Abstract
Objectives
Development of search queries for systematic reviews (SRs) is time-consuming. In this work, we capitalize on recent advances in large language models (LLMs) and a relatively large dataset of natural language descriptions of reviews and corresponding Boolean searches to generate Boolean search queries from SR titles and key questions.
Materials and Methods
We curated a training dataset of 10 346 SR search queries registered in PROSPERO. We used this dataset to fine-tune a set of models to generate search queries based on Mistral-Instruct-7b. We evaluated the models quantitatively using an evaluation dataset of 57 SRs and qualitatively through semi-structured interviews with 8 experienced medical librarians.
Results
The model-generated search queries had median sensitivity of 85% (interquartile range [IQR] 40%-100%) and number needed to read of 1206 citations (IQR 205-5810). The interviews suggested that the models lack both the necessary sensitivity and precision to be used without scrutiny but could be useful for topic scoping or as initial queries to be refined.
Discussion
Future research should focus on improving the dataset with more high-quality search queries, assessing whether fine-tuning the model on other fields, such as the population and intervention, improves performance, and exploring the addition of interactivity to the interface.
Conclusions
The datasets developed for this project can be used to train and evaluate LLMs that map review descriptions to Boolean search queries. The models cannot replace thoughtful search query design but may be useful in providing suggestions for key words and the framework for the query.
Keywords: systematic reviews as topic/methods, artificial intelligence
Background
Literature identification is a vital step in the systematic review process. Traditionally, the review’s population and interventions (or exposures) are translated into a search query by identifying synonyms and controlled vocabulary terms and combining them with the appropriate Boolean operators (OR, AND, or NOT). Then, these queries are executed against a database (eg, PubMed), returning a (typically large) list of candidate citations. Finally, the citations are screened for relevance. Citations deemed relevant during abstract screening are screened in full text, and a final corpus of studies for inclusion in the review is defined.
Many tools have been developed over the last decade that use natural language processing (NLP) to semi-automate search query development and/or abstract screening. Traditional abstract-screening tools that use text mining and machine learning to assist in the screening of records identified from the search query have been shown to rank and classify records accurately, with adequate training.1–4 The CLEF eHealth Lab Series, specifically the Technology Assisted Reviews in Empirical Medicine track,5 has encouraged research into screening prioritization and replacing Boolean search strategies. Studies that leverage deep learning and language models to (semi-) automate database searching have shown improved precision but unacceptable sensitivity.6–9 In addition, these tools suffer from a lack of transparency and replicability, both of which are required by the Cochrane Handbook guidance10 to ensure a high-quality systematic review. Thus, there is a need for tools that produce queries that can be easily evaluated and edited, and can be reported using the standard reporting format, allowing for peer review and improved reproducibility.12,13
There is less evidence for the role of machine learning or artificial intelligence in search query formulation. Most available tools employ text mining to automatically generate terms for query expansion, identify related articles through citation analyses, or translate queries for use in different databases. A recent evaluation of 21 available tools for query development in the Systematic Review Toolbox11 (an online catalogue of tools that support various tasks within the systematic review process) concluded that, although useful tools are available for most steps of the process, all currently available tools struggle with long or complex queries.12 Wang et al13 developed a zero-shot (in that the models were not specifically trained for the task) approach using ChatGPT-3.5, producing very high precision but (unacceptably) low sensitivity results. Chelli et al reported similarly poor sensitivity rates for queries generated by GPT-3.5 (11.9%), GPT-4 (13.7%), and Bard (0%). In addition, they reported hallucination rates (generation of inaccurate or nonsensical text) of 39.6% for GPT-3.5, 28.6% for GPT-4, and 91.4% for Bard.8 There is a clear need to improve the performance of such models before they can be incorporated into the systematic review process. We hypothesize that a model fine-tuned on actual search queries will perform better than zero-shot large language models (LLMs), such as ChatGPT and Bard.
To fine-tune models requires data that incorporate textual information about the review, such as the title and research question(s) it aims to answer, as well as correctly formatted Boolean search queries. Existing data collections, including the CLEF Technology Assisted Review collection,14,15 2 collections from Scells et al16,17 and a collection of systematic review updates,18 are small, including only 25-94 reviews, and are not sufficiently large for fine-tuning a language model to generate queries. All but one of these datasets17 comprise searches in Ovid format only, which limits their utility for fine-tuning a language model for the more widely accessible PubMed interface. The Wang et al dataset17 was deemed appropriate to be incorporated into our evaluation data.
In this work, we capitalize on recent advances in NLP by first developing both training and evaluation datasets, then fine-tuning an LLM to create systematic review search queries that are formatted like the search queries systematic reviewers are accustomed to reading. We then evaluate the models empirically on the evaluation reviews and qualitatively through interviews with experienced medical librarians.
Materials and methods
We assembled 2 datasets of systematic review topics and search queries: 1 for training, representing 10 346 reviews, and 1 for evaluation, representing 57 reviews. From the training set, we separated out 99 of the reviews, which we assessed as of relatively high quality, to use as a validation dataset. We then fine-tuned models on the (reduced) training dataset and, after selecting hyperparameters settings using the validation dataset, evaluated them using the evaluation dataset. Finally, we interviewed 8 medical librarians using semi-structured interviews to establish how to improve the models. Full details of dataset cleaning and evaluation, model training, and qualitative evaluation are given in the supplementary material.
We assembled the training dataset from PROSPERO, an international prospective register of systematic review protocols maintained by the Centre for Reviews and Dissemination at the University of York and funded by the UK National Institute for Health Research.19,20 The PROSPERO team shared the data for the 139 450 records with data in the “search” field, from inception (February 2011) to December 24, 2021. Much of the compilation and annotation of this dataset was done manually. Briefly, we created a cleaned search query by removing methodological filters, translating the query to PubMed syntax if necessary, and fixing any obvious errors. We created 2 separate versions of the dataset: in one, the field tags (such as medical subject heading [MeSH] tags) were standardized; in the other, we removed all field tags, quotation marks, backslashes, and commas. PubMed’s interface automatically matches untagged words to their phrase index and to MeSH terms; thus, it is unlikely that this change would reduce the sensitivity of model-generated queries. However, using untagged words impacts precision, increasing the number of citations that need to be read (NNR) to find one relevant study.
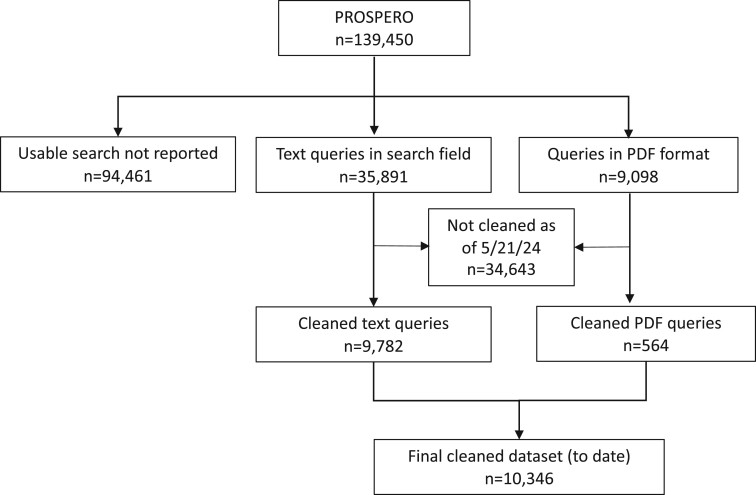
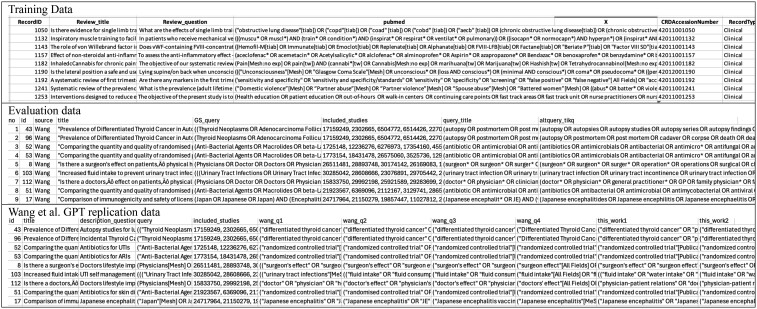
We needed a reasonable number of relatively high-quality queries to use as a validation set for model development and hyperparameter experiments. Thus, we created a subset of 99 search queries, including a random selection of 10 queries from PDFs (which tended to be high-quality queries) and 89 of the better queries from those entered directly in the text field. The flow of the cleaning process to date is shown in Figure 1. Figure 2 (top row) shows the structure of the cleaned training/validation data.
Figure 1.
Flow diagram for PROSPERO queries to date. Each box represents a stage in the data cleaning process. From 139 450 records, 44 989 had usable information in the search fields. Of those, 10 346 have been cleaned.
Figure 2.
Data. The top row shows the structure of the data used to train/validate the models. It includes the title and key question in free text along with a manually curated PubMed search query. The middle row shows the structure of the data used to evaluate the models. It shows the title source (Wang/Adam), the free-text title, the query used in the original review (GS_query), the PMIDs of the included studies, and the 2 model-generated queries, using title only and using title + key questions). The bottom row shows the structure of the data used to replicate Wang et al’s GPT analysis. It shows the free-text title and key questions, the query used in the original review (query), the PMIDs of the included studies, and the model-generated queries, for each prompt.
The training queries are highly skewed with respect to the number of words they contain (median 49; IQR 30-86; range 1-2755), with fewer than 1% containing more than 400 words. This pattern also holds for the number of Boolean operators, which is a proxy for the number of concepts in each search (median 8, IQR 5-8, range 1-276). The total number of records identified by each query is also highly skewed (median 1237; IQR 208-86 682; range 0-23 180 504), with 75% of queries returning fewer than 7000 citations; 597 queries return no records, which suggests that they contain a fatal error. To get a better sense of how much the removal of filters is increasing the size of the returned citation set, as an example, we crossed all searches with the PubMed randomized controlled trials (RCTs) filter. The median number of RCTs retrieved is 57 (IQR 7-368, range 0-609 069).
To develop the evaluation dataset we combined 2 separate subsets: the first contained 38 of the 40 citations in Wang’s seed citation dataset17 (which we will refer to as the “Wang dataset”) used in Wang et al’s investigation of whether ChatGPT® could be used to formulate Boolean search queries.13 This dataset includes the titles, PubMed formatted search queries, and final included studies for 40 Cochrane systematic reviews on a variety of topics. We removed 2 records from this set that retrieved either only one or no citations in PubMed.
To this, we added a second set of 19 reviews known to us (the “Adam dataset”), 18 of which were designed for systematic reviews conducted in the Agency for Healthcare Research and Quality’s (AHRQ) Evidence-based Practice Center (EPC) program21–37 or in the Department of Veterans Affairs (VA).38 These searches were all peer reviewed by another systematic review librarian using the Peer Review Electronic Search Strategies (PRESS) checklist.39 The final record in this set was for a Cochrane review that we were aware of; it is unclear whether this was peer reviewed.40 We did not systematically search for additional reviews. For comparison with model-generated queries, we removed the tags, quotes, and commas from the data. Thus, precision/NNR results are for comparison only and do not represent the actual workload of screening.
The 57 reviews in the evaluation set (38 in the Wang dataset and 19 in the Adam dataset) addressed a wide variety of biomedical topics, with query syntax ranging from simple to complex. The number of studies that were included in the systematic reviews ranged from 4 to 612, with a median of 30. The median sensitivity of the human generated searches was 100% (IQR 88%-100%), with a median NNR of 580 (IQR 161-1466); the median NNR with field tags and punctuation in the original data was 125 (IQR 57-456).
We initially fine-tuned models on both the tagged and stripped datasets, but the dataset stripped of tags performed significantly better in preliminary experiments based on manual evaluation of a subset of the produced strategies in comparison with the initial strategies. Therefore, we proceeded using only this version of the training data. We marked each piece of the training data as belonging to the middle 50% (the middle quartiles) and retained only the data in both middle quartiles, leaving 4324 instances for training. We explored fine-tuning a number of models, including variants of Flan-T541 and BioBART,42 but we found the best performance within our computing resources with a Mistral-Instruct 7 billion parameter chat/instruct LLM.43 All fine-tuning experiments were conducted within the Huggingface library (v4.37.1).44
We used the following instruction to the LLM, which we added to all training samples (ie, each review): “Translate the following into a Boolean search query to find relevant studies in PubMed. Do not add any explanation. Do not repeat terms. Prefer shorter queries.” In separate training rounds, instruction was followed by either the review’s title or its title and objectives/key questions. Due to model size and resource limitations, we fine-tuned this model using Parameter Efficient Fine-Tuning45 specifically with Low Rank Adaptation (LoRA) methods.46 During fine-tuning, we used the adafactor optimizer47 and ran and evaluated a grid search across learning rates (1e−4, 1e−5, 1e−6) and LoRA parameters (rank factor r: 16, 32, 64, 128, 256; scaling factor alpha: 16, r). For each hyperparameter combination, we trained for 10 epochs with a batch size of one (owing to computational constraints), using floating point 16 precision. We used PubMed query results from the validation set to select the best model (for the “title-only” model), choosing the model with the highest sensitivity between the validation set reference query and the model-generated query (0.54). For the “title and key questions” model, we used the hyperparameters established in the best title-only model.
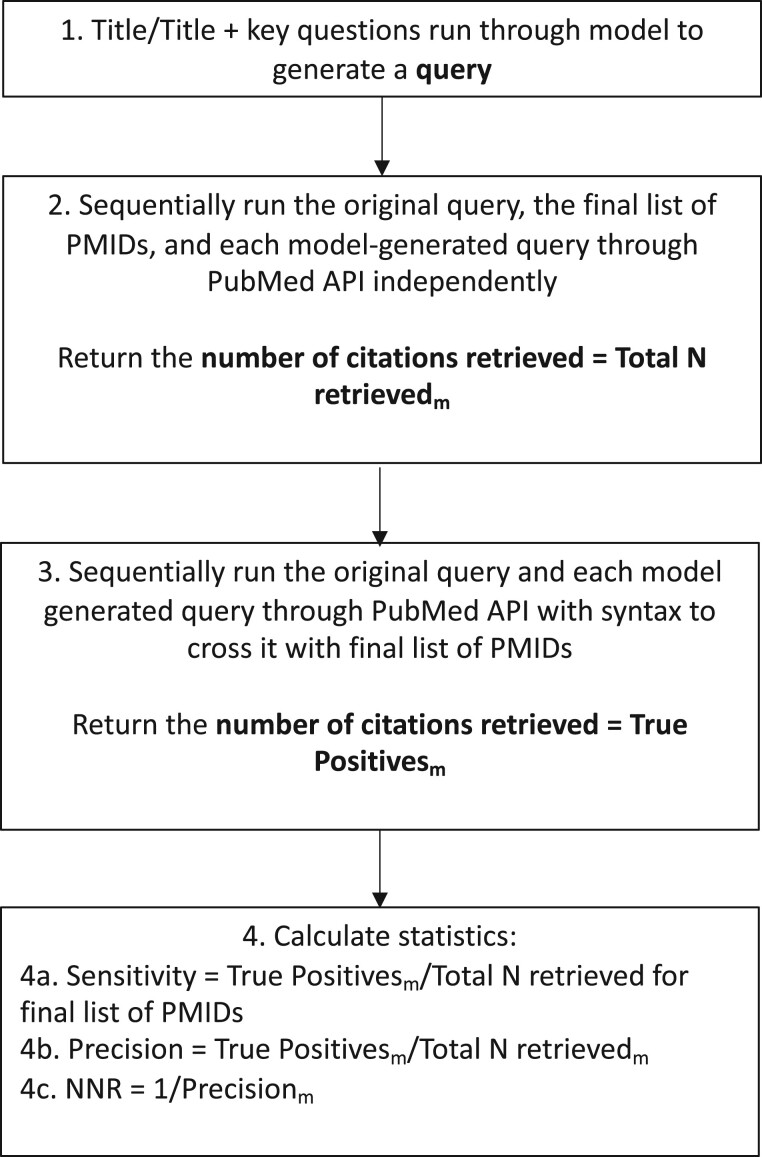
The final models were empirically tested on the evaluation data set. Two search queries were generated for each review in this set, one for each model (title-only and title plus key questions). Figure 2, middle row, shows the structure of the data with the generated queries. These queries were run in PubMed and compared with the reference PMIDs (those included in the corresponding systematic review’s final report) to calculate sensitivity (or recall; the number of included citations correctly identified), precision, and NNR (the number of citations that must be read to identify one relevant citation). Figure 3 illustrates the evaluation process.
Figure 3.
Evaluation flow diagram. Each box represents a stage in the evaluation process. The m subscript indicates that the number belongs to a specific query. Abbreviations: N = total number; PMID = PubMed identifier; NNR = number needed to read.
Further analyses explored whether model performance differed by evaluation review source (Wang or Adam). The search description in the Wang dataset comprises only a few words, while the key questions in the Adam dataset comprise a full description of the questions the review seeks to answer. We also evaluated whether any queries had syntactical errors that lead to errors in PubMed. Finally, we evaluated whether any queries “failed,” defined as retrieving none of the review’s final included PMIDs. Exceedingly high retrieval could also be considered failing. However, in this case, the design of the study led to artificially high NNR, so that was not considered as a failure of the query.
Finally, we re-ran Wang et al’s13 analysis for the first 4 of their original prompts, using GPT-4 to see if the performance had improved. Figure 2, bottom row, shows the structure of the data with the generated queries. Full details of this analysis are in the Supplementary Material.
The models were also qualitatively evaluated through a set of virtual semi-structured interviews with experienced medical librarians who tested the models through a web-based interface,48 hereafter referred to as the query tool. A single researcher (G.P.A.) conducted separate interviews with 8 medical librarians: 4 systematic review librarians affiliated with AHRQ-funded EPCs, 3 university-affiliated academic librarians, and 1 hospital-affiliated librarian. The librarians were asked to share their screens and try out the query tool on a topic for which they had a previously developed a search query. The tool generated 2 queries, one for each model, and the librarians were asked to evaluate one or both generated queries in the PubMed interface and compare them with their manually developed query. As they were doing so, the interviewer encouraged each participant to reflect on their experiences and asked a series of questions in a semi-structured interview format, guided by a topic guide based on the 2015 PRESS Checklist Reviewer Form39 for search strategy peer review.
In analysis, we used a modified version of the framework analysis described by Gale et al.49 To ensure participant anonymity, each librarian was assigned a participant number. The interviewer transcribed all interviews verbatim using the Zoom® automatic transcription tool. Using the qualitative analysis software NVivo (version 1.7.1), the interviewer coded each cleaned transcript, associating each block of text to a set of descriptive or conceptual labels, thus classifying the data and allowing for the collocation of the pieces of text across multiple interviews that addressed the same concepts. Finally, all codes were pulled together, and each was summarized in a few words. This allowed the interviewer to identify a set of 5 themes.
Results
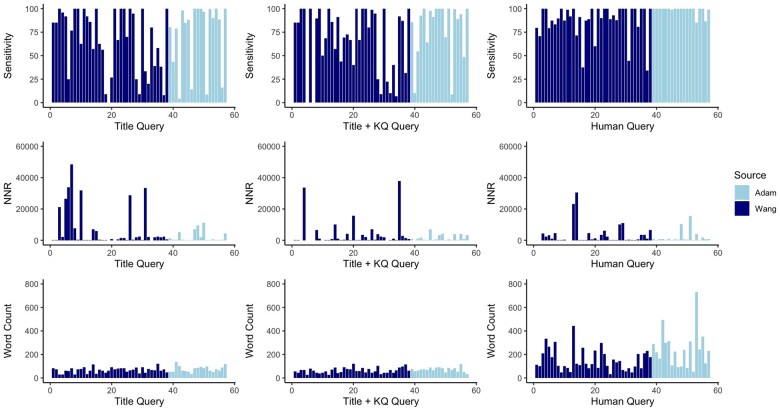
The empirical performance of the queries generated by the title-only and title plus key questions models were similar but varied widely across the reviews in the evaluation set. For 3 reviews, at least one model “failed,” in that it identified none of the relevant citations, while for 20 of the 57 reviews at least one model identified all the relevant citations. Figure 4 shows the sensitivity, NNR, and word count for the queries generated by each model and the human query for each review.
Figure 4.
Performance of models on each review. Each bar represents 1 of the 57 evaluation reviews. The darker lines represent the Wang reviews and the lighter lines represent the Adam reviews. The top row shows sensitivity across search query source, the middle row NNR, and the bottom row word count. Abbreviations: KQ = key question; NNR = number needed to read.
No query had a syntactical error that kept it from running in PubMed or retrieved 0 records. This is not surprising; with the MeSH tags, quotes, commas, and so on stripped, there are few places for the model to introduce syntactical errors, except in the Boolean operators and parentheses. NNRs varied widely, with 7 queries in the title only, 3 in the title and key question model, and 2 in the human having NNR over 2000. All model-generated queries contained fewer words than the human queries. This is likely the result of the instruction provided to the model to prefer shorter queries and training data selection. Table 1 shows the summarized results for each model on the evaluation set, both in total and for each of the 2 evaluation datasets. The model-generated queries performed better on the Adam dataset (which was largely derived from systematic reviews for the AHRQ EPC program) than on the Wang dataset (which was mostly based on Cochrane reviews). In general, topics in EPC reviews tend to be broader and more complex than those in Cochrane reviews. However, it may be that the Wang topics were more challenging, given that the sensitivity of the human generated queries for this dataset was also lower. The precision of the human-generated queries was also worse for the Wang data, although the precision of the model-generated queries in the Wang data was slightly better.
Table 1.
Summarized results for each model on the evaluation set.
| Sensitivity, % | Precision, % | NNR, N | Word count, N | Failed queries, N (%)b | |
|---|---|---|---|---|---|
| Overall | |||||
| Human querya | 100 (88-100) | 0.17 (0.03-0.62) | 580 (161-3466) | 144 (96-240) | 0 (0%) |
| Mistral-Instruct-7b trained on title and key questions query | 86 (51-100) | 0.11 (0.03-0.58) | 908 (171-3906) | 66 (49-83) | 2 (3%) |
| Mistral-Instruct-7b trained on title only query | 85 (40-100) | 0.08 (0.02-0.49) | 1206 (205-5810) | 71 (55-83) | 1 (2%) |
| Wang data | |||||
| Human querya | 91.29 (84.29-100) | 0.19 (0.03-0.74) | 526 (135-3553) | 120 (84-208) | 0 (0%) |
| Mistral-Instruct-7b trained on title and key questions query | 69.23 (40.94-99.74) | 0.07 (0.02-0.65) | 1477 (155-4689) | 65 (44-74) | 2 (3%) |
| Mistral-Instruct-7b trained on title only query | 58.33 (38.34-100) | 0.06 (0.01-0.87) | 1653 (115-7031) | 67 (52-76) | 1 (2%) |
| Adam data | |||||
| Human querya | 100 (100-100) | 0.17 (0.12-0.35) | 590 (282-836) | 230 (116-304) | 0 (0%) |
| Mistral-Instruct-7b trained on title and key questions query | 91.83 (68.09-100) | 0.15 (0.03-0.53) | 661 (188-3226) | 70 (57-84) | 0 (0%) |
| Mistral-Instruct-7b trained on title only query | 88.16 (69.93-99.75) | 0.30 (0.02-0.44) | 331 (227-4588) | 70 (56-90) | 0 (0%) |
All measures median (IQR), except where explicitly noted.
Abbreviations: N = number or count; NNR = number needed to read.
Human queries were the PubMed queries reported in the evaluation dataset that had been developed by the original review team. Note these numbers are the same as the overall numbers in Table 1.
Failed queries retrieved none of the review’s included PubMed identifiers (PMIDs), Word count = number of words in the search queries.
The results of our re-analysis of Wang’s zero-shot study13 in GPT-4 are presented in Table 2. The Wang’s prompts, when run in GPT-4, had worse sensitivity than the GPT-3.5 results reported in their paper. Some prompt engineering produced prompts in GPT-4 with similar sensitivity to those reported in the paper, but nothing close to sufficient for practical use.
Table 2.
Summarized results for each model on the full evaluation set.
| Sensitivity, % | Precision, % | NNR, N | |
|---|---|---|---|
| Wang prompts 1-4, GPT-4 | |||
| Prompt 1 | 0 (0-5.55) | 1.61 (0.50-3.70) | 62 (27-202) |
| Prompt 2 | 0 (0-9.11) | 3.85 (0.50-1.00) | 26 (1-201) |
| Prompt 3 | 0 (0-2.55) | 4.76 (0.61-33.3) | 21 (3-163) |
| Prompt 4 | 0 (0-2.0) | 1.15 (0.27-3.70) | 87 (27-368) |
| Updated prompts, GPT-4 | |||
| New prompt 1 | 12.5 (0-45.4) | 1.61 (0.66-4.35) | 62 (23-151) |
| New prompt 2 GPT-4 | 5.9 (0-27.6) | 3.45 (0.99-8.33) | 29 (12-101) |
| Fine-tuned model | |||
| Mistral-Instruct-7b trained on title and key questions | 86 (51-100) | 0.11 (0.03-0.58) | 908 (171-3906) |
| Mistral-Instruct-7b trained on title only | 85 (40-100) | 0.08 (0.02-0.49) | 1206 (205-5810) |
Results are median (worst performance-best performance).
Abbreviations: N = number or count; NNR = number needed to read.
In the semi-structured interviews, each librarian tested between 1 and 3 topics. The search topics were all health-related but varied widely, including reproductive health, violence, cancer, mental health, pain management, acupuncture, child development, irritable bowel syndrome, cognitive functioning, and medical cannabis. Because the sample of each librarian type was small (1 to 4 librarians), it was not possible to attribute themes to specific librarian types. Thus, the 5 themes shown in Table 3 and described in detail in the Supplementary Material reflect the full set of 8 librarians.
Table 3.
Themes as derived from the coded text.
| Theme | Definition | Example quotations |
|---|---|---|
| Theme 1: Sensitivity | Is the tool-developed query finding the relevant studies? | “I can’t say … that I found a single real article in [the missed citations] that focused on glioblastoma [the search topic]. So think that’s pretty darn good.” [Participant 1] |
| Theme 2: Size of the returned citation set | Is the tool-developed query finding too many or too few citations? | “it’s also retrieving way, way, way, way too many citations.” [Participant 7] |
|
Theme 3: PRESS checklist concepts 3a. Query structure 3b. MeSH Term matching 3c. Key words |
How does the query tool perform in relation to the quality measures included in the most common search query peer review framework? |
3a. “It does a half-decent job with…mapping the question to a basic search, at least identifying what are the different chunks” [Participant 7] 3b. “It’s … overmatching to MeSH terms … [which is] kind of a known issue in PubMed [machine learning models for MeSH term matching].” [Participant 2] 3c. “I really like that. It found all the different … styles of guns. When I first started developing my own search strategy, … it didn't immediately come to me to think of pistols or revolvers or rifles… So that’s pretty helpful.” [Participant 5] |
| Theme 4: Usefulness to searchers | What are the ways in which the query tool is useful to librarians and how do they think it might be useful (or dangerous) for non-expert searchers? | “I guess there’s two different kinds of people, someone who can look at this, and only use it exactly as presented, which isn’t bad. It looks like it covers [this] topic pretty well. … But then there are other people who can look at it and spend a little bit more time, and say, ‘let me take from this what’s useful,’ and I think it would be really helpful in that instance.” [Participant 1] |
|
Theme 5: Future directions 5a: Better training searches 5b: Split by PICO element 5c: Interactivity |
What are some suggestions on how to improve the query tool to make it more useful? |
5a. “[You could use] a search hedge, search filter … but I’m also thinking about your organizations that produce clinical practice guidelines. … That would be a good way to get some really high-quality searches on specific topics.” [Participant 3] 5b. “Break it into sort of two or three basic things … [because] it’s not doing some of the more advanced techniques like nesting…. Let’s say you did break it out. And you had 4 boxes show up based on your PICO. Then would there be another section where it put them all together so that someone could copy and paste it into PubMed.” [Participant 6] 5c. “If there is a way to have it [have] almost a brief interactive reference interview of sorts … and then is able to pull the key concepts out of that” [Participant 6] |
Abbreviations: MeSH = medical subject headings; PICO = population, intervention, comparator, outcome; PRESS = peer review of electronic search strategies.
There were 3 takeaways from the 5 themes: First, the queries developed are sensitive enough to be a good starting place in the search development process for systematic reviews. In general, the queries correctly identified the relevant concepts and produced initial keywords. The comparably large number of citations they return is problematic but may be ameliorated by re-introducing quotation marks, standard vocabulary (MeSH terms), field terms, and filters. Three of the librarians tested this and found that the numbers were reduced, but we did not assess it formally. Additionally, ranking algorithms, such as PubMed’s Best Match algorithm,50 may be useful in prioritizing relevant results. Second, the tool-generated queries are not an end point in and of themselves. They lack both the necessary sensitivity and precision to be used without scrutiny. Each librarian stated that it would be very important to educate users, particularly non-librarian users, about this limitation. It is, however, possible that it could be used as a teaching tool or as a starting place for non-expert searchers. Finally, the librarians made useful suggestions for further development of this technology. These included: training the models on better data, allowing for modular search by concept (for example, population and interventions), adding interactivity to better mimic the reference interview, in which librarians elicit details about the information need, and algorithmic post-processing to assist in the process of re-applying quotation marks, standard vocabulary (MeSH terms), field terms, and filters.
Discussion
The datasets we have compiled should be useful for exploring and testing automation methods for query generation, and perhaps other aspects of search design. To our knowledge, the PROSPERO dataset we generated is the largest dataset of systematic review metadata (titles, objectives, and PICO elements) and corresponding properly formatted search queries. The evaluation data includes basic metadata (title and search description or key questions), the original search query in PubMed format, and the finally included citations for a wide variety of reviews.
The model-generated queries yielded moderate sensitivity but are not ready for unaudited use in searches where the goal is to include all eligible studies. In addition, the NNR was too high for regular use, although the re-introduction of field tags, quotations, and validated filters would reduce the screening burden substantially. Moreover, implementation of machine learning abstract screening tools has been shown to reduce the number of abstracts that must be screened by humans by as much as 50% with minimal loss of relevant records.51 These methods could be combined with automatic query generation to reduce the impact of the higher numbers of records identified during searches.
Limitations
The search strategies in the PROSPERO dataset are likely to be a good representation of final publishable search strategies. But they are imperfect because they were often the initial search for the review published in the protocol rather than the final peer-reviewed query. In addition, they may not always perfectly represent the original query posted in PROSPERO as many were manually or automatically altered in the cleaning process. Finally, we cleaned the queries with pasted search terms first, likely enriching the training dataset with shorter and more generic queries.
Models trained on final searches or on the Population and Intervention/Exposure fields may perform better. We chose to train and test the model on only the review title field and the objectives or key questions field for 3 reasons. First, not everyone is able to break a topic or set of questions down into its component parts, so it is beneficial for a model to be able to automatically parse a topic. Also, not every review follows the standard PICO format (eg, reviews of disease prevalence or research methods). Second, and related, the more the model requires the user to break down the topic and propose PICO terms, the less it actually helps in the process. Third, unlike the title and key questions, these fields vary widely in their content and structure and so may not be sufficiently consistent for fine-tuning.
We evaluated only a single, representative open-source LLM (Mistral-Instruct 7b). Our aim was to establish feasibility of generating search queries automatically via LLMs, rather than to establish which among the now-many LLMs performs “best” at this task. Larger models would likely yield better results, but we did not have the computational resources to fine-tune them. Proprietary models, like ChatGPT, may also be used to aid search design, but these cannot be fine-tuned. Moreover, we are interested in designing and sharing usable, open-source LLMs that can be used as search aids. At the time these experiments began, Mistral-Instruct-7B was outperforming other models by a wide margin. Likewise, we did not perform any extensive prompt engineering experiments. We believe the GPT-4 experiments demonstrate that relatively simple prompting strategies are not likely to succeed, which is why focused on fine-tuning. Additional prompt tuning might be a valuable future research direction.
The qualitative analysis was exploratory and therefore relied on librarians with whom the lead researcher was familiar. The results may, therefore, not be representative of librarians in different settings or countries or with different foci. However, the librarians were experts in the field with many years of experience, which made them uniquely able to provide thoughtful and informed input that is helpful in determining next steps in the effort to leverage AI in the identification of studies for evidence synthesis. Because of the small sample of librarians, we did not explore the ways that different settings (research, academic, and hospital) affected the librarian’s reaction to the query tool, which would be of interest. Finally, due to time and resource constraints, only a single researcher (G.P.A.) conducted interviews and analyzed the qualitative data, potentially introducing flaws and bias.
Conclusion
We have developed a relatively large dataset of natural language descriptions of reviews and corresponding structured Boolean searches, which can be used to train and evaluate LLMs that map the former onto the latter. We demonstrated that an open-source and modestly sized LLM (Mistral-Instruct-7b) can perform this task reasonably well. Although none of the interviewed librarians felt that this query tool was ready to be incorporated into their current workflows, they had useful suggestions for future development based on their expertise in designing search queries and teaching others to do so.
Future research should focus on improving the dataset with more high-quality queries and assessing whether including other fields, such as the population and intervention fields, would produce better performance. In addition to continuing to incorporate expert medical librarians in evaluating the usefulness of query-design tools, future work should reach out to others in the field, including systematic review methodologists and students, with whom librarians work, to include them in the development process and establish how the tools might be most useful to them. Conversations with users can guide implementation of tools, such as how best to incorporate post-processing steps, and whether a tool could be integrated with a database to enable simultaneous query refinement and abstract screening.
Models cannot replace careful and thoughtful search query design, but they can be used to summarize a topic using Boolean logic, providing suggestions for key words and the framework for the query. In conjunction with PubMed’s Best Match algorithm, they may also be a good starting place for searchers either scoping a topic or looking for a non-comprehensive list of relevant articles.
Supplementary Material
Contributor Information
Gaelen P Adam, Center for Evidence Synthesis in Health, Brown University School of Public Health, Providence, RI 02903, United States.
Jay DeYoung, Khoury College of Computer Sciences, Northeastern University, Boston, MA 02115, United States.
Alice Paul, Department of Biostatistics, Brown University School of Public Health, Providence, RI 02903, United States.
Ian J Saldanha, Center for Evidence Synthesis in Health, Brown University School of Public Health, Providence, RI 02903, United States; Center for Clinical Trials and Evidence Synthesis, Johns Hopkins Bloomberg School of Public Health, Baltimore, MD 21205, United States.
Ethan M Balk, Center for Evidence Synthesis in Health, Brown University School of Public Health, Providence, RI 02903, United States.
Thomas A Trikalinos, Center for Evidence Synthesis in Health, Brown University School of Public Health, Providence, RI 02903, United States; Department of Biostatistics, Brown University School of Public Health, Providence, RI 02903, United States.
Byron C Wallace, Khoury College of Computer Sciences, Northeastern University, Boston, MA 02115, United States.
Author contributions
All authors conceived of the study and developed the study design. Gaelen P. Adam, Alice Paul, and Jay Deyoung cleaned the downloaded PROSPERO data. Gaelen P. Adam and Jay Deyoung fine-tuned the model and ran the evaluation. All authors contributed to data analysis, interpretation, and manuscript preparation. All authors have approved its submission and agree to be accountable for all aspects of the work in ensuring that questions related to the accuracy and integrity of any part of the work are appropriately investigated and resolved.
Supplementary material
Supplementary material is available at JAMIA Open online.
Funding
This research did not receive a specific grant from any funding agency in the public, commercial, or not-for-profit sectors.
Conflicts of interest
The authors have no competing interests to declare.
Data availability
Code and data available at 10.5281/zenodo.12802972.
References
- 1. O’Mara-Eves A, Thomas J, McNaught J, Miwa M, Ananiadou S.. Using text mining for study identification in systematic reviews: a systematic review of current approaches. Syst Rev. 2015;4:5. 10.1186/2046-4053-4-5 [published Online First: 2015/01/16]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wallace BC, Small K, Brodley CE, Lau J, Trikalinos TA. Deploying an interactive machine learning system in an evidence-based practice center: abstrackr. In: Proceedings of the 2nd ACM SIGHIT International Health Informatics Symposium. ACM; 2012.
- 3. Carey N, Harte M, Mc Cullagh L.. A text-mining tool generated title-abstract screening workload savings: performance evaluation versus single-human screening. J Clin Epidemiol. 2022;149:53-59. 10.1016/j.jclinepi.2022.05.017 [published Online First: 202200]. [DOI] [PubMed] [Google Scholar]
- 4. Harrison H, Griffin SJ, Kuhn I, Usher-Smith JA.. Software tools to support title and abstract screening for systematic reviews in healthcare: an evaluation. BMC Med Res Methodol. 2020;20:7. 10.1186/s12874-020-0897-3 [published Online First: 2020/01/15]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.CLEF eHealth 2019—Task 2: Technology Assisted Reviews in Empirical Medicine. Secondary CLEF eHealth 2019—Task 2: Technology Assisted Reviews in Empirical Medicine. 2021. Accessed April 24, 2024. https://clefehealth.imag.fr/?page_id=173
- 6. Adam GP, Pappas D, Papageorgiou H, Evangelou E, Trikalinos TA.. A novel tool that allows interactive screening of PubMed citations showed promise for the semi-automation of identification of biomedical literature. J Clin Epidemiol. 2022;150:63-71. 10.1016/j.jclinepi.2022.06.007 [published Online First: 20220620]. [DOI] [PubMed] [Google Scholar]
- 7. Bui DD, Jonnalagadda S, Del Fiol G.. Automatically finding relevant citations for clinical guideline development. J Biomed Inform. 2015;57:436-445. 10.1016/j.jbi.2015.09.003 [published Online First: 2015/09/13]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chelli M, Descamps J, Lavoué V, et al. Hallucination rates and reference accuracy of ChatGPT and bard for systematic reviews: comparative analysis. J Med Internet Res. 2024;26:e53164. 10.2196/53164 [published Online First: 20240522]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wang S, Scells H, Koopman B, Potthast M, Zuccon G. Generating natural language queries for more effective systematic review screening prioritisation. In: Proceedings of the Annual International ACM SIGIR Conference on Research and Development in Information Retrieval in the Asia Pacific Region. Association for Computing Machinery; 2023:73-83.
- 10. Lefebvre CGJ, Briscoe S, Littlewood A, et al. Chapter 4: Searching for and selecting studie. In: Higgins JP, Thomas J, Chandler J, Cumpston M, Li T, Page MJ, Welch VA, eds. Cochrane Handbook for Systematic Reviews of Interventions. London: The Cochrane Collaboration; 2019. [Google Scholar]
- 11. Johnson EE, O’Keefe H, Sutton A, Marshall C.. The systematic review toolbox: keeping up to date with tools to support evidence synthesis. Syst Rev. 2022;11:258. 10.1186/s13643-022-02122-z [published Online First: 20221201]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Paynter RAG, Hedden-Gross A, Twose C, Voisin C.. Systematic Review Search Strategy Development Tools: A Practical Guide for Expert Searchers. Agency for Healthcare Research and Quality; 2024. [Google Scholar]
- 13. Wang S, Scells H, Koopman B, Zuccon G.. 2023. Can ChatGPT write a good Boolean query for systematic review literature search? arXiv, arXiv:2302.03495, preprint: not peer reviewed.
- 14. Wang S, Kanoulas E, Li D, Azzopardi L, Spijker R. CLEF 2017 technologically assisted reviews in empirical medicine overview. In: CEUR Workshop Proceedings. Aachen: CEUR-WS; 2017.
- 15. Wang S, Kanoulas E, Li D, Azzopardi L, Spijker R. CLEF 2018 technologically assisted reviews in empirical medicine overview. In: CEUR Workshop Proceedings. Aachen: CEUR-WS; 2018.
- 16. Scells H, Zuccon G, Koopman B, Deacon A, Azzopardi L, Geva S. A test collection for evaluating retrieval of studies for inclusion in systematic reviews. In: Proceedings of the 40th International ACM SIGIR Conference on Research and Development in Information Retrieval (SIGIR '17). New York, NY: Association for Computing Machinery; 2017:1237-1240. 10.1145/3077136.3080707 [DOI]
- 17. Wang S, Scells H, Clark J, Koopman B, Zuccon G.. 2022. From little things big things grow: a collection with seed studies for medical systematic review literature search. arXiv, arXiv:2204.03096, preprint: not peer reviewed.
- 18. Alharbi A, Stevenson M. A dataset of systematic review updates. In: Proceedings of the 42nd International ACM SIGIR Conference on Research and Development in Information Retrieval. ACM; 2019.
- 19. Booth A, Clarke M, Dooley G, et al. The nuts and bolts of PROSPERO: an international prospective register of systematic reviews. Syst Rev. 2012;1:2-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Schiavo JH. PROSPERO: an international register of systematic review protocols. Med Ref Serv Q. 2019;38:171-180. [DOI] [PubMed] [Google Scholar]
- 21. Adam GP, Di M, Cu-Uvin S, Halladay C, Smith BT, Trikalinos TA.. AHRQ Comparative Effectiveness Technical Briefs. Strategies for Improving the Lives of Women Aged 40 and Above Living With HIV/AIDS. Agency for Healthcare Research and Quality (US; ); 2016. [PubMed] [Google Scholar]
- 22. Balk E, Adam GP, Kimmel H, et al. AHRQ Comparative Effectiveness Reviews. Nonsurgical Treatments for Urinary Incontinence in Women: A Systematic Review Update. Agency for Healthcare Research and Quality (US; ); 2018. [PubMed] [Google Scholar]
- 23. Balk EM, Adam GP, Cao W, Bhuma MR, D’Ambrosio C, Trikalinos TA.. Long-term effects on clinical event, mental health, and related outcomes of CPAP for obstructive sleep apnea: a systematic review. J Clin Sleep Med. 2024;20:895-909. 10.5664/jcsm.11030 [published Online First: 20240201]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Balk EM, Adam GP, Cao W, et al. AHRQ Comparative Effectiveness Reviews. Management of Colonic Diverticulitis. Agency for Healthcare Research and Quality (US; ); 2020. [PubMed] [Google Scholar]
- 25. Balk EM, Adam GP, Langberg V, et al. Omega-3 fatty acids and cardiovascular disease: an updated systematic review. Evid Rep Technol Assess (Full Rep). 2016:1-1252. 10.23970/ahrqepcerta223 [DOI] [PubMed] [Google Scholar]
- 26. Balk EM, Ellis AG, Di M, Adam GP, Trikalinos TA.. AHRQ Comparative Effectiveness Reviews. Venous Thromboembolism Prophylaxis in Major Orthopedic Surgery: Systematic Review Update. Agency for Healthcare Research and Quality (US; ); 2017. [PubMed] [Google Scholar]
- 27. Balk EM, Gazula A, Markozannes G, et al. AHRQ Comparative Effectiveness Reviews. Lower Limb Prostheses: Measurement Instruments, Comparison of Component Effects by Subgroups, and Long-Term Outcomes. Agency for Healthcare Research and Quality (US; ); 2018. [PubMed] [Google Scholar]
- 28. Balk EM, Konnyu KJ, Cao W, et al. AHRQ Comparative Effectiveness Reviews. Schedule of Visits and Televisits for Routine Antenatal Care: A Systematic Review. Agency for Healthcare Research and Quality (US; ); 2022. [PubMed] [Google Scholar]
- 29. Drucker A, Adam GP, Langberg V, et al. AHRQ Comparative Effectiveness Reviews. Treatments for Basal Cell and Squamous Cell Carcinoma of the Skin. Agency for Healthcare Research and Quality (US; ); 2017. [PubMed] [Google Scholar]
- 30. Konnyu KJ, Thoma LM, Bhuma MR, et al. AHRQ Comparative Effectiveness Reviews. Prehabilitation and Rehabilitation for Major Joint Replacement. Agency for Healthcare Research and Quality (US; ); 2021. [PubMed] [Google Scholar]
- 31. Panagiotou OA, Markozannes G, Kowalski R, et al. AHRQ Technology Assessments. Short- and Long-Term Outcomes after Bariatric Surgery in the Medicare Population. Agency for Healthcare Research and Quality (US; ); 2018. [PubMed] [Google Scholar]
- 32. Saldanha IJ, Adam GP, Kanaan G, et al. AHRQ Comparative Effectiveness Reviews. Postpartum Care up to 1 Year After Pregnancy: A Systematic Review and Meta-Analysis. Agency for Healthcare Research and Quality (US; ); 2023. [PubMed] [Google Scholar]
- 33. Saldanha IJ, Cao W, Bhuma MR, et al. Management of primary headaches during pregnancy, postpartum, and breastfeeding: a systematic review. Headache. 2021;61:11-43. 10.1111/head.14041 [published Online First: 20210112]. [DOI] [PubMed] [Google Scholar]
- 34. Steele D, Adam GP, Di M, et al. AHRQ Comparative Effectiveness Reviews. Tympanostomy Tubes in Children with Otitis Media. Agency for Healthcare Research and Quality (US; ); 2017. [PubMed] [Google Scholar]
- 35. Steele DW, Adam GP, Saldanha IJ, et al. Postpartum home blood pressure monitoring: a systematic review. Obstet Gynecol. 2023;142:285-295. 10.1097/aog.0000000000005270 [published Online First: 20230613]. [DOI] [PubMed] [Google Scholar]
- 36. Steele DW, Becker SJ, Danko KJ, et al. AHRQ Comparative Effectiveness Reviews. Interventions for Substance Use Disorders in Adolescents: A Systematic Review. Agency for Healthcare Research and Quality (US; ); 2020. [PubMed] [Google Scholar]
- 37. Guirguis-Blake JM, Evans CV, Perdue LA, Bean SI, Senger CAU.. S. Preventive Services Task Force Evidence Syntheses, Formerly Systematic Evidence Reviews Aspirin Use to Prevent Cardiovascular Disease and Colorectal Cancer: An Evidence Update for the U.S. Preventive Services Task Force. Agency for Healthcare Research and Quality (US; ); 2022. [PubMed] [Google Scholar]
- 38. Jutkowitz E, Hsiao J, Celedon M, et al. VA Evidence-Based Synthesis Program Reports. Accelerated Diagnostic Protocols Using High-Sensitivity Troponin Assays to “Rule In” or “Rule Out” Myocardial Infarction in the Emergency Department: A Systematic Review. Department of Veterans Affairs (US; ); 2023. [PubMed] [Google Scholar]
- 39. McGowan J, Sampson M, Salzwedel DM, Cogo E, Foerster V, Lefebvre C.. PRESS peer review of electronic search strategies: 2015 guideline statement. J Clin Epidemiol. 2016;75:40-46. 10.1016/j.jclinepi.2016.01.021 [published Online First: 2016/03/24]. [DOI] [PubMed] [Google Scholar]
- 40. Woods B, Aguirre E, Spector AE, Orrell M.. Cognitive stimulation to improve cognitive functioning in people with dementia. Cochrane Database Syst Rev. 2012:Cd005562. 10.1002/14651858.CD005562.pub2 [published Online First: 20120215]. [DOI] [PubMed] [Google Scholar]
- 41. Chung HW, Hou L, Longpre S, et al. 2022. Scaling instruction-finetuned language models. arXiv, arXiv:2210.11416, preprint: not peer reviewed.
- 42. Yuan H, Yuan Z, Gan R, Zhang J, Xie Y, Yu S, BioBART. 2022. Pretraining and evaluation of a biomedical generative language model. arXiv, arXiv:2204.03905, preprint: not peer reviewed.
- 43. Jiang AQ, Sablayrolles A, Mensch A, et al. 2023. Mistral 7B. arXiv, arXiv:2310.06825, preprint: not peer reviewed.
- 44. Wolf T, Debut L, Sanh V, et al. 2019. Huggingface’s transformers: state-of-the-art natural language processing. arXiv, arXiv:1910.03771, preprint: not peer reviewed.
- 45.Parameter-Efficient Fine-Tuning Library [program]: HuggingFace. Accessed April 24, 2024. https://huggingface.co/docs/peft/en/index
- 46. Hu EJ, Shen Y, Wallis P, et al. 2021. Lora: low-rank adaptation of large language models. arXiv, arXiv:2106.09685, preprint: not peer reviewed.
- 47. Shazeer N, Stern M. Adafactor: adaptive learning rates with sublinear memory cost. In: International Conference on Machine Learning. PMLR; 2018.
- 48.Anvil [program]. The Tuesday Project Ltd; 2024. Accessed April 24, 2024. https://anvil.works/
- 49. Gale NK, Heath G, Cameron E, Rashid S, Redwood S.. Using the framework method for the analysis of qualitative data in multi-disciplinary health research. BMC Med Res Methodol. 2013;13:117. 10.1186/1471-2288-13-117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Fiorini N, Canese K, Starchenko G, et al. Best match: New relevance search for PubMed. PLoS Biol. 2018;16:e2005343. 10.1371/journal.pbio.2005343 [published Online First: 20180828]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Trikalinos TA, Wallace BC, Jap J, et al. Large scale empirical evaluation of machine learning for semi-automating citation screening in systematic reviews. Med Decis Making. 2019. 10.1177/0272989X19890544 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Code and data available at 10.5281/zenodo.12802972.