaks, suggesting that the different cell lines produced the same soluble CD30 molecule(s). Soluble CD30s eluted after CD30-Fc that forms a disulfide linked homodimer between the two Fc portions. The relative positions of the elution indicate that there is a stable multimer without aggregates. Using molecular weight standards, the molecular sizes of the CD30 and CD30-Fc are estimated to be 317 and 645 kDa, respectively, which possibly agreed with the trimer formation of the tumor necrosis factor receptor (TNFR) (43), a member of the TNFR family to which CD30 belongs.
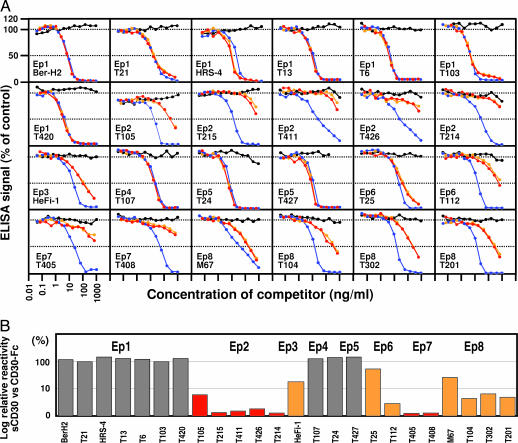
Reactivity of the Anti-CD30 mAbs to Soluble CD30. We assessed the binding of each mAb to soluble CD30 in three different types of experiments. In the first experiment, inhibition by soluble CD30 of the binding of each mAb to CD30-Fc was examined in an ELISA. As shown in Fig. 2A, soluble CD30 produced by L540 cells (red lines) or by Karpas 299 cells (orange lines) inhibited the binding of Ep1, Ep4, Ep5, and Ep6 mAbs in a dose-dependent manner as did CD30-Fc (blue lines). In contrast, very little inhibition by soluble CD30 was observed with Ep2 and Ep7 mAbs. The competitive effects of Ep3 and Ep8 mAbs were intermediate. A control Fc fusion protein, IRTA2-Fc, showed no competition (black lines), indicating that the inhibitory effects are CD30-specific. The cross-reactivity of each anti-CD30 mAb to the soluble CD30 correlated quite well with the topographical epitope, suggesting that the difference in cross-reactivity was based on a structural difference between soluble CD30 and CD30-Fc. For an objective evaluation of the difference in the cross-reactivity, we determined the 50% inhibition concentrations of soluble CD30 (from L540 cells) and compared these concentrations with those of CD30-Fc (Fig. 2B). The epitopes recognized by mAbs whose relative relativities to soluble CD30 were >70% were considered to be conserved epitopes in soluble CD30 (Ep1, Ep4, and Ep5 shown in gray; 124% average cross-reactivity), 5–70% are partially altered epitopes in soluble CD30 (Ep3, Ep6, and Ep8 shown in orange; 16.6% average cross-reactivity), and <5% are considered to be specific to the whole CD30 molecule (Ep2 and Ep7 shown in red; 2.1% average cross-reactivity).
Fig. 2.
Competitive effects of soluble CD30 on the binding of anti-CD30 mAbs to CD30-Fc. (A) The binding of each mAb to the soluble CD30 was examined in a competitive ELISA. Each mAb (20 ng/ml) was mixed with soluble CD30 from L540 cells (red lines), soluble CD30 from Karpas 299 cells (orange lines), CD30-HFc (blue lines), or IRTA2-HFc (black lines) and added to the wells coated with CD30-Fc. The bound mAbs were detected with horseradish peroxidase-labeled rat anti-mouse κ mAb. (B) Relative reactivity of each mAb to soluble CD30 compared with CD30-Fc. The soluble CD30 (from L540) concentrations required for 50% inhibition of the ELISA signal were compared with those for CD30-Fc. The soluble CD30 inhibited the binding of Ep1, Ep4, Ep5, and Ep6 mAbs (gray bars), partially inhibited the binding of Ep3 and Ep8 mAbs (orange bars), and did not inhibit the binding of Ep2 and Ep7 mAbs (red bars).
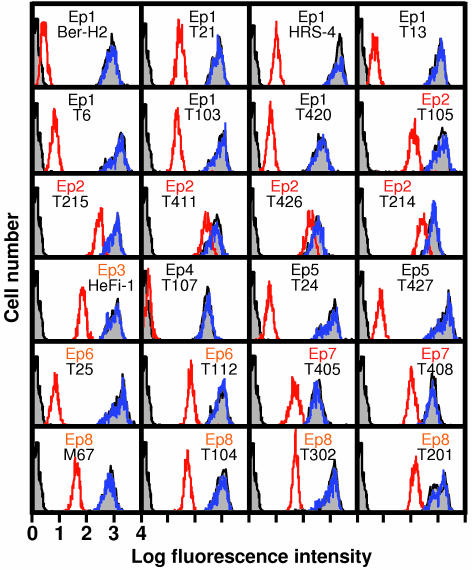
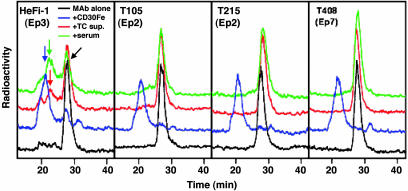
In the second experiment, the binding of each mAb to cell membrane CD30 on L540 cells was examined by FACS in the presence of a 5-fold excess of soluble CD30 (from L540), CD30-Fc, or IRTA2-Fc (Fig. 3). In accordance with the ELISA results, soluble CD30 significantly inhibited the binding of Ep1, Ep4, and Ep5 mAbs to cell membrane CD30, partially inhibited the binding of Ep3, Ep6, and Ep8 mAbs, and inhibited weakly the binding of Ep2 and Ep7 mAbs. The IRTA2-Fc proteins did not show any inhibitory effects on any mAbs. Thus, Ep2 and Ep7 are membrane-specific epitopes. These results also suggest that CD30-Fc and membrane CD30 on L540 cells had a similar structure. To confirm the absence of Ep2 and Ep7 on soluble CD30, selected mAbs were radiolabeled, and their reactivity with soluble CD30 or CD30-Fc was analyzed by size-exclusion chromatography (Fig. 4). As shown in the leftmost panel of Fig. 4, incubation of radiolabeled mAb HeFi-1 with soluble CD30 in the L540 culture supernatant or that in the serum of a mouse bearing a L540 tumor produced immune complexes (red and green arrows) that eluted faster than control mAb HeFi-1 alone (black arrow). CD30-Fc made a larger immune complex (blue arrow) because of the presence of Fc and its homodimerization in the Fc portion. In contrast, the mAbs to the membrane-specific epitopes (Ep2 and Ep7; three rightmost panels in Fig. 4) did not bind to the soluble CD30 but bound to CD30-Fc. These results clearly demonstrate the nonreactivity of Ep2 and Ep7 mAbs to soluble CD30.
Fig. 3.
Binding anti-CD30 mAbs to CD30 on L540 cells in the presence of soluble CD30. Each mAb (100 ng/ml) was mixed with a 5-fold excess of soluble CD30 from L540 cells (red lines) or IRTA2-Fc (blue lines) and then reacted to L540 cells. The two gray peaks in each panel indicated background staining (left, with second Ab only) and CD30 staining without the competitors (right), respectively. Soluble CD30 inhibited the binding of Ep1, Ep4, and Ep5 mAbs, partially inhibited the binding of Ep3, Ep6, and Ep8 mAbs, and did not inhibit the binding of Ep2 and Ep7 mAbs.
Fig. 4.
Size-exclusion chromatography analysis of radiolabeled anti-CD30 mAbs. HeFi-1 (Ep3), T105 (Ep2), T215 (Ep2), and T408 (Ep7) mAbs were labeled with 131I and incubated with appropriate amounts of CD30-HFc (blue lines), L540 culture supernatant containing soluble CD30 (red lines), a pool of sera of mice that had inoculated L540 cells (green lines), or normal mouse serum (black lines). The mixtures were analyzed with the size by running on a Sepharose column. The baseline of each chromatogram is offset for clarification. Arrows show each peak and are explained in the text.
Discussion
Here we show the presence of membrane-specific epitopes (Ep2 and Ep7) on CD30 that are not competed for by soluble CD30. These epitopes are potentially better targets for cancer immunotherapy than other epitopes because they exist only on the malignant cell surface.
Many cell-surface proteins can be cleaved by cellular enzymes to produce soluble proteins (20). If these cell-surface molecules are selected as targets for immunotherapy, the soluble forms will reduce the efficacy of the immunotherapeutic reagents by competition. In general, this drawback has been considered to be unavoidable because the soluble forms are usually entire extracellular domains of membrane proteins and show the same antigenicity as the whole molecule attached to the cell membrane (20). To our knowledge, our results are the first example that a conformational change can occur in the soluble form of a membrane protein that destroys selective epitope structures; these epitopes are, therefore, membrane-specific. The generality of the presence of membrane-specific epitopes on the other targets and the usefulness of targeting membrane-specific epitopes needs to be investigated.
Soluble CD30 has been extensively investigated as a disease marker especially for Hodgkin's lymphoma (3, 19, 25, 26, 40). Its physiological role was also examined in previous studies (44, 45). However, the information about quantitative and biochemical characteristics of soluble CD30 is very limited. Because soluble CD30 has been measured as the immunoreactivity in various sandwich ELISAs with different sets of anti-CD30 mAbs and because there is no standard CD30, the quantity of the soluble antigen was defined in arbitrary units by different investigators. One seminal work that used anti-CD30 mAbs Ki-1 and Ber-H2 in the ELISA roughly estimated 70 pg of CD30 per assay as the quantitation limit using HUT102 cell lysates (19). The same assay was used in another report (25) in which it was found that 48% of patients with Hodgkin's lymphoma produced soluble CD30 in the sera at levels of 15–2,020 unit/ml (corresponding to 1.8–283 ng/ml CD30). Because soluble CD30 will be extensively diluted when it enters the circulation, it is likely that the soluble CD30 level in the microenvironment of CD30-positive tumors can reach levels of micrograms per milliliter. This level of soluble CD30 can neutralize anti-CD30 agents. A precise determination of soluble CD30 levels in the tumors of patients needs to be determined. The formation of immune complexes composed of soluble CD30 and anti-CD30 Fv was reported in clinical studies (11, 12). We have found that the supernatants of L540 cells maintained at 2 × 107 cells per ml in a two-compartment flask contain 2.5 μg/ml soluble CD30. This finding indicates that a level of several micrograms per milliliter of soluble CD30 can be reached in a local environment by a high number of CD30-producing cells.
It has been reported that the soluble CD30 produced by L540 cells migrates at the same size (85–90 kDa) in SDS/PAGE as that prepared from the serum of a patient (40); other reports have shown that soluble CD30 from various cell lines migrates at the 85- to 90-kDa size (19, 22, 41). It is likely that the soluble CD30s produced by different cells are the same, although the precise cleavage site(s) has not been determined (22). We also have shown that soluble CD30s from L540 and Karpas 299 cells have the same antigenic properties, as well as similar sizes.
The membrane-specific epitopes (Ep2 and Ep7) are located near the middle of the extracellular domain of CD30 (unpublished data). They almost correspond to CRD3 and CRD6, amino acids 107–153 and 282–338 of the extracellular domain, which contains amino acids 19–383 (Table 1). Because these epitopes are not accessible to the mAbs after cleavage, a major conformational change of CD30 likely occurs upon shedding and destroys or buries the structure of the epitopes. Both Ep2 and Ep7 are linear epitopes that are recognized by mAbs after reduction in a Western blot (Table 1). Because the sequences of CRD3 and CRD6 are almost identical in the 20 amino acids in the beginning of the domains, Ep2 should be close to the unique region at the end of the CRD3; for the same reason, Ep7 should also be located close to the end of the CRD6. mAbs to these epitopes do not inhibit ligand binding to CD30 (ref. 42 and unpublished data).
In conclusion, we characterized the epitopes on soluble human CD30 and on the full length of CD30 with the goal of the development of better immunotherapies for CD30-positive lymphomas. We found two membrane-specific epitopes. The presence of membrane-specific epitopes and the benefits of targeting them should be extended to other membrane proteins.
Acknowledgments
We thank Dr. Rose G. Mage (National Institutes of Health) for providing pγB1–12,14 plasmid and Anna Mazzuca for editorial assistance.
Author contributions: S.N., T.I., and I.H.P. designed research; S.N., T.I., K.N., and A.R. performed research; S.N., M.O., K.N., A.R., and M.H. contributed new reagents/analytic tools; S.N. and I.H.P. analyzed data; and S.N. and I.H.P. wrote the paper.
Abbreviations: CRD, cysteine-rich domain; Ep, epitope; HFc, human Fc; IRTA2, immunoglobulin superfamily receptor translocation associated 2.
References
- 1.Koon, H. B. & Junghans, R. P. (2000) Curr. Opin. Oncol. 12, 588–593. [DOI] [PubMed] [Google Scholar]
- 2.Stein, H., Gerdes, J., Schwab, U., Lemke, H., Mason, D. Y., Ziegler, A., Schienle, W. & Diehl, V. (1982) Int. J. Cancer 30, 445–459. [DOI] [PubMed] [Google Scholar]
- 3.Horie, R. & Watanabe, T. (1998) Semin. Immunol. 10, 457–470. [DOI] [PubMed] [Google Scholar]
- 4.Wahl, A. F., Klussman, K., Thompson, J. D., Chen, J. H., Francisco, L. V., Risdon, G., Chace, D. F., Siegall, C. B. & Francisco, J. A. (2002) Cancer Res. 62, 3736–3742. [PubMed] [Google Scholar]
- 5.Borchmann, P., Treml, J. F., Hansen, H., Gottstein, C., Schnell, R., Staak, O., Zhang, H. F., Davis, T., Keler, T., Diehl, V., et al. (2003) Blood 102, 3737–3742. [DOI] [PubMed] [Google Scholar]
- 6.Pfeifer, W., Levi, E., Petrogiannis-Haliotis, T., Lehmann, L., Wang, Z. & Kadin, M. E. (1999) Am. J. Pathol. 155, 1353–1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tian, Z. G., Longo, D. L., Funakoshi, S., Asai, O., Ferris, D. K., Widmer, M. & Murphy, W. J. (1995) Cancer Res. 55, 5335–5341. [PubMed] [Google Scholar]
- 8.Francisco, J. A., Cerveny, C. G., Meyer, D. L., Mixan, B. J., Klussman, K., Chace, D. F., Rejniak, S. X., Gordon, K. A., DeBlanc, R., Toki, B. E., et al. (2003) Blood 102, 1458–1465. [DOI] [PubMed] [Google Scholar]
- 9.Matthey, B., Borchmann, P., Schnell, R., Tawadros, S., Lange, H., Huhn, M., Klimka, A., Tur, M. K., Barth, S., Engert, A., et al. (2004) Int. J. Cancer 111, 568–574. [DOI] [PubMed] [Google Scholar]
- 10.Pasqualucci, L., Wasik, M., Teicher, B. A., Flenghi, L., Bolognesi, A., Stirpe, F., Polito, L., Falini, B. & Kadin, M. E. (1995) Blood 85, 2139–2146. [PubMed] [Google Scholar]
- 11.Schnell, R., Staak, O., Borchmann, P., Schwartz, C., Matthey, B., Hansen, H., Schindler, J., Ghetie, V., Vitetta, E. S., Diehl, V., et al. (2002) Clin. Cancer Res. 8, 1779–1786. [PubMed] [Google Scholar]
- 12.Borchmann, P., Schnell, R., Fuss, I., Manzke, O., Davis, T., Lewis, L. D., Behnke, D., Wickenhauser, C., Schiller, P., Diehl, V., et al. (2002) Blood 100, 3101–3107. [DOI] [PubMed] [Google Scholar]
- 13.Nagata, S., Onda, M., Numata, Y., Santora, K., Beers, R., Kreitman, R. J. & Pastan, I. (2002) Clin. Cancer Res. 8, 2345–2355. [PubMed] [Google Scholar]
- 14.Rozemuller, H., Chowdhury, P. S., Pastan, I. & Kreitman, R. J. (2001) Int. J. Cancer 92, 861–870. [DOI] [PubMed] [Google Scholar]
- 15.Kreitman, R. J., Wilson, W. H., White, J. D., Stetler-Stevenson, M., Jaffe, E. S., Giardina, S., Waldmann, T. A. & Pastan, I. (2000) J. Clin. Oncol. 18, 1622–1636. [DOI] [PubMed] [Google Scholar]
- 16.Kreitman, R. J., Wilson, W. H., Bergeron, K., Raggio, M., Stetler-Stevenson, M., Fitzgerald, D. J. & Pastan, I. (2001) N. Engl. J. Med. 345, 241–247. [DOI] [PubMed] [Google Scholar]
- 17.Payne, G. (2003) Cancer Cell 3, 207–212. [DOI] [PubMed] [Google Scholar]
- 18.Durkop, H., Latza, U., Hummel, M., Eitelbach, F., Seed, B. & Stein, H. (1992) Cell 68, 421–427. [DOI] [PubMed] [Google Scholar]
- 19.Josimovic-Alasevic, O., Durkop, H., Schwarting, R., Backe, E., Stein, H. & Diamantstein, T. (1989) Eur. J. Immunol. 19, 157–162. [DOI] [PubMed] [Google Scholar]
- 20.Hooper, N. M., Karran, E. H. & Turner, A. J. (1997) Biochem. J. 321, 265–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hansen, H. P., Dietrich, S., Kisseleva, T., Mokros, T., Mentlein, R., Lange, H. H., Murphy, G. & Lemke, H. (2000) J. Immunol. 165, 6703–6709. [DOI] [PubMed] [Google Scholar]
- 22.Hansen, H. P., Recke, A., Reineke, U., von Tresckow, B., Borchmann, P., von Strandmann, E. P., Lange, H., Lemke, H. & Engert, A. (2004) FASEB J. 18, 893–895. [DOI] [PubMed] [Google Scholar]
- 23.Robb, R. J. & Kutny, R. M. (1987) J. Immunol. 139, 855–862. [PubMed] [Google Scholar]
- 24.Mori, S., Mori, Y., Mukaiyama, T., Yamada, Y., Sonobe, Y., Matsushita, H., Sakamoto, G., Akiyama, T., Ogawa, M., Shiraishi, M., et al. (1990) Jpn. J. Cancer Res. 81, 489–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pizzolo, G., Vinante, F., Chilosi, M., Dallenbach, F., Josimovic-Alasevic, O., Diamantstein, T. & Stein, H. (1990) Br. J. Haematol. 75, 282–284. [DOI] [PubMed] [Google Scholar]
- 26.Nadali, G., Vinante, F., Ambrosetti, A., Todeschini, G., Veneri, D., Zanotti, R., Meneghini, V., Ricetti, M. M., Benedetti, F., Vassanelli, A., et al. (1994) J. Clin. Oncol. 12, 793–797. [DOI] [PubMed] [Google Scholar]
- 27.Cragg, M. S. & Glennie, M. J. (2004) Blood 103, 2738–2743. [DOI] [PubMed] [Google Scholar]
- 28.Spiridon, C. I., Ghetie, M. A., Uhr, J., Marches, R., Li, J. L., Shen, G. L. & Vitetta, E. S. (2002) Clin. Cancer Res. 8, 1720–1730. [PubMed] [Google Scholar]
- 29.Chowdhury, P. S. & Pastan, I. (1999) Nat. Biotechnol. 17, 568–572. [DOI] [PubMed] [Google Scholar]
- 30.Salvatore, G., Beers, R., Margulies, I., Kreitman, R. J. & Pastan, I. (2002) Clin. Cancer Res. 8, 995–1002. [PubMed] [Google Scholar]
- 31.Ho, M., Kreitman, R. J., Onda, M. & Pastan, I. (2005) J. Biol. Chem. 280, 607–617. [DOI] [PubMed] [Google Scholar]
- 32.Horn-Lohrens, O., Tiemann, M., Lange, H., Kobarg, J., Hafner, M., Hansen, H., Sterry, W., Parwaresch, R. M. & Lemke, H. (1995) Int. J. Cancer 60, 539–544. [DOI] [PubMed] [Google Scholar]
- 33.Hecht, T. T., Longo, D. L., Cossman, J., Bolen, J. B., Hsu, S. M., Israel, M. & Fisher, R. I. (1985) J. Immunol. 134, 4231–4236. [PubMed] [Google Scholar]
- 34.Engert, A., Burrows, F., Jung, W., Tazzari, P. L., Stein, H., Pfreundschuh, M., Diehl, V. & Thorpe, P. (1990) Cancer Res. 50, 84–88. [PubMed] [Google Scholar]
- 35.Gruss, H. J., Boiani, N., Williams, D. E., Armitage, R. J., Smith, C. A. & Goodwin, R. G. (1994) Blood 83, 2045–2056. [PubMed] [Google Scholar]
- 36.Schwarting, R., Gerdes, J., Durkop, H., Falini, B., Pileri, S. & Stein, H. (1989) Blood 74, 1678–1689. [PubMed] [Google Scholar]
- 37.Nagata, S., Salvatore, G. & Pastan, I. (2003) J. Immunol. Methods 280, 59–72. [DOI] [PubMed] [Google Scholar]
- 38.Nagata, S., Numata, Y., Onda, M., Ise, T., Hahn, Y., Lee, B. & Pastan, I. (2004) J. Immunol. Methods 292, 141–155. [DOI] [PubMed] [Google Scholar]
- 39.Ise, T., Maeda, H., Santora, K., Xiang, L., Kreitman, R. J., Pastan, I. & Nagata, S. (2005) Clin. Cancer Res. 11, 87–96. [PubMed] [Google Scholar]
- 40.Pfreundschuh, M., Pohl, C., Berenbeck, C., Schroeder, J., Jung, W., Schmits, R., Tschiersch, A., Diehl, V. & Gause, A. (1990) Int. J. Cancer 45, 869–874. [DOI] [PubMed] [Google Scholar]
- 41.Hansen, H. P., Kisseleva, T., Kobarg, J., Horn-Lohrens, O., Havsteen, B. & Lemke, H. (1995) Int. J. Cancer 63, 750–756. [DOI] [PubMed] [Google Scholar]
- 42.Froese, P., Lemke, H., Gerdes, J., Havsteen, B., Schwarting, R., Hansen, H. & Stein, H. (1987) J. Immunol. 139, 2081–2087. [PubMed] [Google Scholar]
- 43.Chan, F. K., Chun, H. J., Zheng, L., Siegel, R. M., Bui, K. L. & Lenardo, M. J. (2000) Science 288, 2351–2354. [DOI] [PubMed] [Google Scholar]
- 44.Hargreaves, P. G. & Al Shamkhani, A. (2002) Eur. J. Immunol. 32, 163–173. [DOI] [PubMed] [Google Scholar]
- 45.Wiley, S. R., Goodwin, R. G. & Smith, C. A. (1996) J. Immunol. 157, 3635–3639. [PubMed] [Google Scholar]