Abstract
Using solely a gene-based procedure, PCR amplification of the 16S ribosomal RNA gene coupled with very deep sequencing of the amplified products, the microbes on 20 human vaginal epithelia of healthy women have been identified and quantitated. The Lactobacillus content on these 20 healthy vaginal epithelia was highly variable, ranging from 0% to 100%. For four subjects, Lactobacillus was (virtually) the only bacterium detected. However, that Lactobacillus was far from clonal and was a mixture of species and strains. Eight subjects presented complex mixtures of Lactobacillus and other microbes. The remaining eight subjects had no Lactobacillus. Instead, Bifidobacterium, Gardnerella, Prevotella, Pseudomonas, or Streptococcus predominated.
Keywords: ribosomal DNA, urogenital bacteria
For more than a century, direct culture methods have been used to identify the microbes in any given ecological niche. A standard culture method involves sampling with a sterile swab, delivering the swab into sterile buffer, dislodging the microbes from the swab into the buffer, and streaking the buffer/microbes across a series of agar plates. The plates contain various nutritious media for selective growth of particular microbes and are incubated under aerobic and anaerobic conditions. However, in the last dozen years, it has become clear that only a small minority of microbes grow and form colonies on agar plates (for recent reviews, see refs. 1 and 2). Previously, the nonculturable microbes went undetected.
In contrast to direct culture methods, gene-based and/or DNA sequence-based technology detects microbes irrespective of whether they can be cultured. We have applied one of these gene-based methods (PCR amplification of a 16S ribosomal RNA gene and very deep sequencing of the PCR products) to the ecological niche of the normal healthy human vaginal epithelium. The state of health of the human female urogenital tract is largely a function of the quantitative mix of microbes present. Therefore, this study provides basic information concerning the ecology of the flora therein and assists in defining the healthy state of the human female urogenital tract.
Materials and Methods
Human Subjects. The use of human subjects in this study had the prior approval of the Stanford University Committee on the Use of Human Subjects in Medical Research. Samples were obtained after voluntary written informed consent. Twenty subjects were recruited for this study. These subjects were at various (recorded) times of their menstrual cycle. All were healthy premenopausal women 27–44 years of age, not taking contraceptive steroids, and without complaints of urogenital symptoms or noticeable infection on physical examination of the urogenital tract. Under direct visualization using a speculum, a sterile cryoloop (Hampton Research, Aliso Viejo, CA) was passed across the vaginal epithelium in the posterior vaginal fornix and was immediately plunged into liquid nitrogen and stored at –80°C until use. Total DNA was prepared by the use of a DNeasy kit, as described by the manufacturer (Qiagen, Valencia, CA). The total DNA from each vaginal epithelium was aliquoted and stored frozen at –80°C until use.
PCR Amplification. The primer pair for the PCR reactions is derived from highly conserved sequences of the Escherichia coli 16S ribosomal RNA gene. We tested (informatically and/or experimentally) most of the published amplification primer pairs for this gene. Based upon our test data (not shown), we chose the primer pair conventionally called 8f and 1492r, because this primer pair produced an amplification product from the widest range of microbial genomic DNA templates and amplified a nearly complete 16S ribosomal RNA gene (>1.4 kb). The forward primer was SDBact0005aS20 (3–5), 5′-AGAGTTTGATCMTGGCTCAG-3′, M = A + C. In the European Ribosomal RNA Database, 8f is called BSF8/20 (www.psb.ugent.be/rRNA). The reverse primer was SDBact1492aA22 (4, 6), 5′-TACGGYTACCTTGTTACGACTT-3′, Y = C +T. The primers were added to an aliquot of the total DNA from the vaginal epithelium from any given subject. PCR amplification was performed with a Takara Bio (Tokyo) PCR kit (catalog no. R011; in the U.S., the distributor is Fisher Scientific), as suggested by the manufacturer: 94°C for 1 min, amplified for 25 cycles where each cycle consisted of 94°C, 30 sec; 55°C, 30 sec; and 72°C, 60 sec. The 1.4-kb amplified product was purified by agarose gel electrophoresis and cloned into a plasmid vector using a TA-Cloning kit, as described by the manufacturer (Invitrogen). That library was transformed into E. coli cells. The plasmid inserts from individual E. coli colonies were PCR-amplified for sequencing by using universal primers equivalent to plasmid vector sequences. The amplified plasmid inserts were sequenced from both ends by using Applied Biosystems BigDye-terminator chemistry and the Applied Biosystems 3730 DNA Analyzer.
Data Processing. Our goal was to collect 2,000 sequence reads (1,000 recombinant plasmids sequenced from both ends of the insert, excluding reads supporting contigs of length <900 bases and all reads of contaminating human DNA) for each of the 20 subjects in our study. Representative data are presented in Tables 1, 2, 3, 4, 5. Summary and detailed data for the remaining subjects can be found in Tables 6–46, which are published as supporting information on the PNAS web site. The net number of reads per subject is given in Table 6. The small number of contaminating human reads per subject is given in Table 7. The individual bases of a sequence read were identified and given a quality score by using the public software phred (7). As per phred, bases with quality scores of 20 or higher are considered to be “good quality” (7). Our average good-quality read length was >700 bases (excluding the 5% failed reads). The reads were processed through our custom software to identify plasmid vector bases (turned to “x” in the sequence) and to trim the usual poor-quality sequence at the 3′ end of each read (bases turned to “n” in the sequence). The public software phrap (7, 8) was used to assemble the reads into contigs and to determine the consensus sequence of each contig. Because our good-quality read length averaged >700 bases, opposing reads from the same plasmid sometimes overlapped. Such overlap assisted the assembly process. The amplification primers were chosen, in part, because they are promiscuous. Therefore, the primer sequences were removed from the contig consensus sequences before comparisons were made to sequences in public databases. Because sequencing errors are most likely to occur at the 5′ and 3′ ends of a read, trimming both ends is useful. In addition, as is well known, Taq polymerase is a relatively faithless enzyme. Therefore, the more reads supporting a contig, the more certain is the contig's consensus sequence. Throughout the text, we downplay the significance of contigs supported by no more than a handful of reads. The consensus sequence of each contig was compared with the sequences in three databases, GenBank, the “arbor” database (9) (ARB), and the Ribosomal Database Project (10, 11). The software clustalw (12–15) and treeview (16) were used for constructing relationship trees among contigs.
Table 1. Summary of subject 01 GenBank microbe matches.
| Microbe | Number of reads | Range of match, % |
|---|---|---|
| Lactobacillus | 1,808 | 94-100 |
| Unidentified γ proteobacterium | 38 | 100 |
| Uncultured bacterium clone 300C-G04 | 9 | 99 |
| Corynebacterium | 4 | 99-100 |
| Pseudomonas | 3 | 97 |
| Anaerococcus | 2 | 97 |
Table 2. Summary of subject 10 GenBank microbe matches.
| Microbe | Number of reads | Range of match, % |
|---|---|---|
| Gardnerella | 1,178 | 98 |
| Gemella | 355 | 99-100 |
| Uncultured bacterium | 57 | 93-100 |
| Acinetobacter | 40 | 99 |
| Streptococcus | 37 | 99-100 |
| Atopobium | 27 | 100 |
| Staphylococcus | 13 | 100 |
| Anaerococcus | 10 | 96-97 |
| Delftia | 6 | 100 |
| Enterococcus | 6 | 100 |
| Janthinobacterium | 6 | 99-100 |
| Peptostreptococcus | 6 | 100 |
| Burkholderia | 5 | 100 |
| Finegoldia | 4 | 99 |
| Stenotrophomonas | 4 | 99 |
| Bromate-reducing bacterium | 3 | 100 |
| Acidovorax | 2 | 100 |
| Agrobacterium | 2 | 100 |
| Brevundimonas | 2 | 100 |
| Clostridium | 2 | 100 |
| Dechloromonas | 2 | 92 |
| Dialister | 2 | 100 |
| Moraxella | 2 | 99 |
| Ochrobactrum | 2 | 100 |
| Prevotella | 2 | 99 |
| Pseudomonas | 2 | 100 |
Table 3. Retrospective simulation of the results for subject 10 assuming ≈400 reads of the experimental length (>700 bases).
| Microbe | Number of reads |
|---|---|
| Gardnerella | 273 |
| Gemella | 70 |
| Acinetobacter | 11 |
| Uncultured bacterium | 9 |
| Atopobium | 8 |
| Streptococcus | 8 |
| Delftia | 6 |
| Enterococcus | 4 |
| Brevibacillus | 3 |
| Anaerococcus | 2 |
| Burkholderia | 2 |
| Clostridium | 2 |
| Finegoldia | 2 |
| Janthinobacterium | 2 |
| Propionibacterium | 1 |
| Uncultured proteobacterium | 1 |
Table 4. Comparison of microbe identification in three databases for the microbe with the most reads for each subject.
| Subject | GenBank best match | ARB best tree position | RDP best match |
|---|---|---|---|
| 01 | Lactobacillus | Lactobacillus | Lactobacillus |
| 02 | Lactobacillus | Lactobacillus | Lactobacillus |
| 03 | Lactobacillus | Lactobacillus | Lactobacillus |
| 04 | Lactobacillus | Lactobacillus | Lactobacillus |
| 05 | Bifidobacterium | Hafnia | Bifidobacterium |
| 06 | Lactobacillus | Lactobacillus | Lactobacillus |
| 07 | Streptococcus | Streptococcus | Streptococcus |
| 08 | Lactobacillus | Lactobacillus | Lactobacillus |
| 09 | Lactobacillus | Lactobacillus | Lactobacillus |
| 10 | Gardnerella | Gardnerella | Gardnerella |
| 11 | Unidentified/uncultured proteobacterium | Pseudomonas | Pseudomonas |
| 12 | Unidentified/uncultured proteobacterium | Pseudomonas | Pseudomonas |
| 13 | Unidentified/uncultured proteobacterium | Pseudomonas | Pseudomonas |
| 21 | Unidentified/uncultured proteobacterium | Pseudomonas | Pseudomonas |
| 22 | Prevotella | Prevotella | Prevotella |
| 41 | Lactobacillus | Lactobacillus | Lactobacillus |
| 42 | Streptococcus | Pseudomonas | Streptococcus |
| 43 | Uncultured bacterium | Eubacterium | Uncultured bacterium |
| 44 | Lactobacillus | Lactobacillus | Lactobacillus |
| 45 | Lactobacillus | Lactobacillus | Lactobacillus |
ARB, the “arbor” small subunit rDNA sequence database; RDP, Ribosomal Database Project.
Table 5. Detailed GenBank microbe matches for subject 01.
| Microbe/GenBank accession no. | Number of reads | Number of bases matched | Percent match, % |
|---|---|---|---|
| Lactobacillus sp. emb|Y16329.1| | 455 | 1,502 | 99 |
| 407 | 1,519 | 100 | |
| 92 | 1,517 | 100 | |
| 7 | 1,428 | 98 | |
| 6 | 1,509 | 100 | |
| 6 | 1,508 | 100 | |
| 5 | 1,491 | 100 | |
| 4 | 1,514 | 99 | |
| 4 | 1,488 | 100 | |
| L. crispatus str. ATCC33820 gb|AF257097.1| | 339 | 1,452 | 100 |
| 188 | 1,504 | 100 | |
| 116 | 1,504 | 100 | |
| L. crispatus emb|V17362.1| | 13 | 1,520 | 100 |
| 10 | 1,414 | 97 | |
| 7 | 1,519 | 99 | |
| 5 | 1,518 | 99 | |
| L. gasseri str. ATCC 33323 gb|AF519171.1| | 20 | 1,496 | 98 |
| 13 | 1,494 | 98 | |
| 8 | 1,526 | 99 | |
| L. gasseri str. KC26 gb|AF243156.1| | 32 | 895 | 100 |
| Unidentified γ proteobacterium dbj|AB015555.1| | 23 | 1,493 | 100 |
| Lactobacillus sp. oral clone CX036 gb|AY005048.1| | 16 | 1,531 | 99 |
| Unidentified γ proteobacterium dbj|AB015581.1| | 12 | 1,493 | 100 |
| Lactobacillus antri gb|AY253659.1| | 7 | 1,428 | 94 |
| Uncultured bacterium clone 300C-G04 gb|AY662007.1| | 7 | 1,486 | 99 |
Results
The 2,000 sequence reads for each subject were assembled separately into contigs. To identify the microbes present on the vaginal epithelium of each of the 20 subjects, the consensus sequence of each contig was compared with the sequences in GenBank. Four of the 20 subjects in this study showed (virtually) only Lactobacillus on the vaginal epithelium (subject 01, Table 1; subjects 02, 03, and 41, Tables 8, 9, and 21, respectively. As an example of the subjects evidencing only Lactobacillus, a summary of the microbes detected on the vaginal epithelium of subject 01 is presented in Table 1. Nine subjects presented complex mixtures of Lactobacillus and many other microbes. These other microbes include, as examples, Bifidobacterium (e.g., subject 06), Gardnerella (e.g., subject 08), Atopobium (e.g., subject 09), Corynebacterium (e.g., subject 44), and Janthinobacterium (e.g., subject 45; Tables 12, 14, 15, 24, and 25, respectively). The remaining seven subjects evidenced no, or virtually no, Lactobacillus on the vaginal epithelium (subject 10, Tables 2 and 3; subjects 05, 11–13, 22, and 42; Tables 11, 16–18, 20, and 22, respectively). As an example, a summary of the microbes detected on the vaginal epithelium of subject 10 is presented in Table 2. As seen in Table 2, the most reads for any microbe on subject 10's vaginal epithelium were for Gardnerella, followed by Gemella. Overall, the percent of Lactobacillus reads on each of the 20 vaginal epithelia in our study ranged from 0% to 100%, with many values in between.
In Table 4, we report the GenBank identification of the microbe supported by the most reads for each of the 20 subjects. In addition to Lactobacillus (Table 1) and Gardnerella (Table 2) as the microbe with the most reads according to GenBank, we found Bifidobacterium (subject 05), Streptococcus (subject 07), Prevotella (subject 22), and “unidentified” and/or “uncultured” proteobacterium and/or bacterium. These last identifications are completely unsatisfactory. Therefore, in addition to GenBank, we analyzed our contig consensus sequences in the ARB (9) and Ribosomal Database Project (10, 11) databases (Table 4). As seen in Table 4, the “unidentified” and “uncultured” GenBank categories are replaced by Pseudomonas in five cases and Eubacterium in one case. Thus, Pseudomonas is supported by the most reads for five subjects. There are two subjects for which microbe identification is different among the three databases: subjects 05 and 42 (Table 4).
A summary of the microbe matches for each subject, as in Tables 1 and 2, presents the big picture but obscures important information, especially clonality or lack thereof. As an example, in Table 5, we present the individual contig matches identified on the vaginal epithelium of subject 01. (The individual contig matches for the remaining 19 subjects are presented in Tables 26–44.) For subject 01 (Table 5), there are contig consensus sequences closely matching seven distinguishable GenBank Lactobacillus sequences, six supported by >10 reads, and the seventh by seven reads. Clearly, the Lactobacillus on the vaginal epithelium of subject 01 is not clonal. There are 10 additional subjects with Lactobacillus contigs supported by >200 reads. For eight (subjects 02–04, 08, 21, 41, 44, and 45), the Lactobacillus is not clonal (Tables 26–28, 32, 38, 40, 43, and 44, respectively). Subject 03 is an example: one contig of L. jensenii supported by 919 reads, one contig of Lactobacillus crispatus supported by 268 reads, and one contig of Lactobacillus gallinarum supported by 239 reads (Table 27). On the other hand, for two subjects (06 and 09), the Lactobacillus appears clonal (Tables 30 and 33, respectively). For subject 06, there is one contig of Lactobacillus gasseri supported by 917 reads (Table 30). In direct contrast, also seen for subject 06 is one contig of Bifidobacterium breve supported by 279 reads, one contig of Bifidobacterium sp. oral strain H6-M4 supported by 57 reads, and one contig of Bifidobacterium urinalis also supported by 57 reads (Table 30). Thus, for subject 06, Lactobacillus appears clonal, whereas Bifidobacterium is not clonal.
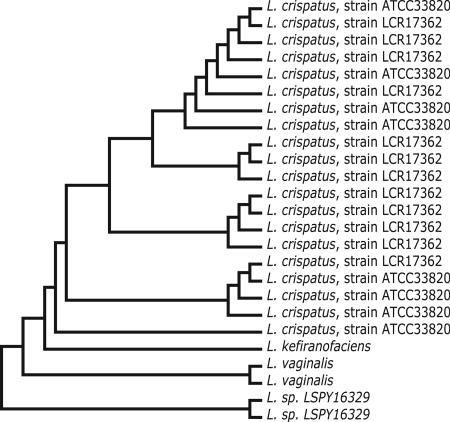
A different, complementary view of the clonality of the Lactobacillus contigs for each subject has been achieved by cluster analysis (12–15). As an example, Fig. 1 presents a relationship “tree” (16) for the Lactobacillus contigs for subject 02. There are 25 distinguishable contigs of at least 1 kb supported by at least four reads (Fig. 1). Twenty-five is the largest number of Lactobacillus contigs for any of the subjects. The 25 contigs form four clusters. Twenty contigs are closely related to each other and to L. crispatus. In fact, the closest GenBank match for 8 of these 20 L. crispatus contigs is ATCC33820. The closest GenBank match for the other 12 L. crispatus contigs is LCR17362. Nevertheless, there are sequence differences that distinguish all 20. In addition to L. crispatus, there are five other Lactobacillus contigs (Fig. 1). In GenBank, one pair of contigs most closely matches “Lactobacillus sp., LSPY16329.” In the ARB database, the best tree position for “Lactobacillus sp., LSPY16329” is Lactobacillus iners. The other pair of contigs most closely matches Lactobacillus vaginalis. Last, there is one contig that most closely matches Lactobacillus kefiranofaciens.
Fig. 1.
Relationship tree of the Lactobacillus contigs for subject 02. Subject 02's Lactobacillus contigs of length at least 1 kb supported by at least four good quality reads were subjected to cluster analysis. The result is presented as a relationship tree with individual contigs identified by their closest GenBank match. There is no “evolutionary distance” scale in the figure, because the lines and connections do not imply quantitative distances. Only qualitative distance is implied.
Discussion
Throughout this report, we have been careful to refer to the number of sequence reads supporting the presence of a microbe and have not equated the number of sequence reads with absolute or relative microbe concentration. As an example, if the presence of bacterium A is supported by 400 sequence reads and the presence of bacterium B is supported by 200 sequence reads, is the concentration of bacterium A twice that of bacterium B? Not necessarily, because, although the number of sequence reads is a function of bacterium concentration, there are several confounding factors. Among the confounding factors are the following: (i) The PCR amplification primer(s) may include one or more base mismatches with any given 16S ribosomal DNA (rDNA) template. The presence of a mismatch between primer and template will reduce the efficiency of priming, especially in the critical early rounds of amplification. Presumably, the end result of such a mismatch between primer and template would be fewer sequence reads than for a perfect primer/template match. (ii) There are substantial differences in the copy number of 16S rDNA genes among bacterial genomes, i.e., different bacterial genomes contain different numbers of 16S rDNA templates (17, 18). The number of sequence reads per template is a function of the concentration of that template. If the genome of bacterium A contains 10 rDNA genes and the genome of bacterium B contains one rDNA gene, then presumably an equimolar mixture of A and B as template will yield more sequence reads for A than for B (if the rDNA genes of A and B both have the same percent match to the primers). (iii) In published studies using this technology, statistically different results have been achieved by changing only the number of PCR amplification cycles (19, 20). Because of these confounding factors, we believe that comparing the number of sequence reads supporting different microbes is only a semiquantitative analysis, but better than no quantitative analysis at all.
We set out to define the microbes on the human vaginal epithelium without culturing the microbes. The 16S ribosomal RNA gene-based technology that we used was adapted from published procedures. We have significantly increased the information derived from the technology by PCR amplifying a large fragment (>1.4 kb) containing nearly all of a 16S ribosomal RNA gene and sequencing the amplified DNA to very high coverage (2,000 reads per subject). That was deliberately exceptionally deep sequencing, and it was very costly in time and money. Because of that double costliness, we have undertaken considerable retrospective analyses of our data to examine the cost-to-benefit ratio. We have modeled the data that we would have achieved from fewer reads (e.g., Table 3), from shorter reads (e.g., Table 45), and from fewer shorter reads (e.g., Table 46). For the number of sequence reads, we chose the last ≈200, ≈400, and ≈900 forward reads for each subject. We chose two good-quality lengths for the sequence reads: bases 50–450 for a 400-base read or the experimental length (>700 bases per read). We assembled the reads into contigs, derived a consensus sequence for each contig, and compared the sequence of each consensus sequence with the sequences in GenBank. One example of the results of this modeling is shown for ≈400 reads of the experimental length for subject 10 (Table 3). As can be seen in Table 3, the fewer reads correctly identify the microbe supported by the most (Gardnerella) and second-most reads (Gemella). There are a handful of reads that suggest the complexity of the mix of microbes. However, even for Gardnerella, there is an insufficient number of reads to address the subject of clonality. With regard to the length of the reads, the cost of the BigDye-terminator reagent (by far the most expensive reagent for DNA sequencing) is the same whether the read length is 400 or 700 bases. Therefore, it makes neither intellectual nor economic sense to sequence a PCR product of length less than the average good-quality read length, which, in our case, is >700 bases. As a result of our retrospective analyses, we conclude that, for the purpose of enumerating the microbes on the human vaginal epithelium while also maximizing the cost-to-benefit ratio, we should achieve 400–500 sequence reads per subject. What will be the difference in microbe detection between 500 and 2,000 reads? We assume that each read is drawn independently from a uniform pool of reads. In this situation, we are sampling from a Poisson distribution. The detection threshold (defined as the minimum frequency required for detection) is proportional to the number of reads. That is, a 4-fold decrease in the number of reads (i.e., from 2,000 to 500 reads per subject) lowers the microbe detection threshold by a factor of 4.
There are three recently published studies using 16S rDNA gene-based technology to identify microbes in the human female urogenital tract (21–23). Although the three published studies took samples from different sites within the vagina and used different primer pairs (yielding different sizes of amplified products), different amplification conditions, different women (from Canada, Belgium, and the U.S., respectively), and at unreported times during the menstrual cycle, these women comprise a reasonable comparison group for our experiments. Burton et al. (21) studied 19 subjects, “premenopausal Caucasian women who had no symptoms or signs of vaginal or urinary tract infection and were otherwise healthy.” Burton et al. (21) amplified 200 bases of the 16S rDNA gene, the smallest fragment of the four studies, purified the fragment(s) from a gel, and sequenced the fragment (but did not report the number of reads per fragment). Of 19 subjects, Burton et al. (21) found that 12 had essentially only Lactobacillus, 3 had Lactobacillus plus Gardnerella, 3 had Gardnerella with other bacteria but without Lactobacillus, and 1 had only Streptococcus. Verhelst et al. (22) studied eight healthy subjects “attending our out-patient clinic for a routine gynecological visit.” Verhelst et al. (22) amplified a 524-base fragment of 16S rDNA; “the 16S rRNA gene was amplified and sequenced from selected clones and from cultured isolates that could not be identified by tDNA-PCR” (tDNA-PCR is a technique that uses one tRNA-based primer for PCR amplification). It is unlikely that rDNA polymorphisms would be detected by this procedure. Verhelst et al. (22) did not report the number of sequence reads per subject. Of the eight subjects, four had essentially only Lactobacillus, one had Atopobium, two had Atopobium plus Prevotella, and one had Peptostreptococcus plus Peptoniphilus. Verhelst et al. (22) recognized that one of their amplification primers had three base mismatches to the Gardnerella 16S rDNA sequence, and that, therefore, the amount of Gardnerella would be underestimated (at the very least). Zhou et al. (23) rigorously excluded subjects with vaginosis or other medical problems in their study group of five subjects; they amplified and cloned a 918-base fragment of 16S rDNA from each of five subjects and sequenced the inserts in both forward and reverse directions from a total of 1,200 plasmids, that is, an average of 240 plasmids per subject. Zhou et al. (23) did not report the number of reads per plasmid. Of the five subjects, two had essentially only Lactobacillus, two had Lactobacillus plus Megasphera, and one had Atopobium.
In comparing the microbes detected on the human vaginal epithelium in our study with those detected in the three recent related published studies, there are interesting similarities and differences. Lactobacillus can be the only, or the majority, microbe on the healthy human vaginal epithelium. That Lactobacillus can be clonal or not. In fact, the totality of the Lactobacillus species/strains on any one vaginal epithelium can be more complex than even recent work has reported (compare refs. 24 and 25). Clearly, the relationship of Lactobacillus clonality and the health of the human vaginal epithelium is a subject that needs further investigation.
Lactobacillus is not always found on the human vaginal epithelium. Gardnerella is commonly found [6 of 19 subjects in the Burton et al. (21) study and 5 of 20 subjects in our study]. Streptococcus is commonly found; 1 of 19 subjects had only Streptococcus in the Burton et al. (21) study, as did 1 of 20 subjects in our study (Table 4). As a direct consequence of our deep sequencing, we found an additional four subjects with a significant number of Streptococcus reads as well as three subjects with a few reads of Streptococcus. Verhelst et al. (22) found no subjects with Streptococcus but one of eight subjects with Peptostreptococcus. We found 4 of 20 subjects with a significant number of Peptostreptococcus reads and another 3 subjects with a few reads of Peptostreptococcus. Atopobium was found in two of eight subjects by Verhelst et al. (22) and was the only microbe in one of five subjects by Zhou et al. (23). Four of our 20 subjects had a small number of Atopobium reads, subjects 03 (3 reads), 05 (41 reads), 09 (53 reads), and 43 (74 reads; Tables 9, 11, 15, and 23, respectively). However, our reverse primer (1492r) has many mismatches with Atopobium 16S rDNA. Therefore, we have probably underestimated the amount of Atpobium present. Prevotella was found in two of eight subjects by Verhelst et al. (22). We found a significant amount of Prevotella on 4 of 20 subjects plus an additional 3 subjects with a few reads of Prevotella. Alone among the four studies, we found Pseudomonas to be a common microbe in the healthy human premenopausal vaginal tract. Pseudomonas had the most reads for 5 of 20 subjects (Table 4); also, an additional 12 subjects evidenced Pseudomonas at some level. The PCR amplification primers used by all four groups of scientists have excellent homology to Pseudomonas 16S rDNA. Therefore, there is no simple molecular explanation as to why we found widespread Pseudomonas, and the other three groups found no Pseudomonas. Obviously, the presence of Pseudomonas in the healthy human female urogenital tract needs further investigation.
The comparisons made in the previous two paragraphs of the microbes in/on the normal healthy human vagina bring us to the clinically important question: the presence and/or absence of which microbes define a healthy vagina? Combining the four studies, only 22 (42%) of 52 subjects evidenced (virtually) only Lactobacillus in the vagina; 16 (31%) subjects evidenced (virtually) no Lactobacillus. These 16 subjects reported no urogenital symptoms. There was no evidence of infection or any other clinical problem upon physical examination of their urogenital tracts. Therefore, we conclude that, alone, the absence of Lactobacillus does not define an unhealthy state, i.e., vaginosis. Complementarily, the presence of solely, or a combination of, Atopobium, Gardnerella, Peptostreptococcus, Prevotella, Pseudomonas, and/or Streptococcus (often noxious bacteria when in/on humans) does not define an unhealthy state.
Our subjects are healthy women consulting at a general gynecology or fertility clinic. Samples were obtained at various recorded times throughout their menstrual cycles. However, the important topic of whether there is hormonal dependence of the endogenous vaginal flora cannot be ascertained from our study [and has not been addressed in other studies using a gene-based approach (21–23)]. Clearly, the influence of hormone concentration on vaginal flora is a subject that needs investigation with a 16S ribosomal RNA gene-based approach.
Conclusion
Although investigations of the microbes in ecological niches using 16S rDNA-based technology have been reported for more than a decade, this technology has been applied only relatively recently to ecological niches defined by the human body. These latter experiments would be even more powerful if investigators of particular human body niches would agree on exactly which E. coli 16S rDNA-based primers and what PCR amplification conditions to use. For example, if Burton et al. (21), Verhelst et al. (22), Zhou et al. (23), and ourselves had used exactly the same primers and PCR amplification conditions, our individual experiments would be more easily comparable and much more powerful. Therefore, we end this manuscript with an invitation. Because we expect to extend our experiments for several years, we invite every scientist using or proposing to use 16S rDNA-based technology to investigate the microbes of the human vagina to contact us in the hope that we will agree on exactly which primers and amplification conditions to use.
Supplementary Material
Acknowledgments
We thank Dr. Paul Lepp, from whose expertise and generosity we have benefited substantially. We also thank Drs. Ruth Lathi, Eva Littman, and Ted Jones for their contributions to this research. This work was supported by a grant from the Ellison Medical Foundation.
Abbreviation: rDNA, ribosomal DNA.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AY958774–AY959212).
References
- 1.Rappe, M. S. & Giovannoni, S. J. (2003). Annu. Rev. Microbiol. 57, 369–394. [DOI] [PubMed] [Google Scholar]
- 2.Schloss, P. D. & Handelsman, J. (2004) Microbiol. Mol. Biol. Rev. 68, 686–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Edwards, U., Rogall, T., Blocker, H., Emde, M. & Bottger, E. C. (1989) Nucleic Acids Res. 17, 7843–7853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wilson, K. H., Blitchington, R. B. & Greene, R. C. (1990) J. Clin. Microbiol. 28, 1942–1946, and erratum (1991) 29, 666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wilmotte, A., Van der Auwera, G. & De Wachter, R. (1993) FEBS Lett. 317, 96–100. [DOI] [PubMed] [Google Scholar]
- 6.Lane, D. J. (1991) in Nucleic Acid Techniques in Bacterial Systematics, eds. Stackebrandt, E. & Goodfellow, M. (Wiley, New York), pp. 115–175.
- 7.Ewing, B. & Green, P. (1998) Genome Res. 8, 186–194. [PubMed] [Google Scholar]
- 8.Ewing, B., Hillier, L., Wendl, M. C. & Green, P. (1998) Genome Res. 8, 175–185. [DOI] [PubMed] [Google Scholar]
- 9.Ludwig, W., Strunk, O., Westram, R., Richter, L., Meier, H., Yadhukumar, N., Buchner, A., Lai, T., Steppi, S., Jobb, G., et al. (2004) Nucleic Acids Res. 32, 1363–1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maidak, B. L., Cole, J. R., Lilburn, T. G., Parker, C. T., Jr., Saxman, P. R., Farris, R. J., Garrity, G. M., Olsen, G. J., Schmidt, T. M. & Tiedje, J. M. (2001) Nucleic Acids Res. 29, 173–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cole, J. R., Chai, B., Farris, R. J., Wang, Q., Kulam, S. A., McGarrell, D. M., Garrity, G. M. & Tiedje, J. M. (2005) Nucleic Acids Res. 33, D294–D296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jeanmougin, F., Thompson, J. D., Gouy, M., Higgins, D. G. & Gibson, T. J. (1998) Trends Biochem. Sci. 23, 403–405. [DOI] [PubMed] [Google Scholar]
- 13.Thompson, J. D., Gibson, T. J., Plewniak, F., Jeanmougin, F. & Higgins, D. G. (1997) Nucleic Acids Res. 24, 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thompson, J. D., Higgins, D. G. & Gibson, T. J. (1994) Nucleic Acids Res. 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Higgins, D. G. & Sharp, P. M. (1988) Gene 73, 237–244. [DOI] [PubMed] [Google Scholar]
- 16.Page, R. D. M. (1996) Comput. Appl. Biosci. 12, 357–358. [DOI] [PubMed] [Google Scholar]
- 17.Farrelly, V., Rainey, F. A. & Stackebrandt, E. (1995) Appl. Environ. Microbiol. 61, 2798–2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Klappenbach, J. A., Dunbar, J. M. & Schmidt, T. M. (2000) Appl. Environ. Microbiol. 66, 1328–1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wilson, K. H. & Blitchington, R. B. (1996) Appl. Environ. Microbiol. 62, 2273–2278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Blaut, M., Collins, M. D., Welling, G. W., Dore, J., van Loo, J. & de Vos, W. (2002) Br. J. Nutr. 87, S203–S211. [DOI] [PubMed] [Google Scholar]
- 21.Burton, J. P., Cadieux, P. A. & Reid, G. (2003) Appl. Environ. Microbiol. 69, 97–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Verhelst, R., Verstraelen, H., Claeys, G., Verschraegen, G., Delanghe, J., Van Simaey, L., De Ganck, C., Temmerman, M. & Vaneechoutte, M. (2004) BMC Microbiol. 4, 16–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou, X., Bent, S. J., Schneider, M. G., Davis, C. C., Islam, M. R. & Forney, L. J. (2004) Microbiology 150, 2565–2573. [DOI] [PubMed] [Google Scholar]
- 24.Pavlova, S. I., Kilic, A. O., Kilic, S. S., So, J. S., Nader-Macias, M. E., Simoes, J. A. & Tao, L. (2002) J. Appl. Microbiol. 92, 451–459. [DOI] [PubMed] [Google Scholar]
- 25.Tarnberg, M., Jakobsson, T., Jonasson, J. & Forsum, U. (2002) APMIS 110, 802–810. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.