Abstract
Nucleobase analogs 5-methylisocytosine (MeisoC) and isoguanine (isoG) form a non-natural base pair in duplex nucleic acids with base pairing specificity orthogonal to the natural nucleobase pairs. Sequencing reactions were conducted with oligodeoxyribonucleotides (ODNs) containing dMeisoC and disoG using modified pyrosequencing and dye terminator methods. Modified dye terminator sequencing was generally useful for the sequence identification of ODNs containing the non-natural nucleobases. The two sequencing methods were also used to monitor nucleotide incorporation and subsequent extension by Family A polymerases used in the sequencing methods with a six-nucleobase system that includes dMeisoC and disoG. Nucleic acids containing the six-nucleobase system could be replicated well, but not as well as natural nucleic acids, especially in regions of high dMeisoC–disoG content. Challenges in replication with dMeisoC–disoG are consistent with nucleobase tautomerism in the insertion step and disrupted minor groove nucleobase pair–polymerase contacts in subsequent extension.
INTRODUCTION
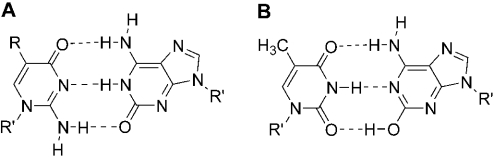
Non-natural nucleobase analogs with base pairing specificity orthogonal to the natural base pairs have been designed to expand the sequence and functional diversity of nucleic acids (1–3). One strategy in the design of additional base pairs has been to work within the Watson–Crick pairing rules of size and hydrogen bonding complementarity. In this approach, nucleobase analogs with carbon/nitrogen ring systems isosteric to natural purines or pyrimidines are used to implement hydrogen bonding functionality arrayed in patterns not found in natural DNA (4). The most thoroughly studied of these non-natural pairs is the 5-methylisocytosine–isoguanine (MeisoC–isoG) pair joined by three hydrogen bonds in duplex nucleic acids (Figure 1) (5–7), and capable of acting as a third base pair in PCR amplification (8). The MeisoC–isoG pair has established technological value in reducing background signal (9) in widely used commercial diagnostic nucleic acid hybridization assays (10,11) approved by the U.S. Food and Drug Administration and other global regulatory authorities. The pair has been used as a component of a real-time quantitative PCR assay (12). Non-natural isoC–isoG or MeisoC–isoG pairs have also been used as mechanistic probes of the fundamental biological processes of template-directed nucleic acid synthesis (13,14), translation (15), protein-mediated strand exchange of DNA (16) and excision repair (17).
Figure 1.
Base pairing with the isoC–isoG non-natural pair. (A) IsoC and isoG form a three hydrogen bond pair. (B) An isoG tautomer is complementary to T in Watson–Crick pairing. 2′-Deoxy-5-methylisocytidine and 2′-deoxyisoguanosine were used in this work (R = CH3, R′ = 2′-deoxyribose).
If nucleic acids containing MeisoC and isoG are to have the utility of natural nucleic acids, the tools and techniques of molecular biology must be available. A powerful tool for characterizing nucleic acids is sequence determination. Non-natural nucleobase positions in nucleic acids have been identified in very limited experiments using various methods, including enzymatic pausing (18), chemical degradation (8,13,14,19) and dye-labeled terminators (20). No reported sequencing method has concurrently identified both nucleobases of a non-natural pair. Here, we describe work to sequence oligodeoxyribonucleotides (ODNs) containing dMeisoC and disoG. We demonstrate that dMeisoC and disoG positions can be unambiguously identified within a single nucleic acid using a dye-labeled terminator method, despite lacking terminators corresponding to the non-natural nucleobases. We have repeatedly used this method to verify synthetic ODN sequences. Another method using pyrosequencing, which detects pyrophosphate generated from the enzymatic addition of a nucleoside triphosphate to a nucleic acid strand, is only partially successful at sequencing ODNs containing dMeisoC and disoG. Development of these sequencing systems with non-natural analogs has afforded the additional benefit of probing polymerase molecular recognition. The two sequencing methods were used to monitor nucleotide incorporation and subsequent extension by polymerases with a six-nucleobase system that includes dMeisoC and disoG.
MATERIALS AND METHODS
Oligodeoxyribonucleotides
Synthetic ODN sequences containing disoG and dMeisoC were synthesized using phosphoramidite chemistry (5,21) and PAGE purified. ODN purity was verified as at least 90%, and nearly always >95%, by capillary electrophoresis analysis (22). The identity of each ODN was confirmed by matrix-assisted laser desorption ionization time-of-flight mass spectrometry and high-performance liquid chromatography (HPLC) analysis of component nucleosides after enzymatic degradation (22).
Pyrosequencing
Pyrosequencing was performed using a PSQ96MA Sequencer (Biotage AB). For optimal results, the concentration of a nucleotide in pyrosequencing reactions should be slightly above the Km of the enzyme for that particular nucleotide. Lower concentrations cause incomplete incorporation and higher concentrations increase misincorporation. Stock solutions of nucleotides are dispensed stepwise from four reservoirs in a pyrosequencing dispensation cartridge. Concentrations of stock solutions of dMeisoCTP and disoGTP required to give nucleotides at the appropriate Km during extension were roughly determined in separate experiments (data not shown) by dispensing a range (10 μM, 50 μM, 500 μM and 2.5 mM) of nucleotide concentrations in pyrosequencing reactions with templates containing either disoG (for dMeisoCTP) or dMeisoC (for disoGTP). The lower concentrations were clearly insufficient and generated less pyrophosphate than expected for complete incorporation. Signal height at 500 μM and 2.5 mM was nearly unchanged for both non-natural nucleotides, suggesting pyrosequencing reservoir stock solutions at 500 μM dispense nucleotide near Km in the reaction solution for the insertion of dMeisoCTP opposite template disoG and disoGTP opposite template dMeisoC. The higher 2.5 mM concentration was chosen for these nucleotides in the cartridge reservoirs to guard against incomplete incorporation at the expense of possibly slightly increasing misincorporation of dMeisoC and disoG. For comparison, standard cartridge concentrations used in pyrosequencing were measured by A260 at 0.3–0.6 mM for dCTP, dGTP and dTTP, and ∼2.5 mM for α-S-dATP.
Most of the pyrophosphate impurities visible in pyrosequencing were removed from dMeisoCTP and disoGTP by HPLC purification with a YMC-Pack ODS-AM column (120 Å, 5 μm, 250 × 4.6 mm). Approximately 0.2 μmol of nucleotide (20 μl) was injected on a Series 1100 HPLC (Hewlett Packard) and purified with a binary gradient (solvent A = 0.2 M triethylammonium acetate, pH 6.8; solvent B = 95% solvent A, 5% acetonitrile) at 1.0 ml/min: 4% solvent B hold for 10 min, then increase solvent B to 100% over 25 min. The eluate containing the dMeisoCTP or disoGTP was collected by monitoring at 260 nm. Eluate containing dMeisoCTP was immediately adjusted to pH 8.3 with triethylamine. The solvent and the volatile buffer were removed under vacuum and the nucleotide was redissolved in water. The purification yielded nucleotides with no detectable impurity peaks upon reinjection and analysis with this HPLC method, and removed most of the pyrophosphate undetectable by UV monitoring, but visible in pyrosequencing.
Fifteen ODN templates were designed to vary the nearest-neighbor positions around dMeisoC and disoG positions. Only three of the four natural nucleobases were used in each template to leave an available reservoir for either dMeisoCTP or disoGTP. Complementary nucleotides were sequentially dispensed for each template. In instances of incomplete incorporation, sequential dispensations of a single nucleotide were used in a subsequent experiment to examine whether extending the time available for incorporation would increase incorporation. One dispensation of an out-of-sequence non-complementary nucleotide was performed with each ODN as a negative control (Supplementary Figure S3). Pyrosequencing data presented are peak heights of emitted light detected and are the average of two replicates.
Preparation of ODNs for cycle sequencing
Synthetic ODNs were ligated with T4 DNA ligase (Amersham Biosciences) to a DNA fragment, which was assembled from three component ODNs (170 nt total, sequences in Supplementary Material). The ODN to be sequenced (1.73 nmol) was ligated to a 5′-phosphorylated 50mer ODN (1.44 nmol) using a reverse complementary linker ODN (2.02 nmol) that formed a 6 nt duplex with each of the ODNs to be ligated. Simultaneously, the 50mer was ligated to a 5′-phosphorylated 57mer (1.20 nmol) through an analogous linker ODN (1.68 nmol), and the 57mer was in turn ligated to a 5′-phosphorylated 63mer (1.00 nmol) through another linker ODN (1.40 nmol). An annealing step was first performed in 1× 100 μl ligation buffer (50 mM Tris–HCl, pH 7.5, 10 mM MgCl2, 2 mM spermidine) by incubating the solution at 55°C (2 min) and reducing the temperature (0.67°C/min) to 22°C, then holding (2 min). To the ODN solution was added 10× ligation buffer (10 μl), 100 mM ATP (4 μl), 500 mM DTT (4 μl), water (28 μl), 50% PEG-8000 (48 μl) and T4 DNA ligase (6 μl, 6 U). The ligation reaction was incubated at 20°C for 14 h. The reaction was quenched with 0.5 M EDTA (6 μl) and the nucleic acid was precipitated by adding pH 4.8 ammonium acetate (137 μl) and ethanol (687 μl) and cooling at −20°C for 1 h. After spinning in a microcentrifuge at 4°C (20 000 r.c.f., 30 min) and rinsing three times with cold 80% ethanol, the pellet was dissolved and the ligation product separated on a 5% polyacrylamide gel. The product band was excised and isolated by electroelution. The ligation product was purified using a NAP-25 column and then ethanol precipitated again.
Cycle sequencing
The template produced by ligation was included in cycle sequencing reactions with dMeisoCTP (21), disoGTP (21), and either BigDye 3.0 (Applied Biosystems) or BigDye 3.1 (Applied Biosystems) kits. Cycle sequencing was performed on a 7700 Sequence Detector (Applied Biosystems) in 9600 emulation mode with 17.6 μl of Ready Reaction Mix using 25 nM template and 160 nM primer in 44 μl reactions for 25 cycles (96°C, 10 s; 60°C, 240 s). The reactions were then purified with DTR spin columns (Edge Biosystems). The sequencing reactions were analyzed on a 310 Genetic Analyzer (Applied Biosystems) using POP-6 polymer gel in a 61 cm × 50 μm uncoated capillary (50°C, 200 V/cm). Concentrations of dMeisoCTP and disoGTP (10–1000 μM) were examined in optimization matrix experiments with the goals of minimal signal attenuation and no mispaired terminator signals opposite dMeisoC or disoG template positions.
RESULTS
Pyrosequencing
In pyrosequencing (23,24), nucleoside triphosphates are singly dispensed into a solution containing primer, template and exo(−) Klenow fragment of DNA polymerase I at 28°C. Incorporation of a complementary nucleotide produces an enzymatically mediated cascade resulting in the generation of visible light. The amount of light generated is proportional to the pyrophosphate produced during incorporation. Excess nucleotide is enzymatically destroyed before subsequent nucleotide dispensations. The ODN templates used here all have natural nucleobases at the first four template positions and these positions always yielded relative signal heights typical of pyrosequencing with natural nucleobases. In evaluating these signals, it is important to note that dispensations of α-S-dATP used in pyrosequencing typically result in peak heights ∼20% higher than the other nucleotides (25). These first four positions form a baseline for comparison of replication performance with non-natural nucleobases. Because of the template design, non-natural positions were challenged with natural nucleotides upon dispensation of the complementary nucleotide opposite the fourth template position of the ODNs containing dMeisoC or disoG positions. Significant incorporation of a natural nucleotide opposite the dMeisoC or disoG at the fifth template position would add to the signal for incorporation opposite the fourth position and give a signal greater than one equivalent.
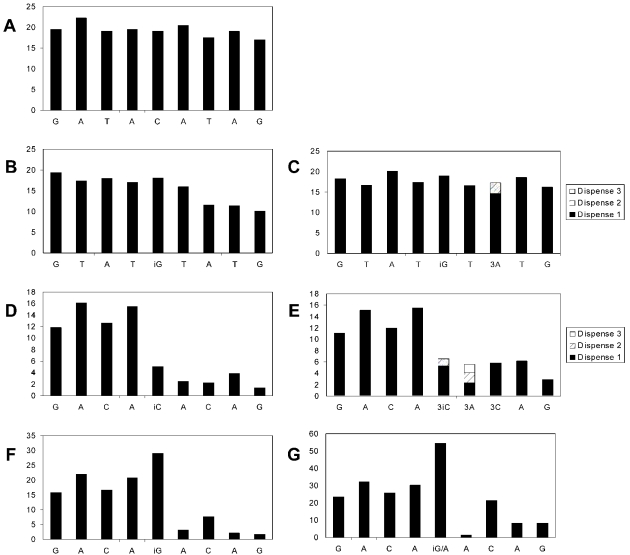
Pyrosequencing reactions performed quite differently depending on whether disoG or dMeisoC was present in the 9 nt template region of the individual ODNs (Figure 2 and Supplementary Figures S1 and S2). Complementary disoG nucleotide was always readily incorporated opposite dMeisoC in the template, while the natural nucleotides were not significantly incorporated opposite dMeisoC (Figure 2B). However, further extension following the dMeisoC–disoG pair was slowed and was often incomplete after a single dispensation of nucleotide. Sequential dispensations of the same complementary nucleotide allowed more cumulative time for incorporation and usually improved the incomplete incorporation observed with a single dispensation at positions following a disoG–dMeisoC pair (Figure 2C). Misincorporation upon dispensing disoGTP was observed when disoGTP was dispensed at dMeisoC positions followed by dT (Figure 2F); more than one equivalent of pyrophosphate was produced, indicating that disoG nucleotide was incorporated opposite dMeisoC and then further incorporated opposite the following dT. When α-S-dATP was mixed 1:1 with disoGTP and dispensed at a template dMeisoC position followed by dT, two equivalents of pyrophosphate were produced and incorporation opposite the position following dT was improved (Figure 2G), suggesting that disoG and dA were incorporated opposite disoC and dT, respectively. This implies that dA is incorporated more readily opposite template dT positions than disoG.
Figure 2.
Pyrosequencing of ODNs containing dMeisoC and disoG. All primer binding regions were identical and a 9 nt template region was varied. Nucleotide dispensations are indicated on the x-axis and relative peak height of light emitted from pyrophosphate release is the y-axis. Consecutive dispensations of a single nucleotide are cumulatively tallied in a single bar. At least one negative control dispensation of a non-complementary nucleotide was made during pyrosequencing of each ODN. These negative control dispensations resulted in no pyrophosphate release and have been omitted for clarity. (A) A template sequence of all natural nucleobases, 3′-CTATGTATC-5′, was sequenced as a positive control. (B) Template sequences containing dMeisoC, such as 3′-CATAiCATAC-5′, displayed complete incorporation of disoG, but extension at subsequent natural nucleobases was inhibited. (C) Allowing more time for incorporation at template positions following dMeisoC through consecutive dispensations of a single nucleotide improved the incomplete incorporation observed in (B). (D) Template sequences containing disoG, such as 3′-CTGTiGTGTC-5′, displayed incomplete incorporation of dMeisoC nucleotide, and extension at subsequent natural nucleobases was inhibited. (E) Consecutive dispensations of dMeisoCTP opposite the template disoG position in (D) incorporated slightly more nucleotide, but still gave less than one equivalent of nucleotide incorporation. Consecutive dispensations of the same nucleotide at positions following disoG improved the incomplete incorporation at these positions observed in (D). (F) Template sequence 3′-CTGTiCTGTC-5′ demonstrated misincorporation of disoG nucleotide opposite template dT following correct incorporation opposite template dMeisoC. (G) Dispensing a 1:1 mixture of disoG and dA nucleotides in place of disoGTP in (F) generated no pyrophosphate at the following dispensation of dA nucleotide and improved incorporation at the second position following the template dMeisoC position, suggesting correct incorporation opposite template dMeisoC and the following dA position.
In contrast, significantly less than one equivalent of pyrophosphate was produced when dMeisoCTP was dispensed at disoG template positions (Figure 2D). The proportion of template disoG paired with dMeisoC was quite variable in different sequence contexts (Supplementary Figure S1). Extending the time available for incorporation through multiple dispensations of dMeisoCTP increased the incorporation of dMeisoC only slightly (Figure 2E). Further extension of the fraction of templates incorporating dMeisoC opposite template disoG was slow and was improved by multiple nucleotide dispensations at positions following dMeisoC–disoG (Figure 2E). Significant misincorporation of any natural nucleotide opposite template disoG was not observed, and dMeisoC nucleotide was not visibly incorporated opposite any of the natural nucleobases.
Natural nucleotides were not significantly incorporated opposite dMeisoC or disoG positions of any template. No misincorporation following the fourth template position is visible in any of the pyrosequencing reactions, which cover all possible natural nucleotide misincorporations opposite non-natural template positions. Interestingly, even dT was not misincorporated opposite template disoG.
Dye terminator sequencing
Sequencing reactions with a thermophilic polymerase and dye terminator chemistry were also conducted with ODNs containing disoG and dMeisoC. Two sequencing kits, BigDye 3.0 and BigDye 3.1 (Applied Biosystems), were used. Amplitaq FS in BigDye 3.0 is a Taq polymerase with two point mutations: the F766Y mutation increases the acceptance of dideoxy nucleotides and G46D eliminates the 5′-exonuclease activity (26). The identity of the polymerase from the BigDye 3.1 kit is undisclosed, but the kit almost certainly includes a Family A polymerase with two analogous mutations (26). The polymerases from the two kits had qualitatively similar performance in sequencing reactions with dMeisoC and disoG.
A series of sequencing reactions with a 59mer template ODN containing 12 non-natural nucleobase positions was performed to determine suitable concentrations of disoGTP and dMeisoCTP for dye terminator sequencing. At low concentrations of disoGTP and dMeisoCTP, ddA and ddT terminators were incorporated opposite dMeisoC and disoG template positions, respectively (Figure 3 and Supplementary Figures S4 and S5). We presume that incorporation of a dideoxy nucleotide opposite a given template position indicates concurrent incorporation of the corresponding deoxynucleotide at a fraction of the template nucleic acid at this position, as in standard dideoxy terminator sequencing. Because disoGTP and dMeisoCTP do not have corresponding fluorescent dideoxy terminators in these reactions, incorporation of these nucleotides lacks an associated dye terminator signal. Therefore, dye signals from ddA vanished opposite dMeisoC template positions as proportionally more disoG nucleotide was incorporated with increasing concentration of disoGTP (Figure 3 and Supplementary Figure S4). Similarly, dye signals from ddT diminished opposite disoG template positions with increasing concentration of dMeisoCTP (Figure 3 and Supplementary Figure S5). Useful concentrations at which incorporation of terminators was substantially suppressed with minimal signal attenuation appear to be 100–200 μM disoGTP and 100–200 μM dMeisoCTP for AmpliTaq FS in the BigDye 3.0 kit. The BigDye 3.1 kits required 200–400 μM disoGTP and 100–400 μM dMeisoCTP for similar results.
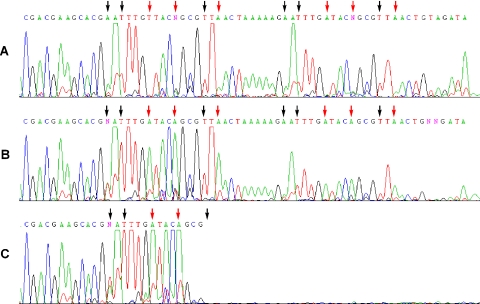
Figure 3.
Dye terminator sequencing (BigDye 3.1 kit) of an ODN containing dMeisoC and disoG used to optimize concentrations of dMeisoCTP and disoGTP. The template sequence in the region shown is 3′-GCTGCTTCGTGCiGTiGAACiCATGiCCGCiGAiC TGATTTTTCiGTiGAACiCATGiCCGCiGAiCTGACATCTA-5′. Changing the concentrations of dMeisoCTP and disoGTP caused differences in dye terminator signals at template disoG (black arrow) and dMeisoC (red arrow) positions. With increasing concentrations of dMeisoCTP, signals from ddT were suppressed opposite template disoG positions. With increasing concentrations of disoGTP, signals from ddA were suppressed opposite dMeisoC template positions. These changes are indicative of competition for incorporation at non-natural template positions between a non-natural nucleotide and a specific natural nucleotide. (A) Sequencing with 100 μM dMeisoCTP and 400 μM disoGTP largely suppressed terminator incorporation opposite the non-natural positions. Modest signal attenuation caused by unwanted strand termination was apparent. (B) As the concentrations of disoGTP and dMeisoCTP were decreased (10 μM dMeisoCTP and 10 μM disoGTP), ddA terminators were incorporated opposite template dMeisoC positions and ddT terminators were incorporated opposite template disoG positions. Signal attenuation caused by unwanted strand termination was increased. (C) In the absence of disoGTP and dMeisoCTP, more ddA was incorporated opposite template disoC and more ddT was incorporated opposite template dMeisoG. Forced misincorporation at all non-natural template positions led to premature termination of the sequencing reaction, unlike in (A) and (B) where it was possible to incorporate some non-natural nucleotide. Similar reactions were performed using the BigDye 3.0 kit (Supplementary Figures S4 and S5).
Sequencing reactions with even very little disoGTP and dMeisoCTP allowed full extension through the 12 non-natural nucleobase positions (Figure 3B), although modest signal attenuation was always observed upon encountering the multiple non-natural nucleobase positions. In contrast, replication of the template was completely terminated in the absence of disoGTP and dMeisoCTP (Figure 3C). These changes in incorporation and extension with varying disoGTP and dMeisoCTP concentrations were primarily a result of the change in nucleotide concentration, and not a generally inhibitory effect, such as an effective reduction in the free Mg2+ concentration. If an increase in disoGTP or dMeisoCTP caused a general inhibition, then the dye signals for the >170 natural nucleobases preceding the non-natural template positions would also be attenuated. General attenuation was observed only at very high concentrations of disoGTP and dMeisoCTP (data not shown).
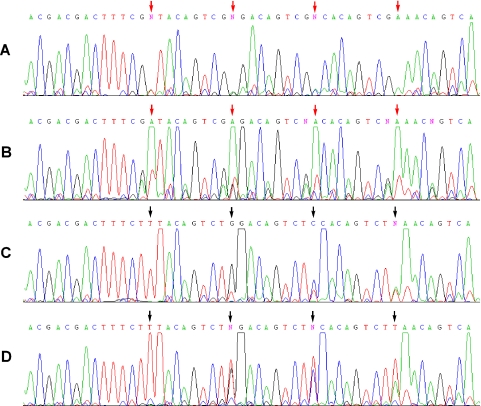
A series of sequencing reactions was conducted to examine the influence of sequence context (Figure 4 and Supplementary Figures S6 and S7). These experiments were performed with 42mer template ODNs containing all 16 natural nucleotide nearest-neighbor contexts possible for dMeisoC and analogous template ODNs for disoG. The ODNs were used as templates in sequencing reactions in the presence or absence of disoGTP and dMeisoCTP. In the absence of disoGTP and dMeisoCTP, extension required misincorporation of natural nucleobases opposite the dMeisoC and disoG template positions in order to proceed; ddA was always incorporated opposite template dMeisoC positions (Figure 4B) and ddT was always incorporated opposite template disoG positions (Figure 4D). In the presence of disoGTP and dMeisoCTP, the complementary non-natural nucleotide was paired opposite dMeisoC and disoG in all sequence contexts, verified by diminished terminator signals opposite the non-natural template positions (Figure 4A and C). Additionally, a noticeable signal that may correspond to the polymerase skipping over a fraction of template disoG positions was often visible opposite disoG (Figure 4C).
Figure 4.
Dye terminator sequencing of two ODNs used to examine dMeisoC and disoG in all possible natural nearest-neighbor contexts (Supplementary Figures S6 and S7 contain additional sequences). An ODN containing four dMeisoC positions (red arrows) was sequenced in a BigDye 3.1 reaction in the presence of (A) 300 μM disoGTP and 0 μM dMeisoCTP or (B) 0 μM disoGTP and 0 μM dMeisoCTP. The template sequence in the region shown is 3′-TGCTGCTGAAAGCiCATGTCAGCiCCTGTCAGCiCGTGTCAGCiCTTTGTCAG-5′. Very large ddA terminator signals were always observed opposite dMeisoC in the absence of disoGTP, perhaps indicating a transition state base pairing geometry different from complementary pairings. An ODN containing four disoG positions (black arrows) was sequenced in a BigDye 3.1 sequencing reaction in the presence of (C) 100 μM dMeisoCTP and 0 μM disoGTP or (D) 0 μM dMeisoCTP and 0 μM disoGTP. The template sequence in the region shown is 3′-TGCTGCTGAAAGAiG ATGTCAGAiGCTGTCAGAiGGTGTCAGAiGTTGTCAG-5′. Terminator ddT was always incorporated opposite disoG in the absence of dMeisoCTP. Extremely large terminator signals were observed at the position following disoG for all conditions. Additionally, a noticeable signal that may correspond to the polymerase skipping over a fraction of template disoG positions was visible opposite disoG. Little, if any, signal attenuation caused by unwanted strand termination was visible upon encountering isolated dMeisoC or disoG positions, either in the presence or absence of complements disoGTP and dMeisoCTP. This contrasts with Figure 3C, in which several proximate non-natural template positions caused premature termination of sequencing reactions lacking disoGTP and dMeisoCTP.
Notable features of the terminator signal intensities were evident in the sequencing reactions. Interestingly, no apparent signal attenuation was visible with these ODN templates containing only isolated non-natural nucleobases, either in the presence or absence of disoGTP and dMeisoCTP. Extremely large terminator signals were always observed at the position following incorporation of either dMeisoC or dT nucleotides opposite template disoG positions, indicating the partitioning of dideoxy and deoxy nucleotides opposite these positions was outside the usual range for natural nucleobase templates. In the absence of disoGTP, extremely large ddA terminator signals were observed opposite dMeisoC positions. The perturbed ratio of dideoxy terminator to deoxynucleotide for incorporation of A nucleotides opposite dMeisoC suggests that transition state base pairing geometry of this pair may be different from complementary pairs.
Sequencing reactions were also conducted to verify the specificity of incorporation of dMeisoC and disoG nucleotides (Supplementary Figure S8). Addition of dMeisoCTP and disoGTP to sequencing reactions with templates lacking dMeisoC and disoG caused no discernable differences in dye terminator patterns from standard sequencing reactions. Addition of dMeisoCTP had no effect on sequencing reactions of templates containing dMeisoC, even in the absence of disoGTP. Similarly, addition of disoGTP had no effect on sequencing reactions of templates containing disoG in the absence of dMeisoCTP. These reactions demonstrate that dMeisoC and disoG were not significantly incorporated opposite natural nucleobases and were not self-paired.
DISCUSSION
We have demonstrated the first generally useful method to determine sequences of nucleic acids containing both constituents of a non-natural nucleobase pair. Our dye terminator method has been routinely used in a single reaction with dMeisoCTP and disoGTP to verify the known sequences of diverse synthetic ODNs containing dMeisoC and disoG. Additionally, the method may have future application in determining unknown sequences of nucleic acids, such as ODNs generated from in vitro selection (27) experiments using a six-nucleobase lexicon with dMeisoC and disoG. More than one reaction is necessary to unambiguously identify dMeisoC and disoG positions in nucleic acids of unknown sequence containing both nucleobases. In a first reaction, dMeisoCTP and isoGTP are present at concentrations sufficient to suppress misincorporation at dMeisoC and disoG positions. In subsequent sequencing reactions, the concentration of dMeisoCTP or disoGTP is reduced (both nucleotide concentrations may also be reduced simultaneously in a single reaction), permitting ddA and ddT nucleotides to be incorporated opposite some of the dMeisoC and disoG positions, respectively. Suppression of specific terminator signals at increased dMeisoCTP or disoGTP concentrations, in addition to the signature large terminator signal following disoG template positions, should allow the identification of the non-natural positions.
The sequencing experiments also demonstrate how replication with the six-nucleobase lexicon falls short of the performance of the natural nucleobases, providing an opportunity to probe features of nucleobases important for polymerase recognition. The pyrosequencing method with dMeisoC and disoG suffered from three defects. First, extension in the positions following dMeisoC–disoG pairs was significantly slowed. Second, dMeisoC nucleotide was not readily incorporated opposite disoG template nucleobases. Third, disoG nucleotide was incorporated opposite template dT positions more readily than natural nucleotides are misincorporated opposite natural nucleobases. The more successful dye terminator sequencing method also had some difficulty with the non-natural nucleobases. Extension at several positions following a dMeisoC–disoG pair was clearly inhibited, leading to modest signal attenuation upon encountering additional proximate disoG and dMeisoC positions.
Tautomerism
Some of the peculiarities in the replication of the dMeisoC–disoG pair may be a consequence of tautomerism. A 2O–H tautomer of isoG (Figure 1B), complementary to T, has long been suspected of confounding replication of isoC–isoG (13,14,28–30). One problem that may result from isoG tautomerism is the difficulty of incorporating dMeisoC nucleotide opposite template disoG positions in the pyrosequencing reactions. Two observations suggest that the deficient incorporation of dMeisoC is indeed the result of interaction between paired nucleobases and not a protein–nucleobase interaction at insertion. First, crystal structures of complexes of Family A polymerases, DNA duplex and dNTP lack direct contacts with nucleobases at the insertion site (31–33). Second, 3-deazaadenine (34) and nonpolar nucleobase analogs, unable to form minor groove hydrogen bonding contacts (35,36), are nonetheless efficiently incorporated opposite template dT by diverse polymerases. Tautomerism of the template isoG is implicated because replication should only be affected by tautomerism at the template nucleobase; an unsuitable tautomer as triphosphate would simply be selectively excluded. It is possible that isoG may not readily interconvert between tautomeric forms at the polymerase active site in the lower temperature pyrosequencing method, leading to problematic dMeisoC incorporation when isoG is locked in an alternate tautomeric form. The evident incorporation of dMeisoC opposite template disoG positions by the thermophilic polymerases may be the result of relatively more rapid interconversion of tautomers in the polymerase active site or a shifted tautomeric equilibrium [although the tautomeric equilibrium of disoG has been reported as unperturbed by variations in this temperature range (30)]. Curiously, if the 2O–H tautomer of isoG was present in the templates, it did not lead to significant dT incorporation opposite disoG in pyrosequencing (Supplementary Figure S1C–F). Pyrosequencing and dye terminator sequencing suggest that the incorporation of dT nucleotide opposite template disoG by Family A polymerases, while apparently facile for a misincorporation event (14,30,37,38), is probably much slower than the incorporation of dMeisoC nucleotide opposite disoG.
In our experiments, the misincorporation of disoG nucleotide opposite template dT occurred much more readily than the misincorporation of dT nucleotide opposite template disoG. The 2O–H tautomer of isoG has also been invoked to explain previously observed incorporation of disoG nucleotide opposite dT template positions (13,14,39,40). Misincorporation of disoG nucleotide opposite dT positions adjacent to template dMeisoC was also apparent in our pyrosequencing reactions. However, this misincorporation was evidently suppressed in the presence of competing dA nucleotide in the pyrosequencing and dye terminator reactions, suggesting a preference for incorporation of disoG over dA opposite template dMeisoC positions (39). The comparative ease of this misincorporation, however, may still lead to relatively high mutation rates in the six-nucleobase system.
Extension following dMeisoC–disoG positions
Another irregularity in dMeisoC–disoG replication is the relatively poor extension following dMeisoC–disoG pairs, reminiscent of slow extension following natural nucleobase mismatches (41). This is the first report of hindered extension following correctly matched dMeisoC–disoG pairs. Hindered extension in dye terminator sequencing of templates with proximate non-natural nucleobases and in pyrosequencing suggests that dMeisoC–disoG pairs, despite adopting a Watson–Crick pairing conformation in duplex nucleic acids (7), do not provide specific contacts necessary for efficient incorporation at subsequent positions. The lone pairs of electrons at N3 on purines and O2 on pyrimidines are symmetrically positioned about a pseudo 2-fold axis of natural base pairs (42) and can act as hydrogen bond acceptors to confirm correct nucleobase pairing. Crystal structures of complexes of polymerase, duplex nucleic acid and dNTP have revealed minor groove hydrogen bonding interactions between the protein and hydrogen bond acceptors on post-insertion nucleobase pairs (31–33,43,44). Mismatched pairs, with associated conformational changes in the base pairing, cannot form these interactions and therefore disrupt the polymerase active site (45). The dMeisoC–disoG pair also cannot satisfy all polymerase minor groove hydrogen bonding sites because dMeisoC lacks the O2 acceptor found in the natural pyrimidines. Slow extension beyond dMeisoC–disoG is likely a result of disruption of the polymerase active site by failure of the non-natural pair to form these contacts. The termination of extension observed at lower non-natural nucleotide concentrations (Figure 3) suggests that the disruption of these contacts hinders polymerase function more severely as the number of mismatched positions near the insertion site increases. Comparison of extension beyond mismatched positions in Figure 3 with Figure 4, in which non-natural nucleobases isolated in templates display no visible signal attenuation in the absence of complementary non-natural nucleotide, indicates that the thermophilic polymerases scanned ≤8 bp of the duplex preceding an insertion site. Our results are consistent with several studies that have found these interactions important in post-insertion extension (34,35,46).
However, despite deficient minor groove protein contacts, the steric equivalence of the dMeisoC–disoG pair to natural nucleobase pairs may provide an advantage in avoiding steric clashes with the protein. Extension by the thermophilic polymerases following complementary dMeisoC–disoG pairs proceeded more successfully than extension following mismatches involving the non-natural bases. Mispairing with dMeisoC or disoG at several proximate template positions caused complete termination of subsequent extension. In contrast, incorporation leading to dMeisoC–disoG pairs at these positions always allowed extension, although accompanied by signal attenuation. Hence, the ability of the thermophilic polymerases to extend several nucleotides following a non-natural pair was not as good as following natural complementary pairs, but better than following the mismatched pairs generated in our experiments.
Nucleobase structure and replication with the dMeisoC–disoG pair
Sequencing with dMeisoC and disoG analogs has highlighted the biological relevance of structural features of nucleobases in polymerase-mediated replication. The analogs reinforce the importance of interactions between polymerase and duplex near the site of insertion during extension. Minor groove interactions observed in natural duplexes with Family A polymerases are unable to form between the protein and dMeisoC–disoG pairs in a duplex and this is a likely cause of the slow extension following dMeisoC–disoG pairs. There may also be a steric screening of base pairs in this region of the duplex, because duplex dMeisoC–disoG pairs unable to form usual minor groove interactions nevertheless allow extension to proceed more smoothly than duplexes containing mismatches of the natural nucleobases with dMeisoC or disoG. Successful utilization of the disoG nucleobase demonstrates that mispairing resulting from nucleobase tautomerism can be minimized to yield workable replication, at least in applications, such as sequencing, which do not demand high fidelity. In addition to helping understand polymerase–nucleic acid interaction, these observations should prove useful in the effective design of nucleobase analogs intended for use in polymerase-mediated replication.
Our sequencing experiments illustrate the utility of replication with a six-nucleobase system that includes dMeisoC and disoG. However, the experiments also reveal potential limitations of the pair. The dMeisoC–disoG pair, while isosteric to natural nucleobase pairs, does not have minor groove hydrogen bond acceptors, and subsequent extension following dMeisoC–disoG pairs, with either dMeisoC or disoG in the template strand, does not proceed as readily as with natural nucleobases. This will likely have undesirable consequences not seen in the sequencing experiments. Polymerases with significant 3′-exonuclease activity may display severe pausing at dMeisoC or disoG template positions. Nucleic acids containing dMeisoC and disoG should suffer relatively high mutation rates compared with high fidelity natural replication systems. In vitro selection experiments may be biased toward yielding nucleic acids with lower dMeisoC–disoG content simply because templates with high dMeisoC–disoG content are less readily replicated. Furthermore, although the dMeisoC–disoG pair demonstrates that it is possible to minimize potential mispairing problems stemming from tautomerism to yield a workable six-nucleobase sequencing system, nucleobases with tautomeric ambiguity may still be problematic as components of systems requiring higher fidelity. Interconversion between tautomers at the polymerase insertion site appears slow, and tautomeric ambiguity will probably slow replication. Appropriate engineering of the environment of the polymerase active site (47) or the carbon–nitrogen heterocycles of disoG (48) may be effective for higher fidelity replication of the dMeisoC–disoG pair.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Supplementary Material
Acknowledgments
The authors thank Prof. C. Ronald Geyer (University of Saskatchewan, Saskatoon, Canada) for helpful discussions. Funding to pay the Open Access publication charges for this article was provided by Bayer HealthCare LLC.
Conflict of interest statement. None declared.
REFERENCES
- 1.Benner S.A., Alleman R.K., Ellington A.D., Ge L., Glasfeld A., Leanz G.F., Krauch T., MacPherson L.J., Moroney S.E., Piccirilli A.J., Weinhold E. Natural selection, protein engineering, and the last riboorganism: rational model building in biochemistry. Cold Spring Harb. Symp. Quant. Biol. 1987;52:53–63. doi: 10.1101/sqb.1987.052.01.009. [DOI] [PubMed] [Google Scholar]
- 2.Ogawa A.K., Wu Y., McMinn D.L., Liu J., Schultz P.G., Romesberg F.E. Efforts toward the expansion of the genetic alphabet: information storage and replication with unnatural hydrophobic base pairs. J. Am. Chem. Soc. 2000;122:3274–3287. [Google Scholar]
- 3.Ishikawa M., Hirao I., Yokoyama S. Synthesis of 3-(2-deoxy-β-d-ribofuranosyl)pyridin-2-one and 2-amino-6-(N,N-dimethylamino)-9-(2-deoxy-β-d-ribofuranosyl)purine derivatives for an unnatural base pair. Tetrahedron Lett. 2000;41:3931–3934. [Google Scholar]
- 4.Piccirilli J.A., Krauch T., Moroney S.E., Benner S.A. Enzymatic incorporation of a new base pair into DNA and RNA extends the genetic alphabet. Nature. 1990;343:33–37. doi: 10.1038/343033a0. [DOI] [PubMed] [Google Scholar]
- 5.Horn T., Chang C.-A., Collins M.L. Hybridization properties of the 5-methyl-isocytidine/isoguanosine base pair in synthetic oligonucleotides. Tetrahedron Lett. 1995;36:2033–2036. [Google Scholar]
- 6.Seela F., Wei C. The base-pairing properties of 7-deaza-2′-deoxyisoguanosine and 2′-deoxyisoguanosine in oligonucleotide duplexes with parallel and antiparallel chain orientation. Helv. Chim. Acta. 1999;82:726–745. [Google Scholar]
- 7.Chen X., Kierzek R., Turner D.H. Stability and structure of RNA duplexes containing isoguanosine and isocytidine. J. Am. Chem. Soc. 2001;123:1267–1274. doi: 10.1021/ja002623i. [DOI] [PubMed] [Google Scholar]
- 8.Johnson S.C., Sherrill C.B., Marshall D.M., Moser M.J., Prudent J.R. A third base pair for the polymerase chain reaction: inserting isoC and isoG. Nucleic Acids Res. 2004;32:1937–1941. doi: 10.1093/nar/gkh522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Collins M.L., Irvine B., Tyner D., Fine E., Zayati C., Chang C.-A., Horn T., Ahle D., Detmer J., Shen L.-P., et al. A branched DNA signal amplification assay for quantification of nucleic acid targets below 100 molecules/ml. Nucleic Acids Res. 1997;25:2979–2984. doi: 10.1093/nar/25.15.2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gleaves C.A., Welle J., Campbell M., Elbeik T., Ng V., Taylor P.E., Kuramoto K., Aceituno S., Lewalski E., Joppa B., et al. Multicenter evaluation of the Bayer VERSANT HIV-1 RNA 3.0 assay: analytical and clinical performance. J. Clin. Virol. 2002;25:205–216. doi: 10.1016/s1386-6532(02)00011-2. [DOI] [PubMed] [Google Scholar]
- 11.Elbeik T., Surtihadi J., Destree M., Gorlin J., Holodniy M., Jortani S.A., Kuramoto K., Ng V., Valdes R., Jr, Valsamakis A., Terrault N.A. Multicenter evaluation of the performance characteristics of the Bayer VERSANT HCV RNA 3.0 assay (bDNA) J. Clin. Microbiol. 2004;42:563–569. doi: 10.1128/JCM.42.2.563-569.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sherrill C.B., Marshall D.J., Moser M.J., Larsen C.A., Daudé-Snow L., Prudent J.R. Nucleic acid analysis using an expanded genetic alphabet to quench fluorescence. J. Am. Chem. Soc. 2003;126:4550–4556. doi: 10.1021/ja0315558. [DOI] [PubMed] [Google Scholar]
- 13.Switzer C., Moroney S.E., Benner S.A. Enzymatic incorporation of a new base pair into DNA and RNA. J. Am. Chem. Soc. 1989;111:8322–8323. [Google Scholar]
- 14.Switzer C.Y., Moroney S.E., Benner S.A. Enzymatic recognition of the base pair between isocytidine and isoguanosine. Biochemistry. 1993;32:10489–10496. doi: 10.1021/bi00090a027. [DOI] [PubMed] [Google Scholar]
- 15.Bain J.D., Switzer C., Chamberlin A.R., Benner S.A. Ribosome-mediated incorporation of a non-standard amino acid into a peptide through expansion of the genetic code. Nature. 1992;356:537–539. doi: 10.1038/356537a0. [DOI] [PubMed] [Google Scholar]
- 16.Rice K.P., Chaput J.C., Cox M.M., Switzer C. RecA protein promotes strand exchange with substrates containing isoguanine and 5-methyl isocytosine. Biochemistry. 2000;39:10177–10188. doi: 10.1021/bi0003339. [DOI] [PubMed] [Google Scholar]
- 17.Moser M.J., Prudent J.R. Enzymatic repair of an expanded genetic information system. Nucleic Acids Res. 2003;31:5048–5053. doi: 10.1093/nar/gkg709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sismour A.M., Lutz S., Park J.-H., Lutz M.J., Boyer P.L., Hughes S.H., Benner S.A. PCR amplification of DNA containing non-standard base pairs by variants of reverse transcriptase from Human Immunodeficiency Virus-1. Nucleic Acids Res. 2004;32:728–735. doi: 10.1093/nar/gkh241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu D., Moran S., Kool E.T. Bi-stranded, multisite replication of a base pair between difluorotoluene and adenine: confirmation by ‘inverse’ sequencing. Chem. Biol. 1997;4:919–926. doi: 10.1016/s1074-5521(97)90300-8. [DOI] [PubMed] [Google Scholar]
- 20.Ohtsuki T., Kimoto M., Ishikawa M., Mitsui T., Hirao I., Yokoyama S. Unnatural base pairs for specific transcription. Proc. Natl Acad. Sci. USA. 2001;98:4922–4925. doi: 10.1073/pnas.091532698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jurczyk S.C., Kodra J.T., Rozzell J.D., Benner S.A., Battersby T.R. Synthesis of oligonucleotides containing 2′-deoxyisoguanosine and 2′-deoxy-5-methylisocytidine using phosphoramidite chemistry. Helv. Chim. Acta. 1998;81:793–811. [Google Scholar]
- 22.Wang C., Jiang J., Battersby T.R. Chemical stability of 2′-deoxy-5-methylisocytidine during oligodeoxynucleotide synthesis and deprotection. Nucleosides Nucleotides Nucleic Acids. 2002;21:417–426. doi: 10.1081/NCN-120014814. [DOI] [PubMed] [Google Scholar]
- 23.Ronaghi M., Karamohamed S., Pettersson B., Uhlén M., Nyrén P. Real-time DNA sequencing using detection of pyrophosphate release. Anal. Biochem. 1996;242:84–89. doi: 10.1006/abio.1996.0432. [DOI] [PubMed] [Google Scholar]
- 24.Ronaghi M. Pyrosequencing sheds light on DNA sequencing. Genome Res. 2001;11:3–11. doi: 10.1101/gr.11.1.3. [DOI] [PubMed] [Google Scholar]
- 25.Pyrosequencing Technical Note 103. 2000. Estimation of SNP allele frequencies.
- 26.Spurgeon S.L., Brandis J.W. New DNA sequencing enzymes. In: Kieleczawa J., editor. DNA Sequencing. Sudbury, MA: Jones and Bartlett; 2004. pp. 35–54. [Google Scholar]
- 27.Tuerk C., Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 28.Roberts C., Bandaru R., Switzer C. Theoretical and experimental study of isoguanine and isocytosine: base pairing in an expanded genetic system. J. Am. Chem. Soc. 1997;119:4640–4649. [Google Scholar]
- 29.Robinson H., Gao Y.-G., Bauer C., Roberts C., Switzer C., Wang A.H.-J. 2′-Deoxyisoguanosine adopts more than one tautomer to form base pairs with thymidine observed by high-resolution crystal structure analysis. Biochemistry. 1998;37:10897–10905. doi: 10.1021/bi980818l. [DOI] [PubMed] [Google Scholar]
- 30.Maciejewska A.M., Lichota K.D., Kuśmierek J.T. Neighbouring bases in template influence base-pairing of isoguanine. Biochem. J. 2003;369:611–618. doi: 10.1042/BJ20020922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Doublié S., Tabor S., Long A.M., Richardson C.C., Ellenberger T. Crystal structure of a bacteriophage T7 DNA replication complex at 2.2 Å resolution. Nature. 1998;391:251–258. doi: 10.1038/34593. [DOI] [PubMed] [Google Scholar]
- 32.Li Y., Korolev S., Waksman G. Crystal structures of open and closed forms of binary and ternary complexes of the large fragment of Thermus aquaticus DNA polymerase I: structural basis for nucleotide incorporation. EMBO J. 1998;17:7514–7525. doi: 10.1093/emboj/17.24.7514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li Y., Waksman G. Crystal structures of ddATP-, ddTTP-, ddCTP-, and ddGTP-trapped ternary complex of Klentaq1: insights into nucleotide incorporation and selectivity. Protein Sci. 2001;10:1225–1233. doi: 10.1110/ps.250101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hendrickson C.L., Devine K.G., Benner S.A. Probing minor groove recognition contacts by DNA polymerases and reverse transcriptases using 3-deazaz-2′-deoxyadenosine. Nucleic Acids Res. 2004;32:2241–2250. doi: 10.1093/nar/gkh542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morales J.C., Kool E.T. Minor groove interactions between polymerase and DNA: more essential to replication than hydrogen bonding? J. Am. Chem. Soc. 1999;121:2323–2324. doi: 10.1021/ja983502+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Morales J.C., Kool E.T. Varied molecular interactions at the active sites of several DNA polymerases: nonpolar nucleoside isosteres as probes. J. Am. Chem. Soc. 2000;122:1001–1007. doi: 10.1021/ja993464+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kamiya H., Ueda T., Ohgi T., Matsukage A., Kasai H. Misincorporation of dAMP opposite 2-hydroxyadenine, an oxidative form of adenine. Nucleic Acids Res. 1995;23:761–766. doi: 10.1093/nar/23.5.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bukowska A.M., Kuśmierek J.T. Miscoding properties of isoguanine (2-oxoadenine) studied in an AMV reverse transcriptase in vitro system. Acta Biochim. Pol. 1996;43:247–254. [PubMed] [Google Scholar]
- 39.Tor Y., Dervan P.B. Site-specific enzymatic incorporation of an unnatural base, N6-(6-aminohexyl)isoguanosine, into RNA. J. Am. Chem. Soc. 1993;115:4461–4467. [Google Scholar]
- 40.Kamiya H., Kasai H. Two DNA polymerases of Escherichia coli display distinct misinsertion specificities for 2-hydroxy-dATP during DNA synthesis. Biochemistry. 2000;39:9508–9513. doi: 10.1021/bi000683v. [DOI] [PubMed] [Google Scholar]
- 41.Huang M.-H., Arnheim N., Goodman M.F. Extension of base mispairs by Taq DNA polymerase: implications for single nucleotide discrimination in PCR. Nucleic Acids Res. 1992;20:4567–4573. doi: 10.1093/nar/20.17.4567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Seeman N.C., Rosenberg J.M., Rich A. Sequence-specific recognition of double helical nucleic acids by proteins. Proc. Natl Acad. Sci. USA. 1976;73:804–808. doi: 10.1073/pnas.73.3.804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kiefer J.R., Mao C., Braman J.C., Beese L.S. Visualizing DNA replication in a catalytically active Bacillus DNA polymerase crystal. Nature. 1998;391:304–307. doi: 10.1038/34693. [DOI] [PubMed] [Google Scholar]
- 44.Hsu G.W., Ober M., Carell T., Beese L.S. Error-prone replication of oxidatively damaged DNA by a high-fidelity DNA polymerase. Nature. 2004;431:217–221. doi: 10.1038/nature02908. [DOI] [PubMed] [Google Scholar]
- 45.Johnson S.J., Beese L.S. Structures of mismatch replication errors observed in a DNA polymerase. Cell. 2004;116:803–816. doi: 10.1016/s0092-8674(04)00252-1. [DOI] [PubMed] [Google Scholar]
- 46.Morales J.C., Kool E.T. Functional hydrogen-bonding map of the minor groove binding tracks of six DNA polymerases. Biochemistry. 2000;39:12979–12988. doi: 10.1021/bi001578o. [DOI] [PubMed] [Google Scholar]
- 47.Chaput J.C., Switzer C. Non-enzymatic transcription of an isoG·isoC base pair. J. Am. Chem. Soc. 2000;122:12866–12867. [Google Scholar]
- 48.Martinot T.A., Benner S.A. Artificial genetic systems: exploiting the ‘aromaticity’ formalism to improve the tautomeric ratio for isoguanosine derivatives. J. Org. Chem. 2004;69:3972–3975. doi: 10.1021/jo0497959. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.