Abstract
Reciprocal effects of adaptive radiations on the evolution of interspecific interactions, like parasitism, remain barely explored. We test whether the recent radiations of European whitefish (Coregonus spp.) across and within perialpine and subarctic lakes promote its parasite Proteocephalus fallax (Platyhelminthes: Cestoda) to undergo host repertoire expansion via opportunity and ecological fitting, or adaptive radiation by specialization. Using de novo genomic data, we examined P. fallax differentiation across lakes, within lakes across sympatric host species, and the contributions of host genetics versus host habitat use and trophic preferences. Whitefish intralake radiations prompted parasite host repertoire expansion in all lakes, whereas P. fallax differentiation remains incipient among sympatric fish hosts. Whitefish genetic differentiation per se did not explain the genetic differentiation among its parasite populations, ruling out codivergence with the host. Instead, incipient parasite differentiation was driven by whitefish phenotypic radiation in trophic preferences and habitat use in an arena of parasite opportunity and ecological fitting to utilize resources from emerging hosts. Whilst the whitefish radiation provides a substrate for the parasite to differentiate along the same water-depth ecological axis as Coregonus spp., the role of the intermediate hosts in parasite speciation may be overlooked. Parasite multiple-level ecological fitting to both fish and crustacean intermediate hosts resources may be responsible for parasite population substructure in Coregonus spp. We propose parasites’ delayed arrival was key to the initial burst of postglacial intralake whitefish diversification, followed by opportunistic tapeworm host repertoire expansion and a delayed nonadaptive radiation cascade of incipient tapeworm differentiation. At the geographical scale, dispersal, founder events, and genetic drift following colonization of spatially heterogeneous landscapes drove strong parasite differentiation. We argue that these microevolutionary processes result in the mirroring of host–parasite phylogenies through phylogenetic tracking at macroevolutionary and geographical scales.
Keywords: speciation, host repertoire expansion, population genetics, species flocks, RADseq, Platyhelminthes
Adaptive species radiations generate stunning diversity of organisms and have broad effects on ecosystems. Yet, we lack understanding of how species radiations affect the evolution of its symbionts through radiation cascades. Diverse habitats support diverse communities of free-living species. Likewise, bursts of host diversities will provide resource-rich habitats favoring symbiont species radiations by specialization, or conversely fostering symbiont expansion into emerging hosts. We tested this hypothesis using a parasitic tapeworm that infects European whitefish, a model system of recent postglacial radiation. Whitefish radiation within lakes provided the substrate for the parasite to differentiate along the same benthic–pelagic ecological axis of lakes, but most likely by specializing in different crustacean intermediate hosts rather than by adaptation to the fish host. Dispersal, founder events, and genetic drift during colonization of heterogeneous landscapes drive parasite differentiation spatially. These microevolutionary processes may be responsible for the mirroring of host–parasite evolutionary histories at the macroevolutionary scale.
Introduction
Adaptive radiations in which organisms diversify rapidly into multitudes of new species have occurred repeatedly throughout the history of life on Earth (Gillespie et al., 2020), often in response to environmental changes that make new resources or niches available (Schluter, 2000). This rise of diversity in ecological roles has broad ecosystem effects (Lundsgaard-Hansen et al., 2017) and may propagate to interacting species resulting in an adaptive radiation cascade (Brodersen et al., 2018; Walsh et al., 2012). Yet, evidence is still scarce, and the mechanisms that drive the evolution of symbiotic and interactive species in radiation cascades remain untested. Analogous to how diverse habitats support diverse communities of free-living species (Lawton, 1983), host diversity can foster parasite community diversity (Johnson et al., 2016; Kamiya et al., 2014) through providing variable and resource-rich habitats promoting symbiont species radiations by specialization (e.g., Abrahamson & Blair, 2008; Feder & Forbes, 2010; Lutzoni et al., 2018; Vanhove et al., 2015). Alternatively, the Stockholm Paradigm (SP), a framework for understanding the evolution of diversity and interspecific associations, postulates that external perturbations changing the environmental conditions of the organisms (hosts or symbionts) will foster them to oscillate from exploitation (specialization) into the exploration (generalization) mode within the limits of their capacity and opportunities (Agosta, 2023; Agosta & Brooks, 2020; Brooks et al., 2019; Nylin et al., 2018). Thus, with hosts undergoing speciation, symbionts can expand their host repertoire (e.g., Gobbin et al., 2021; Kmentová et al., 2020) as a result of ecological opportunity and ecological fitting in sloppy fitness space (Agosta et al., 2010; Braga et al., 2018; Hoberg & Brooks, 2008). Since most symbiont groups remain unstudied (Althoff et al., 2014; Clayton et al., 2015; Ricklefs et al., 2014), more insights are needed before inferring general trends on the effect of host radiations on their symbionts.
Amongst symbionts, parasites are ubiquitous and key components of their host environment (Poulin & Morand, 2004), with whom they engage in reciprocal selection dynamics (Thompson, 2005) and other evolutionary processes that foster micro- and macroevolutionary change (Clayton et al., 2015; Paterson et al., 2010). Metazoan parasite biological characteristics influence their population parameters and/or evolutionary rates in ways uncharacteristic of free-living animals, including their reproductive strategies (e.g., hermaphroditism with self-fertilization and outcrossing, asexual reproduction, or both); short generation times; highly fragmented or isolated populations of endoparasites (each generation reproduces inside a single host individual, regardless of the number of available hosts in the habitat), tendency to seasonal fluctuations due to host death or colonization, resulting in local population extinctions and founder events, respectively, which lead to low effective population sizes and increased mutation fixation rates, favoring their rapid divergence (Huyse et al., 2005). Despite their great potential for speciation research, only a handful of parasitic helminth taxa have been investigated in the context of host radiations (e.g., Beveridge et al., 2002; Rahmouni et al., 2022; Vanhove et al., 2015). These studies are often limited to investigation of parasites with direct (one-host) life cycles, relatively low complexity models allowing to control for important biological attributes (e.g., dispersal). The level of interdependency in parasites with multi-host strategies (e.g., in Rotifera, most Platyhelminthes, Cnidaria, some Nematoda) thus remains unclear (Blasco-Costa & Poulin, 2013; Mazé-Guilmo et al., 2016). Using multiple sequential hosts through the parasite’s development provides heterogeneous selection regimes (Braga et al., 2018; Poulin, 2011) and prolongs generation times—the passage to a downstream host can take even decades (e.g., Curtis, 2003; Moore, 1987)—and thus, parasite’s evolutionary rates might approximate those of its host, contrary to the elevated rates of one-host parasites (e.g., Hafner et al., 1994). Furthermore, most diversification studies of host–parasite associations spanning several million years applied macroevolutionary approaches that can undergo time-dependent bias in the estimation of evolutionary rates (reviewed in Ho et al. (2011)), postspeciation evolution and geological factors [extinctions, bottlenecks (e.g., Drábková et al. (2019)], which can obscure the inferences of diversification/speciation processes. Instead, population genetic studies on parasite recent postglacial differentiation are still rare (Nieberding et al., 2005; Perrot-Minnot et al., 2018), and often based on single mitochondrial markers [but see Nazarizadeh et al. (2023)]. The use of host–parasite models that differentiated more recently may avoid some of these shortcomings and shed new light on the speciation process in parasites.
The European whitefish (Coregonus lavaretus species complex) is a model system for studying rapid speciation and adaptive radiation (Bernatchez, 2004; Hudson et al., 2007) having undergone a complex history of parallel diversifications in large and deep lakes across the Northern Hemisphere after the last glacial maximum, ca 10,000–15,000 BP [reviewed in Hudson et al. (2007)]. Genetically distinct whitefish lineages recolonized Europe from different glacial refugia (Crotti et al., 2021; Hudson et al., 2011; Østbye et al., 2005; Rougeux et al., 2019). Thereafter, species flocks formed through a combination of geographically sympatric and allopatric speciation in high-latitude lakes (Østbye et al., 2005; Praebel et al., 2013), and in perialpine lakes mostly through geographically sympatric speciation from an ancestral hybrid population (Hudson et al., 2011). Intralake diversification occurs along the water-depth axis and is often resource driven, with further subdivision into the littoral and the profundal habitats occasionally (Ingram et al., 2012; Siwertsson et al., 2013). Perialpine European whitefish are monophyletic with respect to their closest relatives outside the region (Hudson et al., 2011). Thus, the lake colonization and sympatric diversification of European whitefish represent a uniquely defined recent radiation model system for parasite evolutionary ecology research.
Tapeworms (Platyhelminthes) are obligatory endoparasites that complete a generation through sequential colonization of several hosts. They are thus suitable models to investigate complex host–parasite associations within the framework of the SP and to test for the adaptive radiation cascade. This study examines the patterns of differentiation within Proteocephalus fallax La Rue, 1911, a specialist tapeworm parasitizing European whitefish, and recently recognized as a species separate from Proteocephalus longicollis (Zeder, 1800) (Brabec et al., 2023). Until now, P. longicollis sensu lato (s.l.) comprised numerous synonyms of species previously described from various salmoniform fishes, including species of Coregonus, Oncorhynchus, Salmo, Salvelinus, and Thymallus (Hanzelová & Scholz, 1999; Scholz & Hanzelová, 1998). Proteocephalus longicolliss.l. (including P. fallax) circulates between two aquatic hosts, an invertebrate (several copepod species) and a vertebrate (salmoniform fishes), whereas its short-lived free-living larval stage disperses by flotation (Scholz, 1999). Thus, parasite’s dispersal is strongly linked to its aquatic hosts. Development of P. longicolliss.l. follows seasonal cycles influenced by abiotic factors (mainly water temperature) and its life cycle takes 1 year to complete (Chubb, 1982). Of all whitefish parasites, its relatively large size offers sufficient DNA quantities to perform state-of-the-art genomic methods to investigate recent divergence with sufficient detail.
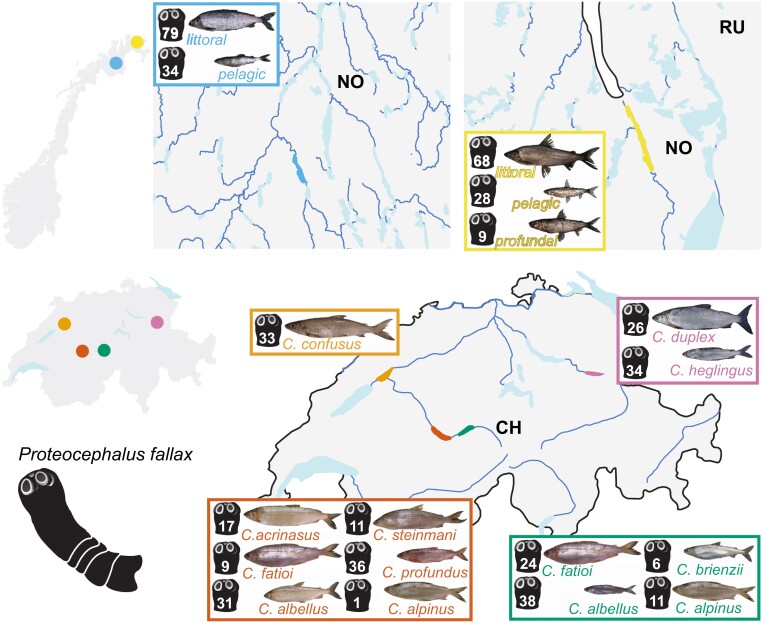
The recent establishment and in situ evolution of the whitefish–P. fallax associations within postglacial lakes make this system an excellent natural experiment suitable to test the adaptive radiation cascade hypothesis and elucidate the mechanisms promoting or preventing parasite differentiation within the SP framework, minimizing large-timescale confounding factors. The novelty lies in investigating population-level differentiation of a complex life cycle parasite in a recently radiated host. We predict that the pressures exerted by the intermediated host and the environment might outweigh the influence of definitive host radiation in this multi-host–parasite differentiation. Using double digest restriction-site associated DNA sequencing (ddRAD) to obtain variable markers for a postglacial radiation scenario, we seek to resolve whether the paradigm “host diversity begets parasite diversity” holds true in the context of whitefish and their tapeworms, by analyzing six replicate lake radiations in two geographically separated areas (Figure 1). Or Conversely, whether the whitefish diversification fosters P. fallax host range expansion by ecological fitting. Our study disentangles the contributions of the fish host genetic and phenotypic diversification (in trophic ecology and habitat use) in driving parasite differentiation. We characterize P. fallax population genomic structure at multiple scales: within lakes across sympatric Coregonus species, and across lakes at regional and continental scales. The study also investigates how lake abiotic characteristics relate to parasite population genetic parameters to shed light on environmental influences on parasite diversity.
Figure 1.
Maps of the two regions studied, Switzerland and northern Norway (insets on the left), with lakes sampled in each region color-coded (enlarged maps: Suohpatjávri, blue; Langfjordvatn, yellow; Bienne, orange; Walen, pink; Thun, dark orange; Brienz, dark green). European whitefish species/ecotypes illustrations are accompanied by black parasite silhouettes showing the number of Proteocephalus fallax specimens genotyped from each host species. Two-letter country codes for reference.
Materials and methods
Fish sampling, identification, and infection statistics
European whitefish were collected from four perialpine lakes in Switzerland and two subarctic lakes in northern Norway (Figure 1) by fishermen or through scientific fishing (permits ZH128/15 and Sak2015/266) between July and December 2017. Fin clips were preserved in ethanol for genotyping and fish species/ecotype identification was done based on genetic results and morphology (see details in Supplementary material). Worms were extracted from the fish intestine and preserved in ethanol.
Parasite whole genome sequencing and assembly
The implementation of genomic analyses of small organisms, like parasitic helminths, is hampered by the limited amount of tissue for obtaining sufficient high-molecular-weight genomic DNA (gDNA) and the risk of contamination with host DNA. Proteocephalus fallax genomic data were generated from a single specimen originating from Lake Geneva (Switzerland) using MinION (Oxford Nanopore Technologies) and NovaSeq 6000 (Illumina) platforms (see Supplementary material for details). Genome assembly was performed on Nanopore raw reads using FLYE v2.8.1(Kolmogorov et al., 2019) with a polishing step integrating Illumina reads and using ntHits and ntEdit (Warren et al., 2019). Genome completeness was assessed using BUSCO v3 (Simão et al., 2015; Warren et al., 2019) with the Metazoa database (metazoa_odb9) including 978 genes.
Parasite DNA isolation, library preparation, and ddRAD sequencing
To extract DNA from small tapeworm individuals and limit sample cross-contamination risks, we adapted the Monarch PCR & DNA Cleanup kit protocol (New England Biolabs). We used 100 μL TL buffer of the E.Z.N.A. Tissue DNA Kit (Omega Bio-tek) and 80 μg Proteinase K (Sigma-Aldrich) to carry out 1-hour tissue lysis at 56 °C followed by manual Monarch PCR & DNA Cleanup following the manufacturer’s protocol (with a volume ratio of binding buffer:sample of 2:1), finalized by two subsequent 7 μl preheated water elutions. A total of 875 gDNA single worm extractions were performed and quantified with Quant-iT PicoGreen dsDNA Assay Kit on Hidex Sense microplate reader, of which 497 produced sufficient DNA amounts and were used for library construction (including 48 technical duplicates).
Our ddRAD library construction protocol followed Peterson et al. (2012), adapted to suggestions of Mastretta-Yanes et al. (2015); (see the step-by-step protocol in Supplementary material). Due to the lack of both prior ddRAD studies and a reference genome of a related tapeworm, we first empirically evaluated the performance of several restriction enzyme pairs (Supplementary material), which led to the selection of two relatively frequent cutters NlaIII and MseI endonucleases as the pair producing preferable gDNA digestion profile. Sixty nanograms gDNA (minimum concentration of 10 ng/μl) was targeted as the starting amount for gDNA double digestion, followed by ligation of adaptors tagged with 24 different barcodes, 20 cycles of PCR amplification with reverse primers tagged with 16 different indexes, and size selection retaining fragments of 320–500 bp. Two equimolar pools of 384 ddRAD libraries each were sequenced at the Lausanne Genomics Technologies Facility using 6 lanes of Illumina HiSeq 2500 and 150 bp paired-end reads.
Reads cleaning, mapping, and SNP calling
Raw ddRAD reads were demultiplexed and trimmed with the first step of ipyrad (Eaton & Overcast, 2020) allowing for one mismatch in barcodes. Read quality was assessed using FastQC (Andrews, 2010). Trimmed reads were mapped to the reference genome using bwa-mem (Li & Durbin, 2009), and reads with mapping quality <20 removed using samtools (Li et al., 2009) to avoid reads with multiple mappings and putative paralogs. Indel realignment was performed using the Picard toolkit (http://broadinstitute.github.io/picard) and IndelRealigner from GATK4 (Van der Auwera et al., 2013). SNP calling was performed using HaplotypeCaller algorithm from GATK4 (Van der Auwera & O'Connor 2020) combining all samples to improve variant detection. SNPs were extracted using VCFTOOLS (Danecek et al., 2011) to keep bi-allelic SNPs with a calling quality >20 and shared by at least 50% of the samples.
Population genetic analysis
SNPs were filtered to keep only a single SNP per ddRAD locus (spaced by at least 200 bp) to minimize the effects of linkage disequilibrium within ddRAD loci. We performed a principal component analysis implemented in the adegenet R package (Jombart & Ahmed, 2011) and a Bayesian admixture analysis using STRUCTURE. We tested K from 1 to 10 with 3 independent Markov chains each, using 200,000 steps, including 10,000 burn-in steps, and verified chains convergence to a stable posterior distribution in each run. The most likely number of clusters was identified using Evanno’s method (Evanno et al., 2005) implemented in Structure Harvester (Earl & vonHoldt, 2012). Genome-wide weighted genetic differentiation (FST) was estimated using Weir and Cockerham’s method (Weir & Cockerham, 1984) implemented in VCFTOOLS. Genetic diversity, heterozygosity, allelic richness, and inbreeding coefficient were estimated using the hierfstat R package (Goudet, 2005). Effective population sizes (Ne) were estimated for each lake using the bias-corrected measure of linkage disequilibrium (Waples & Do, 2010) implemented in NeEstimator v.2.1 (Do et al., 2014) with a minor allele frequency cutoff of 0.05.
Testing Proteocephalus differentiation in replicated whitefish radiations
Pairwise FST were estimated among Proteocephalus populations from distinct sympatric Coregonus spp./ecotypes for each lake, using the hierfstat R package (Goudet, 2005). For each comparison, 999 random permutations were performed to estimate a random FST distribution followed by a Monte-Carlo test to assess whether the observed FST value was significantly higher than the random distribution. A Bonferroni correction of the p-values was applied to correct for multiple comparisons. Pairwise FST among Coregonus species/ecotypes were estimated in GenAlEx (Peakall & Smouse, 2012). Differentiation of Proteocephalus in relation to the genetic differentiation of Coregonus spp. was examined by regressing pairwise FST values of P. fallax populations from sympatric hosts to those of sympatric Coregonus spp./ecotypes using a generalized linear model with a quasibinomial error distribution and a logit link function, implemented in the stats R package.
Spatial and environmental effects on genetic differentiation and diversity indexes
We performed a distance-based redundancy analysis (dbRDA) integrating geographical variables, four quantitative variables on lake characteristics (lake surface, maximum depth, oxygenated lake depth and past highest phosphorous levels), and two quantitative variables on host fish ecology transformed in dummy variables, that is, fish resource preferences (planktivorous = 0, mixed feeding = 1, and benthivorous = 2) and fish habitat occupation (profundal = 0, wide distributed = 1, and shallow = 2), in an individual-based approach (see Laura Benestan https://github.com/laurabenestan/db-RDA-and-db-MEM). In brief, we first created spatial variables using Moran Eigenvector’s Maps (MEMs) implemented in the adespatial R package (Dray et al., 2023). Second, a principal coordinates analysis (PCoA) was performed on the Euclidean genetic distances based on the genotypes of each sample. Finally, a global dbRDA was applied by integrating ecological factors as additional variables to spatial components, and an ANOVA with 1,000 permutations was performed to assess the significance of each variable within the model using the vegan R package (Oksanen et al., 2020). The relationship between the three current environmental lake characteristics mentioned above and the parasite genetic diversity indices was assessed using Spearman’s correlation test for each variable independently using the stats R package. The R computing environment v. 4.1.2 was used (R Development Core Team, 2021).
Results
Population census data of P. fallax among sympatric whitefish and lakes
Tapeworm prevalence (percentage of fish infected among all examined) was high in all lakes, spanning 86–99%, the lowest in Langfjordvatn coinciding with the lowest mean intensity of infection (average number of worms per infected fish), and the highest in Brienz (Table 1). Proteocephalus fallax highest mean intensities of infection were achieved in lakes Bienne and Brienz (range: 1–1,000 individuals; Table 1). Among the perialpine hosts, the highest mean intensities were found in C. confusus (234 ± 52) from lake Bienne, C. fatioi (256 ± 72) and C. albellus (139 ± 23) from lake Thun, and C. brienzii (121 ± 42), C. fatioi (102 ± 14) and C. albellus (94 ± 17) from lake Brienz (Supplementary Table S1), all species typically considered pelagic feeders. In the subarctic lakes, the pelagic ecotypes also carried the highest mean intensities. All other European whitefish species/ecotypes harbored lower mean intensities of infection (6–73 individuals), so that they carry a small proportion of the P. fallax population in each lake.
Table 1.
Summary statistics of genetic diversity per population using 8,072 SNPs and infection parameters per lake. N P. fallax: sample size of parasites, N SNPs: number of SNPs retained per lake, CIs: confidence intervals, Mean Intensity ± S.E.: a measure of population census size indicating the average number of P. fallax specimens and standard error per infected fish, Prevalence of infection: percentage of fish infected from all whitefish examined at each lake, also a measure of population census size.
| Perialpine lakes | Subarctic lakes | |||||
|---|---|---|---|---|---|---|
| Bienne | Brienz | Thun | Walen | Langfjordvatn | Suohpatjávri | |
| N P. fallax | 33 | 93 | 110 | 60 | 105 | 113 |
| N SNPs | 34,843 | 4,758 | 33,488 | 10,152 | 12,375 | 11,611 |
| Allelic richness (Ar) | 1.34 | 1.54 | 1.63 | 1.44 | 1.28 | 1.11 |
| Genetic diversity (Hs) | 0.15 | 0.16 | 0.16 | 0.16 | 0.15 | 0.11 |
| Observed heterozygosity (Ho) | 0.07 | 0.06 | 0.09 | 0.05 | 0.05 | 0.04 |
| Inbreeding coefficient (Fis) | 0.56 | 0.62 | 0.46 | 0.65 | 0.66 | 0.63 |
| Effective Population size (Ne) | 910 | 1,932 | 627 | 450 | 1,253 | 1,170 |
| 95% CIs | 692−1,329 | 780−inf | 600−657 | 352−622 | 954−1,821 | 862–1,814 |
| Mean intensity ± SE | 234 ± 52 | 93 ± 10 | 68 ± 8 | 52 ± 7 | 20 ± 4 | 68 ± 10 |
| Intensity of infection (range) | 3−1,000 | 1 − 749 | 1 − 520 | 1−300 | 1−255 | 1–429 |
| Prevalence of infection (%) | 91 | 99 | 92 | 98 | 86 | 93 |
De novo genome assembly of P. fallax from Lake Geneva (Switzerland)
The Oxford Nanopore run produced 1,680,336 reads with a mean length of 2,697.96 bp and the Illumina sequencing produced 345,501,790 reads and a total of 51.82 Gbp (Table 2). The de novo assembly strategy of the Nanopore reads followed by polishing with Illumina reads yielded the first genome for a representative of the cestode order Onchoproteocephalidea. It comprised 4,060 scaffolds with a scaffold N50 of 419,724 bp and a total assembly length of 131,602,485 bp which is comparable to the k-mer estimate performed on Illumina raw reads (110,347,222 bp; see Table 2). BUSCO metrics showed moderate genome completeness (56.5% complete BUSCO; Table 2) as expected for a nonmodel flatworm representative. For comparison, the same BUSCO analysis performed on the chromosome-level reference genome assembly of Hymenolepis microstoma (GCA_000469805.3) resulted in 65.7% of complete BUSCO. Illumina reads mapping on the final genome was high, 98.08%, suggesting good completeness of the genome assembly.
Table 2.
Reference genome statistics on raw data, genome size estimations, genome assembly, and completeness using BUSCO.
| Statistics | Illumina | Nanopore |
|---|---|---|
| Raw data | ||
| Mean read length (sd) | 150 | 2,697.96 |
| # reads | 345,501,790 | 1,680,336 |
| sum (Gb) | 51.82 | 4.53 |
| Genome size estimation | ||
| Het | 0.0092 | |
| err | 0.0037 | |
| genome size | 110,347,222 | |
| fit | 2.03 | |
| Assembly statistics | ||
| # Scaffolds | 4,060 | |
| N50 scaffold | 419.724 | |
| Mean scaffold size | 32.414 | |
| Longest scaffold | 3,708,885 | |
| %N | 0.0075 | |
| GC content (%) | 43.28 | |
| Total length | 131,602,485 | |
| BUSCO results (%) | ||
| Complete and single-copy BUSCOs | 55.2 | |
| Complete and duplicate BUSCOs | 1.3 | |
| Fragmented BUSCOs | 10.3 | |
| Missing BUSCOs | 33.2 | |
Genomic descriptors of Proteocephalus populations and structure across lakes
A total of 1,283,228,366 quality-trimmed sequence reads were obtained providing an average of 2,496,553 ± 642,993 reads per individual. The mapping step to the newly generated reference genome resulted in an average of 95.27 ± 7.26% mapped reads before cleaning and 72.52 ± 6.78% after cleaning (Supplementary Table S2). The SNP calling identified 8,072 bi-allelic SNPs shared by at least 50% of the samples and an average of 4,495 ± 786 SNPs per sample. A second SNP set was generated keeping only perialpine samples resulting in 26,738 SNPs shared by at least 50% of the perialpine samples. To perform genetic structure analyses, one SNP every 200 bp along the genome was retained, resulting in 2,002 SNP from the dataset generated on all samples and 6,121 SNP from the perialpine dataset. Finally, SNP sets were also generated within each lake using the same parameters as above (Supplementary Table S2).
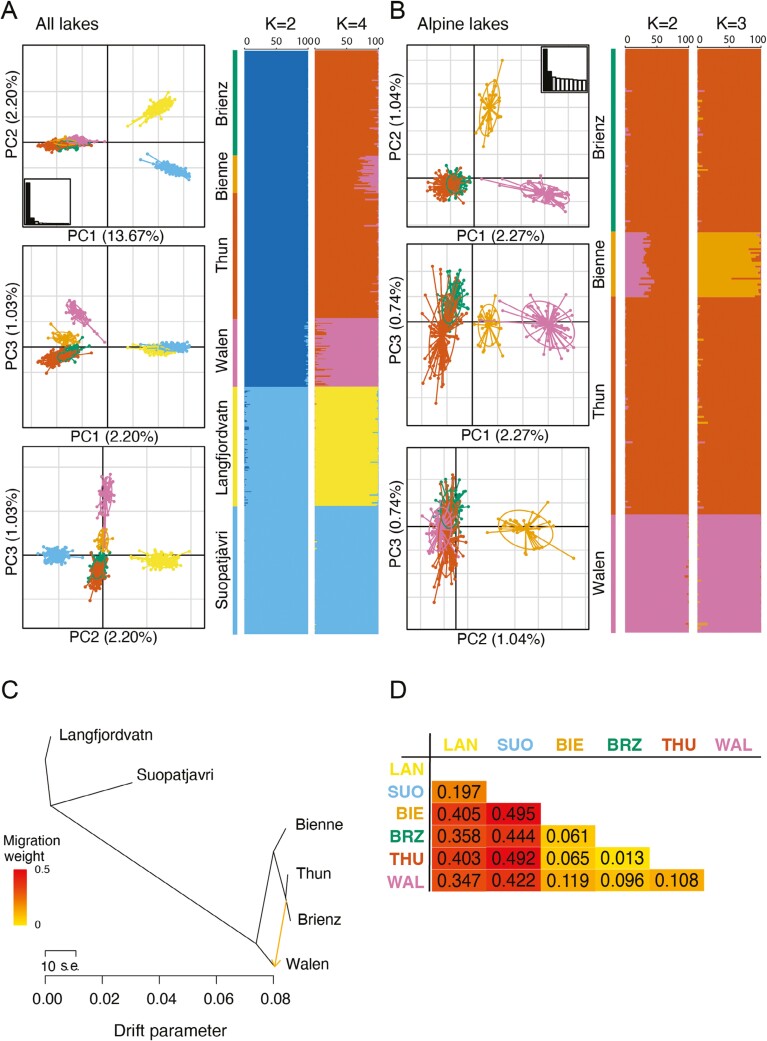
Based on 8,072 SNPs shared among all samples, P. fallax from the subarctic lake Suohpatjávri showed lower observed heterozygosity (HO), gene diversity (HS), and allelic richness (Ar) values, despite its higher mean census population size (approximated from the prevalence and mean intensity of infection data, Table 1) relative to Langfjordvatn. The highest HO, HS, and Ar values were detected in the perialpine region, in Thun (Table 1), which is also the lake with the highest whitefish species richness (six Coregonus spp.). The inbreeding coefficient (FIS) was the highest in Langfjordvatn coinciding with the lowest mean census population size of the subarctic lakes, and the lowest in Thun having the highest mean census population sizes in the perialpine lakes. On the contrary, effective population sizes (Ne) were the largest in the subarctic lakes and the perialpine lake Brienz, whereas the same lakes had low values of HO and high FIS. Walen showed the lowest Ne coinciding with a highly inbred population and the smallest perialpine census population size (Table 1).
Strong genetic structure was detected between the Proteocephalus populations from the subarctic and perialpine regions with clearly separated clusters in the PCA and STRUCTURE plots (Figure 2A, K = 2), and FST values above 0.35 (Figure 2D). Differentiation between populations in the two subarctic lakes was almost as strong as between regions, despite their geographic proximity (Figure 2A,D, K = 4). In the perialpine region, three distinct populations were detected in the lakes Walen, Bienne, and Thun and Brienz (Figure 2B, K = 3). The genetic relationships inferred by the TreeMix analysis (Figure 2C) supported the patterns of genetic differentiation described above. Furthermore, individuals from Thun and Brienz shared a common ancestry and formed a single genetic cluster (FST = 0.01) (Figure 2C,D).
Figure 2.
Proteocephalus fallax population genetic structure across (A) all lakes based on 2,002 shared SNPs, and (B) perialpine lakes based on 6,121 shared SNPs. The best number of genetic clusters (K) is given for each subset. (C) Phylogenetic network of the historical relationships among the populations inferred by Treemix. Putative gene flow is identified by an arrow pointing in the direction of the recipient population and colored proportionally to the gene flow intensity. (D) Pairwise FST matrix for all population comparisons. Colors are proportional to the degree of divergence and correspond to FST values.
Proteocephalus fallax differentiation among hosts in replicated whitefish radiations
Genetic structure of parasite populations infecting sympatric whitefish species was overall weak, but significant in most lakes (pairwise FST, Table 3A). Replicated patterns of differentiation appeared between parasite populations from the littoral and pelagic European whitefish ecotypes in Suohpatjávri, and their equivalents, the shallow/littoral and the deep/pelagic in Walen. In the perialpine region, parasite differentiation was common in the most host-rich lake Thun but less in Brienz, with six and four Coregonus spp., respectively (Table 3A). In Brienz, only the parasite populations associated with C. brienzii and C. albellus showed a significant level of differentiation.
Table 3.
Comparisons of P. fallax population genetic structure (FST) amongst A) sympatric fish species of each lake, B) the host habitat use, and C) the host trophic preference.
| Lake | Comparison | Parasite FST | p-value |
|---|---|---|---|
| (A) | By sympatric fish species | ||
| Brienz | C. brienzii (6) vs. C. fatioi (24) | 0.017 | .051 |
| C. brienzii (6) vs. C. albellus (38) | 0.026 | .007* | |
| C. brienzii (6) vs. C. alpinus (11) | 0.022 | .043 | |
| C. fatioi (24) vs. C. albellus (38) | 0.004 | .074 | |
| C. fatioi (24) vs. C. alpinus (11) | 0.010 | .031 | |
| C. albellus (38) vs. C. alpinus (11) | 0.009 | .042 | |
| Langfjordvatn | pelagic (28) vs. littoral (68) | 0.003 | .047 |
| pelagic (28) vs. profundal (9) | 0.006 | .096 | |
| littoral (68) vs. profundal (9) | 0.005 | .078 | |
| Suohpatjávri | littoral (79) vs. pelagic (34) | 0.003 | .012* |
| Thun | C. acrinasus (17) vs. C. albellus (31) | 0.002 | .121 |
| C. acrinasus (17) vs. C. profundus (36) | 0.002 | .135 | |
| C. acrinasus (17) vs. C. steinmanni (11) | 0.006 | .020 | |
| C. acrinasus (17) vs. C. fatioi (9) | 0.014 | .001** | |
| C. albellus (31) vs. C. profundus (36) | 0.004 | .001** | |
| C. albellus (31) vs. C. steinmanni (11) | 0.005 | .021 | |
| C. albellus (31) vs. C. fatioi (9) | 0.011 | .003* | |
| C. profundus (36) vs. C. steinmanni (11) | 0.006 | .013* | |
| C. profundus (36) vs. C. fatioi (9) | 0.014 | .001** | |
| C. steinmanni (11) vs. C. fatioi (9) | 0.020 | .001** | |
| Walen | C. duplex (26) vs. C. heglingus (34) | 0.006 | .003** |
| (B) | By host habitat use | ||
| Brienz | shallow (11) vs. wide distribution (68) | 0.009 | .030* |
| shallow (28) vs. wide distribution (68) | 0.003 | .037 | |
| Langfjordvatn | shallow (28) vs. profundal (9) | 0.006 | .105 |
| wide distribution (68) vs. profundal (9) | 0.005 | .083 | |
| Suohpatjávri | shallow (34) vs. wide distribution (79) | 0.003 | .014* |
| shallow (18) vs. wide distribution (51) | 0.003 | .050 | |
| Thun | shallow (18) vs. profundal (36) | 0.002 | .098 |
| wide distribution (51) vs. profundal (36) | 0.005 | .001** | |
| Walen | shallow (26) vs. wide distribution (34) | 0.006 | .005** |
| (C) | By host trophic preference | ||
| mix feeding (30) vs. planktivorous (38) | 0.004 | .044 | |
| Brienz | mix feeding (30) vs. benthivorous (11) | 0.010 | .023* |
| planktivorous (38) vs. benthivorous (11) | 0.009 | .030 | |
| Langfjordvatn | benthivorous (77) vs. planktivorous (28) | 0.003 | .038* |
| Suohpatjávri | benthivorous (79) vs. planktivorous (34) | 0.003 | .014* |
| mix feeding (28) vs. planktivorous (40) | 0.003 | .010* | |
| Thun | mix feeding (28) vs. benthivorous (37) | 0.003 | .024* |
| benthivorous (37) vs. planktivorous (40) | 0.005 | .001** | |
| Walen | benthivorous (26) vs. planktivorous (34) | 0.006 | .006** |
p-Values were estimated by performing 999 permutation sets and observed values were compared to a random distribution using a Monte-Carlo test. Significant values (*) indicate observed values were higher than random distribution and p-values were adjusted with Bonferroni correction when more than one comparison in a lake. The P. fallax population from the single host species in Bienne and the single-genotyped parasite specimen from C. alpinus in Thun were excluded from these comparisons, as well as the P. fallax population from the single host species in Bienne. Numbers of genotyped P. fallax specimens are indicated in parentheses.
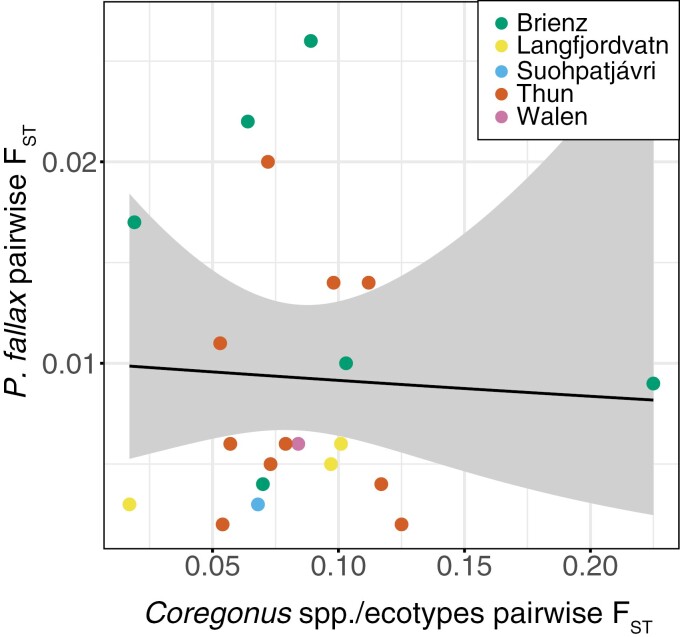
The correlation between P. fallax population differentiation among sympatric fish host species/ecotypes in each lake and the genetic differentiation between the pairs of sympatric Coregonus species/ecotypes was slightly negative (Figure 3), but nonsignificant (GLMFishFST: F1,19 = 0.049, p = .830). In other words, host diversification did not explain the differentiation of its parasite populations, for which genetically well-differentiated host species/ecotypes harbor rather genetically weakly differentiated Proteocephalus populations. The same pattern emerged after excluding the more differentiated C. alpinus/C. albellus pairwise comparison from Lake Brienz (GLMFishFST: F1,18 = 0.098, p = .790).
Figure 3.
Parasite differentiation was unrelated to the level of differentiation between its hosts within lakes. Regression plot of pairwise FST estimates between Proteocephalus fallax populations in sympatric Coregonus hosts (based on SNP data) and pairwise FST between sympatric Coregonus species/ecotypes (based on microsatellite data). Data points are colored according to lakes.
The contribution of host ecology to P. fallax differentiation across replicated radiations
Host ecology played a relevant role in acquiring divergent P. fallax populations (Table 3B,C). The comparisons among sympatric Coregonus spp./ecotypes with distinct habitat use showed that European whitefish species specialized on shallow waters versus those more widespread in the water column exhibit different P. fallax subpopulations in Suohpatjávri, Brienz, and Walen, whereas in the more host species-rich lake of Thun the differentiation within P. fallax occurs between the whitefish species groups with wide distributions in the water column and the profundal specialist, C. profundus (Table 3B). Comparisons of P. fallax between sympatric European whitefish groups with different trophic preferences revealed in almost all cases significant differentiation, with slightly higher differentiation between P. fallax in whitefish with benthivorous versus planktivorous feeding strategies (Table 3C). Thus, the intralake differentiation of the subpopulations of this trophically transmitted parasite in the definitive hosts is associated with phenotypic diversification of Coregonus spp. in habitat segregation and dietary traits.
Spatial environmental associations and abiotic correlations
The distance-based redundancy analysis revealed that the spatial component explained most of the genetic variance across Europe (R2 = .234, p = .002). The contribution of the fish host resource preference and habitat occupation were significant, although much lower (R2 = .039, p = .004 and R2 = .004, p = .002, respectively).
Lake physicochemical characteristics, in particular lake maximal and oxygenation depths showed a significant positive correlation with allelic richness (R2 = .940, p = .017; R2 = .890, p = 0.033, respectively) and genetic diversity of P. fallax (R2 = .930, p = .008; R2 = .930, p = .008), the latter mostly driven by the values in Suohpatjávri (Supplementary Figure S1). Observed heterozygosity showed a strong positive correlation with lake surface area only (R2 = .990, p = .000). Neither allelic richness nor genetic diversity correlated with the number of samples examined or the number of SNPs per lake (Supplementary Figure S2).
Discussion
Our study showed that populations of the tapeworm P. fallax were strongly differentiated spatially among lakes, between, and within the perialpine and subarctic regions. Genomic differentiation within lakes was weak, albeit significant, across replicated radiations of European whitefish in both the Alps and the subarctic. Thus, the European whitefish radiation provided a substrate for the parasites to radiate too, which supports the ‘radiation cascade’ hypothesis, although parasite differentiation was independent from the genetic differentiation of the fish hosts. In sympatry, European whitefish trophic preferences and habitat use had weak, but significant and replicated effects on the differentiation among parasite populations in all lakes. Between lakes, allelic richness and parasite genetic diversity significantly correlated with lake maximum depth and lake oxygenation level, but only heterozygosity correlated with lake surface area. Altogether, our results inform about the mode of evolution of an endoparasitic platyhelminth representative and indicate that P. fallax differentiated spatially in the last 10,000–15,000 years, likely caused by the distinct demographic history of postglacial colonization of the lakes together with its fish hosts. However, genetic differentiation in the tapeworm was constrained in sympatry, being structured along the same water-depth ecological axis of lakes as its Coregonus hosts differentiate, but differentiation remains incipient when compared with the rapid genetic and phenotypic diversification of the fish hosts.
Shared evolutionary history of P. fallax and Coregonus at the continental and regional scales
The comparison of phylogeographic patterns of hosts and symbionts can help elucidate colonization hypotheses and taxon pulses (Galbreath & Hoberg, 2012). Proteocephalus fallax population differentiation between the subarctic and perialpine regions supports the two-refugia origin of European whitefish recolonization of Western Europe (Østbye et al., 2005), the N clade west of the Ural Mountains, and the C clade from near the mouth of the Rhine. Proteocephalus fallax high degree of divergence between Langfjordvatn (Pasvik River system, but isolated from it for ~9,000 YBP), and Suohpatjávri (Alta River catchment), together with high inbreeding values and completely segregated cytochrome c oxidase subunit 1 haplotypes (Brabec et al., 2023) indicate a low number of founders and strong genetic drift as the main drivers of allopatric differentiation. Thus, this recent taxon pulse resulted in the vicariance of both, the tapeworm and the European whitefish (Østbye et al., 2006; Praebel et al., 2013), during the early postglacial recolonization of these lakes. In the Swiss Alps, European whitefish cooccurrence from two glacial refugia led to a hybridogenic ancestral population that underwent rapid adaptive radiations, in at least 5 lake/super-lake systems independently (De-Kayne et al., 2022; Hudson et al., 2011), with three of the major European whitefish genetic clusters being included in our study. Whereas Coregonus spp. from the perialpine lakes Thun-Brienz and Walen are more closely related to each other than either of them to populations from lake Bienne in midland (De-Kayne et al., 2022), the higher divergence of P. fallax detected between Walen and Thun-Brienz speaks to multiple recolonization waves of the perialpine lakes [similar to, e.g., sculpin fish Lucek et al. (2018)]. Though, the influence of historical human-made changes to the connectivity and introductions of whitefish fry (Selz et al., 2020) cannot be excluded. Proteocephalus fallax strong differentiation between the perialpine and the subarctic lakes suggests that the northern European whitefish lineage that recolonized the Alps arrived uninfected, or that the ancestral tapeworm associated with the northern European whitefish lineage was replaced by a different P. fallax lineage that was acquired along the colonization path at intermediate location(s). The loss of parasites, or their lagged arrival, favors reduced parasite pressure (low parasite prevalence) when hosts expand into new regions (Phillips et al., 2010), and predicts an immediate “ecological release” favoring elevated host population growth rates and persistence at the expansion front, followed by an “evolutionary release” (Alexander et al., 2022) for both the host and the parasite. In the context of adaptive radiations through ecological speciation, different mechanisms may contribute to the parasite-driven mediated selection of the host (Karvonen & Seehausen, 2012), but evidence is still limited (Karvonen et al., 2013). We propose that the escape from parasites with the initial “honeymoon phase” sensuPhillips et al. (2010) and “evolutionary release” sensuAlexander et al. (2022) contributed to the initial burst of postglacial whitefish diversification, with a delayed arrival, and simultaneous opportunistic tapeworm host repertoire expansion into emerging Coregonus species flocks. Although today P. fallax may only cause a mild fitness loss to the host by removing nutrients (but notice the high infection levels reported in Results), in a situation of high competition for limited resources in recently formed oligotrophic lakes, escaping infection could confer a considerable advantage. Furthermore, delays in dynamics of colonization and taxon pulses have been observed in other complex host–parasite systems and predicted by models in the framework of the SP (Araujo et al., 2015; Galbreath & Hoberg, 2015; Galbreath et al., 2023; Haas et al., 2020).
Host adaptive radiation results in parasite host–repertoire expansion and subsequent differentiation
Parasite diversification in the context of the adaptive radiation cascade has been sparsely investigated hitherto, with most studies focused on the radiation of cichlids of West African lakes and their direct life cycle monogenean parasites (e.g., Mendlová et al., 2012; Vanhove et al., 2015) that show codifferentiation and predominantly high host specificity. Conversely, we found that gene flow has constrained the differentiation of P. fallax subpopulations among sympatric European whitefish hosts relative to that of its fish hosts during the recent postglacial expansion and within-lake radiations. Even more, the intralake differentiation of this tapeworm was unrelated to the genetic differentiation among sympatric European whitefish hosts, which rules out codivergence with the host (Althoff et al., 2014) and supports the view that codiversification simultaneously with the host is not a common mode of parasite speciation (Agosta & Brooks, 2020; de Vienne et al., 2013). Macroevolutionary studies of typically highly host-specific parasites attribute the lack of codifferentiation to parasite life history traits like having either additional hosts in the life cycle as in the case of P. fallax, stages with short periods of transmission in the environment, possibly facilitating (definitive) host switching (Huyse & Volckaert, 2005; Paterson & Poulin, 1999), or the host ecology and social behavior (Desdevises et al., 2002; Kmentová et al., 2020) among others (Larose & Schwander, 2016). These are all factors that increase the opportunities for encountering alternative hosts. In the context of the SP (Agosta & Brooks, 2020; Nylin et al., 2018), Proteocephalus fallax opportunity and capacity to extend the host range in the European whitefish radiation is facilitated by ecological fitting to whitefish resources via phylogenetic conservatism (Agosta & Klemens, 2008; Brooks & McLennan, 2012) among species flocks at different stages of diversification along the speciation continuum.
By integrating ecological factors that can shape parasite microevolution and underpin macroevolutionary patterns (Blasco-Costa et al., 2021), we revealed that phenotypic diversification in European whitefish exploitation of new ecological niches fosters an incipient nonadaptive radiation cascade in P. fallax. Our results support the oscillation hypothesis (Brooks et al., 2019; Janz & Nylin, 2008), alternating between P. fallax expansion into available European whitefish resources followed by an exploitation (specialization) phase, which progresses with a time-lag as predicted by models (Araujo et al., 2015; Braga et al., 2018). Differentiation along the same water-depth ecological axis of lakes as its Coregonus hosts is most likely associated with the use of different intermediate crustacean hosts for transmission. Three lines of evidence led us to propose this hypothesis. First, parasite intra-lacustrine population substructure, though weak, across European whitefish species/ecomorphs and both, their trophic preference and habitat use, supports a scenario of recent parasite differentiation with gene flow conditional on ecological opportunity (see Hoberg & Brooks, 2008). Second, variation in parasite population genetic parameters correlated with lake depth and oxygenation, both environmental characteristics important for whitefish speciation (typically occurring along depth clines) to the point that changes to the availability of pelagic and profundal habitats have caused speciation reversal (Bhat et al., 2014; Vonlanthen et al., 2012). Third, larval stages of P. longicolliss.l. are known to infect several crustacean taxa in Europe (Scholz, 1999 and references within). Given that salmonid trophic niche variation depends on prey diversity (Sánchez-Hernández et al., 2021), and that European whitefish adaptive radiation was driven by colonization of new habitats (e.g., littoral, pelagic, and profundal) and trophic specialization in postglacial lakes (e.g., Ingram et al., 2012; Siwertsson et al., 2013), it is likely that the differential use of planktonic and benthic crustaceans via ecological fitting in sloppy fitness space (Agosta & Klemens, 2008; Agosta et al., 2010) promotes P. fallax population substructure detected across sympatric whitefish hosts. Thus, P. fallax may represent another example of “multiple-level ecological fitting” (Malcicka et al., 2015) by tracking both fish and intermediate host available resources in the postglacial lakes. These results also highlight the need to stop overlooking the identity and diversity of intermediate hosts if we seek understanding the mechanisms of parasite evolution (Blasco-Costa & Poulin, 2017).
Linking micro- and macroevolutionary research
Differentiation of a parasite is often associated with its most mobile host (Blasco-Costa & Poulin, 2013; Louhi et al., 2010; Prugnolle et al., 2005), but see Mazé-Guilmo et al. (2016). Our findings highlight that dispersal with the fish host during taxon pulses, and founder events associated with the recolonization and establishment of allopatric populations, predispose to high fixation rates (i.e., increased divergence rates) in parasite populations and foster spatial differentiation patterns at micro-, and possibly vicariance later at macroevolutionary time scales. Allopatry predisposed lice to codivergence with its hosts compared to when hosts were found in contact zones (Barker, 1994). In a microsporidian parasite and its aquatic host, shared biogeographic histories and dispersal explained the congruent diversification of both organisms at the regional scale (Park et al., 2020). Taken all together, we propose that host-dependent parasite dispersal during taxon pulses, coupled with parasite founder events during colonizations in range expansions represent important ecological and evolutionary processes generating the pattern of parasite phylogenetic tracking of their hosts in space. In other words, these processes alone could explain some mimicked patterns of codiversification/codifferentiation (correlated diversification between interacting lineages (Clayton et al., 2015)). Microevolutionary research across geographical scales will further reveal processes and underlying mechanisms behind parasite differentiation and eco-evolutionary change.
Supplementary material
Supplementary material is available online at Evolution Letters.
Acknowledgments
We thank two anonymous reviewers for their constructive suggestions. We are thankful to the fish authorities responsible for each lake, Arno Filli, Michael Kugler, Benjamin Gugger and Rolf Schneider, and to the fisherwomen and fishermen who have been essential in the collection of fish specimens. We are also grateful to Laina Dalsbø, Karin Johannessen, Katja Häkli, Janik Pralong, Eloïse Rochat, Tommy Andriolo and colleagues from EAWAG for assistance in the field, lab, or with the analyses, to Alain de Chambrier, Tomáš Scholz and Jean Mariaux for informal discussions and comments, and to the staff of the Lausanne Genomic Technologies Facility.
Contributor Information
Jan Brabec, Department of Invertebrates, Natural History Museum of Geneva, Geneva, Switzerland; Department of Evolutionary Parasitology, Institute of Parasitology, Biology Centre of the Czech Academy of Sciences, České Budějovice, Czech Republic.
Jérémy Gauthier, Department of Invertebrates, Natural History Museum of Geneva, Geneva, Switzerland.
Oliver M Selz, Department of Fish Ecology and Evolution, Centre of Ecology, Evolution and Biogeochemistry (CEEB), Eawag Swiss Federal Institute of Aquatic Science and Technology, Kastanienbaum, Switzerland; Aquatic Restoration and Fisheries section, Federal Office for the Environment (FOEN), Bern, Switzerland.
Rune Knudsen, Department of Arctic Biology, The Arctic University of Norway, Tromsø, Norway.
Julia Bilat, Department of Invertebrates, Natural History Museum of Geneva, Geneva, Switzerland.
Nadir Alvarez, Department of Invertebrates, Natural History Museum of Geneva, Geneva, Switzerland; Department of Genetics and Evolution, University of Geneva, Geneva, Switzerland.
Ole Seehausen, Department of Fish Ecology and Evolution, Centre of Ecology, Evolution and Biogeochemistry (CEEB), Eawag Swiss Federal Institute of Aquatic Science and Technology, Kastanienbaum, Switzerland; Division of Aquatic Ecology & Evolution, Institute of Ecology and Evolution, University of Bern, Bern, Switzerland.
Philine G D Feulner, Department of Fish Ecology and Evolution, Centre of Ecology, Evolution and Biogeochemistry (CEEB), Eawag Swiss Federal Institute of Aquatic Science and Technology, Kastanienbaum, Switzerland; Division of Aquatic Ecology & Evolution, Institute of Ecology and Evolution, University of Bern, Bern, Switzerland.
Kim Præbel, Norwegian College of Fishery Science, UiT The Arctic University of Norway, Tromsø, Norway; Department of Forestry and Wildlife Management, Inland Norway University of Applied Science, Elverum, Norway.
Isabel Blasco-Costa, Department of Invertebrates, Natural History Museum of Geneva, Geneva, Switzerland; Department of Arctic Biology, The Arctic University of Norway, Tromsø, Norway.
Data and code availability
All collapsed and paired-end sequence data for samples sequenced in this study are available in compressed fastq format through NCBI’s BioProject no. PRJNA910576 (ddRAD accessions SAMN32132446–959, reference genome accession SAMN32134074), together with rescaled and trimmed bam sequence alignments against both the nuclear and mitochondrial reference genome. All scripts used to perform the analyses presented in this paper are available through https://github.com/JeremyLGauthier/Scripts_Proteocephalus.
Author contributions
I.B.-C., R.K. conceived the study, I.B.-C., R.K., P.F., O.S. obtained funding, I.B.-C., R.K., J.Br., O.M.S., O.S., J.B., K.P., N.A., P.F. contributed to data collection, J.G., J.Br. I.B.-C. performed analyses and wrote the manuscript. All authors provided feedback and edited the manuscript.
Funding
This work received funding from the Swiss National Science Foundation (SNSF grant 169211 to I.B.-C.).
Conflict of interest: The authors declare no conflicts of interest.
References
- Abrahamson, W. G., & Blair, C. P. (2008). Sequential radiation through host-race formation: Herbivore diversity leads to diversity in natural enemies. In Tilmon K. (Ed.), Specialization, speciation, and radiation: The evolutionary biology of herbivorous insects (pp. 188–202). University of California Press. [Google Scholar]
- Agosta, S. J. (2023). The Stockholm Paradigm explains the eco-evolutionary dynamics of the biosphere in a changing world, including emerging infectious disease. In Gardner S. L., Brooks D. R., Boeger W. A., & Hoberg E. P. (Eds.), An evolutionary pathway for coping with emerging infectious disease (pp. 29–46). Zea E-Books. [Google Scholar]
- Agosta, S. J., & Brooks, D. R. (2020). The major metaphors of evolution. Springer International Publishing. [Google Scholar]
- Agosta, S. J., Janz, N., & Brooks, D. R. (2010). How specialists can be generalists: Resolving the “parasite paradox” and implications for emerging infectious disease. Zoologia (Curitiba), 27(2), 151–162. 10.1590/s1984-46702010000200001 [DOI] [Google Scholar]
- Agosta, S. J., & Klemens, J. A. (2008). Ecological fitting by phenotypically flexible genotypes: Implications for species associations, community assembly and evolution. Ecology Letters, 11(11), 1123–1134. 10.1111/j.1461-0248.2008.01237.x [DOI] [PubMed] [Google Scholar]
- Alexander, J. M., Atwater, D. Z., Colautti, R. I., & Hargreaves, A. L. (2022). Effects of species interactions on the potential for evolution at species’ range limits. Philosophical Transactions of the Royal Society of London, Series B: Biological Sciences, 377(1848), 20210020. 10.1098/rstb.2021.0020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Althoff, D. M., Segraves, K. A., & Johnson, M. T. J. (2014). Testing for coevolutionary diversification: Linking pattern with process. Trends in Ecology & Evolution, 29, 82–89. 10.1016/j.tree.2013.11.003 [DOI] [PubMed] [Google Scholar]
- Andrews, S. (2010). FastQC: A quality control tool for high throughput sequence data. Babraham Bioinformatics, Babraham Institute. [Google Scholar]
- Araujo, S. B. L., Braga, M. P., Brooks, D. R., Agosta, S. J., Hoberg, E. P., Von Hartenthal, F. W., & Boeger, W. A. (2015). Understanding host-switching by ecological fitting. [Article]. PLoS One, 10(10), e0139225. 10.1371/journal.pone.0139225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker, S. C. (1994). Phylogeny and classification, origins, and evolution of host associations of lice. International Journal for Parasitology, 24(8), 1285–1291. 10.1016/0020-7519(94)90195-3 [DOI] [PubMed] [Google Scholar]
- Bernatchez, L. (2004). Ecological theory of adaptive radiation: An empirical assessment from coregonine fishes (Salmoniformes). In Hendry A. P. & Stearns S. C. (Eds.), Evolution illuminated: Salmon and their relatives (pp. 175–207). Oxford Academic. [Google Scholar]
- Beveridge, I., Chilton, N. B., & Spratt, D. M. (2002). The occurrence of species flocks in the nematode genus Cloacina (Strongyloidea: Cloacininae), parasitic in the stomachs of kangaroos and wallabies. Australian Journal of Zoology, 50(6), 597–620. 10.1071/zo02038 [DOI] [Google Scholar]
- Bhat, S., Amundsen, P. -A., Knudsen, R., Gjelland, K., Fevolden, S.-E., Bernatchez, L., & Præbel, K. (2014). Speciation reversal in European whitefish (Coregonus lavaretus (L.)) caused by competitor invasion. PLoS One, 9(3), e91208. 10.1371/journal.pone.0091208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blasco-Costa, I., Hayward, A., Poulin, R., & Balbuena, J. A. (2021). Next-generation cophylogeny: Unravelling eco-evolutionary processes. Trends in Ecology & Evolution, 36(10), 907–918. 10.1016/j.tree.2021.06.006 [DOI] [PubMed] [Google Scholar]
- Blasco-Costa, I., & Poulin, R. (2013). Host traits explain the genetic structure of parasites: A meta-analysis. Parasitology, 140(10), 1316–1322. 10.1017/S0031182013000784 [DOI] [PubMed] [Google Scholar]
- Blasco-Costa, I., & Poulin, R. (2017). Parasite life-cycle studies: A plea to resurrect an old parasitological tradition. Journal of Helminthology, 91(6), 647–656. 10.1017/S0022149X16000924 [DOI] [PubMed] [Google Scholar]
- Brabec, J., Rochat, E. C., Knudsen, R., Scholz, T., & Blasco-Costa, I. (2023). Mining various genomic resources to resolve old alpha-taxonomy questions: A test of the species hypothesis of the Proteocephalus longicollis species complex (Cestoda: Platyhelminthes) from salmonid fishes. International Journal for Parasitology, 53(4), 197–205. 10.1016/j.ijpara.2022.12.005 [DOI] [PubMed] [Google Scholar]
- Braga, M. P., Araujo, S. B. L., Agosta, S., Brooks, D., Hoberg, E., Nylin, S., Janz, N., & Boeger, W. A. (2018). Host use dynamics in a heterogeneous fitness landscape generates oscillations in host range and diversification. Evolution, 72(9), 1773–1783. 10.1111/evo.13557 [DOI] [PubMed] [Google Scholar]
- Brodersen, J., Post, D. M., & Seehausen, O. (2018). Upward adaptive radiation cascades: Predator diversification induced by prey diversification. Trends in Ecology & Evolution, 33(1), 59–70. 10.1016/j.tree.2017.09.016 [DOI] [PubMed] [Google Scholar]
- Brooks, D. R., Hoberg, E. P., & Boeger, W. A. (2019). The Stockholm Paradigm. University of Chicago Press. [Google Scholar]
- Brooks, D. R., & McLennan, D. A. (2012). The nature of diversity: An evolutionary voyage of discovery. University of Chicago Press. [Google Scholar]
- Chubb, J. C. (1982). Seasonal occurrence of helminths in freshwater fishes. Part IV. Adult Cestoda, Nematoda and Acanthocephala. Advances in Parasitology, 20, 1–292. 10.1016/s0065-308x(08)60539-4 [DOI] [PubMed] [Google Scholar]
- Clayton, D. H., Bush, S. E., & Johnson, K. P. (2015). Coevolution of life on hosts: Integrating ecology and history. University of Chicago Press. [Google Scholar]
- Crotti, M., Bean, C. W., Gowans, A. R. D., Winfield, I. J., Butowska, M., Wanzenböck, J., Bondarencko, G., Praebel, K., Adams, C. E., & Elmer, K. R. (2021). Complex and divergent histories gave rise to genome-wide divergence patterns amongst European whitefish (Coregonus lavaretus). Journal of Evolutionary Biology, 34(12), 1954–1969. 10.1111/jeb.13948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis, L. A. (2003). Tenure of individual larval trematode infections in an estuarine gastropod. Journal of the Marine Biological Association of the United Kingdom, 83(5), 1047–1051. 10.1017/s0025315403008257h [DOI] [Google Scholar]
- Danecek, P., Auton, A., Abecasis, G., Albers, C. A., Banks, E., DePristo, M. A., Handsaker, R. E., Lunter, G., Marth, G. T., Sherry, S. T., McVean, G., & Durbin, R.; 1000 Genomes Project Analysis Group. (2011). The variant call format and VCFtools. Bioinformatics, 27(15), 2156–2158. 10.1093/bioinformatics/btr330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vienne, D., Refrégier, G., López‐Villavicencio, M., Tellier, A., Hood, M., & Giraud, T. (2013). Cospeciation vs host‐shift speciation: Methods for testing, evidence from natural associations and relation to coevolution. New Phytologist, 198, 347–385. 10.1111/nph.12150 [DOI] [PubMed] [Google Scholar]
- De-Kayne, R., Selz, O. M., Marques, D. A., Frei, D., Seehausen, O., & Feulner, P. G. D. (2022). Genomic architecture of adaptive radiation and hybridization in Alpine whitefish. Nature Communications, 13(1), 4479. 10.1038/s41467-022-32181-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desdevises, Y., Morand, S., Jousson, O., & Legendre, P. (2002). Coevolution between Lamellodiscus (Monogenea: Diplectanidae) and Sparidae (Teleostei): The study of a complex host-parasite system. Evolution, 56(12), 2459–2471. 10.1111/j.0014-3820.2002.tb00171.x [DOI] [PubMed] [Google Scholar]
- Do, C., Waples, R. S., Peel, D., Macbeth, G. M., Tillett, B. J., & Ovenden, J. R. (2014). NeEstimator v2: Re-implementation of software for the estimation of contemporary effective population size (Ne) from genetic data. Molecular Ecology Resources, 14(1), 209–214. 10.1111/1755-0998.12157 [DOI] [PubMed] [Google Scholar]
- Drábková, M., Jachníková, N., Tyml, T., Sehadová, H., Ditrich, O., Myšková, E., Hypša, V., & Štefka, J. (2019). Population co-divergence in common cuttlefish (Sepia officinalis) and its dicyemid parasite in the Mediterranean Sea. Scientific Reports, 9(1), 14300. 10.1038/s41598-019-50555-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dray, S., Bauman, D., Blanchet, G., Borcard, D., Clappe, S., Guénard, G., Jombart, T., Larocque, G., Legendre, P., Madi, N. & Wagner, H. H. (2023). adespatial: Multivariate multiscale spatial analysis. R package version 0.3-20. https://CRAN.R-project.org/package=adespatial [Google Scholar]
- Earl, D. A., & vonHoldt, B. M. (2012). STRUCTURE HARVESTER: A website and program for visualizing STRUCTURE output and implementing the Evanno method. Conservation Genetics Resources, 4(2), 359–361. 10.1007/s12686-011-9548-7 [DOI] [Google Scholar]
- Eaton, D. A. R., & Overcast, I. (2020). ipyrad: Interactive assembly and analysis of RADseq datasets. Bioinformatics, 36(8), 2592–2594. 10.1093/bioinformatics/btz966 [DOI] [PubMed] [Google Scholar]
- Evanno, G., Regnaut, S., & Goudet, J. (2005). Detecting the number of clusters of individuals using the software structure: A simulation study. Molecular Ecology, 14(8), 2611–2620. 10.1111/j.1365-294X.2005.02553.x [DOI] [PubMed] [Google Scholar]
- Feder, J. L., & Forbes, A. A. (2010). Sequential speciation and the diversity of parasitic insects. Ecological Entomology, 35(s1), 67–76. 10.1111/j.1365-2311.2009.01144.x [DOI] [Google Scholar]
- Galbreath, K. E., & Hoberg, E. P. (2012). Return to Beringia: Parasites reveal cryptic biogeographic history of North American pikas. Proceedings Biological Sciences, 279(1727), 371–378. 10.1098/rspb.2011.0482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galbreath, K. E., & Hoberg, E. P. (2015). Host responses to cycles of climate change shape parasite diversity across North America’s Intermountain West. Folia Zoologica, 64(3), 218–232. 10.25225/fozo.v64.i3.a4.2015 [DOI] [Google Scholar]
- Galbreath, K. E., Makarikov, A. A., Bell, K. C., Greiman, S. E., Allen, J. M., Haas, G. M. S., Li, C., Cook, J. A., & Hoberg, E. P. (2023). Late Cenozoic history and the role of Beringia in assembling a Holarctic cestode species complex. Molecular Phylogenetics and Evolution, 183, 107775. 10.1016/j.ympev.2023.107775 [DOI] [PubMed] [Google Scholar]
- Gillespie, R. G., Bennett, G. M., De Meester, L., Feder, J. L., Fleischer, R. C., Harmon, L. J., Hendry, A. P., Knope, M. L., Mallet, J., Martin, C., Parent, C. E., Patton, A. H., Pfennig, K. S., Rubinoff, D., Schluter, D., Seehausen, O., Shaw, K. L., Stacy, E., Stervander, M., … Wogan, G. O. U. (2020). Comparing adaptive radiations across space, time, and taxa. The Journal of Heredity, 111(1), 1–20. 10.1093/jhered/esz064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gobbin, T. P., Tiemersma, R., Leone, G., Seehausen, O., & Maan, M. E. (2021). Patterns of ectoparasite infection in wild-caught and laboratory-bred cichlid fish, and their hybrids, implicate extrinsic rather than intrinsic causes of species differences in infection. Hydrobiologia, 848(16), 3817–3831. 10.1007/s10750-020-04423-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goudet, J. (2005). hierfstat, a package for r to compute and test hierarchical F-statistics. Molecular Ecology Notes, 5, 184–186. 10.1111/j.1471-8286.2004.00828.x [DOI] [Google Scholar]
- Haas, G. M. S., Hoberg, E. P., Cook, J. A., Henttonen, H., Makarikov, A. A., Gallagher, S. R., Dokuchaev, N. E., & Galbreath, K. E. (2020). Taxon pulse dynamics, episodic dispersal and host colonization across Beringia drive diversification of a Holarctic tapeworm assemblage. Journal of Biogeography, 47(11), 2457–2471. 10.1111/jbi.13949 [DOI] [Google Scholar]
- Hafner, M. S., Sudman, P. D., Villablanca, F. X., Spradling, T. A., Demastes, J. W., & Nadler, S. A. (1994). Disparate rates of molecular evolution in cospeciating hosts and parasites. Science, 265(5175), 1087–1090. 10.1126/science.8066445 [DOI] [PubMed] [Google Scholar]
- Hanzelová, V., & Scholz, T. (1999). Species of Proteocephalus Weinland, 1858 (Cestoda: Proteocephalidae), parasites of coregonid and salmonid fishes from North America: Taxonomic reappraisal. The Journal of Parasitology, 85(1), 94–101. 10.2307/3285708 [DOI] [PubMed] [Google Scholar]
- Ho, S. Y., Lanfear, R., Bromham, L., Phillips, M. J., Soubrier, J., Rodrigo, A. G., & Cooper, A. (2011). Time-dependent rates of molecular evolution. Molecular Ecology, 20(15), 3087–3101. 10.1111/j.1365-294X.2011.05178.x [DOI] [PubMed] [Google Scholar]
- Hoberg, E. P., & Brooks, D. R. (2008). A macroevolutionary mosaic: Episodic host-switching, geographical colonization and diversification in complex host–parasite systems. Journal of Biogeography, 35(9), 1533–1550. 10.1111/j.1365-2699.2008.01951.x [DOI] [Google Scholar]
- Hudson, A. G., Vonlanthen, P., Müller, R., Seehausen, O., Jankun, M., & Brzuzan, P. (2007). Review: The geography of speciation and adaptive radiation in coregonines. Advances in Limnology, 60, 111–146. [Google Scholar]
- Hudson, A. G., Vonlanthen, P., & Seehausen, O. (2011). Rapid parallel adaptive radiations from a single hybridogenic ancestral population. Proceedings Biological Sciences, 278(1702), 58–66. 10.1098/rspb.2010.0925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huyse, T., Poulin, R., & Theron, A. (2005). Speciation in parasites: A population genetics approach. Trends in Parasitology, 21(10), 469–475. 10.1016/j.pt.2005.08.009 [DOI] [PubMed] [Google Scholar]
- Huyse, T., & Volckaert, F. A. M. (2005). Comparing host and parasite phylogenies: Gyrodactylus flatworms jumping from goby to goby. Systematic Biology, 54(5), 710–718. 10.1080/10635150500221036 [DOI] [PubMed] [Google Scholar]
- Ingram, T., Hudson, A. G., Vonlanthen, P., & Seehausen, O. (2012). Does water depth or diet divergence predict progress towards ecological speciation in whitefish radiations? Evolutionary Ecology Research, 14, 487–502. [Google Scholar]
- Janz, N., & Nylin, S. (2008). The oscillation hypothesis of host-plant range and speciation. In Berkeley T. K. J. (Ed.), Specialization, speciation, and radiation: The evolutionary biology of herbivorous insects (pp. 203–215). University of California Press. [Google Scholar]
- Johnson, P. T., Wood, C. L., Joseph, M. B., Preston, D. L., Haas, S. E., & Springer, Y. P. (2016). Habitat heterogeneity drives the host-diversity-begets-parasite-diversity relationship: Evidence from experimental and field studies. Ecology Letters, 19(7), 752–761. 10.1111/ele.12609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jombart, T., & Ahmed, I. (2011). adegenet 1.3-1: New tools for the analysis of genome-wide SNP data. Bioinformatics, 27(21), 3070–3071. 10.1093/bioinformatics/btr521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamiya, T., O’Dwyer, K., Nakagawa, S., & Poulin, R. (2014). What determines species richness of parasitic organisms? A meta-analysis across animal, plant and fungal hosts. Biological Reviews of the Cambridge Philosophical Society, 89(1), 123–134. 10.1111/brv.12046 [DOI] [PubMed] [Google Scholar]
- Karvonen, A., Lundsgaard-Hansen, B., Jokela, J., & Seehausen, O. (2013). Differentiation in parasitism among ecotypes of whitefish segregating along depth gradients. Oikos, 122, 122–128. 10.1111/j.1600-0706.2012.20555.x [DOI] [Google Scholar]
- Karvonen, A., & Seehausen, O. (2012). The role of parasitism in adaptive radiations—when might parasites promote and when might they constrain ecological speciation? International Journal of Ecology, 2012, 1–20. 10.1155/2012/280169 [DOI] [Google Scholar]
- Kmentová, N., Koblmüller, S., Van Steenberge, M., Artois, T., Muterezi Bukinga, F., Mulimbwa N’sibula, T., Muzumani Risasi, D., Masilya Mulungula, P., Gelnar, M., & Vanhove, M. P. M. (2020). Failure to diverge in African Great Lakes: The case of Dolicirroplectanum lacustre gen. nov. comb. nov. (Monogenea, Diplectanidae) infecting latid hosts. Journal of Great Lakes Research, 46(5), 1113–1130. 10.1016/j.jglr.2019.09.022 [DOI] [Google Scholar]
- Kolmogorov, M., Yuan, J., Lin, Y., & Pevzner, P. A. (2019). Assembly of long, error-prone reads using repeat graphs. Nature Biotechnology, 37(5), 540–546. 10.1038/s41587-019-0072-8 [DOI] [PubMed] [Google Scholar]
- Larose, C., & Schwander, T. (2016). Nematode endoparasites do not codiversify with their stick insect hosts. Ecology and Evolution, 6(15), 5446–5458. 10.1002/ece3.2264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawton, J. H. (1983). Plant architecture and the diversity of phytophagous Insects. Annual Review of Entomology, 28(1), 23–39. 10.1146/annurev.en.28.010183.000323 [DOI] [Google Scholar]
- Li, H., & Durbin, R. (2009). Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics, 25(14), 1754–1760. 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H., Handsaker, B., Wysoker, A., Fennell, T., Ruan, J., Homer, N., Marth, G., Abecasis, G., & Durbin, R.; 1000 Genome Project Data Processing Subgroup. (2009). The sequence alignment/map format and SAM tools. Bioinformatics, 25(16), 2078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louhi, K. R., Karvonen, A., Rellstab, C., & Jokela, J. (2010). Is the population genetic structure of complex life cycle parasites determined by the geographic range of the most motile host? Infection, Genetics and Evolution, 10(8), 1271–1277. 10.1016/j.meegid.2010.08.013 [DOI] [PubMed] [Google Scholar]
- Lucek, K., Keller, I., Nolte, A. W., & Seehausen, O. (2018). Distinct colonization waves underlie the diversification of the freshwater sculpin (Cottus gobio) in the Central European Alpine region. Journal of Evolutionary Biology, 31(9), 1254–1267. 10.1111/jeb.13339 [DOI] [PubMed] [Google Scholar]
- Lundsgaard-Hansen, B., Matthews, B., Thierry, A., & Seehausen, O. (2017). The legacy of ecosystem effects caused by adaptive radiation. Copeia, 105, 550–557. 10.1643/ce-16-514 [DOI] [Google Scholar]
- Lutzoni, F., Nowak, M. D., Alfaro, M. E., Reeb, V., Miadlikowska, J., Krug, M., Arnold, A. E., Lewis, L. A., Swofford, D. L., Hibbett, D., Hilu, K., James, T. Y., Quandt, D., & Magallón, S. (2018). Contemporaneous radiations of fungi and plants linked to symbiosis. Nature Communications, 9(1), 5451. 10.1038/s41467-018-07849-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malcicka, M., Agosta, S. J., & Harvey, J. A. (2015). Multi level ecological fitting: Indirect life cycles are not a barrier to host switching and invasion. Global Change Biology, 21(9), 3210–3218. 10.1111/gcb.12928 [DOI] [PubMed] [Google Scholar]
- Mastretta-Yanes, A., Arrigo, N., Alvarez, N., Jorgensen, T. H., Piñero, D., & Emerson, B. C. (2015). Restriction site-associated DNA sequencing, genotyping error estimation and de novo assembly optimization for population genetic inference. Molecular Ecology Resources, 15(1), 28–41. 10.1111/1755-0998.12291 [DOI] [PubMed] [Google Scholar]
- Mazé-Guilmo, E., Blanchet, S., McCoy, K. D., & Loot, G. (2016). Host dispersal as the driver of parasite genetic structure: A paradigm lost? Ecology Letters, 19(3), 336–347. 10.1111/ele.12564 [DOI] [PubMed] [Google Scholar]
- Mendlová, M., Desdevises, Y., Civáňová, K., Pariselle, A., & Šimková, A. (2012). Monogeneans of West African cichlid fish: Evolution and cophylogenetic Interactions. PLoS One, 7(5), e37268. 10.1371/journal.pone.0037268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore, J. (1987). Asexual reproduction and environmental predictability in cestodes (Cyclophyllidea: Taeniidae). Evolution, 35(4), 723–741. 10.2307/2408243 [DOI] [PubMed] [Google Scholar]
- Nazarizadeh, M., Nováková, M., Loot, G., Gabagambi, N. P., Fatemizadeh, F., Osano, O., Presswell, B., Poulin, R., Vitál, Z., Scholz, T., Halajian, A., Trucchi, E., Kočová, P., & Štefka, J. (2023). Historical dispersal and host-switching formed the evolutionary history of a globally distributed multi-host parasite – The Ligula intestinalis species complex. Molecular Phylogenetics and Evolution, 180, 107677. 10.1016/j.ympev.2022.107677 [DOI] [PubMed] [Google Scholar]
- Nieberding, C., Libois, R., Douady, C. J., Morand, S., & Michaux, J. R. (2005). Phylogeography of a nematode (Heligmosomoides polygyrus) in the western Palearctic region: Persistence of northern cryptic populations during ice ages? Molecular Ecology, 14(3), 765–779. 10.1111/j.1365-294X.2005.02440.x [DOI] [PubMed] [Google Scholar]
- Nylin, S., Agosta, S., Bensch, S., Boeger, W. A., Braga, M. P., Brooks, D. R., Forister, M. L., Hambäck, P. A., Hoberg, E. P., Nyman, T., Schäpers, A., Stigall, A. L., Wheat, C. W., Österling, M., & Janz, N. (2018). Embracing colonizations: A new paradigm for species association dynamics. Trends in Ecology & Evolution, 33(1), 4–14. 10.1016/j.tree.2017.10.005 [DOI] [PubMed] [Google Scholar]
- Oksanen, J., Simpson, G., Blanchet, F., Kindt, R., Legendre, P., Minchin, P., O’Hara, R., Solymos, P., Stevens, M., Szoecs, E., Wagner, H., Barbour, M., Bedward, M., Bolker, B., Borcard, D., Carvalho, G., Chirico, M, De Caceres, M., Durand, S., Evangelista, H., FitzJohn, R., Friendly, M., Furneaux, B., Hannigan, G., Hill, M., Lahti, L., McGlinn, D., Ouellette, M., Ribeiro Cunha, E., Smith, T., Stier, A., Ter Braak, C. & Weedon, J. (2020). vegan: Community ecology package. R package version 2.5-7. [Google Scholar]
- Østbye, K., Amundsen, P. A., Bernatchez, L., Klemetsen, A., Knudsen, R., Kristoffersen, R., Naesje, T. F., & Hindar, K. (2006). Parallel evolution of ecomorphological traits in the European whitefish Coregonus lavaretus (L.) species complex during postglacial times. Molecular Ecology, 15(13), 3983–4001. 10.1111/j.1365-294X.2006.03062.x [DOI] [PubMed] [Google Scholar]
- Østbye, K., Bernatchez, L., Naesje, T., Himberg, K. J., & Hindar, K. (2005). Evolutionary history of the European whitefish Coregonus lavaretus (L.) species complex as inferred from mtDNA phylogeography and gill‐raker numbers. Molecular Ecology, 14, 4371–4387. 10.1111/j.1365-294X.2005.02737.x [DOI] [PubMed] [Google Scholar]
- Park, E., Jorge, F., & Poulin, R. (2020). Shared geographic histories and dispersal contribute to congruent phylogenies between amphipods and their microsporidian parasites at regional and global scales. Molecular Ecology, 29(17), 3330–3345. 10.1111/mec.15562 [DOI] [PubMed] [Google Scholar]
- Paterson, A. M., & Poulin, R. (1999). Have chondracanthid copepods co-speciated with their teleost hosts? Systematic Parasitology, 44(2), 79–85. 10.1023/a:1006255822947 [DOI] [PubMed] [Google Scholar]
- Paterson, S., Vogwill, T., Buckling, A., Benmayor, R., Spiers, A. J., Thomson, N. R., Quail, M., Smith, F., Walker, D., Libberton, B., Fenton, A., Hall, N., & Brockhurst, M. A. (2010). Antagonistic coevolution accelerates molecular evolution. Nature, 464(7286), 275–278. 10.1038/nature08798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrot-Minnot, M. -J., Špakulová, M., Wattier, R., Kotlík, P., Düşen, S., & Aydoğdu, A., et al. (2018). Contrasting phylogeography of two Western Palaearctic fish parasites despite similar life cycles. Journal of Biogeography, 45, 101–115. 10.1111/jbi.13118 [DOI] [Google Scholar]
- Peakall, R., & Smouse, P. E. (2012). GenAlEx 6.5: genetic analysis in excel. Population genetic software for teaching and research—an update. Bioinformatics, 28(19), 2537–2539. 10.1093/bioinformatics/bts460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson, B. K., Weber, J. N., Kay, E. H., Fisher, H. S., & Hoekstra, H. E. (2012). Double digest RADseq: An inexpensive method for de novo SNP discovery and genotyping in model and non-model species. PLoS One, 7(5), e37135. 10.1371/journal.pone.0037135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips, B. L., Kelehear, C., Pizzatto, L., Brown, G. P., Barton, D., & Shine, R. (2010). Parasites and pathogens lag behind their host during periods of host range advance. Ecology, 91(3), 872–881. 10.1890/09-0530.1 [DOI] [PubMed] [Google Scholar]
- Poulin, R. (2011). Evolutionary ecology of parasites. Princeton University Press. [Google Scholar]
- Poulin, R., & Morand, S. (2004). Parasite biodiversity. Smithsonian Institution Press. [Google Scholar]
- Praebel, K., Knudsen, R., Siwertsson, A., Karhunen, M., Kahilainen, K. K., Ovaskainen, O., Østbye, K., Peruzzi, S., Fevolden, S.-E. & Amundsen, P.-A. (2013). Ecological speciation in postglacial European whitefish: Rapid adaptive radiations into the littoral, pelagic, and profundal lake habitats. Ecology and Evolution, 3, 4970–4986. 10.1002/ece3.867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prugnolle, F., Théron, A., Pointier, J. P., Jabbour-Zahab, R., Jarne, P., Durand, P., & de Meeûs, T. (2005). Dispersal in a parasitic worm and its two hosts: Consequence for local adaptation. Evolution, 59(2), 296–303. 10.1111/j.0014-3820.2005.tb00990.x [DOI] [PubMed] [Google Scholar]
- Rahmouni, C., Vanhove, M. P. M., Koblmüller, S., & Šimková, A. (2022). Molecular phylogeny and speciation patterns in host-specific monogeneans (Cichlidogyrus, Dactylogyridae) parasitizing cichlid fishes (Cichliformes, Cichlidae) in Lake Tanganyika. International Journal for Parasitology, 52(6), 359–375. 10.1016/j.ijpara.2021.12.004 [DOI] [PubMed] [Google Scholar]
- Ricklefs, R. E., Outlaw, D. C., Svensson-Coelho, M., Medeiros, M. C. I., Ellis, V. A., & Latta, S. (2014). Species formation by host shifting in avian malaria parasites. Proceedings of the National Academy of Sciences, 111(41), 14816–14821. 10.1073/pnas.1416356111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rougeux, C., Gagnaire, P. A., & Bernatchez, L. (2019). Model‐based demographic inference of introgression history in European whitefish species pairs’. Journal of Evolutionary Biology, 32(8), 806–817. 10.1111/jeb.13482 [DOI] [PubMed] [Google Scholar]
- Sánchez-Hernández, J., Finstad, A. G., Arnekleiv, J. V., Kjærstad, G., & Amundsen, P. -A. (2021). Beyond ecological opportunity: Prey diversity rather than abundance shapes predator niche variation. Freshwater Biology, 66, 44–61. 10.1111/fwb.13606 [DOI] [Google Scholar]
- Schluter, D. (2000). The ecology of adaptive radiation. Oxford University Press. [Google Scholar]
- Scholz, T. (1999). Life cycles of species of Proteocephalus, parasites of fishes in the Palearctic Region: A review. Journal of Helminthology, 73(1), 1–19. 10.1017/S0022149X99000013 [DOI] [PubMed] [Google Scholar]
- Scholz, T., & Hanzelová, V. (1998). Tapeworms of the genus Proteocephalus Weinland, 1858 (Cestoda: Proteocephalidae), parasites of fishes in Europe. (Vol. 2/98). Academia. [Google Scholar]
- Selz, O. M., Dönz, C. J., Vonlanthen, P., & Seehausen, O. (2020). A taxonomic revision of the whitefish of lakes Brienz and Thun, Switzerland, with descriptions of four new species (Teleostei, Coregonidae). ZooKeys, 989, 79–162. 10.3897/zookeys.989.32822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simão, F. A., Waterhouse, R. M., Ioannidis, P., Kriventseva, E. V., & Zdobnov, E. M. (2015). BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics, 31(19), 3210–3212. 10.1093/bioinformatics/btv351 [DOI] [PubMed] [Google Scholar]
- Siwertsson, A., Knudsen, R., Praebel, K., Adams, C. E., Newton, J., & Amundsen, P. -A. (2013). Discrete foraging niches promote ecological, phenotypic, and genetic divergence in sympatric whitefish (Coregonus lavaretus). Evolutionary Ecology, 27, 547–564. 10.1007/s10682-012-9607-x [DOI] [Google Scholar]
- R Core Team. (2021). R: A language and environment for statistical computing. [Google Scholar]
- Thompson, J. N. (2005). The geographic mosaic of coevolution. The University of Chicago Press. [Google Scholar]
- Van der Auwera, G. A., Carneiro, M. O., Hartl, C., Poplin, R., del Angel, G., Levy-Moonshine, A., Jordan, T., Shakir, K., Roazen, D., Thibault, J., Banks, E., Garimella, K. V., Altshuler, D., Gabriel, S., & DePristo, M. A. (2013). From FastQ data to high-confidence variant calls: The genome analysis toolkit best practices pipeline. Current Protocols in Bioinformatics, 43, 11.10.11–11.10.33. 10.1002/0471250953.bi1110s43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Auwera, G. A. & O’Connor, B. D. (2020). Genomics in the cloud: using docker, GATK, and WDL in Terra. O’Reilly Media. [Google Scholar]
- Vanhove, M. P. M., Pariselle, A., Van Steenberge, M., Raeymaekers, J. A. M., Hablützel, P. I., Gillardin, C., Hellemans, B., Breman, F. C., Koblmüller, S., Sturmbauer, C., Snoeks, J., Volckaert, F. A. M., & Huyse, T. (2015). Hidden biodiversity in an ancient lake: Phylogenetic congruence between Lake Tanganyika tropheine cichlids and their monogenean flatworm parasites. Scientific Reports, 5(1), 13669. 10.1038/srep13669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vonlanthen, P., Bittner, D., Hudson, A. G., Young, K. A., Mueller, R., Lundsgaard-Hansen, B., Roy, D., Di Piazza, S., Largiader, C. R., & Seehausen, O. (2012). Eutrophication causes speciation reversal in whitefish adaptive radiations. Nature, 482, 357–362. 10.1038/nature10824 [DOI] [PubMed] [Google Scholar]
- Walsh, M. R., DeLong, J. P., Hanley, T. C., & Post, D. M. (2012). A cascade of evolutionary change alters consumer-resource dynamics and ecosystem function. Proceedings Biological Sciences, 279(1741), 3184–3192. 10.1098/rspb.2012.0496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waples, R. S., & Do, C. (2010). Linkage disequilibrium estimates of contemporary N e using highly variable genetic markers: A largely untapped resource for applied conservation and evolution. Evolutionary Applications, 3(3), 244–262. 10.1111/j.1752-4571.2009.00104.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren, R. L., Coombe, L., Mohamadi, H., Zhang, J., Jaquish, B., Isabel, N., Jones, S. J. M., Bousquet, J., Bohlmann, J., & Birol, I. (2019). ntEdit: Scalable genome sequence polishing. Bioinformatics, 35(21), 4430–4432. 10.1093/bioinformatics/btz400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir, B. S., & Cockerham, C. C. (1984). Estimating F-statistics for the analysis of population structure. Evolution, 38(6), 1358–1370. 10.1111/j.1558-5646.1984.tb05657.x. https://www.jstor.org/stable/2408641 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All collapsed and paired-end sequence data for samples sequenced in this study are available in compressed fastq format through NCBI’s BioProject no. PRJNA910576 (ddRAD accessions SAMN32132446–959, reference genome accession SAMN32134074), together with rescaled and trimmed bam sequence alignments against both the nuclear and mitochondrial reference genome. All scripts used to perform the analyses presented in this paper are available through https://github.com/JeremyLGauthier/Scripts_Proteocephalus.