Abstract
Coexpression patterns of gene expression across many microarray data sets may reveal networks of genes involved in linked processes. To identify factors involved in cellulose biosynthesis, we used a regression method to analyze 408 publicly available Affymetrix Arabidopsis microarrays. Expression of genes previously implicated in cellulose synthesis, as well as several uncharacterized genes, was highly coregulated with expression of cellulose synthase (CESA) genes. Four candidate genes, which were coexpressed with CESA genes implicated in secondary cell wall synthesis, were investigated by mutant analysis. Two mutants exhibited irregular xylem phenotypes similar to those observed in mutants with defects in secondary cellulose synthesis and were designated irx8 and irx13. Thus, the general approach developed here is useful for identification of elements of multicomponent processes.
Keywords: Arabidopsis, cell wall, xylem, coexpression
Cellulose, a polymer composed of high molecular weight β-1,4-glucan chains, is a major component of the cell walls of higher plants. Cellulose is synthesized by plasma membrane-localized complexes containing several structurally similar cellulose synthase (CESA) subunits (1, 2). Arabidopsis contains 10 CESA genes. Three of the genes, CESA1, CESA3, and CESA6, corresponding to the mutants rsw1 (3), ixr1 (4), and prc1 (5), respectively, are largely responsible for cellulose production during primary cell wall formation in most tissues. Three other genes, CESA4, CESA7, and CESA8, corresponding to the mutants irx1 (6), irx3 (7), and irx5 (6), are required for cellulose synthesis during secondary cell wall formation in vascular tissues. Where they have been studied, the CESA genes of similar function appear to be coexpressed in the same cells (4, 8, 9). Thus, it has been suggested that at least three different CESA proteins are required for a functional CESA complex (9).
Genetic screens have revealed additional components affecting cellulose biosynthesis (2). The korrigan mutant is deficient in an endo-1,4-β-d-glucanase (10). Mutations in the otherwise anonymous COBRA gene affect the orientation of cell expansion and cause a reduction in cellulose production (11). The brittle culm1 mutation in rice is due to a defect in a COBRA homolog, COBL4 (12). Mutations in an endochitinase-like gene (CTL1) caused ectopic deposition of lignin and cell deformation in pith cells due to a decrease in cellulose (13). The kobito1 (14), knopf (15), and botero1 (16) mutants are also compromised in cellulose biosynthesis.
To identify additional genes required for cellulose synthesis, we have analyzed publicly available microarray data for genes with expression patterns that are highly correlated with those of CESA genes. Similar approaches have previously been used to infer relationships between coexpression and gene function (17–21). Although highly reproducible patterns of genetic coregulation have been reported in some cases (20, 22), several approaches, such as cluster analyses, may cause distortions in coexpression patterns when data from different microarray platforms are included (23). Therefore, we have applied a unique method of analysis based on regression of publicly available Affymetrix ATH1 Arabidopsis microarray data from the Nottingham Arabidopsis Stock Centre (NASC). The approach identified genes previously implicated in cellulose synthesis as well as a vast number of previously uncharacterized genes. Mutations were identified in several of the candidate genes, resulting in cell wall phenotypes characteristic of deficiencies in cellulose synthesis.
Materials and Methods
Microarray Data Sets. Computational analysis was performed on CEL data files purchased from the NASC (24) using the AffyWatch Subscription Service (http://arabidopsis.info). Data sets from 503 Affymetrix 25k ATH1 microarrays (25) were processed by using the robust multiarray analysis (RMA) (26) algorithm in the bioconductor package.
Quality Control Using Deleted Residuals. A quality control analysis of the microarray data sets was performed to identify potential outlier chips using a method based on “deleted residuals.” An assumption underlying most statistical procedures is that data should be independently distributed in order for valid statistical inferences to be made. The deleted residuals method tests whether a data set is drawn from the same distribution as the other data sets in a group. For n genes and m data sets, where n is much larger than m, the deleted residual dij is calculated by
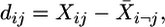 |
where 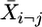 is the mean of gene i for all m data sets excluding the value of data set j. The Studentized deleted residuals are calculated as
is the mean of gene i for all m data sets excluding the value of data set j. The Studentized deleted residuals are calculated as 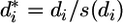 , which obeys t distribution with m - 2 df. For each data set, we have n deleted residuals that follow the expected t distribution. Significant deviation from t distribution of
, which obeys t distribution with m - 2 df. For each data set, we have n deleted residuals that follow the expected t distribution. Significant deviation from t distribution of 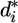 for each data set indicates that the quality of the data set is problematic and provides a criterion for excluding that data set (Fig. 3, which is published as supporting information on the PNAS web site).
for each data set indicates that the quality of the data set is problematic and provides a criterion for excluding that data set (Fig. 3, which is published as supporting information on the PNAS web site).
The Kolmogorov–Smirnov (K–S) goodness-of-fit test is used to decide whether a sample comes from a population with a specific distribution (27). It is based on the empirical distribution function (ECDF). Given N ordered data points X1, X2,..., XN, the ECDF is defined as
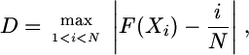 |
where n(i) is the number of points <Xi, the Xi are ordered from smallest to largest value, and F is the theoretical cumulative distribution of the distribution being tested. This ECDF is a step function that increases by 1/N at the value of each ordered data point. The Kolmogorov–Smirnov test statistic is defined as H0 (the data follow a specified distribution) and Ha (the data do not follow the specified distribution).
Rather than using the P value that corresponds to the K–S D statistic, we developed normative standards based on processing all of the Affymetrix chips in Gene Expression Omnibus. The deleted residuals for a total of 10,243 chips were used to develop normative values. We found that between the 80th and 90th percentile (K–S D values of 0.159 and 0.206, respectively), the slope of K–S D curves undergo a large change. Therefore, we used a cutoff for the K–S D of 0.15 to identify chips as potential outliers.
Identification of Potentially Coexpressed Genes by Linear Regression. To identify genes that are coexpressed with CESA genes, linear regression was performed on 408 microarray data sets, randomly subdivided into four subsets, each containing 102 nonoverlapping data sets. The subsets were created to estimate the frequency with which a particular gene was highly correlated with a CESA gene and to ensure that the distribution of the ranked genes was similar for larger subpopulations within the set. We used four subsets because of previous results suggesting that cluster analysis based on 100 data sets produced stable clusters. Regression was performed between a CESA gene and the rest of the genes in the genome. The regression model is described as
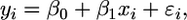 |
where β0 is the intercept, β1 is the regression coefficient, εi is a random error peculiar to the ith observation, xi is the ith observed value of a gene, and yi is the ith observed expression value of a given CESA gene. The estimated regression can be expressed as yi = b0 + b1xi. The fitted values b0 and b1 estimate the true intercept and slope of the regression line.
After regression, all genes were first sorted by the sign of b1 and then by the P values from the regressions between a given CESA gene and each of the other genes. The sign of plus or minus indicates whether the coexpression is positive or negative, respectively. P values indicate the confidence or goodness-of-fit of the regression.
Mapping Coexpressed Genes onto Existing Biological Pathways. For each CESA gene, we obtained four lists of 22,780 P values from the regressions performed with each subset mentioned above. The top 1,000 genes in each list sorted by P values are referred to as coexpressed genes in descending order. The distribution of coexpressed genes with CESA1, 3, and 6 and CESA4, 7, and 8 complexes were mapped onto 186 biological pathways available at The Arabidopsis Information Resource (www.arabidopsis.org/tools/aracyc). A score reflecting the extent of coexpression was assigned to each pathway by using the following formula
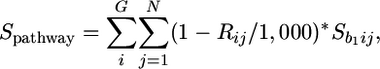 |
where G is the number of genes in a pathway present in the top 1,000 of all lists, and N is the number of lists in which gene i is present in the top 1,000 genes. Rij is the rank of the ith gene of the specified pathway in a list. Sb1ij is the sign of the regression coefficient of gene i with any one of CESA1, 3, or 6 or CESA4, 7, or 8 in list j, where Sb1ij = 1 if b1 > 0 and Sb1ij = -1 for b1 < 0.
Plant Material and Genetic Analysis. Arabidopsis thaliana (Col-0) plants were germinated on standard MS medium under continuous light (140–220 μmol·m-2·sec-1) at 23°C. Seedlings were transferred to soil and grown in greenhouse chambers under 16 h light/8 h dark conditions at 23°C.
Insertion lines (28) were obtained from the Arabidopsis Biological Resource Center (http://arabidopsis.org). The lines used were SALK_055713 and SA IL_545_A07 (At3g16920), SALK_137109 and SAIL_1186_A05 (At4g27435), SALK_014026 and SA IL_603_G02 (At5g54690), and SA LK_046976 (At5g03170). PCR primer sequences (Table 4, which is published as supporting information on the PNAS web site) were generated against the genomic regions flanking the insert and a standard primer for the 3′ end of the insertion sequence. RT-PCR was used to test for the presence of transcripts (primers are listed in Table 4). RNA was isolated by using an RNeasy plant mini kit (Qiagen, Valencia, CA), and a Qiagen Onestep RT-PCR kit was used for first-strand synthesis and subsequent PCR steps.
Because only one insertion line was obtained for At5g03170 (SALK_046976), the line was backcrossed to Columbia WT, and homozygous plants were reassessed for stem and Fourier transform infrared (FTIR) phenotypes.
Microscopy. Hand-cut stem sections (≈200 μm in thickness) were stained in 0.02% toluidine blue O (Sigma) for 5 min and then rinsed, mounted in water, and viewed with a compound microscope (Leitz DMRB, Leica, Deerfield, IL).
Cell Wall Analyses. Plants were placed in the dark overnight to deplete starch. Stems from 10 individual plants for each line were ground in liquid nitrogen by using a mortar and pestle. Noncovalently bound proteins were extracted by 5 min of homogenization in 0.5 M potassium phosphate, pH 7.0/1% SDS. The material was centrifuged at 2,000 × g for 10 min, washed five times with water, and extracted with chloroform:methanol (1:1) and acetone. The cell wall material was air-dried at room temperature overnight and then ground to a fine powder by ball-milling for 1–2 h. The powder was dried at 30°C overnight, mixed with KBr, and pressed into 13-mm pellets. Fifteen FTIR spectra for each line were collected on a Thermo Nicolet Nexus 470 spectrometer over the range 4,000–400 cm-1. For each spectrum, 32 scans were coadded at a resolution of 8 cm-1 for Fourier transform processing and absorbance spectrum calculation by using omnic software (Thermo Nicolet). Spectra were corrected for background by automatic subtraction and saved in JCAMP.DX format for further analysis. Using win-das software (Wiley, New York), spectra were baseline-corrected and area-normalized and analyzed by using the principal component (PC) analysis covariance matrix method (29).
The cellulose content of ball-milled material was determined as described by Updegraff (30).
Results
Microarray Analysis for Stably and Closely Coexpressed Genes. To minimize the effects of experimental artefacts that may arise during preparation of RNA or processing of Affymetrix arrays, all experimental data were filtered by using deleted residuals. Of the 503 ATH1 microarray data sets that were available, 95 were found to have a K–S D of 0.15 or greater (Fig. 3) and were removed from this analysis. The resulting 408 data sets were analyzed by using a linear regression approach (Fig. 4, which is published as supporting information on the PNAS web site).
To investigate whether the individual CESA subunits in either primary or secondary cellulose biosynthesis were coexpressed, RMA signal values corresponding to the 10 CESA genes from the 408 quality data sets were plotted against each other (Fig. 5, which is published as supporting information on the PNAS web site). Not surprisingly, the CESA genes involved in cellulose biosynthesis during primary and secondary cell wall formation exhibited a high degree of coexpression based on the correlation coefficients (Fig. 5). By contrast, plotting RMA signal values for CESA1, 3, or 6 against CESA4, 7, or 8 did not show coexpression (data not shown). These results are consistent with direct experimental data indicating that CESA 1, 3, and 6 and CESA 4, 7, and 8 are coexpressed during primary and secondary cell wall formation, respectively (4, 6, 9).
To test the utility of the method for identification of genes affecting cellulose production, several genes involved in cellulose biosynthesis (i.e., CESA1, 2, 3, 4, 5, 6, 7, 8, and 9, COBRA, CTL1, and KORRIGAN) were used as reference points during regression analyses. The expression level for each candidate gene was regressed on all of the other 22,780 genes for the 408 chips. Genes were subsequently sorted by the signs of b1 and by the average rank of P values. Table 1 is a list of the 40 most highly ranked genes for CESA1, 3, and 6. Not surprisingly, the CESA genes involved in primary cell wall cellulose formation were ranked among the top candidate genes (Table 1). In addition, the gene for the glycosylphosphatidylinositol-anchored protein COBRA, which affects cellulose deposition (11), is highly coregulated with the primary cell wall cellulose complex. KORRIGAN and CTL1, both implicated in cellulose deposition during primary cell wall formation (10, 13), were also highly correlated (Table 1). Interestingly, the CESA2 gene appears to be highly coexpressed with CESA1, 3, and 6, indicating a potential role for the gene in primary cell wall cellulose deposition. Most of the other proteins in Table 1 have no specific functional annotation and cannot be associated with the overall process of cellulose synthesis on the basis of current knowledge.
Table 1. Most highly coregulated genes for CESA1, 3, and 6.
| Arabidopsis Genome Initiative no. | Protein homology | Score | P value* |
|---|---|---|---|
| 5G05170 | Cellulose synthase, CESA3 | 6 | 1.6E-75 |
| 4G32410 | Cellulose synthase, CESA1 | 7 | 5.9E-60 |
| 5G64740 | Cellulose synthase, CESA6 | 8 | 5.9E-60 |
| 5G60920 | COBRA | 9 | 9.5E-64 |
| 1G76670 | Transporter-related | 45 | 4.1E-35 |
| 1G04430 | Dehydration-responsive like | 47 | 2.5E-36 |
| 1G05850 | Chitinase-like protein 1 (CTL1) | 47 | 5.9E-31 |
| 4G26690 | Glycerophosphoryl diester phosphodiesterase family protein | 49 | 1.2E-35 |
| 1G29470 | Dehydration-responsive like | 64 | 1.6E-33 |
| 4G39350 | Cellulose synthase, CESA2 | 69 | 1.3E-28 |
| 1G12500 | Phosphate translocator-related | 79 | 2.3E-29 |
| 5G35160 | Endomembrane protein 70 | 106 | 2.1E-31 |
| 3G62660 | Glycosyl transferase family 8 protein | 130 | 1.1E-26 |
| 4G39840 | Expressed protein | 149 | 9.3E-27 |
| 2G41770 | Expressed protein | 171 | 4.4E-23 |
| 1G58440 | Squalene monooxygenase | 174 | 4.3E-25 |
| 4G31590 | Glycosyl transferase family 2 protein | 179 | 7.2E-24 |
| 4G18030 | Dehydration-responsive protein | 185 | 4.3E-26 |
| 5G01460 | LMBR1 integral membrane protein | 188 | 1.3E-24 |
| 4G03390 | Leucine-rich repeat protein kinase | 194 | 3.0E-24 |
| 1G45688 | Expressed protein | 198 | 8.2E-22 |
| 2G42880 | Mitogen-activated protein kinase | 220 | 6.9E-24 |
| 3G07330 | Glycosyl transferase family 2 protein | 232 | 8.2E-23 |
| 5G06700 | Expressed protein | 241 | 1.4E-24 |
| 5G19780 | Tubulin α-3 (TUA5) | 260 | 2.3E-22 |
| 1G12850 | Phosphoglycerate/bisphosphoglycerate mutase protein | 271 | 9.5E-20 |
| 5G12850 | Zinc finger family protein | 274 | 3.5E-23 |
| 3G05070 | Expressed protein | 280 | 8.2E-20 |
| 3G17390 | S-adenosylmethionine synthetase | 284 | 1.2E-22 |
| 1G14670 | Endomembrane protein 70, putative | 300 | 5.1E-22 |
| 4G27430 | COP1-interacting protein 7 (CIP7) | 322 | 1.2E-23 |
| 3G26700 | Protein kinase family protein | 335 | 2.6E-18 |
| 5G49720 | Endo-1,4-β-glucanase (KORR) | 356 | 2.6E-18 |
| 2G22125 | C2 domain-containing protein | 371 | 4.7E-20 |
| 3G11745 | Expressed protein | 382 | 3.7E-21 |
| 5G17920 | Methyltetrahydropteroyltriglutamatehomocysteine methyltransferase | 385 | 3.7E-19 |
| 5G53500 | WD-40 repeat family protein | 385 | 3.4E-20 |
| 1G06850 | bZIP transcription factor, putative | 392 | 2.6E-20 |
| 1G12750 | Rhomboid family protein | 393 | 5.2E-21 |
| 3G28180 | Glycosyl transferase family 2 protein | 400 | 8.9E-20 |
The score indicates the average coexpressed rank of the gene for CESA1, 3, and 6. The P value indicates the level of linear regression fitness.
For example, 1.6E-75 = 1.6 × 10-75
The 40 most highly ranked genes for CESA4, 7, and 8 are listed in Table 2. CESA4, 7, and 8 were among the top ranked candidates, confirming tight coregulation of the three CESA subunits. Interestingly, both the COBRA homolog, COBL4, and a CTL1 homolog (At3g216920) are ranked among the 10 most highly coexpressed genes for CESA 4, 7, and 8 (Table 2). The presence of a number of lignin-related genes in Table 2 (i.e., laccases and phenylalanine ammonia lyase) is consistent with the fact that lignin synthesis is associated with the final stages of secondary wall synthesis in vascular tissues. Overall, 16 of the top 40 proteins in Table 2 are known to be involved in, or are good candidates for, cell wall synthesis or modification (i.e., glycosyltransferases, arabinogalactan proteins, epimerases, polygalacturonases, laccases, and glycoside hydrolases), and 10 have no functional annotation. Of the remaining proteins on the list, none can be excluded as having a role in cellulose synthesis.
Table 2. Most highly coregulated genes for CESA4, 7, and 8.
| Arabidopsis Genome Initiative no. | Protein homology | Score | P value* |
|---|---|---|---|
| 5G44030 | Cellulose synthase (IRX5) | 3 | 8.4E-144 |
| 4G18780 | Cellulose synthase (IRX1) | 4 | 4.8E-123 |
| 5G54690 | Glycosyl transferase family 8 | 5 | 2.6E-125 |
| 5G17420 | Cellulose synthase (IRX3) | 5 | 4.8E-123 |
| 2G38080 | Laccase, putative | 5 | 3.0E-118 |
| 3G16920 | Glycoside hydrolase (CTL1-like) | 6 | 5.3E-123 |
| 5G15630 | COBRA homolog COBL4 | 6 | 1.8E-120 |
| 5G03170 | Fasciclin-like AGP (FLA11) | 7 | 7.6E-124 |
| 2G37090 | Glycosyl transferase family 43 | 9 | 5.3E-116 |
| 3G18660 | Glycogenin glucosyltransferase like | 10 | 3.2E-94 |
| 4G27435 | Expressed protein | 10 | 1.6E-105 |
| 5G60720 | Expressed protein | 12 | 1.2E-99 |
| 3G62020 | Germin-like protein (GLP10) | 13 | 2.9E-94 |
| 4G28500 | No apical meristem (NAM) family | 15 | 7.4E-80 |
| 5G60020 | Laccase, putative | 17 | 1.8E-81 |
| 5G60490 | Fasciclin-like AGP (FLA12) | 18 | 5.8E-72 |
| 1G79620 | Leucine-rich repeat kinase | 19 | 4.8E-75 |
| 1G27440 | Exostosin family protein | 21 | 1.9E-61 |
| 1G09610 | Expressed protein | 22 | 6.2E-69 |
| 1G79420 | Expressed protein | 22 | 6.1E-73 |
| 3G50220 | Expressed protein | 22 | 4.1E-67 |
| 1G54790 | GDSL-motif lipase/hydrolase | 26 | 2.7E-62 |
| 4G18640 | Leucine-rich repeat protein kinase | 27 | 1.3E-65 |
| 1G08340 | Rac GTPase activating protein | 27 | 6.4E-62 |
| 4G23496 | Expressed protein | 28 | 2.9E-65 |
| 2G41610 | Expressed protein | 29 | 2.3E-57 |
| 1G72230 | Plastocyanin-like domain-containing protein | 30 | 1.2E-59 |
| 2G29130 | Laccase, putative | 31 | 5.9E-61 |
| 3G15050 | Calmodulin-binding family protein | 32 | 1.6E-54 |
| 2G28760 | NAD-dependent epimerase | 32 | 4.7E-52 |
| 1G62990 | Homeodomain protein (KNAT7) | 32 | 7.1E-62 |
| 5G01190 | Laccase, putative | 38 | 5.2E-52 |
| 5G01360 | Expressed protein | 40 | 2.5E-54 |
| 1G27380 | p21-rho-binding domain protein | 40 | 2.5E-52 |
| 1G73640 | Ras-related GTP-binding family | 40 | 7.9E-52 |
| 1G25530 | Lysine/histidine transporter | 40 | 3.1E-56 |
| 2G47500 | Kinesin motor protein-related | 41 | 4.1E-51 |
| 5G16600 | Expressed protein | 43 | 2.8E-55 |
| 3G42950 | Glycoside hydrolase family 28 | 44 | 1.3E-47 |
| 2G40480 | Expressed protein | 44 | 4.3E-49 |
The score indicates the average coexpressed rank of the gene for CESA4, 7, and 8. The P value indicates the level of linear regression fitness.
For example, 8.4E-144 = 8.4 × 10-144
As shown above, the CESA genes for respective CESA complexes are highly coregulated. Therefore, one might expect that genes encoding other factors required for cellulose synthesis should be highly coregulated with each of the three CESA genes individually. Fig. 1 shows a Venn diagram representing the level of coregulatory linkage of the 100 highest ranked genes for the individual CESA genes. Sixteen of the 100 highest ranked genes are shared among the CESA1, 3, and 6 subunits (Fig. 1). Furthermore, 64 of the 100 highest ranked genes are shared among the CESA4, 7, and 8 subunits (Fig. 1). The lower coregulatory linkage score for CESA1, 3, and 6 may be attributed to the overall high expression levels of CESA1, 3, and 6. Recent reports have shown that greater perturbations in expression changes are common among highly expressed genes, referred to as the law of “richer travel more” (31), which may contribute to an increase in positively coexpressed genes of unrelated function and a decreased number in linked coexpressed genes for CESA1, 3, and 6.
Fig. 1.
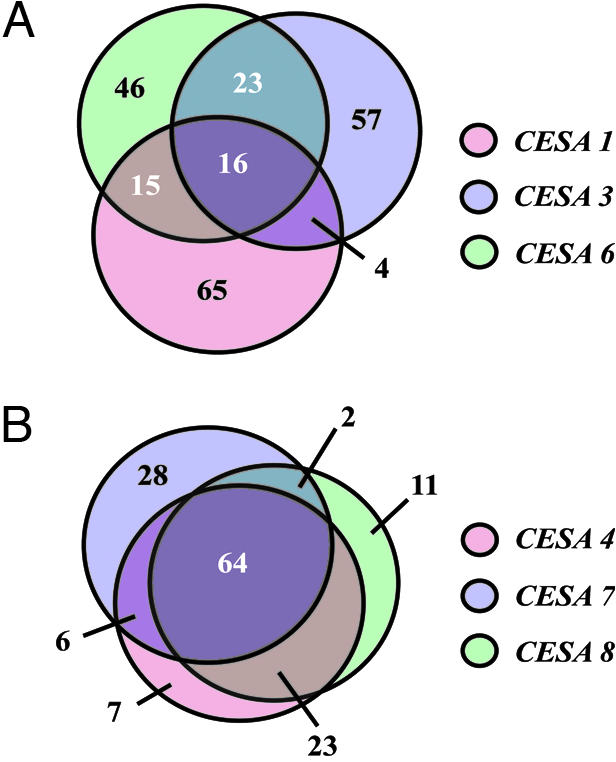
Venn diagrams show overlapping coexpressed genes for the 100 highest ranked genes with the individual CESA genes for the primary (A) and secondary (B) CESA complexes.
Physiological Implications of Genes Coregulated with CESA 4, 7, and 8. To test the biological significance of the apparent coexpression of the genes in Tables 1 and 2 with the CESA genes, we selected a subset of genes (At5g54690, At4g27435, At5g03170, and At3g16920) that was implicated as being coexpressed with CESA4, 7, and 8 for functional analysis. The reason for selecting genes coexpressed with CESA4, 7, and 8, rather than CESA1, 3, and 6, was based on the overall higher linkage scores in CESA4, 7, and 8 coexpression and because secondary cellulose mutations are not lethal, whereas defects in primary cellulose synthesis may be lethal. All of the genes showed a high level of coexpression with CESA4, 7, and 8 in different tissues in Arabidopsis. Furthermore, when plotting RMA signal values for the selected genes against CESA4, 7, and 8, the regression scores were very similar to scores obtained for regression analyses of the individual CESA genes (compare Figs. 5 and 6, which are published as supporting information on the PNAS web site).
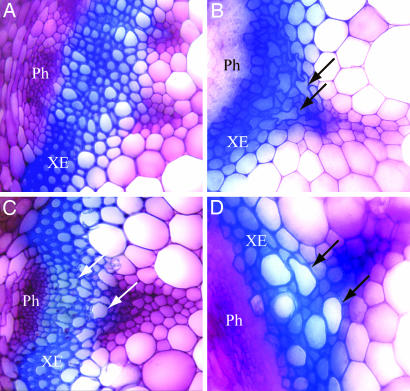
Insertion lines for the subset of genes above were obtained from the collection of sequence-indexed transferred DNA (T-DNA) insertions (28). Except for At5g03170, where only one mutation was available, two homozygous insertion lines were identified for each of the genes. The vascular cell morphology was examined by light microscopy of hand-cut stem sections from 7-week-old insertion line plants stained with toluidine blue. Whereas the insertion lines corresponding to At5g54690 exhibited severe deformations in the xylem cell morphology, the xylem phenotype for the At5g03170 insertion line was subtle (Fig. 2). No xylem deformations were evident in At3g16920 and At4g27435. With the exception of the deformations in xylem cell morphology, no discernable differences in stem organization were apparent. The phenotypes for At5g54690 and At5g03170 were similar to those observed for mutations in CESA4, 7, and 8, which result in an irregular or collapsed xylem (irx) phenotype (32). We have recently learned that Simon Turner and colleagues have obtained similar results by using a different approach (S. Turner, personal communication). In accordance with their nomenclature, we have named the mutants with xylem deformations irx8 (At5g54690) and irx13 (At5g03170).
Fig. 2.
Cross sections of stem vascular bundles. Stem sections were stained with toluidine blue O and viewed under a compound microscope. (A) WT. (B) irx1. (C) irx8. (D) irx13. Arrows indicate deformed xylem elements.
To assess whether the mutants exhibited an alteration in cell wall composition, FTIR analyses were performed. Spectra from stem cell walls of 7-week-old plants were collected and analyzed by using PC analysis. Fig. 7, which is published as supporting information on the PNAS web site, shows plots of PCs 1 and 2 for the mutants and WT. The spectra for the mutant lines show a clear separation from WT spectra based on PC 1, indicating alterations in the chemical composition of the mutant cell walls. The corresponding loadings show that spectra from irx8 have a negative correlation with peaks at 985, 1,043, and 1115 cm-1, corresponding to potential alterations in noncellulosic polymers compared with WT (Fig. 7 and ref. 34). Similar patterns can be seen for At4g27435, At3g16920, irx13, and irx1. The PC plot indicates both a cellulose signature (987, 1,060, and 1,168 cm-1) and differences in noncellulosic polymers for irx1 and irx8 compared with WT (Fig. 7). The loading for At4g27435 and At3g16920 vs. WT also suggests an alteration in cellulose (987, 1,035, and 1,060 cm-1) and pectin (1,022, 1,087, 1,143, and 1,749 cm-1) for the mutants (Fig. 7).
The cellulose contents of 7-week-old stem material from the mutants was measured by a colorimetric method (Fig. 8, which is published as supporting information on the PNAS web site). The mutants irx8 and irx13 exhibited a significant reduction in cellulose, whereas At3g16920 and At4g27435 did not show any significant alteration in cellulose contents compared with WT (Fig. 8).
Mapping Coexpressed Genes to Biological Pathways. To examine similarities and differences in coexpressed gene patterns for the two CESA complexes, coregulated genes were assigned to the 186 existing biological pathways (Table 3; see also Tables 5 and 6, which are published as supporting information on the PNAS web site). Not surprisingly, cellulose biosynthesis exhibited very high pathway scores for both the primary and secondary wall CESA genes. Several other pathways, and homologs within the pathways, were also linked with both types of CESA genes (Tables 3 and 5). These pathways include both cell wall-associated pathways (e.g., homogalacturonan degradation) and potential precursor pathways (e.g., glycolysis and galactose, and galactoside and glucose catabolism). However, coexpression of genes involved in several pathways differed significantly between the two types of CESA genes (e.g., brassinosteroid biosynthesis, lignin biosynthesis, and dTDP-rhamnose biosynthesis pathways) (Table 3). Genes associated with brassinosteroid synthesis exhibited a high coregulation with CESA1, 3, and 6 but not with CESA4, 7, and 8. Brassinosteroids affect morphogenesis and cell expansion of plant cells, processes involving rearrangements of the primary wall matrix (for review, see ref. 35). Lignin biosynthesis genes, however, showed a significantly higher coexpression with the secondary cell wall genes than with CESA1, 3, and 6 (Table 3). Lignin accumulates during vascular differentiation and may reinforce the cell walls during formation of the vascular bundles (36). Pathways that exhibited coregulation with only one of the two CESA types are listed in Table 6.
Table 3. Selected pathway components that are coregulated with both CESA1, 3, and 6 and CESA4, 7, and 8.
| Pathway | Score | CESAs |
|---|---|---|
| Similarly coregulated | ||
| Cellulose biosynthesis | 76.3 | CESA136 |
| 41.9 | CESA478 | |
| Homogalacturonan degradation | 15.7 | CESA136 |
| 37.3 | CESA478 | |
| Galactose, galactoside, and glucose catabolism | 5.9 | CESA136 |
| 11.0 | CESA478 | |
| Gluconeogenesis | 8.9 | CESA136 |
| 7.4 | CESA478 | |
| Serine-isocitrate lyase pathway | 6.3 | CESA136 |
| 5.5 | CESA478 | |
| Aerobic glycerol catabolism | 8.4 | CESA136 |
| 6.8 | CESA478 | |
| Differentially coregulated | ||
| Brassinosteroid biosynthesis | 19.1 | CESA136 |
| 3.3 | CESA478 | |
| Lignin biosynthesis | 3.6 | CESA136 |
| 13.8 | CESA478 | |
| dTDP-rhamnose biosynthesis | 0.4 | CESA136 |
| 9.3 | CESA478 |
Score indicates congruence (i.e., the extent of coexpression between CESA1, 3, and 6 or CESA4, 7, and 8 complexes and the genes in the specified pathway).
Discussion
Results presented here provide evidence that a number of genes are coregulated to varying degrees with two functionally distinct types of CESA genes. The regression analyses reduce drawbacks encountered by classic cluster analyses and their derivative approaches, such as problems in establishing numbers of clusters to use, instability of the clusters when exploring different data sets, and the inability to identify negative correlations (37).
Microarray experiments are liable to a large number of nonbiological sources of error (38). In general, there are few defined metrics available for assigning the quality of microarray data in public data sets. The method used here facilitates a quantitative assessment of each data set compared with a large body of other microarray data sets. The method further allows for quality control within a subset of microarray experiments, as opposed to across all data sets from many tissues, which may decrease the utility of other metrics.
The regression method was used to assess coregulatory networks of genes for cellulose biosynthesis in Arabidopsis. The notion that three CESA subunits are required to assemble into a functional CESA complex implies coexpression of the corresponding genes. Analysis of the levels of mRNA for the known CESA genes in barley was consistent with this idea (21). Indeed, the genes for the three CESA subunits involved in primary and secondary cell wall cellulose biosynthesis were highly coexpressed over the 408 microarray data sets analyzed here. Additionally, genes such as COBRA and CTL1, which have previously been implicated in cellulose synthesis by genetic analysis, were highly coexpressed with CESA1, 3, and 6. Similarly, the COBRA homolog COBL4, which has been implicated in cellulose synthesis in rice (12), was highly coregulated with CESA4, 7, and 8. A CTL1 homolog (At3g16920) was found here to be highly coexpressed with CESA4, 7, and 8, and disruption of the gene caused an FTIR phenotype indicative of alterations in the cell wall composition. Several other pairs of homologous genes are also present in Tables 1 and 2 (i.e., several ADP-glucose pyrophosphorylases and glycosyltransferases). Thus, it appears that the two types of CESA complexes have specialized homologs of other proteins that are involved in synthesis and assembly of the cell wall.
Three other genes, At5g54690 (IRX8), At5g03170 (IRX13), and At4g27435 that were coexpressed with CESA4, 7, and 8, were analyzed genetically to assess their potential functions in cell wall synthesis. Stem sections from transferred DNA (T-DNA) insertion mutations in the IRX8 and IRX13 genes displayed deformations in xylem cells, similar to established phenotypes for CESA4, 7, and 8 mutants (32). Cellulose analyses of cell wall preparations from stems revealed reduction in cellulose for irx8 and irx13, and PC analysis of FTIR spectra further suggested alterations in noncellulosic polymers for these mutants. IRX8 encodes an apparent family 8 glycosyltransferase with sequence similarity to QUASIMODO1, an endomembrane-localized glycosyltransferase implicated in pectin synthesis (33). IRX13 and At3g16920 are similar to an arabinogalactan protein and a chitinase-like protein, respectively, whereas At4g27435 is a gene with no similarity to known proteins. The identification of these four genes substantially increases the list of genes implicated in secondary cellulose synthesis. It seems likely that mutant analysis of other genes listed in Tables 1 and 2 will yield additional genes of interest in this context.
The mapping of coexpressed genes onto biological pathway schemes provides a more comprehensible way of displaying the data. Unfortunately, only a limited number of pathways are presently accessible through the Gene Ontology annotation guide and AraCyc at The Arabidopsis Information Resource. The majority of genes listed in Tables 1 and 2 are, therefore, not included in the pathway assembly. Nevertheless, the results revealed a complex pattern across many biological pathways, indicating that cell wall synthesis is coordinated with several other biological processes. Not surprisingly, Table 3 indicates that both types of CESA genes appear to be coordinated with several shared pathways. However, although the genes in these shared pathways often are functional homologs, they are very rarely the same gene, suggesting the existence of dual biosynthetic networks for the two CESA complexes.
Several pathways differed significantly in coregulation for the CESA complexes (e.g., brassinosteroid, dTDP-rhamnose, and lignin biosynthesis pathways). A connection between brassinosteroids and the cell wall matrix was previously suggested by a reduction of transcription of KORRIGAN observed in det2, a mutant deficient in brassinosteroid synthesis (10). In addition, brassinosteroids modulate the transcript levels of cell wall-related genes involved in cell elongation and morphogenesis (39). Because secondary wall synthesis takes place when cell expansion has ceased, it makes sense that expression of the CESA4, 7, and 8 genes are not linked to brassinosteroid synthesis. Lignin deposition, however, is largely associated with secondary wall formation (36). Genes encoding lignin monomer-polymerizing laccases and lignin monomer synthesis are among the 50 most closely coexpressed genes for CESA 4, 7, and 8 (Table 2). In addition, genes linked to the lignin-related pathway for suberin synthesis are highly coexpressed with CESA4, 7, and 8 (Tables 5 and 6).
The analyses performed here can be readily extended to identify additional coregulatory networks. The coexpression approach may also be used to identify previously unknown coregulatory patterns in sets of genes with known functions to decipher underlying networks among the genes. The results presented here indicate that the integration of multiple data sets for linking coexpressed genes is practical and that the use of simple linear regression is feasible. Since completing this work, it has come to our attention that Simon Turner and colleagues have used a proprietary data set and different statistical methods to identify genes that are coexpressed with secondary cell wall CESA genes (S. Turner, personal communication). More than half of the 25 genes identified as being coregulated with IRX3 were common to both studies.
Supplementary Material
Acknowledgments
We thank Mehta Tapan for help in running RMA in the Cahaba cluster at the Department of Mechanical Engineering, University of Alabama at Birmingham. S.P. was a recipient of a fellowship from the Carl Tryggers Foundation (CTS03:258). This work was supported by United States Department of Energy Grant DE-FG02-03ER20133 and National Science Foundation Grant NSF MCB 0114562.
Author contributions: S.P., H.W., J.M., G.P.P., and C.R.S. designed research; S.P., H.W., and J.M. performed research; S.P., H.W., J.M., and G.P.P. contributed new reagents/analytic tools; S.P., H.W., J.M., G.P.P., and C.R.S. analyzed data; and S.P., H.W., J.M., G.P.P., and C.R.S. wrote the paper.
Abbreviations: CESA, cellulose synthase; FTIR, Fourier transform infrared; RMA, robust multiarray analysis; K–S, Kolmogorov–Smirnov; PC, principal component.
References
- 1.Doblin, M. S., Kurek, I., Jacob-Wilk, D. & Delmer, D. P. (2002) Plant Cell Physiol. 43, 1407-1420. [DOI] [PubMed] [Google Scholar]
- 2.Somerville, C., Bauer, S., Brininstool, G., Facette, M., Hamann, T., Milne, J., Osborne, E., Paredez, A., Persson, S., Raab, T., et al. (2004) Science 306, 2206-2211. [DOI] [PubMed] [Google Scholar]
- 3.Arioli, T., Peng, L., Betzner, A. S., Burn, J., Wittke, W., Herth, W., Camilleri, C., Höfte, H., Plazinski, J., Birch, R., et al. (1998) Science 279, 717-720. [DOI] [PubMed] [Google Scholar]
- 4.Scheible, W. R., Eshed, R., Richmond, T., Delmer, D. & Somerville, C. (2001) Proc. Natl. Acad. Sci. USA 98, 10079-10084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fagard, M., Desnos, T., Desprez, T., Goubet, F., Refregier, G., Mouille, G., McCann, M., Rayon, C., Vernhettes, S. & Höfte, H. (2000) Plant Cell 12, 2409-2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Taylor, N. G., Laurie, S. & Turner, S. R. (2000) Plant Cell 12, 2529-2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Taylor, N. G., Scheible, W. R., Cutler, S., Somerville, C. R. & Turner, S. R. (1999) Plant Cell 11, 769-780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tanaka, K., Murata, K., Yamazaki, M., Onosato, K., Miyao, A. & Hirochika, H. (2003) Plant Physiol. 133, 73-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Taylor, N. G., Howells, R. M., Huttly, A. K., Vickers, K. & Turner, S. R. (2003) Proc. Natl. Acad. Sci. USA 100, 1450-1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nicol, F., His, I., Jauneau, A., Vernhettes, S., Canut, H. & Höfte, H. (1998) EMBO J. 17, 5563-5576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schindelman, G., Morikami, A., Jung, J., Baskin, T. I., Carpita, N. C., Derbyshire, P., McCann, M. C. & Benfey, P. N. (2001) Genes Dev. 15, 1115-1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li, Y., Qian, Q., Zhou, Y., Yan, M., Sun, L., Zhang, M., Fu, Z., Wang, Y., Han, B., Pang, X., et al. (2003) Plant Cell 15, 2020-2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhong, R., Kays, S. J., Schroeder, B. P. & Ye, Z. H. (2002) Plant Cell 14, 165-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pagant, S., Bichet, A., Sugimoto, K., Lerouxel, O., Desprez, T., McCann, M., Lerouge, P., Vernhettes, S. & Höfte, H. (2002) Plant Cell 14, 2001-2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gillmor, C. S., Poindexter, P., Lorieau, J., Palcic, M. M. & Somerville, C. (2002) J. Cell Biol. 156, 1003-1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bichet, A., Desnos, T., Turner, S., Grandjean, O. & Höfte, H. (2001) Plant J. 25, 137-148. [DOI] [PubMed] [Google Scholar]
- 17.Eisen, M. B., Spellman, P. T., Brown, P. O. & Botstein, D. (1998) Proc. Natl. Acad. Sci. USA 95, 14863-14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kemmeren, P., van Berkum, N. L., Vilo, J., Bijma, T., Donders, R., Brazma, A. & Holstege, F. C. (2002) Mol. Cell 9, 1133-1143. [DOI] [PubMed] [Google Scholar]
- 19.Stuart, J. M., Segal, E., Koller, D. & Kim, S. K. (2003) Science 302, 249-255. [DOI] [PubMed] [Google Scholar]
- 20.Lee, H. K., Hsu, A. K., Sajdak, J., Qin, J. & Pavlidis, P. (2004) Genome Res. 14, 1085-1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Burton, R. A., Shirley, N. J., King, B. J., Harvey, A. J. & Fincher, G. B. (2004) Plant Physiol. 134, 224-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dabrowski, M., Aerts, S., Van Hummelen, P., Craessaerts, K., De Moor, B., Annaert, W., Moreau, Y. & De Strooper, B. (2003) J. Neurochem. 85, 1279-1288. [DOI] [PubMed] [Google Scholar]
- 23.Kuo, W. P., Jenssen, T. K., Butte, A. J., Ohno-Machado, L. & Kohane, I. S. (2002) Bioinformatics 18, 405-412. [DOI] [PubMed] [Google Scholar]
- 24.Craigon, D. J., James, N., Okyere, J., Higgins, J., Jotham, J. & May, S. (2004) Nucleic Acids Res. 32, D575-D577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Redman, J. C., Haas, B. J., Tanimoto, G. & Town, C. D. (2004) Plant J. 38, 545-561. [DOI] [PubMed] [Google Scholar]
- 26.Irizarry, R. A., Hobbs, B., Collin, F., Beazer-Barclay, Y. D., Antonellis, K. J., Scherf, U. & Speed, T. P. (2003) Biostatistics 4, 249-264. [DOI] [PubMed] [Google Scholar]
- 27.Chakravarti, I. M., Laha, R. G. & Roy, J. (1967) Handbook of Methods of Applied Statistics (Wiley, New York), Vol. 1, pp. 392-394. [Google Scholar]
- 28.Alonso, J., Stepanova, A., Leisse, T., Kim, C., Chen, H., Shinn, P., Stevenson, D., Zimmerman, J., Barajas, P., Cheuk, R., et al. (2003) Science 301, 653-657. [DOI] [PubMed] [Google Scholar]
- 29.Kemsley, E. K. (1998) Discriminant Analysis and Class Modelling of Spectroscopic Data (Wiley, New York), p. 179.
- 30.Updegraff, D. M. (1969) Anal. Biochem. 32, 420-424. [DOI] [PubMed] [Google Scholar]
- 31.Ueda, H. R., Hayashi, S., Matsuyama, S., Yomo, T., Hashimoto, S., Kay, S. A., Hogenesch, J. B. & Iino, M. (2004) Proc. Natl. Acad. Sci. USA 101, 3765-3769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Turner, S. R. & Somerville, C. R. (1997) Plant Cell 9, 689-701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bouton, S., Leboeuf, E., Mouille, G., Leydecker, M. T., Talbotec, J., Granier, F., Lahaye, M., Hofte, H. & Truong, H. N. (2002) Plant Cell 14, 2577-2590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mouille, G., Robin, S., Lecomte, M., Pagant, S. & Höfte, H. (2003) Plant J. 35, 393-404. [DOI] [PubMed] [Google Scholar]
- 35.Nemhauser, J. L. & Chory, J. (2004) J. Exp. Bot. 55, 265-270. [DOI] [PubMed] [Google Scholar]
- 36.Demura, T., Tashiro, G., Horiguchi, G., Kishimoto, N., Kubo, M., Matsuoka, N., Minami, A., Nagata-Hiwatashi, M., Nakamura, K., Okamura, Y., et al. (2002) Proc. Natl. Acad. Sci. USA 99, 15794-15799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gibbons, F. D. & Roth, F. P. (2002) Genome Res. 12, 1574-1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Page, G. P., Edwards, J. W., Barnes, S., Weindruch, R. & Allison, D. B. (2003) Nutrition 19, 997-1000. [DOI] [PubMed] [Google Scholar]
- 39.Goda, H., Shimada, Y., Asami, T., Fujioka, S. & Yoshida, S. (2002) Plant Physiol. 130, 1319-1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.