Abstract
Principles of modular design are evident in signaling networks that detect and integrate a given signal and, depending on the organism in which the network module is present, transduce this signal to affect different metabolic or developmental pathways. Here we report a global transcriptional analysis of an oxygen sensory/signaling network in Caulobacter crescentus consisting of the sensor histidine kinase FixL, its cognate response regulator FixJ, the transcriptional regulator FixK, and the kinase inhibitor FixT. It is known that in rhizobial bacteria these proteins form a network that regulates transcription of genes required for symbiotic nitrogen fixation, anaerobic and microaerobic respiration, and hydrogen metabolism under hypoxic conditions. We have identified a positive feedback loop in this network and present evidence that the negative feedback regulator, FixT, acts to inhibit FixL by mimicking a response regulator. Overall, the core circuit topology of the Fix network is conserved between the rhizobia and C. crescentus, a free-living aerobe that cannot fix nitrogen, respire anaerobically, or metabolize hydrogen. In C. crescentus, the Fix network is required for normal cellular growth during hypoxia and controls expression of genes encoding four distinct aerobic respiratory terminal oxidases and multiple carbon and nitrogen metabolic enzymes. Thus, the Fix network is a conserved sensory/signaling module whose transcriptional output has been adapted to the unique physiologies of C. crescentus and the nitrogen-fixing rhizobia.
Keywords: Caulobacter, genetic circuit, hypoxia, signal transduction, two component
A central tenet in the design of complex systems is that individual components of the system are modular. Recent work has demonstrated that modularity is a fundamental design principle not only in human engineered systems, but in biological systems as well with transcriptional (1), biochemical (2), and protein–protein (3) interaction networks exhibiting modular properties. Here, we present a global transcriptional analysis of an oxygen-responsive sensory/signaling system. The core of this system is comprised of the sensor histidine kinase FixL, its cognate response regulator FixJ, the transcriptional regulator FixK, and the kinase regulatory protein FixT. These Fix proteins form a hypoxia-induced signaling module that serves to regulate both common and distinct transcriptional outputs in related species of bacteria.
FixL histidine kinases of rhizobial species bind a single molecule of iron protoporphyrin IX (heme b) through their amino-terminal PAS domain (4), which, in the presence of molecular oxygen, forms a ferrous (Fe2+) oxyheme complex that inhibits kinase activity (5). Under low oxygen concentrations, oxygen dissociates from the iron center of heme leading to up-regulation of kinase activity, transphosphorylation of the FixJ response regulator, and increased transcription of genes required for nitrogen fixation (6). Indeed, the chemistry of biological nitrogen fixation can only occur in microaerobic/anaerobic environments, and the FixLJ two-component system ensures that genes required for this process are only expressed under such conditions. FixLJ has an additional function in the soybean symbiont, Bradyrhizobium japonicum, where it also regulates genes required for anaerobic nitrate respiration (7) and symbiotic hydrogenase activity (8). Here, we demonstrate that Caulobacter crescentus FixL (gene no. CC0759) is a heme-binding histidine kinase that has identical spectral properties to rhizobial FixL kinases. In C. crescentus, which does not fix nitrogen, respire nitrate, or metabolize hydrogen, FixL is required for normal cellular growth under low dissolved oxygen concentrations and regulates the transcription of a range of genes, including four distinct respiratory terminal oxidase complexes and multiple carbon and nitrogen metabolic enzymes.
Global transcriptional analyses of C. crescentus strains carrying a deletion of fixL and its cotranscribed response regulator, which we have annotated fixJ (gene no. CC0758), identified two additional conserved components of this network: an FNR/CAP family transcription factor homologous to the rhizobial transcription factor FixK (gene no. CC0752) (9) and a gene encoding a homolog of the rhizobial kinase regulatory protein FixT (gene no. CC0753), which acts as an inhibitor of FixL kinase activity (10). Protein sequence identity between C. crescentus FixJ, FixK, and FixT and its rhizobial counterparts was sufficiently low (48–53% for FixJ, 36–44% for FixK, and 25% for FixT) that none of these genes were annotated as members of the Fix signaling network. Nevertheless, biochemical analysis of the putative FixL protein and transcriptional analyses of deletion strains of each of the putative fix genes demonstrate that these proteins form a conserved Fix signaling module that utilizes both positive and negative feedback controls.
Materials and Methods
PAS Domain Identification and Cofactor-Binding Motif Search. The Caulobacter genome was scanned for PAS domains by using the NCBI Conserved Domain Database (CDD) (www.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml). PAS domains with a cutoff score <1 were selected and subjected to a focused blast search for structurally and biochemically defined cofactor-binding motifs. Heme–PAS hsitidine kinases were identified by scanning PAS domains in the CDD for the presence of the conserved histidine residue that ligates the heme iron (residue 200 in B. japonicum FixL) and an arginine essential for signal transduction of FixL (residue 214 in B. japonicum FixL).
Cloning, Expression, and Purification of Caulobacter and Bradyrhizobium Heme-PAS Domains. Residues 116–245 of C. crescentus FixL (GenBank accession no. NP.419576) were amplified from C. crescentus genomic DNA with KOD Hot Start DNA Polymerase (Novagen) using a standard PCR protocol. The PCR product was cloned into the EcoRI site of pET28c (Novagen), and transformed into Rosetta(DE3)pLysS (Novagen). This expression strain was grown to OD600 = 0.4 and induced with 500 μM isopropyl-β-d-thiogalactopyranoside (IPTG). Cells were harvested after 12 h and lysed, and the protein was purified by affinity chromatography using Zn2+ Chelating Sepharose Resin (Amersham Pharmacia). Fractions were run on SDS/PAGE to confirm purity. Heme-PAS domain residues 141–270 of B. japonicum FixL (GenBank accession no. P23222) were expressed from a vector and purified as described (11).
Visible Absorption Spectroscopy. Absorption spectra of C. crescentus FixL-PAS (residues 116–245) and B. japonicum FixL-PAS (residues 141–270) were collected on a Hewlett Packard 8452A diode array absorption spectrophotometer using a quartz cuvette with a 1-cm path length. The unliganded met state of FixL-PAS was prepared by dialyzing pure protein fractions against 50 mM Tris, pH 8.0/150 mM NaCl to remove residual imidazole. The ferrous (Fe2+) oxy-heme state of C. crescentus and B. japonicum FixL-PAS were prepared by reduction of the heme iron with 10 mM sodium ascorbate and subsequent addition of air-saturated Tris, pH 8.0/150 mM NaCl buffer. A ferrous carbon monoxideheme complex of FixL-PAS was prepared by adding carbon monoxide-saturated buffer to protein that had been reduced to the Fe2+ state with 10 mM sodium ascorbate.
Construction of Deletion Strains. Gene deletions of fixL (CC0759), fixJ (CC0758), fixK (CC0752), fixT (CC0753), ftrA (CC3367), and ftrB (CC1410) were generated to test their function in Caulobacter. All gene deletions described in this study were constructed in C. crescentus strain CB15. fixJ was the only deletion that was not in-frame, as it contained an Ω-cassette insertion (12). Two PCR products extending ≈500 bp from either side of the gene to be deleted were amplified from genomic DNA by using KOD Hot Start Polymerase (Novagen) and cloned into pNPTS138 (see Table 3, which is published as supporting information on the PNAS web site). The Ω cassette was subsequently cloned into the BamHI site of the pNPTS138-fixJ deletion construct. The pNPTS138 deletion vectors were transformed into wild-type Caulobacter and gene replacement was performed by a two-step sacB counterselection procedure (13). Colony PCR using a set of primers flanking each deleted gene was used to screen for the presence of the deletion.
Growth Assays Under Controlled Oxygen Conditions. C. crescentus strains CB15 (wild-type) and CB15 ΔfixL (LS3728) were grown in peptone–yeast extract medium (PYE) supplemented with 500 μM CaCl2 (14) at 30°C. Cells were cultured in a New Brunswick BioFlo 3000 (New Brunswick Scientific) at pH 7.0. Oxygen levels were measured by using a dissolved oxygen (DO) probe. The probe output was a relative value that was calibrated to medium that had been continuously stirred and bubbled with air for 30 min. DO concentration in this air-saturated medium was set as 100%. Medium that had been continuously bubbled with nitrogen for 30 min was set as 0% DO. Dissolved oxygen in the medium was controlled by adjusting the agitation rate and the ratio of air to nitrogen that was bubbled through the culture. Generation times for the wild-type and deletion strains were calculated from batch culture that was inoculated to an initial density of 0.05 OD at 660 nm.
RNA Isolation and Microarray Protocols. For microarray experiments testing the global expression profiles of deletion strains versus wild-type Caulobacter, cells were grown in PYE broth at 30°C and shaken at 250 rpm. Four cultures of each strain were grown in parallel to 0.3 OD600, pelleted at 8,000 × g in a microcentrifuge, and allowed to stand in the pellet for 10 min to deplete oxygen (to activate the FixLJ two-component system).
For a comprehensive description of the RNA extraction, microarray, and array analysis protocols used in these experiments, see ref. 15.
Results
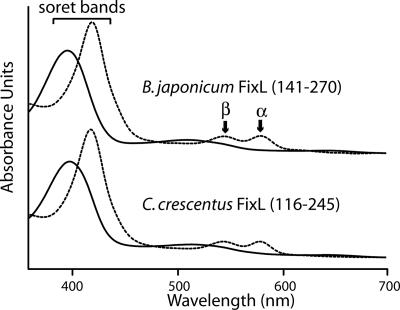
C. crescentus FixL Is a Heme-Binding Histidine Kinase. C. crescentus FixL has four predicted transmembrane helices at its amino terminus and is part of a two-gene operon with a DNA-binding response regulator sharing ≈50% sequence identity with rhizobial FixJ proteins. To determine whether C. crescentus FixL is indeed a heme-binding kinase, it was necessary to express and purify the protein. Heterologous overexpression of full-length FixL and FixL minus its amino-terminal transmembrane domain did not yield any soluble protein. However, the PAS domain of FixL (residues 116–245) was cloned, overexpressed, and purified to yield soluble protein that was suitable for spectroscopic analysis. Visible absorption spectroscopy of purified C. crescentus FixL-PAS and B. japonicum FixL-PAS (residues 141–270) (11) yielded qualitatively identical spectra (Fig. 1). C. crescentus FixL-PAS exhibits a canonical hemoprotein spectrum with a soret band at 396 nm in the ferric (Fe3+) “met” state. Reduction of the heme cofactor to its physiological ferrous (Fe2+) state permits formation of a complex with O2 with a soret band at 417 nm and α/β bands at 578 and 543 nm, respectively. A comparison of λmax values for various liganded states of C. crescentus FixL-PAS and B. japonicum FixL-PAS is shown in Table 4, which is published as supporting information on the PNAS web site. These spectroscopic data show that C. crescentus FixL, like its rhizobial homologs, is a heme-binding histidine kinase that can serve as a direct sensor of molecular oxygen.
Fig. 1.
Absorption spectrum of FixL heme-PAS domains. Visible absorption spectrum of the heme-PAS domain of C. crescentus FixL (116–245) compared to the heme-PAS domain of B. japonicum FixL (141–270). The unliganded met (Fe3+) state is shown as a solid line; the ferrous (Fe2+) state bound to molecular oxygen is shown as a dashed line.
FixL Is Necessary for Normal Growth of C. crescentus Under Hypoxia. We tested the effect of decreased oxygen concentration on growth of wild-type and ΔfixL strains by using a gas probe to monitor DO in the medium. At DO levels of 90% (222 μM) and 8% (20 μM), relative to air-saturated medium, there was no significant difference in the generation times between wild-type and ΔfixL. However, lowering DO to 0.1% (238 nM), the detection limit of our culture system, resulted in a 30% increase in generation time in ΔfixL as compared to the wild-type strain (128 ± 6 versus 97 ± 4 min) (Table 1). We predict this growth defect at DO = 0.1% would be accentuated if the wild-type and mutant strains were cultured under even lower oxygen concentrations (beyond the capacity of our apparatus). Indeed, prolonged growth of wild-type C. crescentus under oxygen-limited conditions more extreme than those tested here leads to a severe phenotype in which cells elongate up to 8-fold and exhibit cell division defects (16).
Table 1. Generation times (in minutes) of wild-type Caulobacter strain CB15 and a CB15::ΔfixL in-frame deletion strain grown under decreasing DO concentrations.
| Strain | 90% DO (222 μM) | 8% DO (20 μM) | 0.1% DO (238 nM) |
|---|---|---|---|
| Wild type | 84 ± 2 | 87 ± 4 | 97 ± 4 |
| ΔfixL | 78 ± 2 | 80 ± 3 | 128 ± 6 |
Molarity of oxygen (in parentheses) was calculated according to Henry's Law (35).
Deletion of fixL and fixJ Affects Transcription of Genes Controlling Aerobic Respiration. To determine the downstream target genes of FixL and its response regulator, FixJ, we compared the global transcriptional profiles of strains carrying gene deletions of either fixL (ΔfixL) or fixJ (ΔfixJ) to a wild-type strain by using DNA microarrays. In ΔfixL and ΔfixJ strains, which cannot respond to oxygen, genes that are activated or repressed by FixLJ were determined by depleting oxygen and comparing gene expression to a wild-type strain that has been subjected to identical hypoxic conditions.
Consistent with a model of FixL and FixJ acting together as a two-component system, microarray cluster analysis (17) identified a set of genes that are regulated in a consistent manner by these sensor/signaling proteins during hypoxia (Fig. 2A and Tables 5 and 6, which are published as supporting information on the PNAS web site). Specifically, transcription of 79 genes in ΔfixL and 71 genes in ΔfixJ exhibited significant change relative to wild-type C. crescentus (Table 5) (see Materials and Methods for microarray significance cutoff parameters). Thirty-seven of these genes changed in an equivalent manner in both deletion strains. Among the largest changes in expression (up to 12-fold) in this common set of genes are operons encoding components of three of the four respiratory terminal oxidases in C. crescentus: Cytbb3 (genes CC1401–1408), Cytbd (CC0762–0763), and Cytbo3 (CC1767–1773). An operon encoding the fourth terminal oxidase complex in C. crescentus, Cytaa3 (CC3402–3407), shows evidence of weak repression by FixLJ and is discussed below.
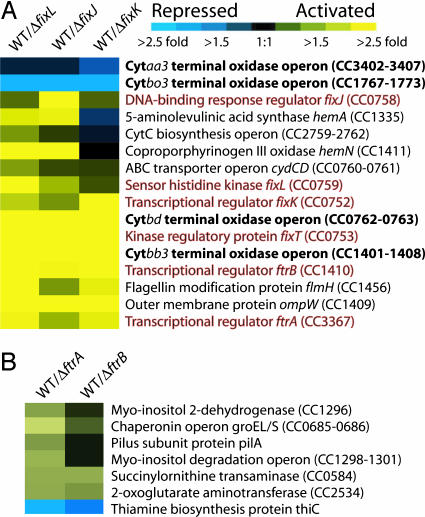
Fig. 2.
Microarray cluster presenting expression of genes in fixL, fixJ, fixK, ftrA, and ftrB deletion backgrounds relative to wild-type C. crescentus strain CB15. Relative expression of genes in wild-type C. crescentus versus mutant strains was calculated by dividing average wild-type expression by average mutant expression values (n = 4). All strains (deletion and wild-type reference) were subjected to oxygen depletion to induce the FixLJ two-component system. Data are clustered and shown for wild type versus ΔfixL, ΔfixJ, and ΔfixK (A) and ΔftrA and ΔftrB (B) strains. Yellow indicates genes that are positively regulated by a given transcription regulator (higher expression in wild-type cells versus mutant), whereas blue indicates genes that are repressed by a given transcriptional regulator (higher expression in mutant cells versus wild-type). A color scale is included above A. The respiratory terminal oxidase operons are highlighted in bold text, whereas the sensory and transcriptional regulatory components of the FixLJ two-component network are in red text. A complete list of all genes that showed statistically significant change can be found in Table 4.
Transcription of the Cytbb3 and Cytbd operons is strongly activated by FixLJ in the absence of oxygen, suggesting that these terminal oxidases are used for respiration by C. crescentus under low DO concentrations. Indeed, Cytbb3 and Cytbd terminal oxidase complexes in B. japonicum and Escherichia coli, respectively, have an extremely high affinity for O2 (Km = 7–20 nm) (18, 19). Conversely, FixLJ acts to repress transcription of the Cytbo3 operon upon oxygen depletion (Fig. 2 A and Tables 4 and 5). Measurements on bo3 and aa3 complexes from other species revealed a 20- to 50-fold lower affinity for oxygen than Cytbb3 and Cytbd (20, 21) and demonstrated that these terminal oxidase complexes function most efficiently under high DO concentrations (19, 22). Other respiration-related genes including several involved in heme biosynthesis, cytochrome c biogenesis, and electron transport are also positively regulated by FixLJ during hypoxia (Fig. 2 A and Table 5).
Multiple Downstream Regulatory Factors Are Under the Control of FixLJ. At least three transcription factors (TFs) are part of the FixLJ regulon (Figs. 2 A and 3). These include an FNR/CAP family TF (gene no. CC0752) sharing ≈40% identity with rhizobial fixK, an AraC-family TF (gene no. CC3367) we have named fix transcriptional regulator ftrA, and an FNR/CAP-family TF (gene no. CC1410) ftrB, which all exhibited 3- to 10-fold higher expression in wild-type cells compared to either ΔfixL or ΔfixJ (Table 5). Additionally, gene no. CC0753, which is annotated as a hypothetical protein but shares weak sequence homology with several response regulators, is strongly up-regulated by FixLJ, with 20-fold higher expression in wild-type cells than in ΔfixL or ΔfixJ. The regulatory role of this protein will be discussed below.
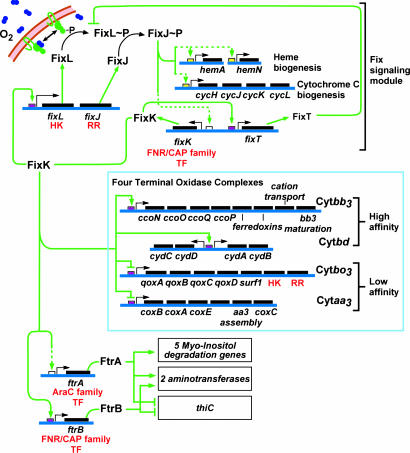
Fig. 3.
Primary topology and regulation of the FixLJ–FixK–FixT system in C. crescentus. Activity of FixL is inhibited by oxygen. Like its rhizobial homologs, we propose that, in the absence of oxygen, FixL autophosphorylates at the membrane and initiates the signaling cascade via transphosphorylation of FixJ and transcriptional activation of fixK (via the FixJ binding sequence shown in white). FixJ either directly or indirectly regulates the uncharacterized promoters (in yellow) of hemA, hemN, and the cycHJKL operon. FixK serves to regulate the promoters of all four respiratory terminal oxidases, likely through the upstream FixK box (in magenta). In addition, FixK serves as a positive regulator of fixLJ transcription and a positive regulator of fixT, which encodes a protein homologous to canonical response regulator receiver domains and serves as an inhibitor of FixLJ-mediated signaling. The transcriptional regulators ftrA and ftrB are downstream of FixK. FtrA and FtrB both serve as either direct or indirect transcriptional repressors of thiC and activators of two aminotransferases (CC0584 and CC2534). Additionally, FtrA activates transcription of genes required for myo-inositol degradation. FtrB contains an upstream FixK box (magenta), whereas the promoter of FtrB is uncharacterized (yellow). Solid lines represent direct interactions based on the presence of defined promoter sequences; dashed lines indicate interactions that may be direct or indirect and for which no common promoter element has been defined. HK, histidine kinase; RR, response regulator; TF, transcription factor.
FixK Is a Global Regulator of Terminal Oxidase Transcription. Deletion of fixK in C. crescentus (ΔfixK) results in aberrant transcription of a similar set of genes as is seen in ΔfixL and ΔfixJ (Fig. 2 A). Forty-four genes are under the regulation of FixK, 25 of which appear in the ΔfixL and ΔfixJ data sets (Table 5). Transcription of all four terminal oxidase operons are regulated by FixK under conditions of hypoxia, with the direction of regulation the same as is seen in Fig. 2 A and Table 5. Notably, the low-affinity Cytaa3-type oxidase operon is significantly repressed by fixK during hypoxia, providing evidence that transcription of all four respiratory terminal oxidase complexes is controlled by FixLJ-FixK. Bioinformatic analysis of several bacterial genomes had previously predicted the gene encoding FixK to be a transcription factor that regulates Cytbb3 transcription in C. crescentus (23). Our results demonstrate that FixK is a global regulator of respiration in C. crescentus, acting as a positive regulator of the predicted high-affinity oxidases, Cytbb3 and Cytbd, and a negative regulator of the low-affinity oxidases, Cytbo3 and Cytaa3.
We searched for cis-acting elements in the promoters of genes regulated by FixK using MAST/MEME (24) and found a conserved motif (TTGA-N6-TCAA) upstream of several genes. This motif closely resembles the binding motif for FixK in rhizobia (6, 9) and FNR in E. coli (25) (Table 2). The FixK motif is present in the upstream region of all four terminal oxidase operons, and upstream of the transcription factor ftrB, the predicted flagellar acetyltransferase/glycosyltransferase flmH (26), fixLJ, and a putative homolog of the rhizobial kinase regulatory gene, fixT (gene no. CC0753) (see Fig. 3 and Table 2). The presence of this motif upstream of fixLJ, combined with the transcriptional profiles of ΔfixL, ΔfixJ, and ΔfixK, is consistent with a model in which FixK acts to up-regulate the transcription of fixLJ (see Fig. 3). Indeed, deletion of fixK leads to a decrease (1.5-fold) in transcription from the fixLJ operon at low oxygen levels as measured on DNA microarrays, providing evidence for a positive feedback loop between FixK and fixLJ. This positive feedback was confirmed by measuring the expression of lacZ fused to the fixLJ promoter in both wild-type and ΔfixK backgrounds under low oxygen (expression was 1.7-fold higher in a wild-type background compared to ΔfixK) (Fig. 6A, which is published as supporting information on the PNAS web site).
Table 2.
Predicted FixK-binding sites in the promoter regions of C. crescentus are homologous to the FixK-binding sites of rhizobia (6) and FNR-binding sites of E. coli (21)
| Regulator | Consensus sequence | Genes/operons preceded by consensus |
|---|---|---|
| FixK (C. crescentus) | TTGAC-C-GATCAA-GC | cydAB (Cytbd), ccoNOQP (Cytbb3), qoxABCD (Cytbo3), coxAB (Cytaa3), fixT, ftrB, flmH, fixLJ |
| FixK (rhizobia) | TTGA-C--GATCAA-G- | fixNOQP (Cytbb3), fixT, nifA, rpoN1, fixLJ, fixT |
| FNR (E. coli) | AAA-TTGAT----ATCAA-TTT | cydAB (Cytbd), fumB, narX, nirB |
Selected FixK consensus sequences are all found <150 bases from the translational start site. Nucleotides that are at least 60% conserved are shown. Common names of the cytochrome terminal oxidase complexes are shown in parentheses after the names of the genes which encode them.
CC0753 Encodes FixT, a Negative Regulator of FixLJ-Mediated Signaling. In S. meliloti, the protein FixT is positively regulated by FixK and acts to inhibit the formation and/or accumulation of phosphoryl-FixL under hypoxic conditions (10). Thus, FixT is a negative regulator of FixLJ-mediated signaling. In C. crescentus, the hypothetical gene CC0753 is weakly homologous to S. meliloti FixT (25% identity) and is strongly up-regulated by FixLJ/FixK under oxygen limitation (Fig. 2 A and Table 5). To determine whether this gene has any regulatory function in the FixLJ/FixK signaling circuit, we generated an in-frame deletion and tested the expression of the fixK promoter fused to lacZ in the deletion and wild-type backgrounds. In the wild-type background under high-oxygen conditions, fixK-lacZ expression is low (Fig. 6B). In the CC0753 deletion background under high oxygen, expression of fixK-lacZ is derepressed (3.1-fold higher) (Fig. 6B). Thus, the regulatory role of C. crescentus CC0753 is consistent with this protein, like rhizobial FixT, acting as an inhibitor of FixL sensor kinase activity. Although sequence homology with S. meliloti FixT is low, we propose that C. crescentus gene CC0753 is a functional homolog of rhizobial FixT, and we have annotated this gene as such.
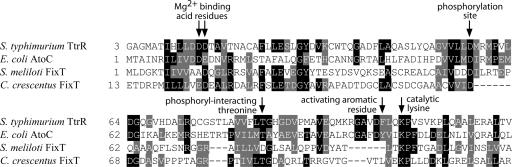
Previous studies on S. meliloti FixT reported that this protein exhibits no known homology to any protein (10, 27). However, we discovered weak homology between S. meliloti and C. crescentus FixT and the TtrR and AtoC response regulator receiver domain families (CDD score <10–4). Among the amino acids conserved between FixT and these canonical receiver domains are all of the residues that have been reported to form the catalytic active site (28), including two-acidic residues at the amino terminus that chelate Mg2+, the catalytic lysine, and the aspartic acid residue that serves as the phosphoacceptor during phosphotransfer from a cognate histidine kinase (Fig. 4). Moreover, a conserved threonine that has been demonstrated to interact with and stabilize the phosphoryl group of phosphoaspartate (28) is also conserved. The homology between FixT and canonical response regulator receiver domains, including all of the catalytic residues responsible for removing a phosphoryl group from a histidine kinase, suggests that FixT acts as an inhibitor of FixL-mediated signaling by competing for FixL phosphate. This result raises the interesting possibility that FixT is simply a response regulator mimic whose sole function is to inhibit FixL signaling via phosphatase-like activity.
Fig. 4.
Alignment of C. crescentus and S. meliloti FixT proteins against the receiver domains of the AtoC response regulator of E. coli and the TtrR response regulator of S. typhimurium. Residues that are required for histidine-to-aspartate phosphotransfer and subsequent response regulator transcriptional activity are highlighted with arrows.
FtrA and FtrB Are Regulators of Carbon and Amino Acid Metabolism. The expression of two genes encoding the transcriptional regulators, ftrA and ftrB, is strongly up-regulated by FixLJ/FixK under oxygen limitation (Fig. 2 A and Table 4). FtrA has close homologs (>90% sequence identity) of unknown function in Burkholderia mallei and B. cepecia. The closest homolog of FtrB is NarR of Paracoccus pantotrophus (44% identity), a positive regulator of respiratory nitrate reductase (narKGHJI) (29). As discussed above, ftrB contains a FixK-binding motif in its promoter region and is predicted to be a direct target of FixK; ftrA does not contain a FixK motif in its promoter. The role of FtrA and FtrB in the Fix regulon was explored by preparing strains carrying deletions of each of these genes (ΔftrA and ΔftrB) and comparing the global transcriptional profiles to wild-type cells by using DNA microarrays.
Under oxygen starvation, FtrA and FtrB regulate a small set of genes that are primarily involved in carbon and amino acid metabolism (Fig. 2B and Table 7, which is published as supporting information on the PNAS web site). A total of 26 genes in the ftrA deletion and 7 genes in the ftrB deletion exhibit significant transcriptional differences from wild-type; three of these genes are common to the two microarray data sets. In both mutant strains, the gene whose transcription is most strongly repressed is thiC (CC2029), which is expressed as a single gene. ThiC is part of an alternative segment of the thiamine biosynthesis pathway that utilizes 5-aminoimidazole ribotide, an intermediate in the de novo synthesis of purine nucleotides, as a thiamine building block. Intermediates in the purine biosynthetic pathway have been demonstrated to have a regulatory role in aerobic respiration. Specifically, 5-aminoimidazole-4-carboxamide ribonucleotide inhibits transcription of the Cytbb3 terminal oxidase complex in S. meliloti via FixK (30). In this way, the repression of thiC may modulate crosstalk between the purine biosynthetic pathway and the FixLJ/FixK signaling pathway.
Other genes that are positively regulated by FtrA include the myo-inositol degradation operon (iolBCDE) (Fig. 2B and Table 5). Notably, strains of Rhizobium leguminosarum carrying mutations in the myo-inositol catabolism operon are at a 100-fold competitive disadvantage in the environment of the root nodule (31). This finding raises the interesting question of what benefit myo-inositol catabolism may confer to these organisms under microaerobic conditions.
Discussion
Positive and Negative Feedback in Fix Signaling Systems. A series of global transcriptional analyses on deletion strains, beginning with a gene that we have demonstrated to encode a heme-binding histidine kinase, has enabled the identification and topological characterization of an entire Fix signaling system in C. crescentus. The protein components and regulatory topology of the Fix signaling systems of C. crescentus and of the nitrogen-fixing rhizobia are similar. The core signaling circuit shown in Fig. 3 has two important feedback pathways. FixJ up-regulates the transcription of fixK either directly or indirectly. FixK then acts as a positive feedback regulator of the fixLJ two-component operon and a negative regulator of its own transcription via up-regulation of the FixT inhibitor of the FixL kinase. Although this paper provides evidence for positive feedback between FixK and fixLJ, putative FixK binding sequences are present upstream of fixLJ promoters in B. japonicum, M. loti, and S. meliloti (6), suggesting that positive feedback control is a general feature of the Fix sensory/signaling module.
The presence of positive feedback in this signaling circuit likely serves to increase the response rate (i.e., slope of the hypoxia dose–response curve) of this system (32). If the magnitude of positive feedback between FixK and the fixLJ promoter is large enough, it could create bistable/switch-like behavior in this circuit (32). Positive feedback has the additional consequence of shifting the regulatory range of this system to higher oxygen concentrations by generating more FixL, FixJ, and FixK molecules per cell. To avoid unchecked positive feedback that would lead to runaway expression of fixLJ and fixK, FixT damps system output by acting as a negative regulator of FixLJ-mediated signaling.
We interpret this arrangement of positive and negative feedback controls in the FixLJ–FixK–FixT network as a mechanism that enables C. crescentus to quickly and stably adapt to changes across a wide range of oxygen concentrations. Because C. crescentus is found in freshwater ecosystems where the concentration of oxygen varies depending on depth in the water column and on the titer of other organisms, this oxygen response system likely serves to optimize the respiratory capacity of C. crescentus within a particular oxygen microenvironment. This adaptation is particularly important for stalked C. crescentus cells, which remain sessile once they have attached to a surface and are unable to move to richer oxygen environments.
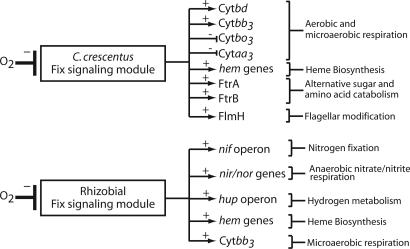
Functional Modularity in Fix Signaling Systems. Importantly, the Fix signaling systems of C. crescentus and the rhizobia show differences in the functions regulated by FixK. C. crescentus contains a FixK motif upstream of several genes, including those encoding all four of its respiratory terminal oxidase complexes. S. meliloti, B. japonicum, and M. loti possess operons encoding these same four aerobic terminal oxidases, but FixK has only been shown to up-regulate transcription of the high-affinity Cytbb3 complex (fixNOQP) in these species (33). Indeed, a search of the promoter regions of the other terminal oxidases in these species reveals no FixK binding sites upstream of any of these operons except for the Cytbo3 operon of S. meliloti. However, previous work by Trzebiatowski and colleagues (34) has shown that the FixLJ/FixK signaling system in S. meliloti does not affect transcription of the Cytbo3 operon during oxygen limitation. Rather, rhizobial FixK serves to directly or indirectly up-regulate genes encoding nitrogenase, nitrogenase-associated genes, respiratory nitrate/nitrite reductase, and hydrogenase (6). All of these genes are absent from the genome of C. crescentus. These results suggest that the Fix signaling system can be thought of as a sensory/signaling module whose sensory features, protein components, and topology is conserved. However, differing placement of the FixK cis-regulatory sequences in the genomes of C. crescentus and the rhizobial bacteria lead to differing transcriptional outputs from this system in response to the same environmental stimulus. In other words, the Fix sensory/signaling module is “wired” in into the genome in a way that best suits the physiology of the organism. A schematic overview of genes under the control of the Fix signaling module in C. crescentus and the rhizobia is shown in Fig. 5.
Fig. 5.
Transcriptional output under the control of the Fix signaling module of C. crescentus and the rhizobia. The circuit topology of the Fix signaling module is shown in Fig. 3. Here, the Fix module is treated as a “black box” that detects and integrates molecular oxygen by using the same core circuitry in both C. crescentus and rhizobial species. However, the transcriptional output of this sensory/signaling module is different in these related bacteria, having been adapted to suit the distinct physiologies of these species.
High-throughput genome sequencing efforts have identified numerous heme-PAS histidine kinases in prokaryotic species of widely varying physiologies. FixL-like histidine kinases are primarily found in the α-proteobacteria, although heme-PAS kinases exist in the β and γ proteobacteria as well as the cyanobacteria and flavibacteria. Species containing heme-PAS kinases occupy a range of physiological niches but all share the ability to respire oxygen. We predict that the great majority of heme-PAS kinases will be found to directly or indirectly regulate the efficiency of aerobic respiration across a range of oxygen concentrations. It will be interesting to compare of the regulatory circuitry built around these heme-PAS kinases to the Fix signaling module described in this manuscript. System-wide comparisons of these oxygen-regulated networks is certain to reveal many similarities (and differences) in network architecture and target gene transcription.
Supplementary Material
Acknowledgments
We thank Alfred Spormann for use of the chemostat. Alison Hottes and Maliwan Meewan provided valuable assistance with microarray preparation and analysis. Jason Key shared reagents and equipment while collecting spectroscopic data on the FixL heme-PAS domains. This work benefited from insightful discussion with Martin Thanbichler. S.C. is a National Institutes of Health–Kirschstein postdoctoral fellow. P.T.M. acknowledges support from the Stanford Human Genome Training Program (National Institutes of Health/National Human Genome Research Institute). C.S. is supported by National Science Foundation Grant MCB-0317037. This study was partly supported by Department of Energy Grant DE-FG03-01ER63219 (to H.H.M. and L.S.), National Institutes of Health Grant GM32506 (to L.S.), and Office of Naval Research Grant N00014-02-0538 (to H.H.M.).
Author contributions: S.C. designed research; S.C. and C.S. performed research; S.C. contributed new reagents/analytic tools; S.C., P.T.M., H.H.M., and L.S. analyzed data; and S.C. and L.S. wrote the paper.
Abbreviation: DO, dissolved oxygen.
References
- 1.McAdams, H. H. & Shapiro, L. (2003) Science 301, 1874–1877. [DOI] [PubMed] [Google Scholar]
- 2.Papin, J. A., Reed, J. L. & Palsson, B. O. (2004) Trends Biochem. Sci. 29, 641–647. [DOI] [PubMed] [Google Scholar]
- 3.Reichmann, D., Rahat, O., Albeck, S., Meged, R., Dym, O. & Schreiber, G. (2005) Proc. Natl. Acad. Sci. USA 102, 57–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gilles-Gonzalez, M. A., Ditta, G. S. & Helinski, D. R. (1991) Nature 350, 170–172. [DOI] [PubMed] [Google Scholar]
- 5.Gilles-Gonzalez, M. A., Gonzalez, G., Perutz, M. F., Kiger, L., Marden, M. C. & Poyart, C. (1994) Biochemistry 33, 8067–8073. [DOI] [PubMed] [Google Scholar]
- 6.Fischer, H. M. (1994) Microbiol. Rev. 58, 352–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Anthamatten, D. & Hennecke, H. (1991) Mol. Gen. Genet. 225, 38–48. [DOI] [PubMed] [Google Scholar]
- 8.Durmowicz, M. C. & Maier, R. J. (1998) J. Bacteriol. 180, 3253–3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Batut, J., Daveranmingot, M. L., Jacobs, M. D. J., Garnerone, A. M. & Kahn, D. (1989) EMBO J. 8, 1279–1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garnerone, A. M., Cabanes, D., Foussard, M., Boistard, P. & Batut, J. (1999) J. Biol. Chem. 274, 32500–32506. [DOI] [PubMed] [Google Scholar]
- 11.Gong, W. M., Hao, B., Mansy, S. S., Gonzalez, G., Gilles-Gonzalez, M. A. & Chan, M. K. (1998) Proc. Natl. Acad. Sci. USA 95, 15177–15182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Prentki, P. & Krisch, H. M. (1984) Gene 29, 303–313. [DOI] [PubMed] [Google Scholar]
- 13.Stephens, C., Reisenauer, A., Wright, R. & Shapiro, L. (1996) Proc. Natl. Acad. Sci. USA 93, 1210–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ely, B. (1991) Methods Enzymol. 204, 372–384. [DOI] [PubMed] [Google Scholar]
- 15.Holtzendorff, J., Hung, D., Brende, P., Reisenauer, A., Viollier, P. H., McAdams, H. H. & Shapiro, L. (2004) Science 304, 983–987. [DOI] [PubMed] [Google Scholar]
- 16.Cohen-Bazire, G., Kunisawa, R. & Poindexter, J. (1966) J. Gen. Microbiol. 42, 301–308. [DOI] [PubMed] [Google Scholar]
- 17.Eisen, M. B., Spellman, P. T., Brown, P. O. & Botstein, D. (1998) Proc. Natl. Acad. Sci. USA 95, 14863–14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.D'mello, R., Hill, S. & Poole, R. K. (1996) Microbiology 142, 755–763. [DOI] [PubMed] [Google Scholar]
- 19.Preisig, O., Zufferey, R., Thony-Meyer, L., Appleby, C. A. & Hennecke, H. (1996) J. Bacteriol. 178, 1532–1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.D'mello, R., Hill, S. & Poole, R. K. (1995) J. Bacteriol. 177, 867–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Millar, A. H., Bergersen, F. J. & Day, D. A. (1994) Plant Physiol. Biochem. 32, 847–852. [Google Scholar]
- 22.Gennis, R. B. & Stewart, V. (1996) Respiration (Am. Soc. Microbiol. Press, Washington, DC).
- 23.Cosseau, C. & Batut, J. (2004) Arch. Microbiol. 181, 89–96. [DOI] [PubMed] [Google Scholar]
- 24.Bailey, T. L. & Gribskov, M. (1998) Bioinformatics 14, 48–54. [DOI] [PubMed] [Google Scholar]
- 25.Eiglmeier, K., Honore, N., Iuchi, S., Lin, E. C. C. & Cole, S. T. (1989) Mol. Microbiol. 3, 869–878. [DOI] [PubMed] [Google Scholar]
- 26.Leclerc, G., Wang, S. P. & Ely, B. (1998) J. Bacteriol. 180, 5010–5019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Foussard, M., Garnerone, A. M., Ni, F., Soupene, E., Boistard, P. & Batut, J. (1997) Mol. Microbiol. 25, 27–37. [DOI] [PubMed] [Google Scholar]
- 28.Robinson, V. L., Buckler, D. R. & Stock, A. M. (2000) Nat. Struct. Biol. 7, 626–633. [DOI] [PubMed] [Google Scholar]
- 29.Wood, N. J., Alizadeh, T., Bennett, S., Pearce, J., Ferguson, S. J., Richardson, D. J. & Moir, J. W. (2001) J. Bacteriol. 183, 3606–3613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Soberon, M., Morera, C., Kondorosi, A., Lopez, O. & Miranda, J. (2001) Mol. Plant–Microbe. Interact. 14, 572–576. [DOI] [PubMed] [Google Scholar]
- 31.Fry, J., Wood, M. & Poole, P. S. (2001) Mol. Plant–Microbe. Interact. 14, 1016–1025. [DOI] [PubMed] [Google Scholar]
- 32.Xiong, W. & Ferrell, J. E. (2003) Nature 426, 460–465. [DOI] [PubMed] [Google Scholar]
- 33.Mesa, S., Bedmar, E. J., Chanfon, A., Hennecke, H. & Fischer, H. M. (2003) J. Bacteriol. 185, 3978–3982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Trzebiatowski, J. R., Ragatz, D. M. & de Bruijn, F. J. (2001) Appl. Environ. Microbiol. 67, 3728–3731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Atkins, P. W. (1993) The Elements of Physical Chemistry (Oxford Univ. Press, Oxford).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.