Abstract
Insulin stimulates glucose uptake in fat and muscle by mobilizing Glut4 glucose transporters from intracellular membrane storage sites to the plasma membrane. This process requires the trafficking of Glut4-containing vesicles toward the cell periphery, docking at exocytic sites, and plasma membrane fusion. We show here that phospholipase D (PLD) production of the lipid phosphatidic acid (PA) is a key event in the fusion process. PLD1 is found on Glut4-containing vesicles, is activated by insulin signaling, and traffics with Glut4 to exocytic sites. Increasing PLD1 activity facilitates glucose uptake, whereas decreasing PLD1 activity is inhibitory. Diminished PA production does not substantially hinder trafficking of the vesicles or their docking at the plasma membrane, but it does impede fusion-mediated extracellular exposure of the transporter. The fusion block caused by RNA interference-mediated PLD1 deficiency is rescued by exogenous provision of a lipid that promotes fusion pore formation and expansion, suggesting that the step regulated by PA is late in the process of vesicle fusion.
INTRODUCTION
Insulin-stimulated uptake of glucose by fat and muscle and the maintenance of glucose homeostasis are primarily mediated by the Glut4 glucose transporter (Bryant et al., 2002). Glut4 cycles between the plasma membrane and cytoplasmic storage sites, with most of the transporter residing intracellularly in the absence of insulin signaling because the basal rate of endocytosis exceeds the basal rate of exocytosis. Insulin signaling greatly stimulates the rate of exocytosis, leading to recruitment of up to 50% of the transporter to the cell surface where it facilitates glucose uptake. Each of the major elements of this regulated exocytosis process—vesicle mobilization, trafficking to the plasma membrane, docking, and fusion—have been shown to be rate limiting under different circumstances, and the mechanisms that regulate them remain under investigation.
Phospholipase D (PLD), a membrane-associated enzyme regulated by agonist stimulation (reviewed in Frohman and Morris, 1999), has been proposed to function at many different steps in vesicle trafficking, including activation of signaling networks (Andresen et al., 2002), budding of vesicles from the trans-Golgi (Chen et al., 1997), and vesicle fusion (reviewed in McDermott et al., 2004). PLD generates the lipid phosphatidic acid (PA), which has been shown to activate phosphatidylinositol 4-phosphate 5-kinase (Honda et al., 1999) and thus increase the levels of phosphatidylinositol 4,5-bisphosphate, a lipid critically required for exocytosis (Di Paolo et al., 2004). PA also has been reported to serve as a membrane anchor for a growing number of protein targets (Manifava et al., 2001), including yeast and mammalian components of the soluble N-ethylmaleimide-sensitive factor attachment protein receptor (SNARE) complex (Wagner and Tamm, 2001; Nakanishi et al., 2004), and it has been proposed to act as a fusogenic lipid in biophysical modeling studies by lowering the activation energy for membrane bending during generation and expansion of fusion pores (Kozlovsky et al., 2002; Kooijman et al., 2003).
PLD has been proposed to play a role at an undefined step in insulin-stimulated Glut4 translocation to the plasma membrane in fat cells (Emoto et al., 2000). Emoto et al. (2000) demonstrated that injection of bacterial PLD into adipocytes increased their insulin sensitivity with respect to Glut4 translocation. However, bacterial PLD is cytosolic, constitutively active, uses as substrate multiple species of phospholipids, and seems to preferentially increase PA levels on perinuclear membranes (Delon et al., 2004). In contrast, mammalian PLD is membrane associated, regulated by signal transduction events, and restricted to acting on phosphatidylcholine. Thus, it was difficult to draw definitive conclusions regarding the physiological significance of PA generation in the Glut4 translocation process. Moreover, another report suggested that PLD does not play a role in insulin-stimulated glucose uptake, based on the use of alcohol to decrease PA production (Millar et al., 2000).
The lack of highly effective PLD inhibitors and lack of information regarding the endogenous enzyme has limited the ability to clarify the role of PLD in Glut4 trafficking. Using newly developed antisera capable of imaging endogenous PLD1, RNA interference (RNAi) to examine the role of the endogenous protein in Glut4 translocation, and exogenous rescue of the RNAi phenotype by using a lipid that facilitates the terminal steps in fusion, we report here that PLD1 activation plays a rate-limiting step late in the process of insulin-stimulated fusion of Glut4-containing storage vesicles into the plasma membrane and suggest that this may be a general role for PLD1 in regulated exocytosis.
MATERIALS AND METHODS
General Reagents
DMEM and cell culture supplements were from Invitrogen (Carlsbad, CA); fetal bovine serum was from Hyclone Laboratories (Logan, UT); 2-deoxy-d-glucose, [1,2-3H] was from MP Biomedicals (Irvine, CA); palmitic acid, [9,10-3H(N)] was from American Radiolabeled Chemicals (St. Louis, MO); palmitoyl-lysophosphatidylcholine was from Avanti Polar Lipids (Alabaster, AL); and insulin and phorbol 12-myristate 13-acetate (PMA) were from Sigma-Aldrich (St. Louis, MO). All other reagents were of analytical grade unless otherwise specified.
Antibodies
Rabbit anti-PLD1 and rat anti-PLD2 antisera have been described previously (Du et al., 2004; Zhang et al., 2004). The anti-PLD1 antisera were affinity purified and recognized only one band by Western blot analysis. The band, and the PLD1 immunofluorescent signal, are decreased by targeting of PLD1 via RNAi. Other antibodies included anti-c-myc (9E10; Sigma-Aldrich), -hemagglutinin (HA) (3F10; Roche Diagnostics, Indianapolis, IN), -tubulin (Sigma-Aldrich), and -transferrin receptor (Zymed Laboratories, South San Francisco, CA). Antibodies against glycogen synthase kinase 3β (GSK-3β) and caveolin-1 were obtained from BD Biosciences Transduction Laboratories (Lexington, KY), and anti-phospho-GSK-3β from Cell Signaling Technology (Beverly, MA). Rhodamine-conjugated wheat germ agglutinin (WGA) and Alexa 647-conjugated secondary antibody were from Molecular Probes (Eugene, OR). Cy3-conjugated secondary antibodies were from Jackson ImmunoResearch Laboratories (West Grove, PA).
Cell Culture and Transfection of 3T3-L1 Adipocytes
3T3-L1 and myc7-Glut4-GFP (Bogan et al., 2001) fibroblasts were cultured and differentiated into adipocytes for 8–12 d and electroporated according to established protocols (Thurmond et al., 1998). After electroporation, the cells were allowed to adhere to collagen-coated coverslips for 24 h before serum starvation and insulin treatment.
Adenovirus Infection
Adenovirus vectors for expression of wild-type and catalytically inactive (dominant negative) mutants of PLD1 (Sung et al., 1997) were generated using the AdEasy system (He et al., 1998). 3T3-L1 adipocytes were infected at a multiplicity of infection of 100 plaque-forming units/cell in DMEM with 0.5% bovine serum albumin overnight. These media were replaced with DMEM containing 10% serum the next morning. All the experiments were performed 48 h after infection. This protocol resulted in an infection efficiency of at least 80%.
Retroviral Infection and Generation of PLD1-RNAi Stable Cell Lines
The PLD1 sequence CTGGAAGATTACTTGACAA (nt 547–565 of the open reading frame) was subcloned into pSuper (Brummelkamp et al., 2002) and shuttled into a modification of the Retroviral GeneSuppressor System (Imgenex, San Diego, CA) in which the U6 promoter had been replaced by the H1-promoter. Retrovirus was produced in human embryonic kidney 293 cells and used to infect myc7-Glut4-GFP fibroblasts according to the manufacturer's instructions. Stably infected fibroblasts were selected 48 h later in medium containing G418 (1 mg/ml; Geneticin), and single colonies were picked for Western blot analysis and PLD assay. One control and two PLD1-RNAi sublines were chosen for subsequent experiments.
For rescue assays, hPLD1 wobble mutations (A552G, T555C, and T559C) were generated using QuikChange (Stratagene, La Jolla) and confirmed by sequencing. PLD1-RNAi adipocytes were electroporated with 200 μg of pCGN-vector (HA tagged) or pCGN-PLD1-wobble constructs and the Myc-Glut4-EGFP translocation assay conducted as described below.
Myc-Glut4-EGFP and HA-Glut4-EGFP Translocation Assays
Myc-Glut4-EGFP and HA-Glut4-EGFP translocation assays were conducted as described in Bogan et al. (2001). In rescue experiments, adipocytes were immunostained at 4°C with anti-myc antibody for 1 h, washed with cold phosphate-buffered saline (PBS), fixed with 4% paraformaldehyde for 10 min, permeabilized with 0.1% Triton X-100 for 10 min, immunostained with anti-HA antibody for 1 h, and visualized using fluorescent dye-conjugated secondary antibodies. In K+-depletion experiments, myc7-Glut4-GFP cells were incubated in K+-free buffer for 2 h before detection of cell surface myc (Li et al., 2001). Lysophosphatidylcholine (LPC) (palmitoyl; Avanti Polar Lipids) was added to media 15 min before insulin stimulation. Unless otherwise stated, all stimulations were performed using 100 nM insulin for 5 min. Quantitative analysis was performed using either Leica TCS2 confocal imaging or NIH Image software. All images were acquired using a 40× 1.25 numerical aperture oil immersion objective and represent single z-sections of height 0.25 μm. Cell surface versus total Glut4 was calculated by measuring cyanin 3 (Cy3)/green fluorescent protein (GFP) fluorescence ratio. The Cy3 and GFP fluorescence intensities were summed over all cells in a field, and the Cy3/GFP ratio determined as an average of five fields.
Microinjection and Plasma Membrane Sheet Assay
3T3-L1 adipocytes were grown on coverslips, and microinjection carried out as described previously (Watson et al., 2004). In brief, cells were impaled using an Eppendorf model 5171 micromanipulator, and the nuclei were injected with 50 μg/ml MBP-Ras expression plasmid plus 200 μg/ml vector or Exo70-N expression plasmid by using an Eppendorf model 5246 transinjector. The cells were allowed to recover for 24 h, and plasma membrane sheets prepared as described previously (Robinson et al., 1992). In myc7-Glut4-GFP cells, Glut4 translocation was visualized by GFP, and total plasma membrane was visualized using WGA (Bose et al., 2001). The data were quantified by counting the number of GFP-positive plasma membrane sheets from WGA staining-positive cells in five randomly selected fields.
PLD Activity Assay
PLD activities were determined using in vivo transphosphatidylation to measure the accumulation of phosphatidylbutanol in intact cells (Morris et al., 1997). For insulin-stimulated PLD activity, adipocytes were incubated with 100 nM insulin for 5 min before addition of 0.3% 1-butanol for 15 min.
2-Deoxyglucose Uptake
3T3-L1 adipocytes were seeded in 12-well plates and 2-deoxyglucose uptake determined as described previously (Kanzaki and Pessin, 2001). In brief, 3T3-L1 adipocytes were serum starved for at least 3 h before assay. Cells were then washed twice with KRPH buffer (5 mM Na2HPO4, 20 mM HEPES, pH 7.4, 1 mM MgSO4, 1 mM CaCl2, 136 mM NaCl, 4.7 mM KCl, 0.1% [wt/vol] BSA) and stimulated with or without insulin for 5 min. Glucose uptake was measured by incubation with 0.1 mM 2-deoxyglucose containing 1 μCi/ml 2-deoxy-d-glucose, [1,2-3H] at 4°C for 5 min. Transport was terminated by washing the cells three times with ice-cold PBS. Cells were solubilized with 1% Triton X-100, and the radioactivity was detected by scintillation counting. Nonspecific deoxyglucose uptake was measured in the presence of 20 μM cytochalasin B and was subtracted from each determination to obtain specific uptake. Data are representative of three experiments, and each value was corrected for protein content.
Western Blotting
Immunoreactive bands were visualized with SuperSignal West Pico Chemiluminescent Substrate (Pierce Chemical, Rockford, IL). Phosphorylation of GSK-3β was detected with a phospho-GSK-3β (Ser9) antibody (1:1000), and total GSK-3β (1:2000) was measured as a loading control. The images were quantified using a Bio-Rad model GS-700 imaging densitometer with Image-Quant 5.1 software. Protein concentration was determined by the Bradford method, and equal amounts loaded for each sample.
RESULTS
PLD1 Is Expressed in Adipocytes, Colocalizes with Glut4 before and after plasma membrane translocation, and Is Activated by Insulin Signaling
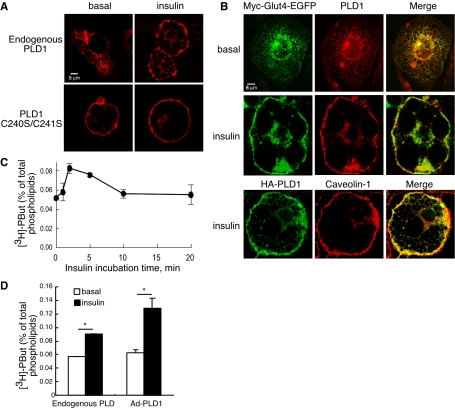
To explore the potential role of PLD-generated PA in glucose uptake, we first examined localization of the endogenous protein in 3T3-L1 mouse adipocytes using a highly sensitive polyclonal anti-PLD1 antiserum (Zhang et al., 2004). Under basal conditions, endogenous (Figure 1, A and B, and Supplemental Figure 1) and overexpressed (our unpublished data) PLD1 localizes to dispersed small intracellular membrane vesicles; upon insulin challenge, translocation to the plasma membrane was observed (Figure 1, A and B, and Supplemental Figure 1) in the same time frame in which Glut4 translocates (2–5 min). The punctate localization pattern suggests that PLD1 concentrates at specific plasma membrane sites. These sites may represent rosettes of clustered lipid raft domains (Kanzaki and Pessin, 2002), which are important for PLD1 reentry (Du et al., 2003), because expression of a mutant PLD1 allele unable to enter into lipid rafts (Du et al., 2003) yielded the uniform plasma membrane localization pattern characteristic of non-raft-localizing proteins (Figure 1A). PLD1 colocalized with Glut4 to a substantial degree before and after insulin stimulation (Figure 1B), suggesting that PA generated by PLD1 activation could accumulate in Glut4-containing vesicles and play a role in signaling or trafficking.
Figure 1.
PLD1 colocalizes with Glut4 and is activated by insulin signaling. (A) Endogenous PLD1 translocates to the plasma membrane in response to insulin stimulation. 3T3-L1 adipocytes were either untreated or electroporated with the HA-tagged mutant PLD1 allele C240S/C241S. Basal and insulin-stimulated cells were then stained with anti-PLD1 antibody to detect endogenous PLD1 or with anti-HA to detect the mutant PLD1 allele, followed by the appropriate Cy3-conjugated secondary antibody. Images were acquired by confocal microscopy. (B) PLD1 substantially colocalizes with Glut4 before and after insulin-induced translocation. Basal and insulin-stimulated myc7-Glut4-GFP–expressing adipocytes were stained with anti-PLD1 antibody followed by Alexa 647-conjugated secondary antibody. Glut4 was detected using its GFP tag. Caveolin-1 was used as a maker for the plasma membrane (Watson et al., 2001). (C) PLD activity peaks rapidly after insulin stimulation. Individual culture wells were supplemented with 0.3% butanol at the indicated times after stimulation with insulin, cultured for an additional 2 min, and then processed for PLD activity as described in Materials and Methods. The data represent mean ± SD of triplicate measurements. The experiment is representative of three independent experiments. (D) Insulin stimulates endogenous PLD and overexpressed PLD1 activity in 3T3-L1 adipocytes. PLD activity was measured as described in Materials and Methods. Overexpression of PLD1 was accomplished using an adenoviral delivery system. The data represent mean ± SD of triplicate measurements. The experiment is representative of three independent experiments. *, significant difference, p < 0.001. Ad, adenovirus, Pbut, phosphatidylbutanol.
Whether insulin stimulates endogenous PLD has remained controversial, potentially because the alcohol substrate required to detect PLD activity decreases insulin receptor activation (Seiler et al., 2000; Xu et al., 2003). We found that insulin-stimulated endogenous PLD activation could be detected using a sensitive in vivo assay when the insulin receptor is preactivated before the addition of butanol (Figure 1, C and D; 60% increase in activity) and that this activity increases after exposure of the cells to PLD1-expressing adenovirus (which achieved an infection rate of ∼80%; Supplemental Figure 2). The bulk of PLD activity occurs 2–10 min after insulin stimulation, as determined through addition of butanol for a fixed period of time (2 min) at successive intervals after insulin stimulation (Figure 1C). These data demonstrate that PLD1 is insulin responsive in the time frame in which Glut4 translocation takes place and that PLD1 overexpression can be used to investigate the role of PA in regulation of Glut4 translocation.
Increased PLD1 Activation Facilitates Insulin-stimulated Translocation of Glut4 to the Plasma Membrane
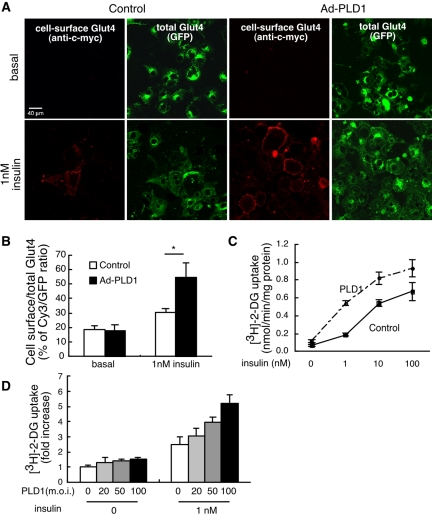
We explored PLD1's role by using three independent approaches based on gain and loss of function. In the first approach, gain-of-function effects were examined using adenoviral overexpression of wild-type PLD1 in 3T3-L1 adipocytes. Glut4 translocation was monitored using an allele doubly tagged with GFP at its C terminus to visualize all of the transporter, and with seven copies of c-myc in its first extracellular loop. The latter permitted visualization of just the transporter on the cell surface by using anti-c-myc immunostaining on nonpermeabilized cells (Bogan et al., 2001). PLD1 overexpression did not significantly alter the amount of Glut4 at the plasma membrane under basal conditions, but it did increase insulin-stimulated Glut4 plasma membrane translocation at submaximal levels of insulin (1 nM) as determined by fluorescence visualization (Figure 2A) and quantitation (Figure 2B). As a second and independent means of assessment, PLD1 overexpression increased [3H]glucose uptake over a range of PLD1 and insulin concentrations (Figure 2, C and D). PLD1 overexpression had little to no effect on basal glucose uptake, but it increased uptake 2–3-fold at submaximal (1 nM) levels of insulin stimulation. These results demonstrate that increasing PLD1 activity promotes insulin-stimulated Glut4 translocation.
Figure 2.
Increasing PLD1 activity promotes insulin-stimulated Glut4 translocation. (A) Overexpression of PLD1 facilitates Glut4 translocation. myc7-Glut4-GFP adipocytes were infected with wild-type PLD1-expressing adenoviruses. Cells were serum starved, stimulated with insulin for 5 min, chilled, and stained without fixation or permeabilization to detect cell surface Glut4 through visualization of the externalized Myc epitope tag, whereas total Glut4 was visualized using the GFP tag. Images are representative of three independent experiments. (B) Quantification of surface-to-total distribution of myc7-Glut4-GFP. The Cy3 and GFP fluorescence intensities were summed over all cells in a field, and the Cy3/GFP ratio determined as an average of five different fields. *, significant difference, p < 0.0006. The experiment is representative of three independent experiments. (C and D) Overexpression of PLD1 increases glucose uptake. Differentiated adipocytes were infected with control or PLD1-expressing adenoviruses and stimulated with insulin. The rate of [3H]2-deoxyglucose (2-DG) uptake was determined. Results are expressed as the mean ± SD of triplicate determinations. Representative of five independent experiments.
Expression of a Catalytically Inactive PLD1 Allele Inhibits Insulin-stimulated Translocation of Glut4 to the Plasma Membrane
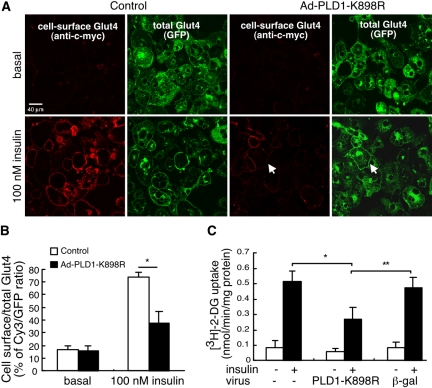
In the second approach, loss-of-function effects were examined by expressing PLD1-K898R, a catalytically inactive mutant allele (Sung et al., 1997) that has been shown to exhibit dominant negative activity (Shen et al., 2001; Vitale et al., 2001, 2002; Choi et al., 2002). Overexpression of PLD1-K898R inhibited insulin-stimulated Glut4 translocation by 62 ± 8.7%, even at maximal levels (100 nM) of insulin stimulation (Figure 3, A and B). Similarly, PLD1-K898R inhibited insulin-stimulated glucose uptake by 50 ± 9.1% (Figure 3C). Adenoviral expression in itself was not inhibitory, because viral delivery of β-galactosidase had no significant effect on insulin-stimulated Glut4 translocation (Figure 3C).
Figure 3.
Competitive interference with PLD1 function via expression of a catalytically inactive mutant allele inhibits insulin-stimulated Glut4 translocation. (A–C) Overexpression of catalytically-inactive PLD1 (PLD1-K898R) inhibits Glut4 translocation and glucose uptake. myc7-Glut4-GFP adipocytes were infected with control (β-gal) or PLD1-K898R-expressing adenoviruses and analyzed as in Figure 2. *, significant difference, p < 0.001; **, p < 0.01. 2-DG, 2-deoxyglucose. Arrows indicate cell in which vesicles translocated to the plasma membrane but did not undergo fusion. See text (Figure 5) for details. Representative of five independent experiments.
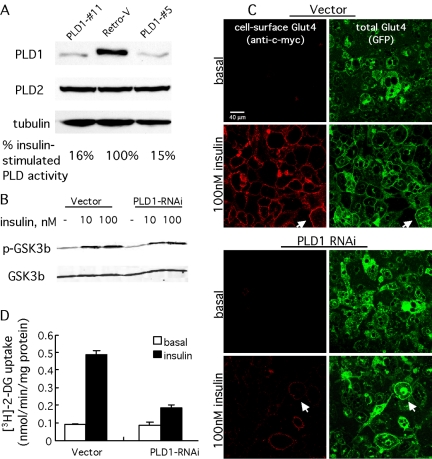
RNAi-mediated PLD1 Down-Regulation Inhibits Insulin-stimulated Translocation of Glut4 to the Plasma Membrane but Not Basal Glut4 Cycling or Insulin-stimulated Transferrin Receptor Trafficking
For the third approach, we used RNAi to examine the role of endogenous PLD1 and PA production. A retroviral-based system was used to establish 3T3-L1 cell lines in which 85–90% of PLD1 expression had been eliminated (Figure 4A). PLD1 down-regulation had no readily apparent effects on morphology, production of Glut4-containing vesicles, rates of cell proliferation, or adipocyte differentiation (our unpublished data) and did not elicit up-regulation of PLD2, the second mammalian PLD isoform, which also is expressed in 3T3-L1 cells (Figure 4A). Insulin stimulation revealed that the PLD1-RNAi cell lines exhibited an 85% decrease in PLD activity in comparison with the control (Retro-V) line (Figure 4A). Similar results were obtained using PMA to activate PLD (our unpublished data). A portion of the residual PLD activity presumably ensues from PLD2 activation.
Figure 4.
RNAi-mediated ablation of PLD1 inhibits insulin-stimulated Glut4 translocation. (A) Isoform-specific targeting of PLD1 by RNAi. Immunoblot analysis of endogenous PLD1 in myc7-Glut4-GFP adipocytes stably transformed with control (empty) or PLD1-RNAi retrovirus. Cell lysates from control (Retro-V) and two PLD1-shRNA cell lines (clones 5 and 11) were analyzed by immunoblotting with anti-PLD1, anti-PLD2, and anti-tubulin antibodies. Insulin-stimulated PLD activity was measured in control and PLD1 RNAi cells; 100% corresponds to a [3H]phosphatidylbutanol (percentage of total lipids) value of 0.091 ± 0.010. Note that some of the residual activity derives from the nontargeted PLD2 isoform. (B) Insulin receptor signaling is not altered in PLD1-RNAi cells. GSK-3β phosphorylation was examined in vector- and PLD1-RNAi cells as a function of insulin stimulation. An antibody that recognizes GSK-3β independent of phosphorylation was used as a loading control. (C) Endogenous PLD1 is required for insulin-stimulated Glut4 translocation. Glut4 translocation in control (Retrovector) and PLD1-RNAi adipocytes was analyzed by immunofluorescence staining of externalized Myc epitope after 5 min of insulin stimulation. Arrows indicates control (vector-transformed) cells in which vesicles translocated to the plasma membrane and underwent fusion, and PLD1-RNAi cells in which vesicles translocated to the plasma membrane but did not undergo fusion. See text (Figure 5) for details. Additional control, not shown: changes in insulin-stimulated Glut4 translocation and fusion were not observed in cells expressing an irrelevant short hairpin RNA. (D) Endogenous PLD1 is required for insulin-stimulated glucose uptake. Insulin-stimulated [3H]2-deoxyglucose (2-DG) uptake in control and PLD1-RNAi adipocytes. Data represent mean ± SD of triplicate measurements. All experiments were performed at least three times.
PLD2 has been reported to function in insulin signaling pathways (Rizzo et al., 1999). To investigate whether PLD1 functioned similarly, insulin-stimulated phosphorylation of GSK-3β, a direct downstream target of AKT, was examined in control and PLD1-RNAi cell lines. No differences in activation-dependent phosphorylation were observed, suggesting that PLD1 activity is not required for signaling events upstream of GSK-3β (Figure 4B). Similar results were obtained for AKT and mitogen-activated protein kinase (our unpublished data).
The PLD1-RNAi cell lines exhibited resistance to insulin stimulation with decreases in both Glut4 translocation (decrease of 71 ± 3.8%; Figure 4C) and glucose uptake (decrease of 75 ± 1.8%; Figure 4D). In addition to Glut4, insulin also stimulates a less dramatic translocation of the transferrin receptor (TfnR) from more typical recycling endosomes. TfnR translocation was unaffected in PLD1-RNAi cells (Supplemental Figure 3, A and B), suggesting that PLD1 activity is required only for the more specialized high-output Glut4 translocation pathway. A low level of Glut4 cycling occurs under basal (nonstimulated) conditions, although the origin of this Glut4 (recycling endosomes vs. specialized vesicles) remains controversial (reviewed in Simpson et al., 2001). The basal movement of Glut4 can be assessed using depletion of extracellular potassium to block endocytic return of the Glut4 from the plasma membrane to storage vesicles. This maneuver traps basally trafficking Glut4 at the plasma membrane, allowing it to be visualized and quantitated. Accumulation of Glut4 at the plasma membrane via depletion of extracellular potassium was unaltered in PLD1-RNAi cells (Supplemental Figure 3C), suggesting that PLD1 activity is not required and supporting the proposal that the basal cycling occurs via a pathway distinct from the insulin-stimulated pathway (Simpson et al., 2001).
Glut4-containing Membrane Vesicles Dock at Exocytic Sites but Exhibit Delayed Fusion into the Plasma Membrane
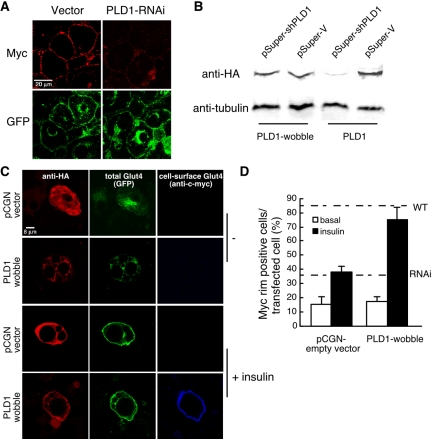
The most striking finding of the RNAi-PLD1 (Figures 4C and 5A) and PLD1-K898R (Figure 3A) experiments was that the appearance of the exofacial c-myc tag was markedly reduced (23 ± 8% of control value in Figure 5A; Figures 3A and 4C, arrows) in comparison with the extent of translocation of the GFP tag into a “rim” pattern at the plasma membrane of many cells (Figures 3A, see cells marked by arrows, and 4B). This suggests that although PLD1 could play roles at many steps, the most obvious consequence of PLD1 reduction is not on insulin signaling, Glut4-containing vesicle mobilization, or translocation of the vesicles to the plasma membrane, but rather on a distal event such as docking (tethering) or fusion. To confirm that the RNAi-mediated phenotype was specific, an HA-tagged rescue plasmid was generated by mutating the PLD1 cDNA sequence at wobble codons within the RNAi-targeted region. The PLD1 protein generated from the rescue cDNA exhibited a level of activity identical to PLD1 expressed from a wild-type cDNA (our unpublished data), was resistant to RNAi-mediated degradation (Figure 5B), colocalized with Glut4 before and after insulin stimulation (Figure 5C), increased the level of exofacial Glut4 in cells exhibiting GFP rim fluorescence (Figure 5C), and increased the number of cells exhibiting exofacial Glut4 in response to insulin nearly to wild-type levels (Figure 5D).
Figure 5.
PLD1-generated PA facilitates fusion of Glut4 vesicles into the plasma membrane. (A) Higher magnification image of insulin-stimulated control and PLD1-RNAi adipocytes reveals a discrepancy between the equal extents of Glut4 GFP (green) plasma membrane rim fluorescence in control and PLD1-RNAi cells versus exofacial c-myc epitope detection (red), which is almost absent in the RNAi-targeted cells. (B) A wobble codon-mutated PLD1 cDNA is immune to PLD1 RNAi. HeLa cells were cotransfected with 0.5 μg of pCGN-PLD1 or pCGN-PLD1-wobble and 1 μg of pSuper constructs as indicated. Twenty-four hours later, Western blot analysis was performed, by using tubulin as a loading control. (C) PLD1-wobble expression rescues exofacial Glut4 exposure in PLD1-RNAi cells. pCGN or pCGN-PLD1-wobble were transfected into PLD1-RNAi–expressing adipocytes by electroporation, cultured for 24 h, stimulated with 100 nM insulin for 5 min, and fixed for immunostaining. Representative cells are shown. Note that the pCGN empty vector generates a small, HA-tagged cytosolic peptide. (D) Quantitation of rescue. Fifty cells were counted for each condition. Data represent mean ± SD of three independent experiments. Dotted lines indicate frequencies of myc-rim–positive cells observed for untransfected insulin-stimulated RNAi and wild-type 3T3-L1 cells.
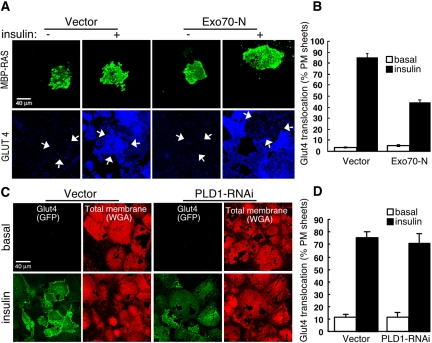
To further delineate the step at which PLD1/PA was required, we used the plasma membrane sheet assay to examine docking, reasoning that docked vesicles should be retained on the plasma membrane sheets even if fusion had not taken place, whereas undocked vesicles should be washed away. Microinjection of an expression plasmid for Exo70-N, a dominant-interfering allele of the exocyst component Exo70 that blocks Glut4-containing vesicle docking (Inoue et al., 2003), was performed to confirm that vesicles that have translocated to the plasma membrane but have not successfully docked are washed away under the assay conditions used (Figure 6A). An ∼50% decrease in the number of insulin-stimulated Glut4-positive plasma membrane sheets was observed in Exo70-N–injected cells (Figure 6B), which is consistent with the decrease in exofacial exposure reported previously by Inoue et al. (2003). In contrast, the fraction of plasma membrane sheets positive for Glut4 was not significantly decreased in PLD1-RNAi cells: control and PLD1-RNAi adipocytes adhering to coverslips were disrupted before or after insulin stimulation, after which the plasma membranes sheets adhering to the coverslips were washed to remove undocked vesicles and imaged. Glut4 was detected at the plasma membrane in control plasma membrane sheets only in response to insulin stimulation (Figure 6C); Glut4 in insulin-stimulated PLD1-RNAi cells associated with the plasma membrane sheets at control frequencies (Figure 6D), demonstrating that the Glut4 vesicles were tightly adhered to the plasma membrane. Together, these results indicate a role for PLD1 at a step distal to docking in the exocytic pathway.
Figure 6.
PLD1-derived PA is not required for vesicle docking. (A) Exo70-N prevents vesicle docking in the plasma membrane sheet assay. Differentiated 3T3-L1 nuclei were microinjected with MBP-RAS and vector or Exo70-N expression plasmids. MBP-Ras served as a membrane marker to indicate the cells injected during subsequent analysis steps. The cells were allowed to recover for 24 h, stimulated with 100 nM insulin, and used to prepare plasma membrane sheets. To prepare plasma membrane sheets, the adipocytes were disrupted using sonication, the coverslips were washed to remove unattached vesicles, and the residual plasma membrane sheets adhering to the coverslips were fixed and stained. Glut4 was imaged using anti-Glut4 antibody. Arrows indicate MBP-Ras–positive plasma membrane sheets. (B) Quantification of Glut4-positive plasma membrane sheets. Seventy to 80 MBP-Ras–positive sheets were counted for each condition. Data represent mean ± SD of three independent experiments. (C) PLD1-RNAi does not block vesicle docking. Basal and insulin-stimulated (5 min) control and PLD1-RNAi adipocytes were disrupted using sonication, the coverslips were washed to remove unattached vesicles, and the residual plasma membrane sheets adhering to the coverslips were fixed and stained to visualize the plasma membrane sheets by using rhodamine-conjugated WGA, a nonspecific membrane marker. Glut4 was visualized using its GFP tag. (D) Quantification of Glut4-positive plasma membrane sheets. One hundred cells were counted for each condition. Data represent mean ± SD of three independent experiments. Analysis also was examined by performing confocal quantitation of the GFP fluorescence and normalizing to WGA fluorescence. No significant difference was observed between the insulin-stimulated vector and PLD1-RNAi sheets.
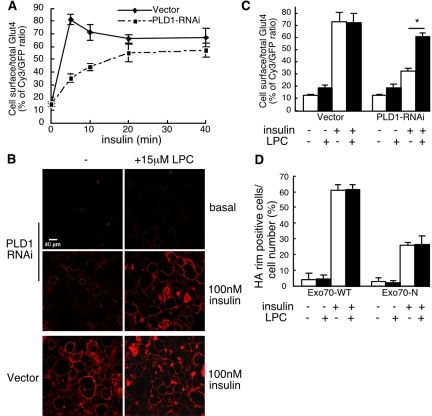
Kinetic analysis was used to examine the rate of fusion in PLD1-RNAi cells. As shown in Figure 7A, wild-type adipocytes exhibit a brief elevated peak of cell-surface Glut4 before settling to lower steady-state levels. This is thought to ensue from mobilization of a highly insulin-responsive intracellular compartment that is depleted upon insulin addition, after which the steady-state exocytosis rate for Glut4 becomes limited at some other step in the recycling pathway (Yeh et al., 1995; Bogan et al., 2001). Glut4 plasma membrane insertion is slowed down more than 10-fold in PLD1-RNAi cells, and no overshoot was observed (Figure 7A; our unpublished data); PA production would accordingly seem to be a rate-limiting component underlying fusion efficiency in insulin-stimulated Glut4 translocation. The remaining low rate of fusion could either reflect action of the residual PLD1 in the RNAi cells or indicate that fusion can proceed nonetheless, albeit at a decreased rate, in the absence of PLD1-generated PA.
Figure 7.
The PLD1-RNAi fusion block is rescued by exogenous provision of LysoPC, whereas the Exo70N docking block is not. (A) Kinetics of insulin-stimulated Glut4 translocation in PLD1-RNAi adipocytes. Measurements were performed in pentuplicate as described in Figure 2 and in Materials and Methods; representative of three experiments. Insulin (100 nM) used for stimulation. (B) Cell surface myc-Glut4 was visualized with or without addition of 15 μM LPC 15 min before insulin stimulation (5 min) in control and PLD1-RNAi cells. (C) Cy3/GFP fluorescence intensity was quantified and averaged for five separate fields. *, significant difference, p < 0.0001. (D) The docking block caused by overexpression of dominant negative Exo70 (Exo70-N; Inoue et al., 2003) cannot be rescued by exogenous addition of LysoPC. Wild-type 3T3-L1 adipocytes were transfected with 50 μg of HA-Glut4-EGFP plus 200 μg of Exo70-WT or Exo70-N. Cells were treated with or without addition of LPC 15 min before 100 nM insulin stimulation and stained with anti-HA antibody followed by Cy3 goat anti-rat IgG to detect cell surface expression of HA-Glut4 (see Supplemental Figure 4 for representative images). Fifty HA-Glut4-EGFP–overexpressing cells from each culture condition were scored for detectable HA plasma membrane rim fluorescence; data represent mean ± SD of four independent experiments.
Decreased Production of PA on the Inner Leaflet of the Plasma Membrane Can Be Rescued by Provision of the Positive Curvature Lipid Lysophosphatidylcholine to the Outer Leaflet of the Plasma Membrane
Once a docked vesicle has become juxtaposed to the plasma membrane, there is progressive formation of a vesicle: plasma membrane stalk, a hemifusion diaphragm, and a micropore, followed by pore expansion (Chernomordik and Kozlov, 2003; Jahn et al., 2003). Grote et al. (2000) previously reported that replacement of SNARE protein transmembrane domains with COOH-terminal geranylgeranylation signals resulted in otherwise normal-seeming, membrane-associated SNARE complexes that are unable to catalyze fusion of the distal leaflets of secretory vesicles and the plasma membrane. Supportive evidence for the proposal that the fusion step was blocked at the relatively late pore formation/expansion step was that the inhibition could be rescued by addition of the positive curvature-inducing lipid LPC to the outside of the cell (Grote et al., 2000). Addition of LPC is thought to lower the energy required to bend the outer membrane leaflet inwards and thus to promote pore formation/expansion. To examine whether the fusion block resulting from PA deficiency occurred at early steps in the fusion process or at the step impacted by geranylgeranylated SNAREs, we challenged PLD1-RNAi cells with LPC and found that the Glut4 fusion block was rescued (Figure 7, B and C; without LPC exposure, 33 ± 2.4% of control; with LPC exposure, 78 ± 3.2% of control). In contrast, Exo-70 dominant negative-transfected cells in which the Glut4-containing vesicles had translocated to the inner surface of the plasma membrane but had not docked (Inoue et al., 2003) could not be rescued by LPC addition (Figure 7D and Supplemental Figure 4). This signifies that the step hindered by PA absence is sufficiently late in the fusion process that an agent that promotes pore formation/expansion is able to compensate for the phenotypic consequence resulting from PLD1 deficiency.
DISCUSSION
Together, the findings reported here provide evidence for an important role for PLD1-generated PA in the final steps of Glut4-containing vesicle fusion into the plasma membrane in response to insulin. The findings constitute the first examination of the endogenous enzyme's localization in adipocytes and physiological role using an RNAi-based loss-of-function model system.
Czech and colleagues previously reported that PLD plays a role in Glut4 translocation based on the findings that overexpressed, epitope-tagged human PLD1 localized to Glut4-containing vesicles and that injection of bacterial or plant PLD into 3T3-L1 adipocytes facilitated insulin-stimulated Glut4 movement to the plasma membrane (Emoto et al., 2000). However, they also concluded that PLD1 played a passive (constitutive) role in Glut4 trafficking because they could not detect PLD activation in response to insulin signaling. One limitation to this work was that the bacterial and plant PLD enzymes used for the functional studies are cytosolic, constitutively active, promiscuous (i.e., they hydrolyze many types of phospholipids, not just phosphatidylcholine), and raise PA levels primarily on perinuclear membranes (Delon et al., 2004). In contrast, mammalian PLD1 is targeted to and cycles through specific membrane locations via the action of pleckstrin homology, Phox domain, and phosphatidylinositol 4,5-bisphosphate membrane-interacting domains (Du et al., 2003); is regulated in a complex manner by agonist signaling pathways (McDermott et al., 2004); and specifically hydrolyzes phosphatidylcholine. However, overexpression of PLD1 can lead to its mislocalization (Freyberg et al., 2001), and another study concluded that PLD does not have a role in insulin-stimulated glucose uptake based on the use of butanol as an inhibitor to divert the catalytic activity of PLD toward generation of phosphatidylbutanol at the expense of PA production (Millar et al., 2000).
Our current study largely supports and extends the observations reported by Emoto et al. (2000). Demonstration that PLD1 is expressed endogenously in 3T3-L1 cells and colocalizes with Glut4 before and after insulin stimulation validates the use of the overexpressed recombinant enzyme as a tool for functional studies. Overexpression of human PLD1, which is targeted and activated physiologically correctly, does facilitate insulin-stimulated Glut4 trafficking and glucose uptake, and we show here that interference with PLD1 function, via expression of a catalytically inactive allele or via selective loss of PLD1 through RNAi targeting, leads to insulin resistance with respect to Glut4 trafficking and glucose uptake. However, because we can detect PLD1 activation by insulin, we conclude that PLD1 plays an active role rather than a passive one in insulin-stimulated Glut4 translocation.
In contrast, our conclusions differ from those of Millar et al. (2000). Skippen et al. (2002) have reported that the concentrations of butanol typically used do not divert PA production sufficiently to block PLD stimulation of phosphatidylinositol 4,5-bisphosphate production via phosphatidylinositol 4,5-kinase. Thus, it is possible that PA generation was not eliminated to the degree required to inhibit Glut4 vesicle fusion at the plasma membrane in the prior study. Alternately, because we observed a delay in Glut4 fusion but not a complete block, differences in the specific assays used or analysis after longer period of insulin stimulation may have made this delay less apparent.
Where and how is PLD1 activated by insulin? Romero and colleagues (Li et al., 2003) have reported that insulin stimulation of PLD is mediated by ARF and its activator ARNO. Vitale et al. (2002) have reported that ARNO localizes to the plasma membrane in PC12 cells, whereas ARF6 localizes to translocating vesicles (Vitale et al., 2002), suggesting that activation takes place as the vesicles approach the plasma membrane. On the other hand, in situ activation of PLD1 on intracellular vesicles also has been demonstrated (Hughes et al., 2002). In the experiments we describe here, the PLD1-generated PA seems to function at the plasma membrane, that is, it is not required for trafficking of the vesicles to the plasma membrane, and thus PLD1 does not need to activated until the vesicles arrive there, but we cannot rule out its activation at an earlier time.
What might the role of PLD1/PA be in late fusion? The ability of LPC to rescue the PLD1-RNAi phenotype suggests as one possibility that PA may play a role in formation of the fusion pore or in its expansion. Theoretical models suggest that the presence of PA at sites of inward membrane curvature should lessen the energy requirements of the curvature process and promote the lipid zippering process that generates the hemifusion diaphragm and provides the force for micropore formation and expansion (Kozlovsky et al., 2002). Accordingly, PA might function via a biophysical mechanism to act in synergy with the SNARE complex fusogenic machinery to generate and expand fusion pores during exocytosis. Alternately, localization and function of the yeast SNARE protein Spo20 is regulated by phosphatidic acid (Coluccio et al., 2004; Nakanishi et al., 2004), and Syntaxin 1A encodes an acidic phospholipid binding motif important for its function (Wagner and Tamm, 2001). Thus, PA might additionally serve as an anchor or recruitment site for components of the fusogenic protein machinery. PA also could function by increasing levels of phosphatidylinositol 4,5-bisphosphate through stimulation of its rate limiting enzyme (Honda et al., 1999; Skippen et al., 2002) or by recruiting any of the growing number of reported PA-binding traffic-related proteins (Manifava et al., 2001). Alternately, PA could be converted by phosphatidic acid phosphohydrolases to diacylglycerol, which itself is a highly fusogenic lipid (Jun et al., 2004).
Finally, our findings are potentially relevant to the many examples of regulated secretion in which PLD activation has been proposed to be involved, including insulin secretion (Lawrence and Birnbaum, 2003; Hughes et al., 2004).
Supplementary Material
Acknowledgments
We thank J. Bogan for the myc-Glut4-EGFP plasmids and cell lines; M. Czech, M. Kanzaki, and J. Smith for sharing protocols and reagents; A. Saltiel for providing the Exo70-N plasmid; C. Alonso for assistance with confocal analysis; and J. Castle, S. Tsirka, and members of the Frohman laboratory for critical discussions. This work was supported by grant DK64166 from the American Diabetes Association and the National Institutes of Health.
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E04–12–1124) on March 16, 2005.
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
References
- Andresen, B. T., Rizzo, M. A., Shome, K., and Romero, G. (2002). The role of phosphatidic acid in the regulation of the Ras/MEK/Erk signaling cascade. FEBS Lett. 531, 65–68. [DOI] [PubMed] [Google Scholar]
- Bogan, J. S., McKee, A. E., and Lodish, H. F. (2001). Insulin-responsive compartments containing Glut4 in 3T3–l1 and chinese hamster ovary cells: regulation by amino acid concentrations. Mol. Cell. Biol. 21, 4785–4806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bose, A., Cherniack, A. D., Langille, S. E., Nicoloro, S.M.C., Buxton, J. M., Park, J. G., Chawla, A., and Czech, M. P. (2001). Galpha11 Signaling through ARF6 Regulates F-actin Mobilization and GLUT4 Glucose Transporter Translocation to the Plasma Membrane. Mol. Cell. Biol. 21, 5262–5275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brummelkamp, T. R., Bernards, R., and Agami, R. (2002). A system for stable expression of short interfering RNAs in mammalian cells. Science 296, 550–553. [DOI] [PubMed] [Google Scholar]
- Bryant, N. J., Govers, R., and James, D. E. (2002). Regulated transport of the glucose transporter GLUT4. Nat. Rev. Mol. Cell. Biol. 3, 267–277. [DOI] [PubMed] [Google Scholar]
- Chen, Y. G., Siddhanta, A., Austin, C. D., Hammond, S. M., Sung, T. C., Frohman, M. A., Morris, A. J., and Shields, D. (1997). Phospholipase D stimulates release of nascent secretory vesicles from the trans-Golgi network. J. Cell Biol. 138, 495–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernomordik, L. V., and Kozlov, M. M. (2003). Protein-Lipid interplay in fusion and fission of biological membranes. Annu. Rev. Biochem. 72, 175–207. [DOI] [PubMed] [Google Scholar]
- Choi, W. S., Kim, Y. M., Combs, C., Frohman, M. A., and Beaven, M. A. (2002). Phospholipase D1 and 2 regulate different phases of exocytosis in mast cells. J. Immunol. 168, 5682–5689. [DOI] [PubMed] [Google Scholar]
- Coluccio, A., Malzone, M., and Neiman, A. M. (2004). Genetic evidence of a role for membrane lipid composition in the regulation of soluble NEM-sensitive factor receptor function in Saccharomyces cerevisiae. Genetics 166, 89–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delon, C., Manifava, M., Wood, E., Thompson, D., Krugmann, S., Pyne, S., and Ktistakis, N. T. (2004). Sphingosine kinase 1 is an intracellular effector of phosphatidic Acid. J. Biol. Chem. 279, 44763–44774. [DOI] [PubMed] [Google Scholar]
- Di Paolo, G., et al. (2004). Impaired PtdIns(4,5)P2 synthesis in nerve terminals produces defects in synaptic vesicle trafficking. Nature 431, 415–422. [DOI] [PubMed] [Google Scholar]
- Du, G., Huang, P., Liang, B. T., and Frohman, M. A. (2004). Phospholipase D2 localizes to the plasma membrane and regulates angiotensin II receptor endocytosis. Mol. Biol. Cell 15, 1024–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du, G., Huang, P., Vitale, N., Chasserot-Golaz, S., Altshuller, Y. M., Morris, A. J., Bader, M.-F., and Frohman, M. A. (2003). Regulation of Phospholipase D subcellular cycling through coordination of multiple membrane association motifs. J. Cell Biol. 62, 305–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emoto, M., Klarlund, J., Waters, S., Hu, V., Buxton, J., Chawla, A., and Czech, M. (2000). A role for phospholipase D in GLUT4 glucose transporter translocation. J. Biol. Chem. 275, 7144–7151. [DOI] [PubMed] [Google Scholar]
- Freyberg, Z., Siddhanta, A., Sweeney, D., Bourgoin, S., Frohman, M. A., and Shields, D. (2001). Intracellular localization of phospholipase D1 in mammalian cells. Mol. Biol. Cell 12, 943–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frohman, M. A., and Morris, A. J. (1999). Phospholipase D structure and regulation. Chem. Phys. Lipids 98, 127–140. [DOI] [PubMed] [Google Scholar]
- Grote, E., Baba, M., Ohsumi, Y., and Novick, P. J. (2000). Geranylgeranylated SNAREs are dominant inhibitors of membrane fusion. J. Cell Biol. 151, 453–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, T. C., Zhou, S., da Costa, L. T., Yu, J., Kinzler, K. W., and Vogelstein, B. (1998). A simplified system for generating recombinant adenoviruses. Proc. Nat. Acad. Sci. USA 95, 2509–2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda, A., et al. (1999). Phosphatidylinositol 4-phosphate 5-kinaseα is a downstream effector of the small G protein ARF6 in membrane ruffle formation. Cell 99, 521–532. [DOI] [PubMed] [Google Scholar]
- Hughes, W. E., Elgundi, Z., Huang, P., Frohman, M. A., and Biden, T. J. (2004). Phospholipase D1 regulates secretagogue-stimulated insulin release in pancreatic b-cells. J. Biol. Chem. 279, 27534–27541. [DOI] [PubMed] [Google Scholar]
- Hughes, W. E., Larijani, B., and Parker, P. J. (2002). Detecting protein-phospholipid interactions. Epidermal growth factor-induced activation of phospholipase D1b in situ. J. Biol. Chem. 277, 22974–22979. [DOI] [PubMed] [Google Scholar]
- Inoue, M., Chang, L., Hwang, J., Chiang, S. H., and Saltiel, A. R. (2003). The exocyst complex is required for targeting of Glut4 to the plasma membrane by insulin. Nature 422, 629–633. [DOI] [PubMed] [Google Scholar]
- Jun, Y., Fratti, R. A., and Wickner, W. (2004). Diacylglycerol and its formation by phospholipase C regulate Rab- and SNARE-dependent yeast vacuole fusion. J. Biol. Chem. 279, 53186–53195. [DOI] [PubMed] [Google Scholar]
- Jahn, R., Lang, T., and Sudhof, T. C. (2003). Membrane fusion. Cell 112, 519–533. [DOI] [PubMed] [Google Scholar]
- Kanzaki, M., and Pessin, J. E. (2001). Insulin-stimulated GLUT4 translocation in adipocytes is dependent upon cortical actin remodeling. J. Biol. Chem. 276, 42436–42444. [DOI] [PubMed] [Google Scholar]
- Kanzaki, M., and Pessin, J. E. (2002). Caveolin-associated filamentous actin (Cav-actin) defines a novel F-actin structure in adipocytes. J. Biol. Chem. 277, 25867–25869. [DOI] [PubMed] [Google Scholar]
- Kooijman, E. E., Chupin, V., de Kruijff, B., and Burger, K. N. (2003). Modulation of membrane curvature by phosphatidic acid and lysophosphatidic acid. Traffic 4, 162–174. [DOI] [PubMed] [Google Scholar]
- Kozlovsky, Y., Chernomordik, L. V., and Kozlov, M. M. (2002). Lipid intermediates in membrane fusion: formation, structure, and decay of hemifusion diaphragm. Biophys. J. 83, 2634–2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence, J. T., and Birnbaum, M. J. (2003). ADP-ribosylation factor 6 regulates insulin secretion through plasma membrane phosphatidylinositol 4,5-bisphosphate. Proc. Nat. Acad. Sci. USA 100, 13320–13325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, D., Randhawa, V. K., Patel, N., Hayashi, M., and Klip, A. (2001). Hyperosmolarity reduces GLUT4 endocytosis and increases its exocytosis from a VAMP2-independent pool in l6 muscle cells. J. Biol. Chem. 276, 22883–22891. [DOI] [PubMed] [Google Scholar]
- Li, H. S., Shome, K., Rojas, R., Rizzo, M. A., Vasudevan, C., Fluharty, E., Santy, L. C., Casanova, J. E., and Romero, G. (2003). The guanine nucleotide exchange factor ARNO mediates the activation of ARF and phospholipase D by insulin. BMC Cell Biol. 4, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manifava, M., Thuring, J. W., Lim, Z. Y., Packman, L., Holmes, A. B., and Ktistakis, N. T. (2001). Differential binding of traffic-related proteins to phosphatidic acid- or phosphatidylinositol (4,5)-bisphosphate-coupled affinity reagents. J. Biol. Chem. 276, 8987–8994. [DOI] [PubMed] [Google Scholar]
- McDermott, M., Wakelam, M. J., and Morris, A. J. (2004). Phospholipase D. Biochem. Cell Biol. 82, 225–253. [DOI] [PubMed] [Google Scholar]
- Millar, C. A., Meerloo, T., Martin, S., Hickson, G. R., Shimwell, N. J., Wakelam, M. J., James, D. E., and Gould, G. W. (2000). Adipsin and the glucose transporter GLUT4 traffic to the cell surface via independent pathways in adipocytes. Traffic 1, 141–151. [DOI] [PubMed] [Google Scholar]
- Morris, A. J., Frohman, M. A., and Engebrecht, J. (1997). Measurement of phospholipase D activity. Anal. Biochem. 252, 1–9. [DOI] [PubMed] [Google Scholar]
- Nakanishi, H., de los Santos, P., and Neiman, A. M. (2004). Positive and negative regulation of a SNARE protein by control of intracellular localization. Mol. Biol. Cell 15, 1802–1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzo, M. A., Shome, K., Vasudevan, C., Stolz, D. B., Sung, T.-C., Frohman, M. A., Watkins, S. C., and Romero, G. (1999). Phospholipase D and its product, phosphatidic acid, mediate agonist-dependent Raf-1 translocation to the plasma membrane and the activation of the MAP kinase pathway. J. Biol. Chem. 274, 1131–1139. [DOI] [PubMed] [Google Scholar]
- Robinson, L. J., Pang, S., Harris, D. S., Heuser, J., and James, D. E. (1992). Translocation of the glucose transporter (GLUT4) to the cell surface in permeabilized 3T3–L1 adipocytes: effects of ATP insulin, and GTP gamma S and localization of GLUT4 to clathrin lattices. J. Cell Biol. 117, 1181–1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seiler, A. E., Henderson, A., and Rubin, R. (2000). Ethanol inhibits insulin receptor tyrosine kinase. Alcohol Clin. Exp. Res. 24, 1869–1872. [PubMed] [Google Scholar]
- Shen, Y., Xu, L., and Foster, D. (2001). Role for phospholipase D in receptor-mediated endocytosis. Mol. Cell. Biol. 21, 595–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson, F., Whitehead, J. P., and James, D. E. (2001). GLUT4–at the cross roads between membrane trafficking and signal transduction. Traffic 2, 2–11. [DOI] [PubMed] [Google Scholar]
- Skippen, A., Jones, D. H., Morgan, C. P., Li, M., and Cockcroft, S. (2002). Mechanism of ADP ribosylation factor-stimulated phosphatidylinositol 4,5-bisphosphate synthesis in HL60 cells. J. Biol. Chem. 277, 5823–5831. [DOI] [PubMed] [Google Scholar]
- Sung, T. C., Roper, R. L., Zhang, Y., Rudge, S. A., Temel, R., Hammond, S. M., Morris, A. J., Moss, B., Engebrecht, J., and Frohman, M. A. (1997). Mutagenesis of phospholipase D defines a superfamily including a trans-Golgi viral protein required for poxvirus pathogenicity. EMBO J. 16, 4519–4530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurmond, D. C., Ceresa, B. P., Okada, S., Elmendorf, J. S., Coker, K., and Pessin, J. E. (1998). Regulation of insulin-stimulated GLUT4 translocation by Munc18c in 3T3L1 adipocytes. J. Biol. Chem. 273, 33876–33883. [DOI] [PubMed] [Google Scholar]
- Vitale, N., Caumont-Primus, A.-S., Chasserot-Golaz, S., Du, G., Wu, S., Sciorra, V. A., Morris, A. J., Frohman, M. A., and Bader, M.-F. (2001). Phospholipase D 1, a key factor for the exocytotic machinery in neuroendocrine cells. EMBO J. 20, 2424–2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitale, N., Chasserot-Golaz, S., Bailly, Y., Morinaga, N., Frohman, M. A., and Bader, M.-F. (2002). Calcium-regulated exocytosis of dense core vesicles requires the activation of ARF6 by ARNO at the plasma membrane. J. Cell Biol. 159, 79–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner, M. L., and Tamm, L. K. (2001). Reconstituted syntaxin1a/SNAP25 interacts with negatively charged lipids as measured by lateral diffusion in planar supported bilayers. Biophys. J. 81, 266–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson, R. T., Khan, A. H., Furukawa, M., Hou, J. C., Li, L., Kanzaki, M., Okada, S., Kandror, K. V., and Pessin, J. E. (2004). Entry of newly synthesized GLUT4 into the insulin-responsive storage compartment is GGA dependent. EMBO J. 23, 2059–2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson, R. T., Shigematsu, S., Chiang, S. H., Mora, S., Kanzaki, M., Macara, I. G., Saltiel, A. R., and Pessin, J. E. (2001). Lipid raft microdomain compartmentalization of TC10 is required for insulin signaling and GLUT4 translocation. J. Cell Biol. 154, 829–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, J., Yeon, J. E., Chang, H., Tison, G., Chen, G. J., Wands, J., and de la Monte, S. (2003). Ethanol impairs insulin-stimulated neuronal survival in the developing brain: role of PTEN phosphatase. J. Biol. Chem. 278, 26929–26937. [DOI] [PubMed] [Google Scholar]
- Yeh, J. I., Verhey, K. J., and Birnbaum, M. J. (1995). Kinetic analysis of glucose transporter trafficking in fibroblasts and adipocytes. Biochem. 34, 15523–15531. [DOI] [PubMed] [Google Scholar]
- Zhang, Y., Huang, P., Du, G., Kanaho, Y., Frohman, M. A., and Tsirka, S. E. (2004). Increased expression of two phospholipase D isoforms during experimentally induced hippocampal mossy fiber outgrowth. Glia 46, 74–83. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.