Abstract
Recent X-ray crystallographic studies on the human telomere sequence d[AGGG(TTAGGG)3] revealed a unimolecular, parallel quadruplex structure in the presence of potassium ions, while earlier NMR results in the presence of sodium ions indicated a unimolecular, antiparallel quadruplex. In an effort to identify and isolate the parallel form in solution, we have successfully ligated into circular products the single-stranded human telomere and several modified human telomere sequences in potassium-containing solutions. Using these sequences with one or two terminal phosphates, we have made chemically ligated products via creation of an additional loop. Circular products have been identified by polyacrylamide gel electrophoresis, enzymatic digestion with exonuclease VII and electrospray mass spectrometry in negative ion mode. Optimum pH for the ligation reaction of the human telomere sequence ranges from 4.5 to 6.0. Several buffers were also examined, with MES yielding the greatest ligation efficiency. Human telomere sequences with two phosphate groups, one each at the 3′ and 5′ ends, were more efficient at ligation, via pyrophosphate bond formation, than the corresponding sequences with only one phosphate group, at the 5′ end. Circular dichroism spectra showed that the ligation product was derived from an antiparallel, single-stranded guanine quadruplex rather than a parallel single-stranded guanine quadruplex structure.
INTRODUCTION
The structural elaboration and complexity of telomeric DNA sequences, as well as their potential importance for the design of anticancer drugs, have received much attention in the past decade (1–9). Wang and Patel reported that a human telomeric sequence, d(AG3(T2AG3)3), formed a unimolecular antiparallel basket-type guanine quadruplex in Na+-containing solution (10). Parkinson and colleagues (11) described a strikingly different crystal structure of the same sequence in the presence of K+ ions, a parallel quadruplex which could readily be incorporated into a higher-order DNA architecture and whose three loops splayed out like a propeller from the main body of the quadruplex. This remarkable structural feature could also facilitate the necessary folding and unfolding of stacked quadruplexes during chromosome replication since there is no formation of knots in the structure (12). Interest in the type of unimolecular conformations formed by human telomere sequences in solution, as a function of ionic conditions, continues to grow and some new results have been recently proposed.
In a study using platinum cross-linking, only the antiparallel basket-type conformation was detected in both Na+ and K+ solutions (13). Ying and co-workers also published a paper concerning parallel and antiparallel conformations using single-molecule fluorescence energy transfer and proposed that the two distinct conformations could coexist and interconvert under near-physiological conditions (14). More recently, He et al. suggested for the first time that there was a third unimolecular antiparallel chair-type quadruplex for this sequence in solution in the presence of either Na+ or K+ ions with 125I-radioprobing (15). Although some of the methods above cannot distinguish fine structural details among the three different types of unimolecular quadruplex, parallel propeller-type, antiparallel chair-type or basket-type of conformation, these informative papers suggest that additional studies on the flexibility and polymorphism of telomeric oligodeoxynucleotides in solution are warranted.
In this paper, we apply chemical ligation of linear human telomere sequences into circular oligodeoxynucleotides, based on unimolecular guanine quadruplex formation of the unligated species, to explore the various solution conformations exhibited by these sequences. This approach to circularization of short oligonucleotides has been recently applied to a variety of sequences (16–19). Circular oligodeoxynucleotides are promising as molecular tools for hybridization, diagnostics and inhibition of gene expression since they possess higher DNA-binding affinity, greater sequence selectivity, stronger exonuclease resistance and higher stability than the corresponding linear sequences (17,20–24). Furthermore, many other non-telomeric guanine-rich DNA sequences have also been found in a number of important regions, such as the promoter region of the c-myc gene (25,26), the switch regions of immunoglobulin genes (27) and the region upstream of the insulin gene (28). Quadruplex structures are also important as potential therapeutic agents, e.g. the thrombin binding aptamer (29) and an inhibitor of HIV integrase (30). At present, some circular products of oligodeoxynucleotides based on unimolecular quadruplex or hairpin quadruplex structures have been successfully synthesized, all having their ligation position within the quartet stack (18). Here, we report, for the first time, that chemical ligation can occur at loop positions of unimolecular quadruplexes by pyrophosphate bond formation between two phosphate groups located at the 3′ and 5′ ends.
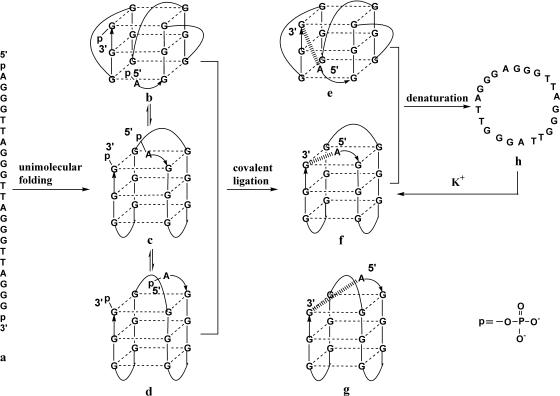
Our strategy for constructing a circular quadruplex through covalent ligation of the human telomere sequence a at a loop position is shown in Figure 1. The parallel, b and two antiparallel, c and d, conformations may coexist at equilibrium, in varying amounts in either K+-, Na+-, Pb2+- or Ba2+-containing solution. Once one conformation among them is ligated into the corresponding circular quadruplex, the dynamic equilibrium may adjust by repopulating the species thus depleted. Continued ligation should result in conversion of a substantial amount of DNA to the ligated form.
Figure 1.
Schematic diagram of possible unimolecular guanine quadruplex conformations of human telomere and our strategy for constructing circular quadruplexes through covalent ligation at the loop position. Hashed lines represent pyrophosphate bond formation; arrows indicate 5′ → 3′ direction.
The likelihood of ligation occurring depends strongly on the geometry of the quadruplex conformation. The linear distance between the 3′ and 5′ ends of the parallel human telomere quadruplex, determined by X-ray crystallography (11), is 6.9 Å (Figure 1, b and e), while that of the antiparallel basket-type quadruplex, determined by NMR (10), is 19.75 Å (Figure 1, d and g). The distance of covalent ligation formed through one phosphodiester bond between adjacent bases in an oligodeoxynucleotide is usually ∼3.5 Å. Therefore, we speculated that the ligation distance of a pyrophosphate bond formed by two phosphate groups located at the 3′ and 5′ ends, ∼7.0 Å, would closely approximate the distance separating the two ends in the parallel quadruplex, and thus would be favorable for ligation of b to form the circular parallel quadruplex e. Pyrophosphate bond formation involves ligation of two free phosphate groups and has been shown to have little effect on DNA duplex conformation (31). Since the two ends in the antiparallel conformation c are located on the same side of the top quartet, we may expect that ligation would occur readily here as well. Obviously, the much longer separation of the two ends in the basket-type quadruplex d would be unlikely to lead to the formation of circular product g.
In consideration of the fact that ends at loop positions are more flexible than those within the quartet stack, extension by one base at the 5′ end or one base each at both 3′ and 5′ ends was designed to increase the collision and ligation efficiency. Even so, it can be imagined that the diagonal loop in conformation d probably prevents ligation by steric hindrance. Additionally, in order to confirm the possibility of ligation of the chair-type quadruplex, we investigate covalent ligation of the extended thrombin binding aptamer (TBA), d(pTG2T2G2TATG2T2G2p), which we assume possesses a chair-type conformation similar to c (32). On the other hand, a basket-type quadruplex, d(pTG4(T4G4)3p), as determined by NMR (33), resembling d, is utilized to examine further the possibility of forming a circular basket-type quadruplex, g. Our results suggest that the unimolecular chair-type quadruplex c is the main conformation giving rise to the formation of circular product under our experimental ligation conditions.
MATERIALS AND METHODS
Oligodeoxyribonucleotides and reagents
All oligodeoxyribonucleotides in the study (Table 1) were purchased from Integrated DNA Technologies, Inc (Coralville, Iowa, USA). Concentrations of oligodeoxyribonucleotides were determined by UV spectrophotometry (Cary 100, Varian, Palo Alto, CA) using calculated extinction coefficients (34), also included in this table. N-cyanoimidazole was synthesized from imidazole and cyanogen bromide (35), purified by sublimation and stored under argon at −20°C prior to use. Covalent ligation products were analyzed via 20% denaturing polyacrylamide gel electrophoresis (PAGE) and visualized by stains-all. CD spectra were obtained on an Aviv model 215 spectrophotometer (Aviv Biomedical, Inc, NJ).
Table 1.
Oligodeoxyribonucleotides used in this study
| Designation | Sequence | ɛ260a |
|---|---|---|
| HT | d(AGGGTTAGGGTTAGGGTTAGGG) | 236 880 |
| HT1 | d(pAGGGTTAGGGTTAGGGTTAGGG) | 236 880 |
| HT2 | d(pAGGGTTAGGGTTAGGGTTAGGGp) | 236 880 |
| HT3 | d(pAGGGTTAGGGTTAGGGTTAGGGT) | 245 160 |
| HT4 | d(pAGGGTTAGGGTTAGGGTTAGGGTp) | 245 160 |
| HT5 | d(pTAGGGTTAGGGTTAGGGTTAGGGT) | 253 380 |
| HT6 | d(pTAGGGTTAGGGTTAGGGTTAGGGTp) | 253 830 |
| HT7 | d(pGTTAGGGTTAGGGTTAGGGTTAGG) | 254 660 |
| HT8 | d(pGTTAGGGTTAGGGTTAGGGTTAGGp) | 254 660 |
| TBA1 | d(pTGGTTGGTGTGGTTGG) | 155 160 |
| TBA2 | d(pTGGTTGGTGTGGTTGGp) | 155 160 |
| Oxy1 | d(pTGGGGTTTTGGGGTTTTGGGGTTTTGGGGp) | 281 420 |
| T20 | d(TTTTTTTTTTTTTTTTTTTT) |
aExtinction coefficients, l/(mol cm), at 260 nm.
Covalent ligation reaction
The selected sequence (40 pmol) was dissolved in 10 μl of 200 mM MES (2-(N-morpholino) ethanesulfonic acid monohydrate) buffer containing 20 mM KCl. Incubation at 4°C for 2 h allowed for the formation of unimolecular guanine quadruplexes (Figure 1, b–d). The 5′- and 3′-terminal phosphates were activated by adding 1 μl of 1 M N-cyanoimidazole and 1 μl of 1 M MnCl2 for a total volume of 20 μl (36–38). The reaction mixture was kept at 4°C for 12 h to form a circular product generated by pyrophosphate bond formation between two phosphates. In the case of sequences with one phosphate at the 5′ end, ligation was carried out via formation of a phosphodiester bond between the 5′-phosphate and adjacent 3′-hydroxyl group. Reactions were terminated by addition of loading buffer and ligation products were subsequently analyzed by PAGE under denaturing conditions.
Structural identification of circular products
Exonuclease VII hydrolysis
Reaction mixtures containing 10 μl exonuclease VII buffer (200 mM Tris–HCl, 200 mM potassium phosphate, 33.2 mM EDTA and 40 mM 2-mercaptoethanol, pH 7.9), 4 μl of 10 U/μl exonuclease VII (Amersham Phamercia Biotech, USA) (39) and 6 μl of purified ligation reaction solution in a total volume of 20 μl was incubated at 37°C for 2 h. Reaction products were then analyzed by PAGE under denaturing conditions.
Mass spectrometry
Sequences were analyzed by nano-electrospray ionization mass spectrometry (ESI) on an LTQ (Thermo Finnigan Corporation, USA) spectrometer operating in negative ion mode at the School of Pharmacy Mass Spectrometry Facility, UCSF.
RESULTS AND DISCUSSION
Formation of covalent ligation circular products
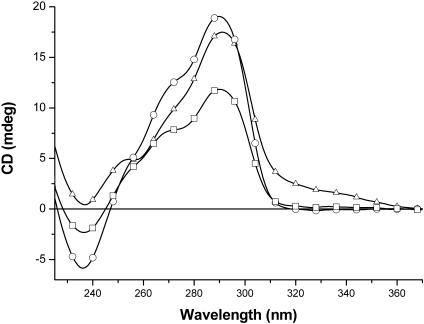
As shown in Figure 2, circular dichroism reveals that solutions of the human telomere sequence (HT) or its modification with one phosphate (HT1) or two phosphates (HT2) consist of a mixture of parallel and antiparallel quadruplexes because each spectrum exhibits a major peak near 290 nm, consistent with antiparallel quadruplexes, along with a minor peak near 265 nm, consistent with parallel quadruplexes. Relying solely on CD to assign strand alignment is not foolproof (40), but in most cases has proved to be a useful guide. Recent studies of vertebrate telomere repeats indicate that parallel quadruplex structures have at least the 265 nm peak, and possibly a secondary peak near 290 nm (41). Several NMR reports have appeared demonstrating that 2-fold repeats of the human telomere, or related, sequences can take on both parallel and antiparallel conformations (42). Thus, it appears reasonable to infer the presence of a parallel structure (possibly along with antiparallel structures) when a significant CD peak near 265 nm is present and an antiparallel structure when only the 290 nm peak appears. Furthermore, CD spectra cannot distinguish between unimolecular and four-stranded parallel quadruplexes, nor between basket and chair antiparallel quadruplexes. However, native gels show no evidence for four-stranded quadruplexes (data not shown), and thus we infer that the peak near 265 nm most likely arises from a unimolecular parallel conformation. Sequence HT2 was accordingly chosen for most of our studies, as it resulted in higher yields of circular products than HT1 (see below).
Figure 2.
CD spectroscopy of HT (triangles, 5 μM), HT1 (circles, 6 μM) and HT2 (squares, 4 μM) in 10 mM of Li-cacodylate buffer and 10 mM of KCl.
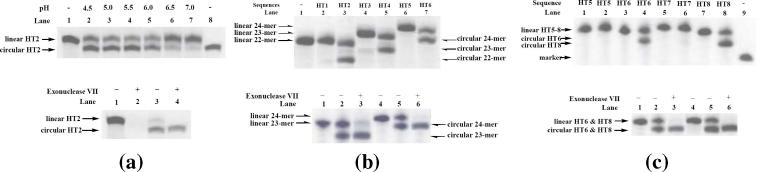
HT2 was first incubated in a pH 5.5 buffer to allow the desired quadruplex structure to form. Both 3′- and 5′-terminal phosphates within the unimolecular complex were next activated by addition of N-cyanoimidazole to facilitate the formation of a pyrophosphate bond between 3′- and 5′-phosphates (Figure 1, b → e and c → f). Analysis of the reaction products was subsequently carried out through denaturing polyacrylamide gel electrophoresis. As shown in Figure 3a, top, lane 4, a new product was generated from this ligation reaction with greater mobility than that of its corresponding linear precursor HT2 (lane 1). Based on this information, the newly formed product was identified tentatively as the circular oligodeoxyribonucleotide generated from its corresponding linear precursor, HT2. The efficiency of these ligation reactions was found to be strongly dependent on pH. Circularization increased with the increase of pH from 4.5 to 5.5 (Figure 3a, top, lanes 2–4), with an optimum at pH 5.5 (lane 4). The amount of circular product decreased with higher pH, and at pH 7.0 (lane 7) only a small amount of product was observed. A linear 20mer, d(T20) (lane 8), was used as a molecular weight marker.
Figure 3.
(a) (Top) Effect of pH on circularization of linear oligodeoxyribonucleotide HT2. Covalent ligation reactions were performed as described in Materials and Methods. The circularization reaction proceeded at pH 4.5 (lane 2), 5.0 (lane 3), 5.5 (lane 4), 6.0 (lane 5), 6.5 (lane 6) and 7.0 (lane 7), respectively. Lane 1: same reaction as the one loaded in lane 4, except for the absence of N-cyanoimidazole. Lane 10: the 20mer d(T20) as a molecular weight marker. (Bottom) Hydrolysis of HT2 ligation product by exonuclease VII. Lane 1: linear HT2 alone. Lane 2: linear HT2 after treatment with 20 U of exonuclease VII at 37°C for 2 h. Lane 3: ligation reaction mixture alone. Lane 4: ligation reaction mixture after treatment with 20 U of exonuclease VII at 37°C for 2 h. (b) (Top) Loop size and phosphate group number dependency of the covalent ligation of human telomere sequences. Same reaction as the one loaded in lane 4 in Figure 3a, except for replacing sequence HT2 with HT1, d(pAG3T2AG3T2AG3T2AG3), HT3, d(pAG3T2AG3T2AG3T2AG3T), HT4, d(pAG3T2AG3T2AG3T2AG3Tp), HT5, d(pTAG3T2AG3T2AG3T2AG3T) or HT6, d(pTAG3T2AG3T2AG3T2AG3Tp), respectively. Lane 1: HT1 alone. Lanes 2–7 contain reaction mixtures of HT1 (lane 2), HT2 (lane 3), HT3 (lane 4), HT4 (lane 5), HT5 (lane 6) and HT6 (lane 7), respectively. (Bottom) Hydrolysis of ligation product of HT4 and HT6 by exonuclease VII. Lane 1: HT4 alone. Lane 2: ligation reaction mixture of HT4. Lane 3: ligation reaction mixture of HT4 after treatment with 20 U of exonuclease VII at 37°C for 2 h. Lane 4: HT6 alone. Lane 5: ligation reaction mixture of HT6. Lane 6: ligation reaction mixture of HT6 after treatment with 20 U of exonuclease VII at 37°C for 2 h. (c) (Top) End position dependency of the covalent ligation of human telomeres. Same reaction as the one loaded in lane 4 in Figure 3a, top, except for replacing sequence HT2 with HT5, d(pTAG3T2AG3T2AG3T2AG3T), HT6, d(pTAG3T2AG3T2AG3T2AG3Tp), HT7, d(pGT2AG3T2AG3T2AG3T2AG2) and HT8, d(pGT2AG3T2AG3T2AG3T2AG2p), respectively. Lanes 1, 3, 5 and 7: same reaction as the one loaded in lane 4, except for the absence of N-cyanoimidazole. Lanes 2, 4, 6 and 8 contain reaction mixtures of HT5, HT6, HT7 and HT8, respectively. Lane 9: the 20mer of 5′-d(T20)-3′ as a molecular weight marker. (Bottom) Hydrolysis of ligation product of HT6 and HT8 by exonuclease VII. Lane 1: HT6 alone. Lane 2: ligation reaction mixture of HT6. Lane 3: ligation reaction mixture of HT6 after treatment with 20 U of exonuclease VII at 37°C for 2 h. Lane 4: HT8 alone. Lane 5: ligation reaction mixture of HT8. Lane 6: ligation reaction mixture of HT8 after treatment with 20 U of exonuclease VII at 37°C for 2 h.
In order to verify the circular nature of HT2 with a pyrophosphate bond, the ligation reaction mixture, containing circular product and linear precursor, was digested with exonuclease VII, an enzyme that hydrolyzes nucleotides from both 3′ and 5′ ends of single-stranded deoxyribonucleic acids. Circular oligodeoxyribonucleotides are known to resist degradation by this enzyme (21,39) due to the absence of open termini within their structures. As shown in Figure 3a, bottom, only the linear precursor was susceptible to exonuclease digestion (lane 4). As a control, the hydrolysis reaction of unreacted HT2 with exonuclease VII was also carried out, which consequently led to lower molecular-weight products in quantitative yield (lane 2). The complete resistance of our ligation product to hydrolysis by this exonuclease demonstrates the absence of open termini within its unimolecular structure.
With the aim of further confirming the nature of the unimolecular ligation product, we purified it by PAGE and then analyzed it by electrospray mass spectrometry in negative ion mode. The expected mass for the ligated product is 7108 Da, and we observed a peak at a mass of 709.55 Da, which corresponds to an ion with 10 negative charges and a molecular mass of 709.55 × 10 + 10 = 7105.5 Da, in good agreement with the expected mass. Similarly, for the linear precursor, we observed a peak at 711.45 Da, corresponding to an ion with 10 negative ions and a molecular mass of 711.45 × 10 + 10 = 7124.5 Da. This is consistent with the expected molecular mass of 7126.5 Da. It should be noted that we observed a peak at 695.45 Da for HT (same sequence as HT2 but without any phosphate group at its ends) processed by the same circularization and purification procedures as HT2. This corresponds to an ion with 10 negative charges and a mass of 695.45 × 10 + 10 = 6964.5 Da, in close agreement with its expected molecular weight of 6966.5. This result demonstrates that the presence of terminal phosphate groups is a prerequisite for circularization of the sequence. Taken together, these results provide additional evidence for the formation of circular, ligated product.
Influence of loop size and number of phosphate groups on covalent ligation
The results presented above demonstrate that circular product was indeed formed by our ligation procedure. However, what is not clear is which kind or kinds of quadruplex conformation generated the circular product. From our previous structural considerations, conformations b and c in Figure 1 should allow for circular product formation, while conformation d is unlikely to form the corresponding circular product g because of the large distance (19.75 Å) separating the ends. Assuming that conformation d exists in solution, we examined the effect of one or two extra terminal bases, decreasing the end-to-end distance, on covalent ligation. Figure 3b, top, presents the results from these experiments.
The increase of loop size did not improve the ligation reaction, even for the sequences with two phosphates such as HT4 and HT6. There are probably two geometric factors controlling linkage efficiency: the end-to-end distance and the end flexibility, the latter becoming important once the former is satisfied. We can imagine that the longer the loop size, the greater the flexibility, resulting in a lower frequency of effective collisions. For conformations b and c in Figure 1, elongation of the loop would increase the end flexibility, while for conformation d, although a longer loop may better satisfy the end-to-end distance requirement, the existing diagonal loop would still sterically hinder one end from coming into close proximity of the other end, as required for chemical ligation.
One of the interesting phenomena observed with sequences containing only one phosphate, such as HT1, HT3 and HT5, is a far smaller amount of ligation product than with two phosphates, such as HT2, HT4 and HT6. This may be due to the fact that sequences containing two phosphates possess higher reactant collision frequencies leading to circular products than those sequences with one phosphate and one hydroxyl group, because there are several available oxygen atoms in each phosphate, while there is only one oxygen atom available in the terminal hydroxyl group. In addition, nucleophilic attack by a hydroxyl group is weaker than by an oxygen anion.
Finally, ligation products from HT4 and HT6 were confirmed by exonuclease VII digestion in the same way as HT2. As shown in Figure 3b, bottom, the linear precursors in the ligation reaction mixture of HT4 or HT6 were digested, and their corresponding gel bands have disappeared (lanes 3 and 6) while circular products remained unaffected. These results reveal again that covalent ligation at loop sites is highly efficient under our reaction conditions.
Influence of end positions on covalent ligation
In order to compare the feasibility of the ligation reaction at different positions within the guanine quadruplex, HT5 and HT6 with loop ends, as well as HT7 and HT8 with columnar ends (i.e. ends within the quartet core), were subject to the circularization reaction. As shown in Figure 3c, top, sequences HT5 (lane 2) and HT7 (lane 6) with only one phosphate at the 5′ end led to undetectable amounts of circular products, while sequences HT6 and HT8 with two phosphate groups, one each at the 3′ and 5′ ends, possess better ligation yields whether the end position was within a loop (lane 4) or quartet stack (lane 8). Moreover, the yield of circular product for HT8 with columnar ends appears higher than that of HT6 with loop ends. This result indicates again that human telomere sequences with two phosphates lead to higher yields for covalent ligation than those with one phosphate. As in the case of DNA duplexes (31), pyrophosphate formation does not appear to significantly distort the quadruplex conformation. The circular products from HT6 and HT8 were also confirmed by exonuclease VII digestion in the same way as HT2. The results are shown in Figure 3c, bottom.
Influence of metal ions and buffers on covalent ligation
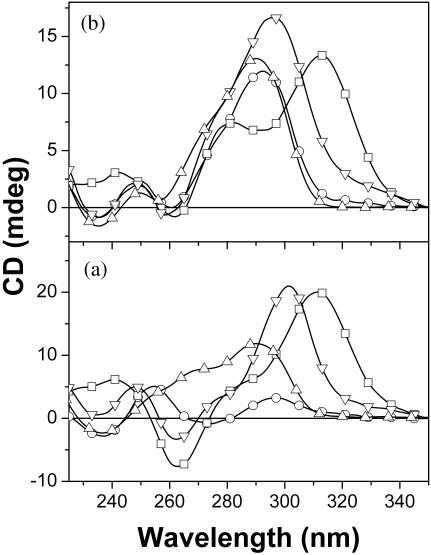
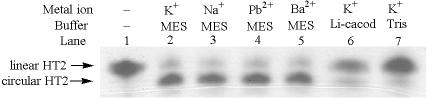
Nucleic acid quadruplexes based on the guanine quartet are stabilized not only by monovalent cations such as K+, Na+ and (43), but also by certain divalent cations such as Pb2+ (44), Sr2+ (45,46) and Ba2+ (43). With the purpose of exploring the effect of different metals on the unimolecular quadruplex conformations of the human telomere sequence, we determined CD spectra of HT2 in K+, Na+, Pb2+ or Ba2+ ions (Figure 4a). The peaks at 295 nm in Na+, 302 nm in Ba2+ and 312 nm in Pb2+ clearly indicate that HT2 forms antiparallel quadruplex structures although, again, we cannot distinguish chair-type from basket-type conformations. In contrast, the presence of K+ leads to a significant contribution near 265 nm, arising from a parallel quadruplex conformation, as seen in Figure 2. The effect of different cations was investigated further by carrying out covalent ligation reactions, as described above. The results of these studies are presented in Figure 5.
Figure 4.
CD spectroscopy of HT2 (4 μM) in KCl (triangles, 20 mM), NaCl (circles, 20 mM), Pb(NO3)2 (squares, 4 μM), BaCl2 (inverted triangles, 4 μM) and 10 mM of Li-cacodylate buffer (pH 5.5) (a) or 200 mM of MES buffer (pH 5.5) (b).
Figure 5.
Metal ion and buffer dependency of the covalent ligation of human telomere HT2. Lane 1: Same as lane 1 in Figure 3a, top. Lane 2: Same as lane 4 in Figure 3a, top. Lanes 3–5: reactions were carried out in the same way as the one loaded in lane 4 in Figure 3a, top, except for replacing KCl with 20 mM NaCl (lane 3), 20 μM Pb2+ (lane 4), 20 μM Ba2+ (lane 5). Lanes 6–7: Same as lane 4 in Figure 3a, top, except for replacing MES with 10 mM Li-cacodylate (lane 6) or 10 mM Tris–HCl (lane 7).
As illustrated in Figure 5, conversion of HT2 to covalently ligated product is seen for all cations examined (lanes 1–5) when MES is used as buffer. The result for HT2 in Li-cacodylate buffer indicates that only small amounts of circular product were formed whether in K+ (lane 6) or in Na+, Pb2+ or Ba2+ solution (data not shown). The same experiments were also carried in tris–HCl buffer and K+-, Na+-, Pb2+- or Ba2+-containing solutions. The circular products obtained in all metals in tris–HCl buffer were almost the same as illustrated in lane 7, Figure 5. It is not clear whether the different amounts of circular products obtained in MES compared to the other buffers reflect differences in DNA conformation in these buffers or differences in ligation efficiency or a combination of both. Nonetheless, Figure 4b reveals some degree of buffer impact on the quadruplex conformation prior to covalent ligation. Specifically, in MES buffer, there is relatively little evidence of a peak at 265 nm in the CD spectra, indicating a relatively small amount of parallel quadruplex is formed. The fairly high yields of circular products for HT2 in MES buffer suggest that the chair-type antiparallel conformation is present in significant amounts under these conditions.
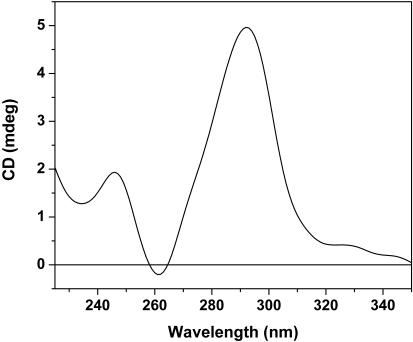
Quadruplex conformation of the circular product
The HT2 circular product was purified by column chromatography and its CD spectrum was determined under native conditions. As shown in Figure 6, the spectrum is characteristic of antiparallel quadruplexes and thus reflects the major component observed in K+-MES buffer prior to covalent ligation (Figure 4b). Based on geometric factors discussed above, the circular product most likely derives from the antiparallel chair conformation (Figure 1, c and f). There are several possible explanations for the lack of covalently ligated, parallel quadruplex products. First, based on the CD spectrum in Figure 4b, there is only a small amount, at best, of parallel structure present prior to ligation, which may result in undetectable amounts of covalent product. A second explanation may be that the structural details of the unimolecular, parallel quadruplex in solution differ from those observed in the crystal structure (11), such that the terminal phosphate groups are too far apart for efficient ligation.
Figure 6.
CD spectroscopy of purified circular product of HT2 (1.2 μM) in 200 mM MES buffer (pH 5.5) containing 20 mM KCl.
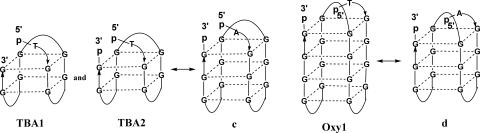
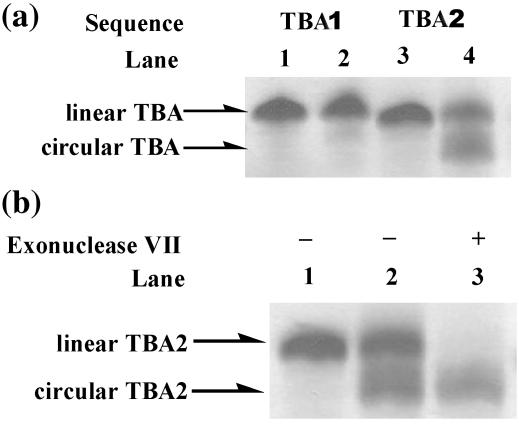
In order to gain further insight into the possible solution conformations susceptible to covalent ligation, we examined the cyclization behavior of several other quadruplex-forming sequences. The conformation of the TBA has been shown to be the antiparallel chair-type quadruplex in the presence of K+ ions by NMR (29,47). We selected TBA to confirm the feasibility of covalent ligation for the chair-type quadruplex conformation of the human telomere sequence (Figure 7, c), to further support our interpretation that the circular product of HT2 results from this particular antiparallel conformation. Thus, we subjected several TBA-like sequences to covalent ligation, both containing an extra T at the 5′ end, TBA1, with one terminal phosphate and TBA2, with two terminal phosphates (Figure 7). As shown in Figure 8a, both TBA1 (lane 4) and TBA2 (lane 2) can be ligated into circular products, with a higher yield for TBA2 than TBA1. The ligation product from TBA2 was also confirmed by enzymatic digestion with exonuclease VII. As shown in Figure 8b, the linear precursor in the ligation reaction mixture of TBA2 was digested (lane 2) while the circular product remained unaffected.
Figure 7.
Schematic illustration of certain structural features of guanine quadruplexes comparing thrombin binding aptamer, oxytricha telomeric and human telomeric sequences.
Figure 8.
(a) Circularization of thrombin binding aptamer (TBA). Lane 1: TBA1 alone. Lane 2: TBA1 ligated in the same reaction as the one loaded in lane 4 in Figure 3a, top. Lane 3: TBA2 alone. Lane 4: TBA2 ligated in the same reaction as the one loaded in lane 4 in Figure 3a, top, except for ligation reaction at 25°C. (b) Hydrolysis of ligation product of TBA2 by exonuclease. Lane 1: TBA2 alone. Lane 2: mixture of ligation reaction of TBA2. Lane 3: mixture of ligation reaction of TBA2 after treatment with 20 U of exonuclease VII at 37°C for 2 h.
These results demonstrate that covalent ligation can occur from the antiparallel, chair conformation. Furthermore, they indicate again that sequences with two phosphate groups, one each at the 3′ and 5′ ends, form a pyrophosphate bond more readily than the corresponding sequences with only one phosphate group at the 5′ end formed a phosphodiester bond.
On the other hand, the sequence d(G4(T4G4)3), based on the Oxytricha telomere repeat, has been shown to possess a basket-type guanine quadruplex conformation (33). Its modified sequence d(pTG4(T4G4)3p), Oxy1 (Figure 7) was utilized to examine the feasibility of covalent ligation for a sequence with the structural feature similar to Figure 1, d. Our experimental result revealed that no ligation product was obtained (data not shown). This observation further supports our conclusion that the circular product obtained from covalent ligation of human telomere sequences originated from an antiparallel chair-type guanine quadruplex conformation.
If covalent ligation cannot proceed directly from a given quadruplex conformation, such as the basket-type antiparallel conformation, then prior to ligation, molecules in such a conformation would have to undergo a conformational change to one that allows ligation to proceed. For example, using the structures in Figure 1, the only way molecules existing in a basket-type conformation (d) could lead to covalent product (f) would be via a mechanism such as d ⇌ c → f. The obligate change in conformation would then have significant consequences for the kinetics of product formation.
CONCLUSIONS
Our studies indicate that single-stranded circular guanine quadruplexes based on the human telomere repeat can be synthesized by covalent ligation reaction at loop positions, and that the resulting product possesses an antiparallel folding topology. Human telomere sequences with two phosphate groups, one each at the 3′ and 5′ ends, cyclize more readily than the corresponding sequences with only one phosphate group at the 5′ end. The covalent ligation reactions can be successfully carried out in MES buffer containing any one of the ions K+, Na+, Pb2+ or Ba2+. MES buffer is more efficient for the chemical ligation than Li-cacodylate or tris–HCl buffers. In addition, the pH and the number of loop oligonucleotides at the sequence termini can affect ligation efficiency. These circularization reactions are highly efficient under our standard reaction conditions not only for human telomere but also for the TBA.
Acknowledgments
We would like to thank Drs Martin Shetlar and Stephen Kahl for providing the equipment in the synthesis of the N-cyanoimidazole and Dr Dan Minor for providing access to his circular dichroism instrument. Mass spectrometry results were kindly provided by Dr David Maltby at the UCSF Mass Spectrometry Facility (A.L. Burlingame, Director) supported by the Biomedical Research Technology Program of the National Center for Research Resources, NIH NCRR RR01614. This work has been supported by grant GM067607 from the National Institute of General Medical Sciences, NIH, DHHS. Funding to pay the Open Access publication charges for this article was provided by NIH grant GM067607.
Conflict of interest statement. None declared.
REFERENCES
- 1.Blackburn E.H. Switching and signaling at the telomere. Cell. 2001;106:661–673. doi: 10.1016/s0092-8674(01)00492-5. [DOI] [PubMed] [Google Scholar]
- 2.Cech T.R. Beginning to understand the end of the chromosome. Cell. 2004;116:273–279. doi: 10.1016/s0092-8674(04)00038-8. [DOI] [PubMed] [Google Scholar]
- 3.de Lange T. T-loops and the origin of telomeres. Nat. Rev. Mol. Cell Biol. 2004;5:323–329. doi: 10.1038/nrm1359. [DOI] [PubMed] [Google Scholar]
- 4.Gellert M., Lipsett M.N., Davies D.R. Proceedings of the National Academy of Sciences of the United States of America. 1962;48:2013–2018. doi: 10.1073/pnas.48.12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sen D., Gilbert W. Novel DNA superstructures formed by telomere-like oligomers. Biochemistry. 1992;31:65–70. doi: 10.1021/bi00116a011. [DOI] [PubMed] [Google Scholar]
- 6.Horvath M.P., Schweiker V.L., Bevilacqua J.M., Ruggles J.A., Schultz S.C. Crystal structure of the Oxytricha nova telomere end binding protein complexed with single strand DNA. Cell. 1998;95:963–974. doi: 10.1016/s0092-8674(00)81720-1. [DOI] [PubMed] [Google Scholar]
- 7.Griffith J.D., Comeau L., Rosenfield S., Stansel R.M., Bianchi A., Moss H., de Lange T. Mammalian telomeres end in a large duplex loop. Cell. 1999;97:503–514. doi: 10.1016/s0092-8674(00)80760-6. [DOI] [PubMed] [Google Scholar]
- 8.Schaffitzel C., Berger I., Postberg J., Hanes J., Lipps H.J., Pluckthun A. In vitro generated antibodies specific for telomeric guanine-quadruplex DNA react with Stylonychia lemnae macronuclei. Proc. Natl Acad. Sci. USA. 2001;98:8572–8577. doi: 10.1073/pnas.141229498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hurley L.H. DNA and its associated processes as targets for cancer therapy. Nat. Rev. Cancer. 2002;2:188–200. doi: 10.1038/nrc749. [DOI] [PubMed] [Google Scholar]
- 10.Wang Y., Patel D.J. Solution structure of the human telomeric repeat d[AG3(T2AG3)3] G-tetraplex. Structure. 1993;1:263–282. doi: 10.1016/0969-2126(93)90015-9. [DOI] [PubMed] [Google Scholar]
- 11.Parkinson G.N., Lee M.P., Neidle S. Crystal structure of parallel quadruplexes from human telomeric DNA. Nature. 2002;417:876–880. doi: 10.1038/nature755. [DOI] [PubMed] [Google Scholar]
- 12.Patel D.J. Structural biology: a molecular propeller. Nature. 2002;417:807–808. doi: 10.1038/417807a. [DOI] [PubMed] [Google Scholar]
- 13.Redon S., Bombard S., Elizondo-Riojas M.A., Chottard J.C. Platinum cross-linking of adenines and guanines on the quadruplex structures of the AG3(T2AG3)3 and (T2AG3)4 human telomere sequences in Na+ and K+ solutions. Nucleic Acids Res. 2003;31:1605–1613. doi: 10.1093/nar/gkg259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ying L., Green J.J., Li H., Klenerman D., Balasubramanian S. Studies on the structure and dynamics of the human telomeric G quadruplex by single-molecule fluorescence resonance energy transfer. Proc. Natl Acad. Sci. USA. 2003;100:14629–14634. doi: 10.1073/pnas.2433350100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.He Y., Neumann R.D., Panyutin I.G. Intramolecular quadruplex conformation of human telomeric DNA assessed with 125I-radioprobing. Nucleic Acids Res. 2004;32:5359–5367. doi: 10.1093/nar/gkh875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen J., Liu D., Lee A.H., Qi J., Chan A.S., Li T. G-quadruplex as a new class of structural entities for directing the formation of circular oligodeoxyribonucleotides. Chem. Commun. (Camb.) 2002:2686–2687. doi: 10.1039/b208075n. [DOI] [PubMed] [Google Scholar]
- 17.Li T., Liu D., Chen J., Lee A.H., Qi J., Chan A.S. Construction of circular oligodeoxyribonucleotides on the new structural basis of i-motif. J. Am. Chem. Soc. 2001;123:12901–12902. doi: 10.1021/ja011401x. [DOI] [PubMed] [Google Scholar]
- 18.Zhou T., Chen G., Wang Y., Zhang Q., Yang M., Li T. Synthesis of unimolecularly circular G-quadruplexes as prospective molecular probes. Nucleic Acids Res. 2004;32:e173. doi: 10.1093/nar/gnh162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu D., Chen J., Lee A.H., Chow L.M., Chan A.S., Li T. Small circular oligodeoxynucleotides achieved from self-assembling entities. Angew Chem. Int. Ed. Engl. 2003;42:797–799. doi: 10.1002/anie.200390211. [DOI] [PubMed] [Google Scholar]
- 20.Kool E.T. Recognition of DNA, RNA, and proteins by circular oligonucleotides. Acc. Chem. Res. 1998;31:502–510. doi: 10.1021/ar9602462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kool E.T. Circular oligonucleotides: new concepts in oligonucleotide design. Annu. Rev. Biophys. Biomol. Struct. 1996;25:1–28. doi: 10.1146/annurev.bb.25.060196.000245. [DOI] [PubMed] [Google Scholar]
- 22.Escude C., Garestier T., Helene C. Padlock oligonucleotides for duplex DNA based on sequence-specific triple helix formation. Proc. Natl Acad. Sci. USA. 1999;96:10603–10607. doi: 10.1073/pnas.96.19.10603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fire A., Xu S.Q. Rolling replication of short DNA circles. Proc. Natl Acad. Sci. USA. 1995;92:4641–4645. doi: 10.1073/pnas.92.10.4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuhn H.D., Vadim V., Frank-Kamenetskii M.D. Topological links between duplex DNA and a circular DNA single strand. Angew. Chem. Int. Ed. Engl. 1999;38:1446–1449. doi: 10.1002/(SICI)1521-3773(19990517)38:10<1446::AID-ANIE1446>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 25.Simonsson T., Pecinka P., Kubista M. DNA tetraplex formation in the control region of c-myc. Nucleic Acids Res. 1998;26:1167–1172. doi: 10.1093/nar/26.5.1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rangan A., Fedoroff O.Y., Hurley L.H. Induction of duplex to G-quadruplex transition in the c-myc promoter region by a small molecule. J. Biol. Chem. 2001;276:4640–4646. doi: 10.1074/jbc.M005962200. [DOI] [PubMed] [Google Scholar]
- 27.Sen D., Gilbert W. Formation of parallel four-stranded complexes by guanine-rich motifs in DNA and its implications for meiosis. Nature. 1988;334:364–366. doi: 10.1038/334364a0. [DOI] [PubMed] [Google Scholar]
- 28.Hammond-Kosack M.C., Kilpatrick M.W., Docherty K. Analysis of DNA structure in the human insulin gene-linked polymorphic region in vivo. J. Mol. Endocrinol. 1992;9:221–225. doi: 10.1677/jme.0.0090221. [DOI] [PubMed] [Google Scholar]
- 29.Macaya R.F., Schultze P., Smith F.W., Roe J.A., Feigon J. Thrombin-binding DNA aptamer forms a unimolecular quadruplex structure in solution. Proc. Natl Acad. Sci. USA. 1993;90:3745–3749. doi: 10.1073/pnas.90.8.3745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.de Soultrait V.R., Lozach P.Y., Altmeyer R., Tarrago-Litvak L., Litvak S., Andreola M.L. DNA aptamers derived from HIV-1 RNase H inhibitors are strong anti-integrase agents. J. Mol. Biol. 2002;324:195–203. doi: 10.1016/s0022-2836(02)01064-1. [DOI] [PubMed] [Google Scholar]
- 31.Purmal A.A., Shabarova Z.A., Gumport R.I. A new affinity reagent for the site-specific, covalent attachment of DNA to active-site nucleophiles: application to the EcoRI and RsrI restriction and modification enzymes. Nucleic Acids Res. 1992;20:3713–3719. doi: 10.1093/nar/20.14.3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schultze P., Macaya R.F., Feigon J. Three-dimensional solution structure of the thrombin-binding DNA aptamer d(GGTTGGTGTGGTTGG) J. Mol. Biol. 1994;235:1532–1547. doi: 10.1006/jmbi.1994.1105. [DOI] [PubMed] [Google Scholar]
- 33.Wang Y., Patel D.J. Solution structure of the Oxytricha telomeric repeat d[G4(T4G4)3] G-tetraplex. J. Mol. Biol. 1995;251:76–94. doi: 10.1006/jmbi.1995.0417. [DOI] [PubMed] [Google Scholar]
- 34.Gray D.M., Hung S.H., Johnson K.H. Absorption and circular dichroism spectroscopy of nucleic acid duplexes and triplexes. Methods Enzymol. 1995;246:19–34. doi: 10.1016/0076-6879(95)46005-5. [DOI] [PubMed] [Google Scholar]
- 35.Luebke K.J., Dervan P.B. Nonenymatic ligation of double-helical DNA by alternate-strand triple helix formation. Nucleic Acids Res. 1992;20:3005–3009. doi: 10.1093/nar/20.12.3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Luebke K.J., Dervan P.B. Nonenzymatic sequence-specific ligation of double-helical DNA. J. Am. Chem. Soc. 1991;113:7447–7448. [Google Scholar]
- 37.Kanaya E., Yanagawa H. Template-directed polymerization of oligoadenylates using cyanogen bromide. Biochemistry. 1986;25:7423–7430. doi: 10.1021/bi00371a026. [DOI] [PubMed] [Google Scholar]
- 38.Ferris J.P., Huang C.H., Hagan W.J., Jr N-cyanoimidazole and diimidazole imine: water-soluble condensing agents for the formation of the phosphodiester bond. Nucleosides Nucleotides. 1989;8:407–414. doi: 10.1080/07328318908054184. [DOI] [PubMed] [Google Scholar]
- 39.Dolinnaya N.G., Blumenfeld M., Merenkova I.N., Oretskaya T.S., Krynetskaya N.F., Ivanovskaya M.G., Vasseur M., Shabarova Z.A. Oligonucleotide circularization by template-directed chemical ligation. Nucleic Acids Res. 1993;21:5403–5407. doi: 10.1093/nar/21.23.5403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hazel P., Huppert J., Balasubramanian S., Neidle S. Loop-length-dependent folding of G-quadruplexes. J. Am. Chem. Soc. 2004;126:16405–16415. doi: 10.1021/ja045154j. [DOI] [PubMed] [Google Scholar]
- 41.Rujan I.N., Meleney J.C., Bolton P.H. Vertebrate telomere repeat DNAs favor external loop propeller quadruplex structures in the presence of high concentrations of potassium. Nucleic Acids Res. 2005;33:2022–2031. doi: 10.1093/nar/gki345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Phan A.T., Patel D.J. Two-repeat human telomeric d(TAGGGTTAGGGT) sequence forms interconverting parallel and antiparallel G-quadruplexes in solution: distinct topologies, thermodynamic properties, and folding/unfolding kinetics. J. Am. Chem. Soc. 2003;125:15021–15027. doi: 10.1021/ja037616j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hardin C.C., Perry A.G., White K. Thermodynamic and kinetic characterization of the dissociation and assembly of quadruplex nucleic acids. Biopolymers. 2001;56:147–194. doi: 10.1002/1097-0282(2000/2001)56:3<147::AID-BIP10011>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 44.Smirnov I., Shafer R.H. Lead is unusually effective in sequence-specific folding of DNA. J. Mol. Biol. 2000;296:1–5. doi: 10.1006/jmbi.1999.3441. [DOI] [PubMed] [Google Scholar]
- 45.Chen F.M. Sr2+ facilitates intermolecular G-quadruplex formation of telomeric sequences. Biochemistry. 1992;31:3769–3776. doi: 10.1021/bi00130a006. [DOI] [PubMed] [Google Scholar]
- 46.Nagesh N., Chatterji D. Ammonium ion at low concentration stabilizes the G-quadruplex formation by telomeric sequence. J. Biochem. Biophys. Methods. 1995;30:1–8. doi: 10.1016/0165-022x(94)00057-k. [DOI] [PubMed] [Google Scholar]
- 47.Wang K.Y., McCurdy S., Shea R.G., Swaminathan S., Bolton P.H. A DNA aptamer which binds to and inhibits thrombin exhibits a new structural motif for DNA. Biochemistry. 1993;32:1899–1904. doi: 10.1021/bi00059a003. [DOI] [PubMed] [Google Scholar]