Abstract
The non-ORC protein, TIF1, recognizes sequences in the Tetrahymena thermophila ribosomal DNA (rDNA) minichromosome that are required for origin activation. We show here that TIF1 represses rDNA origin firing, but is required for proper macronuclear S phase progression and division. TIF1 mutants exhibit an elongated macronuclear S phase and diminished rate of DNA replication. Despite this, replication of the rDNA minichromosome initiates precociously. Because rDNA copy number is unaffected in the polyploid macronucleus, mechanisms that prevent reinitiation appear intact. Although mutants exit macronuclear S with a wild-type DNA content, division of the amitotic macronucleus is both delayed and abnormal. Nuclear defects are also observed in the diploid mitotic micronucleus, as TIF1 mutants lose a significant fraction of their micronuclear DNA. Hence, TIF1 is required for the propagation and subsequent transmission of germline chromosomes. The broad phenotypes associated with a TIF1-deficiency suggest that this origin binding protein is required globally for the proper execution and/or monitoring of key chromosomal events during S phase and possibly at later stages of the cell cycle. We propose that micro- and macronuclear defects result from exiting the respective nuclear S phases with physically compromised chromosomes.
INTRODUCTION
The initiation of eukaryotic DNA replication is regulated by protein-DNA interactions that occur within defined chromosomal domains, termed replicators or replicons. Eukaryotic replicators are modular and contain binding sites for the conserved six-subunit origin recognition complex (ORC; Bell and Stillman, 1992) and non-ORC DNA binding proteins (Marahrens and Stillman, 1992). ORC plays a central role, recruiting proteins involved in replication initiation and elongation to form the prereplicative complex (pre-RC). These proteins include a replicative helicase—the minichromosome maintenance (MCM) complex (Ishimi, 1997; Labib et al., 2000) and factors that regulate origin activation, such as Cdc6 and Cdt1 (Nishitani et al., 2000; Oehlmann et al., 2004).
Although ORC binding sites are plentiful in the Saccharomyces cerevisiae (Sc) genome, only a fraction are routinely engaged in replication initiation (Theis and Newlon, 1993; Wyrick et al., 2001). Genetic studies indicate that ScORC binding is necessary, but not sufficient, to confer replicator status to a chromosomal domain. Although ScORC binds DNA in a sequence-specific manner, metazoan ORCs exhibit no obvious sequence-specificity, displaying a preference for degenerate, asymmetric A:T-rich sequences in vitro (Austin et al., 1999; Chesnokov et al., 2001; Vashee et al., 2001) and in vivo (Kong et al., 2003; Vashee et al., 2003). This relaxed specificity is similar to Schizosaccharomyces pombe (Sp) ORC, which binds DNA via an unusual A-T hook domain in its Orc4 subunit (Chuang and Kelly, 1999; Kong and DePamphilis, 2001). Although metazoan ORCs lack AT hooks, they still associate with specific replicator domains in vivo (Austin et al., 1999; Abdurashidova et al., 2004).
The contribution of non-ORC DNA binding proteins to replication initiation is less well understood. Several proteins have been shown to impart replicator status to a given chromosomal site. For example, S. cerevisiae ABF1 appears to function primarily as a physical barrier that prevents nucleosomes from invading the adjacent ORC binding site (Venditti et al., 1994; Lipford and Bell, 2001). By comparison, localization of DmORC to the chromosome 3 chorion gene locus may be facilitated by interactions with a sequence-specific DNA binding complex (Beall et al., 2002). Genetic experiments indicate that this myb-containing complex facilitates chorion gene amplification, but represses replication at sites in the genome in terminally differentiated follicle cells (Beall et al., 2004). Consequently, the selective activation of chorion gene replicons involves additional layers of regulation.
Similar to metazoan replicators, the Tetrahymena thermophila ribosomal DNA (rDNA) minichomosome contains dispersed cis-acting replicator elements, including essential determinants that either colocalize to replication initiation sites or act at a distance. The 1.9-kb 5′ nontranscribed spacer (5′ NTS) is necessary and sufficient for developmentally programmed amplification and cell cycle–regulated replication of the macronuclear rDNA minichromosome (reviewed in Kapler et al., 1996). Origin-proximal and distal type I elements act in concert to control replication that initiates within two nucleosome-free regions (Figure 1A; Larson et al., 1986; Blomberg et al., 1997; Zhang et al., 1997; Gallagher and Blackburn, 1998; Reischmann et al., 1999). In addition to their role in replication initiation, type I elements induce the pausing of replication forks at adjacent pause site elements (PSEs; MacAlpine et al., 1997). Promoter-proximal type I elements are also required for rRNA transcription (Gallagher and Blackburn, 1998). Separation-of-function alleles support a model in which replication and transcription are regulated by different type I element binding factors. Whether these factors compete or cooperate to regulate origin firing is unknown.
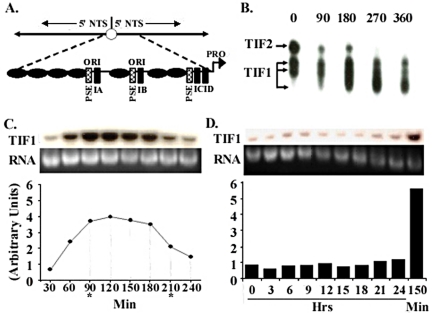
Figure 1.
TIF1 DNA binding activity and mRNA levels peak in S phase. (A) Schematic of the 21-kb palindromic rDNA minichromosome and 1.9-kb 5′ nontranscribed spacer (5′NTS; expanded diagram), including TIF1-binding sites (PSE and type I elements) and positioned nucleosomes (black ovals), and replication origins (ori) that reside in the 230 base pairs nucleosome-free regions that are part of an imperfect 430-base pair tandem duplication. (B) TIF1 DNA binding activity is cell cycle regulated. Gel shift analysis of extracts prepared from vegetative cultures synchronized by starvation and refeeding. The probe, ssA37, corresponds to the A-rich strand of the type IB element (Saha and Kapler, 2000). (C) TIF1 mRNA levels are cell cycle regulated. Northern blot analysis with an intron-spanning TIF1 coding region probe on mRNA prepared from cells synchronized by starvation and refeeding (Mohammad et al., 2000). Hybridization signals were normalized to ethidium bromide staining of the rRNA and plotted as a function of time. Asterisks demarcate the approximate beginning and end of macronuclear S phase. (D) TIF1 mRNA levels are constant throughout development. TIF1 Northern blot analysis on mRNA prepared from cells at various time points during conjugation. Developmental landmarks: 3 h: premeiotic S, 7–8 h: postzygotic S, 10–24 h: macronuclear development, including rDNA gene amplification. T = 150 min: peak TIF1 mRNA level in synchronized vegetative cell cultures.
Type I elements are recognized by four distinct DNA binding activities in vitro, designated type I factors (TIF1, TIF2, TIF3, and TIF4; Umthun et al., 1994; Mohammad et al., 2000). In contrast to ORC and non-ORC replicator proteins in other systems, all four TIFs associate exclusively with single-stranded DNA in vitro (Mohammad et al., 2000, 2003; Saha and Kapler, 2000). Consistent with an in vivo role for these sequence-specific single-stranded DNA binding proteins (ss-SSBs), in vivo footprinting studies indicate that the Tetrahymena rDNA origin and promoter regions are naturally unwound in native chromosomes (Saha et al., 2001). The multisubunit TIF4 complex (MW ∼550 kDa) shares several biochemical properties with ScORC (Mohammad et al., 2003). Like ScORC, TIF4 recognizes a single known target that colocalizes with the site of replication initiation. TIF4 binding to the type I element T-rich strand, is both ATP-dependent and sequence-specific. TIF4 contains an Orc2-related subunit, Tt-p69, which cross-reacts with antibodies that recognize yeast, Drosophila and human Orc2 proteins. Tt-p69 associates with DNA in a cell cycle–regulated manner, similar to the metazoan Orc1 and Orc2 subunits (Kreitz et al., 2001; Fugita et al., 2002; Li and DePamphilis, 2002). Tt-p69 relocalizes from the cytoplasm to macronucleus during vegetative S phase (Mohammad et al., 2003). Furthermore, Tt-p69 is restricted to replicating nuclear compartments during development and has been implicated in rDNA gene amplification.
The TIF1 homotetramer (subunit MW 21 kDa) exhibits a more relaxed sequence specificity than TIF4, binding to the A-rich or T-rich strand of type I elements or adjacent PSEs, but not to the respective duplex DNA substrates. PSEs map to the 5′ border of the nucleosome-free origin regions (MacAlpine et al., 1997) and are required for replication of the rDNA minichromosome (Saha et al., 2001). Footprinting studies revealed that TIF1 modulates the occupancy of origin- and promoter-proximal PSE and type I elements in vivo (Saha et al., 2001). Remarkably, TIF1 affects the footprint on the A-rich strand at the origin and T-rich strand at the promoter. Consequently, TIF1 might facilitate TIF4 binding to the T-rich origin strand or function at a later step to regulate origin firing.
An important event in the Tetrahymena life cycle is the programmed amplification of rDNA minichromosomes. The rDNA minichromosome is formed as part of a genetic program that transforms a transcriptionally silent, diploid (germline) micronucleus into a transcriptionally active polyploid (somatic) macronucleus. The single copy 10.3-kb rRNA gene is excised from its parental chromosome by site-specific fragmentation, rearranged into a palindromic 21-kb minichromosome and amplified to ∼9000 copies in a single S phase (Yao et al., 1974). The remainder of the five germline chromosomes are fragmented into ∼280 segments that attain a copy number of 45. During subsequent vegetative growth, macronuclear chromosomes replicate once (on average) per cell cycle. Although these chromosomes lack centromeres, genic balance is somehow maintained in the macronucleus (Doerder, 1979; Preer and Preer, 1979; Pan and Blackburn, 1995). In contrast, micronuclear chromosomes segregate by conventional mitosis and meiosis.
Here we assess the role of the non-ORC rDNA origin binding protein, TIF1, in DNA replication. We demonstrate that TIF1 is a negative regulator of rDNA replication. TIF1-deficient cells replicate the rDNA minichromosome precociously, but exhibit a delay in macronuclear S phase progression that is associated with a diminished rate of DNA replication. TIF1 mutants also undergo aberrant macronuclear division, despite the fact that they exit S phase with a normal DNA content. TIF1's role is not restricted to macronuclear functions, as the mutant also fails to faithfully propagate chromosomes in the mitotic, diploid germline micronucleus. Thus, TIF1 plays an important role in the replication and transmission of chromosomes in these two distinct nuclear compartments.
MATERIALS AND METHODS
Tetrahymena Strains and DNA Transformation
Tetrahymena thermophila strains were cultured as previously described (Kapler and Blackburn, 1994). The wild-type strain CU428 was used for comparative studies with TIF1 gene replacements. The TIF1 gene disruption plasmid (pTIF1::neo) was generated by replacing the TIF1 coding region with a β-tubulin promoter-driven neomycin phosphotransferase (neo) gene. The plasmid insert was released from the vector backbone by restriction digestion for homologous targeting in the micro- or macronucleus.
Mating cultures (strains CU427 × B2086) were transformed by biolistic bombardment at 3.5 h after mixing to generate germline transformants that underwent targeted homologous recombination in the micronucleus (Cassidy-Hanley et al., 1997). Cells that expressed the transforming neo gene were selected for resistance to 150 μg/ml paromomycin (pm). Heterozygous germline transformants were subsequently cultured in the absence of drug selection. Gene replacements were initially verified by PCR using TIF1 and neo-coding region primers. The genotypes of heterozygous germline transformants (TXh102 and TXh106) and additional strains were verified by PCR, Southern blot analysis, and/or mating to wild-type testers as previously described (Kapler et al., 1994).
TIF1:neo heterokaryons were generated by mating sexually mature heterozygous TIF1::neo micronuclear transformants with the functionally-amicronucleate A* strains (mating type III or V). Because A* strains contribute no genetic information to progeny, a process termed Round 1 genomic exclusion generates progeny that are homozygous at all loci in the micronucleus, but retain the macronucleus from the A* or transformant parent (Allen, 1967). Pm-sensitive progeny that expressed the mating type of the A* parent and contained the TIF1::neo allele in the micronucleus were isolated. A single homozygous null TIF1 strain, TXk202, was subsequently generated by mating homozygous TIF1 germline knockout heterokaryons (TXa28 and TXa42) to one another and selecting for pm-resistant progeny. Micronuclear genome stability of homozygous TIF1:neo heterokaryons (TXa28 and TXa42) and the homozygous TIF1 knockout (TXk202) was assessed by mating these mutants with heterokaryon strains (CU354, CU357, CU361, and CU371) that are nullisomic for micronuclear chromosome 2, 3, 4, or 5, respectively, but contain a wild-type macronucleus.
Transformation of the vegetative macronucleus was achieved by biolistic transformation of starved CU428 cells. Transformant strains TXh48 and TXh29 were cultured in increasing concentrations of pm (150–4500 μg/ml) to select for cells with decreasing amounts of the wild-type TIF1 gene in the macronucleus. Random assortment of amitotic macronuclear chromosomes (∼45C) was exploited in an attempt to generate complete macronuclear gene replacements (reviewed in Turkewitz et al., 2002).
Molecular Biology Techniques
Standard molecular biology techniques, including Southern blotting, Northern blotting, electrophoretic mobility shift assays (EMSA), and RT-PCR were performed as described (Mohammad et al., 2000; Saha et al., 2001). DNA and RNA hybridization signal were quantified on a Bio-Rad Molecular Imager FX PhosphoImager (Richmond, CA). Quantitation of rDNA chromosome copy number was achieved by determining the ratio of the rDNA hybridization signal to two non-rDNA probes derived from either a large (>1000 kb) or small (50 kb) macronuclear chromosome in wild-type and TIF1-deficient strains. RNA samples were prepared using an RNAeasy mini-kit (Qiagen, Chatsworth, CA) according to the manufacturer's recommendations.
Cell Cycle Synchronization
Cell cycle synchronization was achieved by starvation for at least 8 h and refeeding or by modification of a stationary phase synchronization protocol in which saturated cultures are placed in starvation medium for 8 h before dilution into growth medium at a density of 0.6 × 105 cells/ml (Mohammad et al., 2003). Cells were collected at each time point and incubated with 100 μg/ml bromo-deoxyuridine (BrdU; Sigma Chemical, St. Louis, MO) for 15 min to assess micro- and macronuclear DNA synthesis by immunofluorescence microscopy (see below). Alternatively, cells were radiolabeled for 15 min at 30-min intervals with tritiated thymidine (Perkin Elmer-Cetus, Norwalk, CT; 79Ci/mmol) at a final concentration of 5 μCi/ml. Thymidine incorporation was measured by liquid scintillation counting of trichloroacetic acid precipitates.
Two-dimensional Gel Electrophoresis of rDNA Replication Intermediates
DNA samples were prepared from refed stationary phase synchronized cultures harvested at 30 min intervals. For each time point, twenty micrograms of HindIII-digested total genomic DNA was resolved by two-dimensional gel electrophoresis and hybridized to an rDNA 5′ NTS probe (Zhang et al., 1997).
Immunofluorescence Studies
For mating experiments, wild-type strains (CU427, CU428, and B2086) were distinguished from TIF1 knockout (TXk202) and knockdown (TXh48, TXh29) strains by incorporation of Mitotracker Green FM or Red-CMXRos dyes (Molecular Probes, Eugene, OR) during overnight starvation of premating (single strain) cultures. Reciprocal labeling experiments revealed that these dyes do not alter the phenotypes described in Results. Cell preparation and fluorescence microscopy were performed essentially as previously described (Marsh et al., 2000). One-milliliter mating cultures were harvested at selected developmental time points, washed sequentially with 1 ml of distilled water, 50% methanol, 70% methanol, and 70% methanol/15% acetic acid fixative. Cells were resuspended in 100 μl of methanol/acetic acid fixative, dropped onto microscope slides from a height of 30–60 cm, and air-dried. Slides were sequentially dipped in 95% ethanol (15 s), 0.1 μg/ml 4′,6′-diamidino-2-phenylidole (DAPI, Sigma Chemical) in 70% ethanol/300 mM NaCl (1 min), 70% ethanol (15 s), 35% ethanol (15 s), and examined by fluorescence microscopy. Live cells were stained with Apofluor (0.001% acridine orange and 5 μg/ml Hoescht 33342/ml) and examined by fluorescence microscopy.
For cell cycle analysis of DNA replication, BrdU-labeled cells (15-ml cultures) were harvested by centrifugation, washed with 10 mM Tris (pH 7.4), and incubated in 2 ml of PHEM buffer (60 mM PIPES, 25 mM HEPES, 10 mM EGTA, 2 mM MgCl2, pH 6.9) + 0.5% Triton X-100 for 3 min. Cells were recentrifuged, fixed in 1 ml PHEM + 3% paraformaldehyde for 30–60 min at 4°C, and washed three times with phosphate-buffered saline (PBS; Marsh et al., 2000). Fixed cells were then placed in PBT blocking buffer (PBS + 3% bovine serum albumin/0.1% Tween 20) for >1 h at 4°C. Cells were sequentially incubated at room temperature (RT) for 20 min with PBT + 5% normal donkey serum (Jackson ImmunoResearch Laboratories, West Grove, PA), 1 h with anti-bromo-deoxyuridine antibody (mouse monoclonal, Amersham-Pharmacia Biotech, Piscataway, NJ) according to the manufacturer's specifications and 1 h with rhodamine-conjugated goat anti-mouse antibody (Jackson ImmunoResearch Laboratories, 1:100 dilution). Cells were resuspended in 0.1 μg/ml DAPI (Sigma) for 10 min and washed twice with PBS, and mounted onto slides in glycerol:phosphate-buffered saline (9:1) for microscopy examination. The percentage of BrdU-labeled nuclei was determined for 300–500 cells per time point.
Flow Cytometry
Vegetative wild-type (CU428) and TIF1 KO (TXk202) cultures were starved overnight and refed at a density of 1.0 × 105 cells/ml. Cells were harvested at 1 h intervals, resuspended in 0.5 ml PBS + 4.5 ml of 70% ethanol, and incubated for 2 h at 4°C. Samples were washed at RT with 0.5 ml PBS followed by staining for 30 min in PBS + 0.1% Triton X-100, 0.02 mg/ml propidium iodide, 0.2 mg/ml RNAseA (Darzynkiewicz et al., 2003). Cell fluorescence was measured on a Becton Dickinson (Mountain View, CA; FACScalibur) flow cytometer.
RESULTS
TIF1 binds to dispersed, genetically defined replicator sequence elements in vitro (Figure 1A, type I elements and PSEs) and modulates the in vivo footprint of the rDNA replication origins and promoter in native chromosomes (Saha et al., 2001). To better understand the biological role of TIF1, we examined vegetative cell division and development in strains carrying partial or complete replacements of the wild-type TIF1 gene.
TIF1 DNA Binding Activity and mRNA Levels are Cell Cycle Regulated
The ability of TIF1 to alter the in vivo occupancy of the rDNA origin suggests a role for this protein in the initiation of DNA replication. Because many S phase–specific genes are subjected to cell cycle regulation at the RNA or protein level, we examined TIF1 steady state mRNA and protein (in vitro DNA binding activity) levels in synchronized vegetative cultures. Affinity-purified TIF1 generates three distinct DNA:protein complexes that can be resolved by electrophoretic mobility shift analysis (EMSA) in a Tris:glycine buffer (Saha et al., 2001). The EMSA profile of extracts prepared from starved/refed vegetative cultures revealed that TIF1 DNA binding activity is cell cycle regulated (Figure 1B), producing a profile similar to that obtained for the TIF4 origin binding complex (Mohammad et al., 2003).
Northern blotting was performed to monitor TIF1 gene expression. This analysis revealed that TIF1 mRNA levels are also regulated across the cell cycle (Figure 1C). Maximal signals were obtained at the 90–180-min interval, which includes the distinct periods for macro- and micronuclear DNA replication (Mohammad et al., 2003). A pronounced decline in TIF1 mRNA abundance was evident before cytokinesis (240 min). Although tritiated thymidine labeling revealed experimental variation in the lag period before S phase (unpublished data), TIF1 mRNA levels reproducibly rose before the onset of macronuclear DNA synthesis. The maximal TIF1 mRNA signal was 4–7-fold greater than that observed in pre-S phase cells (Figure 1C, graph).
Northern blotting was also used to monitor TIF1 mRNA levels during development. Pre- and postmeiotic micronuclear replication precedes the formation of a new macronucleus in progeny cells (3–8 h), with the Orc2-related TIF4 subunit specifically localizing to nuclei that are actively engaged in DNA replication (Mohammad et al., 2003). Selective amplification of the rDNA occurs between 10 and 20 h. In contrast to cycling vegetative cells, TIF1 mRNA levels were relatively constant throughout development. Signal intensities were comparable to that seen in starved cell cultures (Figure 1D, 0–24 h), and much lower than S phase vegetative cells (Figure 1D, right lane: 150 min). Despite the low level of TIF1 mRNA, previous EMSA experiments showed an increase in TIF1 DNA binding activity in cells undergoing macronuclear development (Mohammad et al., 2000). The basis for this difference is unknown.
Partial and Complete Macronuclear Replacement of the Wild-type TIF1 Gene
Partial inhibition of TIF1 mRNA translation using an antisense ribosome strategy previously revealed that TIF1 modulates the occupancy of PSE and type I elements in vivo and differentially marks the rDNA origin and promoter regions (Saha et al., 2001). To further investigate the role of TIF1 in rDNA replication and assess whether TIF1's role is restricted to macronuclear rDNA functions, we generated strains that were partially depleted for TIF1 or that lacked the TIF1 gene entirely (Cassidy-Hanley et al., 1997).
Because macronuclear chromosomes segregate randomly, TIF1:neo transformants that retain more copies of the disruption allele can be obtained by pm-selection (reviewed in Turkewitz et al., 2002). Transformation of the vegetative macronucleus with a TIF1 disruption construct (Figure 2A, left panel) produced strains TXh48 and TXh29. Phenotypic assortment of the wild-type TIF1 gene was used to titrate TIF1 to a rate-limiting dosage. Primary transformants were sequentially cultured in increasing concentrations of pm (from 100 to 4500 μg/ml) to select for cells that harbored a higher percentage of the TIF1::neo disruption allele in the polyploid amitotic macronucleus. Cells that were resistant to intermediate and high levels of pm grew more slowly than wild type (unpublished data), suggesting that selection against the TIF1 gene was being counteracted by a slower rate of cell division. Southern blot analysis of two macronuclear transformants, resistant to 4500 μg/ml pm, revealed that the wild-type TIF1 gene had not been completely replaced (Figure 2A, middle panel, TXh48 and TXh29). PhosphorImager quantitation indicated that the copy number of the intact TIF1 gene in lines 48 and 29 was reduced to ∼25 and 50% of wild-type, respectively (after normalization to the β-tubulin loci, btu1 and btu2). Northern blotting revealed a comparable decrease in TIF1 mRNA abundance (unpublished data). The inability to completely replace the wild-type TIF1 gene in the macronucleus argues that there is strong selective pressure to retain a threshold level of TIF1 in the cell.
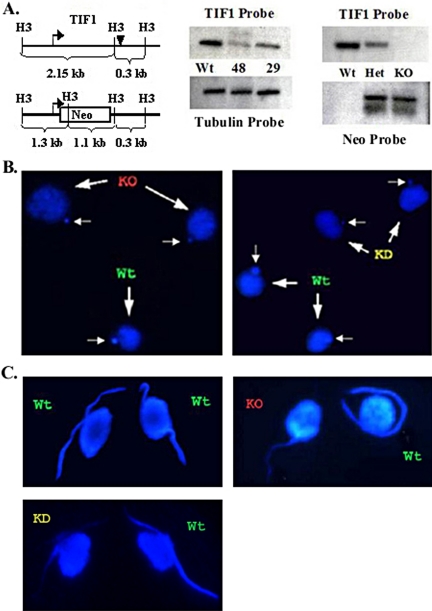
Figure 2.
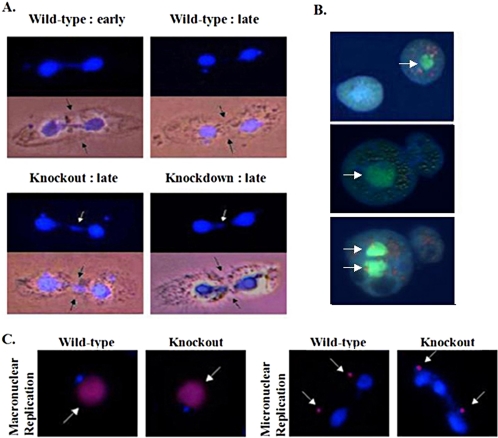
Molecular and cytological analysis of TIF1 knockout and knockdown strains. (A) Restriction map of the wild-type TIF1 gene and TIF1::neo replacement allele (Neo). Bent arrow, TIF1 initiator methionine; filled triangle, TIF1 stop codon. The TIF1 coding region probe recognizes an ∼2.15-kb HindIII fragment, whereas the neo probe hybridizes to 1.3- and 1.1-kb fragments. Middle panel: Southern blot analysis of a macronuclear TIF1::neo transformants (TXh48 and TXh29: resistant to 4500 μg/ml paromomycin) and the wild-type strain, CU428. The TIF1 signal was normalized to β-tubulin to estimate the macronuclear TIF1 gene copy number. Right panel: Southern blot analysis of micronuclear (germline) TIF1::neo transformants. Het: heterozygous germline TIF1::neo replacement, TXh102. KO: homozygous TIF1::neo gene replacement, TXk202 (knockout). (B) DAPI staining of vegetative wild-type (Wt), TIF1::neo germline knockout (KO, TXk202) and macronuclear knockdown (KD, TXh48) strains. Wild-type and mutant cells were prelabeled with different mitochondrial dyes and mixed before DAPI staining for comparative analysis. Macronuclei (large arrows); micronuclei (small arrows). (C) Diminished DAPI staining in premeiotic micronuclear crescents in TIF1-deficient cells. DAPI (nuclear) staining of mating pairs in wild-type (CU427 × CU428) and wild-type (CU428) × mutant (KO, TXk202; KD, TXh48) crosses. Before meiotic S phase, the micronucleus elongates into a characteristic crescent shape. The mitochondrial dyes used to identify individual cells in mating pairs had no effect on DAPI staining intensity.
Germline transformation was used to determine if TIF1 is essential for cell viability. Biolistic transformation of premeiotic mating cells was used to generate heterozygous germline knockout strains, in which the micronuclear TIF1 coding region was replaced with the neomycin phosphotransferase gene (neo). Primary transformants were heterozygous for the disruption in the transcriptionally silent micronucleus and contain approximately equivalent amounts of the wild-type and disrupted TIF1 alleles in the new progeny macronucleus (Figure 2A: TXh102 (HET); TXh106, unpublished data).
Heterozygous transformants were grown to sexual maturity (∼80–100 fissions) in the absence of selection for the disrupted allele to minimize potentially deleterious effects of depleting TIF1. Heterokaryon strains were subsequently generated by Round 1 genomic exclusion in crosses with functionally amicronucleate strains, A* mating type III or A* mating type V (Allen, 1967). Twenty-five percent of the progeny derived from this abortive developmental program should be homozygous for the TIF1::neo disruption allele in the micronucleus and contain a wild-type macronucleus derived from the A* parental strain. Clonal isolates were genotyped to identify progeny that met these criteria. These strains were then mated to one another to generate homozygous TIF1 null progeny that completely lack the TIF1 gene in the transcriptionally active macronucleus. A single homozygous null line, TXk202, was obtained (Figure 2A, right panel, KO). Although there is strong selective pressure to retain the TIF1 gene in the macronucleus (Figure 2A, middle panel), our ability to isolate a homozygous null line indicates that TIF1 is not absolutely required for vegetative (macronuclear) functions.
TIF1 Is Required for Micronuclear Genome Stability
Although we were able to generate a homozygous null line, TXk202 exhibited defects in macronuclear DNA replication and division during vegetative propagation (see below) and eventually senesced. Repeated attempts to generate new homozygous null mutants failed, even when freshly derived heterozygous germline knockout transformants were mated immediately upon reaching sexual maturity. These observations raised the possibility that TIF1 might be required for long-term vegetative propagation of the micronucleus.
To determine if TIF1 is required for micronuclear genome stability, the homozygous null strain (TXk202) and two homozygous TIF1::neo heterokaryons (TXa28 and TXa42) were mated with nullisomic heterokaryon strains that lacked one of the five micronuclear chromosomes. In contrast to wild-type controls, all of the TIF1 mutants failed to generate viable progeny in these crosses, suggesting that the chromosomal composition of the micronucleus was compromised during vegetative cell divisions. Similar results were obtained when these mutants were mated to wild-type tester strains.
To assess whether a TIF1-deficiency led to degeneration of the micronuclear genome, we visualized nuclei in asynchronous vegetative cultures with DAPI. Partial (TXh48) and complete (TXk202) TIF1-deficient mutants exhibited two consistent differences from wild-type cells at early stages of the cell cycle (in cells containing one micronucleus and one macronucleus). First, the mutant macronucleus was somewhat enlarged, producing fainter, punctate DAPI staining (Figure 2B, large arrows). Second, the micronucleus was reproducibly smaller than wild-type (Figure 2B, small arrows). Cell-to-cell variation in the size of mutant micronuclei was observed, along with an overall diminution in DAPI staining intensity over time.
More revealing information on the micronucleus was obtained from sexually mature mating cells (after 80+ fissions). Before mating, cells were prelabeled with fluorescent mitochondrial dyes to identify the wild-type (green) and mutant partner (red) in each mating pair. During the developmental stage that precedes meiosis I, the micronucleus detaches from the macronucleus and elongates into a crescent (reviewed in Karrer, 2000). Control crosses between two wild-type strains (CU427 and CU428) produced mating partners with crescent nuclei of comparable length and DAPI staining intensity (Figure 2C, top left panel). In contrast, the intensity of the micronuclear DAPI crescent was markedly diminished in the homozygous TIF1 knockout (TXk202; Figure 2C, top right panel). Crescent formation was not simply delayed in the mutant, because the staining intensity did not increase at later time points (unpublished data). Similar results were obtained with heterozygous TIF1: neo germline transformants that contained partial macronuclear gene replacements (Figure 2C, TXh48: bottom left panel; TXh29: unpublished data) and homozygous TIF1 knockout heterokaryons (unpublished data). Although the mutant macronucleus is replaced with a wild-type (TIF1+) macronucleus in heterokaryons before conjugation, diminished micronuclear crescent staining was still observed. Consequently, the apparent loss of micronuclear DNA must occur during vegetative cell divisions.
Precocious Replication of the rDNA Minichromosome in TIF1-depleted Cells
Because TIF1 recognizes rDNA replication determinants (type I elements and PSEs) in vitro and contributes to the in vivo footprint at these sites, replication of the rDNA minichromosome was examined in TIF1-deficient cells. Cells were synchronized to examine the timing of bulk macronuclear DNA replication and rDNA origin firing. A stationary phase/starvation/refeeding protocol was used to synchronize cells (Mohammad et al., 2003) and BrdU pulse-labeling was used to monitor cell cycle progression. Wild-type (CU428) and TIF1-deficient strains (TXh48) entered S phase with similar kinetics; however, the mutant reproducibly exhibited an elongated macronuclear S phase (2.5 h vs. 2.0 h; Figure 3A, TXh48, solid line; CU428, dashed line). BrdU labeling was first detected 1.5–2 h after refeeding, with a modestly higher percentage of BrdU-positive mutant cells (2–3%) being observed throughout the first cell cycle.
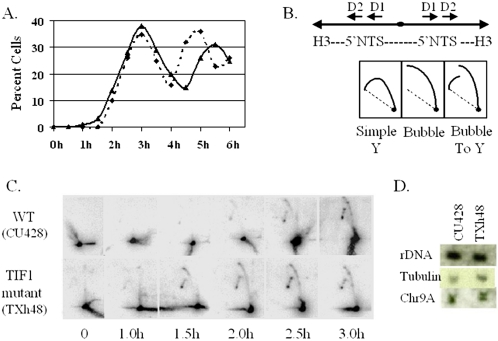
Figure 3.
Precocious rDNA replication in TIF1-deficient cells. (A) BrdU-labeling profiles of cells synchronized by a stationary phase/starvation protocol. The graph depicts the percent BrdU-positive cells in TIF1-deficient (TXh48, solid line) and wild-type (CU428) cultures examined at 30-min intervals after refeeding. (B) Top panel: restriction map of the 4.2 kb HindIII fragment that spans the two inverted copies of the rDNA 5′ NTS. Replication origins were previously localized within the tandem 430-base pair duplications, designated Domains 1 and 2 (D1, D2; Zhang et al., 1997). Bottom panel: expected 2D gel arc profiles for passive replication of the rDNA 5′ NTS (simple Y), replication from a centrally positioned origin (bubble), and replication from an asymmetrically position origin (D1 or D2; bubble-to-Y). (C) 2D gel analysis of HindIII-digested DNA from samples from the 0–3-h refeeding interval depicted in A. About 20 μg of total genomic DNA was loaded in each lane. (D) Quantitation of rDNA and non-rDNA chromosome abundance in wild-type (CU428) and TIF1-deficient (TXh48) cells. Southern blot analysis of total genomic DNA with probes specific for the rDNA 5′ NTS, β-tubulin genes (BTU1 and BTU2), and the 51-kb macronuclear chromosome, Chr9A (TIGR sequence scaffold 1172176).
2D gel analysis was performed on DNA samples prepared from time points in the first cell cycle to assess origin utilization, replication fork arrest at PSE elements, and the timing of rDNA replication. Total genomic DNA was digested with HindIII to generate a palindromic fragment containing both (inverted) copies of the 5′ NTS (Figure 3B). Wild-type and mutant strains produced identical (bubble-to-Y) replication arc profiles with an rDNA 5′ NTS probe, indicating that the mutant initiated replication exclusively from 5′ NTS origins (Figure 3C). Furthermore, the patterns of stalled intermediates were indistinguishable, indicating that replication fork pausing at PSE elements was not perturbed in the mutant. However, rDNA replication intermediates were detected earlier in TIF1-deficient cells. Extremely faint signals, corresponding to accumulated replication intermediates that had stalled at PSE elements, were detected at 1.0 h in the mutant and 1.5 h in wild-type cells. Clear replication intermediate arcs were detected at 1.5 h in the mutant and at 2.0–2.5 h in wild-type cells. We conclude that TIF1 functions in a negative regulatory manner to repress initiation at rDNA replication origins.
Quantitative Southern blotting was used to assess whether the rDNA was overreplicated in TIF1-deficient cells. No significant change in the abundance of macronuclear rDNA was detected relative to two non-rDNA chromosomes of differing length (Figure 3D; Chr 9A: 50 kb, β-tubulin: ∼1000 kb). Thus, it seems unlikely that TIF1 regulates rDNA copy number control in the amitotic macronucleus (Pan and Blackburn, 1995).
TIF1 Is Required for Normal S Phase Progression and Cytokinesis
Similar to the TIF1 knockdown strain, TXh48 (Figure 3A), the TIF1 null mutant, TXk202, exhibited a reduced growth rate compared with wild-type cells (Figure 4A). Microscopic analysis revealed a statistically significant increase in the percentage of mutant cells undergoing cytokinesis, suggesting that the null mutation perturbs a late step in cell division (Figure 4B). Cells were synchronized to evaluate the temporal relationship between S phase progression and cytokinesis. The TIF1 null strain reproducibly exhibited an elongated macronuclear S phase, indistinguishable from that observed for the TIF1 knockdown mutant (Figure 4C, dashed black line: wild-type (CU428) and solid black line: mutant (TXk202); see Figure 3A for comparison). On exiting S phase, the TIF1 null strain showed a further delay in cytokinesis (Figure 4C, dashed gray line: wild-type and solid gray line: mutant). Although the peak for macronuclear BrdU labeling typically occurred 30 min later than wild-type, the peaks for cytokinesis was delayed an additional 30 min. Furthermore, whereas wild-type cells divided with good synchrony, the mutant division profile was extremely broad. We conclude that TIF1 is required, either directly or indirectly, for the normal temporal progression of at least two cell cycle–regulated processes, DNA replication and cytokinesis.
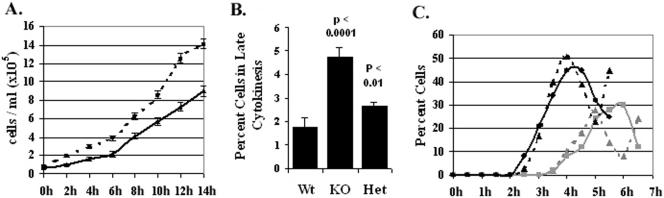
Figure 4.
Cell cycle defects in the TIF1 knockout mutant. (A) Growth curves for the wild-type (CU428, dashed line) and homozygous TIF1 knockout (TXk202, solid line) mutant were generated by averaging six hemocytometer cell counts per time point (n = 5 experiments). (B) Delayed cell division in the TIF1 germline knockout. Asynchronous vegetative cultures were visually examined for dividing cells (presence of a cleavage furrow). Knockout: homozygous TIF1::neo germline replacement (TXk202). Heterozygote: heterozygous TIF1::neo germline replacement (TXh102). The percentage of cells in late cytokinesis was determined by averaging the results from six to seven independent experimental analyses. (C) Elongated macronuclear S phase and delayed cytokinesis in TIF1-deficient cells. Wild-type and homozygous TIF1 knockout strains were synchronized with a stationary/starvation/refeeding protocol (Mohammad et al., 2003). Refed cultures were pulse-labeled with BrdU for a 15 min at 30-min intervals. Indirect immunoflourescence was used to quantify the percentage of BrdU-positive cells; dashed black line: wild-type (CU428); solid black line: TIF1 knockout (TXk202). Cytokinesis (dashed gray line: wild-type; solid gray line: TIF1 knockout) was scored by microscopic detection of a cleavage furrow.
Macronuclear Division and Cytokinesis Are Temporally Uncoupled in TIF1 Mutants
Asynchronous wild-type and TIF1 knockout cultures were stained with DAPI to investigate whether the delay in cytokinesis was associated with a defect in macronuclear division. Macronuclear division reproducibly occurred well before cytokinesis in wild-type controls (Figure 5A, top panels). Consistent with previous cytological studies, a trailing signal of DAPI staining material is associated with dividing macronuclei, and daughter nuclei are well separated before extensive constriction at the cleavage furrow in 90–95% of dividing cells. In contrast, TIF1 knockout cells (TXk202) often contained residual DNA at the cleavage furrow at late stages in cell division (Figure 5A, bottom left panel). The frequency of dividing cells that were simultaneously undergoing macronuclear division and cytokinesis was 4–8-fold higher in the mutant, fluctuating between 30 and 50% in asynchronous log phase cultures. Identical results were obtained with the partial macronuclear replacement strain, TXh48 (Figure 5A, bottom right panel). In the vast majority of cases, aberrantly dividing macronuclei were symmetrically positioned across the cleavage furrow, suggesting that TIF1 does not play a significant role in nuclear migration. Amacronucleate cells or cells with two macronuclei were observed less frequently in the mutant (Figure 5B; ∼1% of aberrant cell divisions). Asymmetric cell division was also observed (Figure 5B, bottom two panels). Although rare, the incidence of these events was elevated in the TIF1 mutant; no such cells were seen in a comparable sampling of wild-type cells (n = >5000).
Figure 5.
Aberrant macronuclear division and cytokinesis in TIF1-deficient strains. (A) Nuclear division and cytokinesis were examined in asynchronous wild-type (CU428), homozygous TIF1 knockout (TXk202), and macronuclear TIF1 knockdown (TXh48) strains after fixation of asynchronous log phase cultures. Light images of representative predivisional wild-type cells show the typical relationship between the extent of cleavage furrow invagination (black arrows) and position of daughter macronuclei (DAPI) at early and late stages of cytokinesis. Residual macronuclear DNA at the cleave furrow in mutant cells (white arrow, late stage cytokinesis) was observed in 30–50% of mutant cell divisions. (B) Less frequent cell division phenotypes in TIF1-deficient cells. Apoflour-stained live cells were photographed immediately after cell division. Top micrograph: macronuclear division failure associated with normal cytokinesis (TXh48; arrow: macronucleus). Center micrograph: macronuclear division failure associated with asymmetric cytokinesis. Bottom micrograph: macronuclear segregation failure associated with asymmetric cytokinesis. The daughter cell on the left has two macronuclei and the one on the right has none. (C) BrdU pulse-labeling of wild-type (CU428) and homozygous TIF1 knockout (TXk202) strains. Cells were pulse-labeled for 15 min with BrdU, fixed, and stained with DAPI (blue stain) and anti-BrdU antibodies (red stain, white arrow) to examine macronuclear and micronuclear DNA replication. BrdU labeling of the macronucleus was not observed in TIF1-deficient cells undergoing aberrant macronuclear division (right micrograph).
The global macronuclear defects associated with the loss of TIF1-prolonged S phase and delayed nuclear division raised two possibilities: that dividing macronuclei had not completed S phase or that macronuclei exited S, but were unable to undergo normal nuclear division. To distinguish between these possibilities, log phase cultures were briefly pulse-labeled with BrdU and examined by immunofluorescence with a BrdU-specific antibody. DAPI analysis was simultaneously performed to identify cells undergoing aberrant macronuclear division. Both dividing and predivisional cells were examined. Predivisional wild-type and mutant cells (cells with a single macronucleus) incorporated BrdU into their micronucleus (Figure 5C, left panels) or macronucleus (Figure 5C, right panels), but not both. Thus, the relative timing of macro- and micronuclear replication was not altered in the mutant. BrdU labeling was restricted to the micronucleus in dividing wild-type cells, whose daughter macronuclei were well separated (Figure 5C, third panel). Similarly, only the micronucleus was labeled in aberrantly dividing TIF1 mutants (Figure 5C, right panel); no BrdU was detected in daughter macronuclei or residual nuclear material at the cleavage furrow. Thus, TIF1-deficient cells that undergo abnormal macronuclear division exit macronuclear S phase, albeit later than normal.
DNA Replication Occurs at a Slower Rate in TIF1-deficient Cells
Sytox staining of log phase vegetative cultures detected a significant population of predivisional mutant cells with a single prominent macronucleus and extranuclear vesicles that contained DNA (Figure 6A, bottom left panel (TIF1 homozygous knockout, TXk202); frequency ∼25%). Although these vesicles could be “chromatin extrusion bodies” (CEBs) that form when macronuclear DNA content exceeds an upper limit (Bodenbender et al., 1992), the loss of micronuclear DNA during vegetative propagation (Figure 2, B and C) is inconsistent with a simple overreplication model. Alternatively, these structures could be generated by mechanical shearing of undivided macronuclei during cytokinesis. Extranuclear sytox staining was not detected in starved or predivisional (post-S phase) cells, suggesting that these DNA-containing vesicles either fuse with the macronucleus or are degraded.
Figure 6.
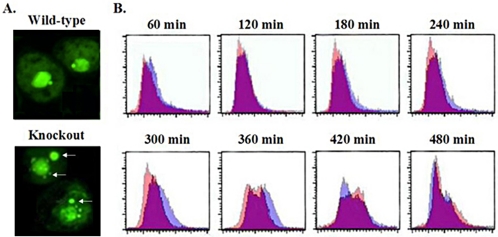
TIF1-deficient cells synthesize DNA more slowly than wild type, but maintain a normal macronuclear DNA content. (A) Sytox (DNA) staining of log phase vegetative cultures of wild-type (CU428) and homozygous TIF1::neo knockout (TXk202) strains. White arrows point to extranuclear DNA-staining bodies in TIF1-deficient cells. These structures were absent in cells transferred into starvation media (unpublished data). (B) Flow cytometry of synchronized wild-type and TIF1 knockout strains reveals a diminished rate of DNA synthesis in TIF1-deficient cells. Stationary phase/starved cultures were refed and stained with propidium iodide at 1-h intervals for FACS analysis. The FACS profiles for synchronized wild-type (CU428, purple) and TIF1 knockout (TXk202, pink) cultures were overlaid, the area of overlap appearing as magenta.
Flow cytometry was used to assess whether the prolonged macronuclear S phase in TIF1-deficient cells is associated with a diminished rate of DNA synthesis or overreplication of the macronuclear genome. DNA content and cell cycle progression were evaluated by fluorescent activated cell sorting (FACS) of propidium iodide–stained cells. Synchronized cultures were assayed at 1-h intervals over an 8-h period. To facilitate comparisons, the FACS profiles for wild-type (purple) and mutant (pink) cells were overlaid. Magenta areas correspond to overlapping cell populations with the same DNA content. The DNA content (peak widths) of wild-type and mutant strains were in good agreement over the first 4 h (Figure 6B, 60–240 min) and were consistent with BrdU-labeling experiments presented above (Figure 4C), which showed that wild-type and mutant cells enter S phase with similar kinetics. However, the wild-type DNA peak shifted to the right first, suggesting a slower rate of DNA replication (initiation and/or elongation) in the mutant.
The difference in wild-type and mutant replication rates was more apparent in the 5–8 h interval (Figure 6B, 300–480 min). Although wild-type cells generated a symmetric peak at 300 min with a 2N DNA content, the mutant peak was not only asymmetric, but contained a predominance of cells with lower DNA content. At the 360-min time point, two peaks were detected in both strains. The 1N peak in wild-type cells corresponds to cells that have divided and entered the second cell cycle. Cells in the mutant 2N peak had not achieved the maximal DNA content of wild-type cells at this time. However, the 2N DNA content was achieved at 420 min in the mutant, and a 1N peak appeared at the appropriate position 60 min later. Within the limits of resolution, the mutant appears to have replicated its entire macronuclear genome. Importantly, there was no evidence for gross over- or under-replication.
DISCUSSION
TIF1 was previously shown to bind to essential replication determinants in the T. thermophila rDNA minichromosome (Umthun et al., 1994; Saha and Kapler, 2000) and generate in vivo marks that distinguish the sites for replication and transcription initiation (Saha et al., 2001). In the work presented here we provide evidence that TIF1 regulates rDNA origin firing. We also show that TIF1 serves a more global role during macronuclear S phase. A deficiency in TIF1 produces opposite effects on replication of the rDNA minichromosome and bulk macronuclear DNA. Mutant cells precociously activate rDNA replication origins, but require additional time to replicate the remainder of their macronuclear genome. Consequently, TIF1 delays rDNA replication, but promotes S phase progression. We uncovered additional cellular processes that were perturbed in the TIF1 mutant. Cytokinesis was delayed and frequently associated with aberrant macronuclear division. Furthermore, TIF1 mutants failed to faithfully propagate their micronuclear genome during vegetative cell divisions and consequently, were sterile. Because micronuclear chromosomes contain centromeres and macronuclear chromosomes do not, it seems unlikely that TIF1 regulates a common pathway for chromosome segregation.
Regulation of the T. themophila rDNA Replicon
TIF1 is one of four single-stranded DNA binding activities that specifically recognize type I elements in vitro (Mohammad et al., 2000, 2003). As such, these distinct biochemical entities might compete or cooperate in vivo for binding to these reiterated, essential replication determinants. Although TIF1 shares homology with a sequences-specific, single-stranded DNA binding protein that functions as a transcription factor, sequence and structural similarity is confined to a segment required for homo-tetramerization (Desveaux et al., 2000, 2002; Saha et al., 2001; 3D Possum analysis, GMK, unpublished data). Although TIF1 lacks motifs found in transcriptional activator proteins, we cannot rule out a role in transcription. Directly or indirectly, TIF1 serves an important function during DNA replication and subsequent transmission of chromosomes.
We show here that TIF1 regulates the initiation of DNA replication, specifically affecting the timing of rDNA origin firing. Although TIF1-deficient cells exhibit a prolonged macronuclear S phase (TIF1 mutant, 2.5 h; wild-type, 2.0 h), rDNA origins begin firing ∼30–60 min earlier than wild-type cells. Our unexpected discovery that TIF1 depletion accelerates the timing of rDNA origin activation rather than delaying or eliminating it indicates that TIF1 is a negative regulator of rDNA replication. Epigenetic mechanisms have been shown to influence the temporal pattern of origin firing in other eukaryotes (reviewed in McNairn and Gilbert, 2003). A link between histone acetylation and origin activation has been recently demonstrated (Pasero et al., 2002; Aggarwal and Calvi, 2004, Aparicio et al., 2004; Kemp et al., 2005). Because we have not detected substantive change in histone H3 acetylation in the rDNA 5′ NTS of TIF1 mutants (Yakisich and Kapler, unpublished results), TIF1 probably regulates rDNA replication timing by a different mechanism.
We previously showed that TIF1 contributes to the type I element A-rich strand footprint at the rDNA origin (Saha et al., 2001). Consequently, a potential target for TIF1 regulation is the T strand-specific, type I element binding factor TIF4. Cytological studies have implicated the TIF4 Orc2-related subunit, Tt-p69, in global DNA replication in the micro- and macronucleus, as well as selective amplification of the rDNA minichromosome (Mohammad et al., 2003). The biochemical properties of TIF4 raise the possibility that this multiprotein complex is Tetrahymena ORC. Our recent discovery of Orc and MCM subunit orthologues in the T. thermophila genome sequence database (http://www.tigr.org; Kapler, unpublished results) not only indicates that the fundamental constituents of the prereplicative complex are conserved, but provides new avenues for exploring the relationship between TIF4 and ORC.
Global Roles for TIF1 in Macro- and Micronuclear Chromosomal Processes
To a first approximation TIF1 mutants fully replicate their macronuclear genome during S phase, albeit more slowly than wild-type. Mutants exit macronuclear S, but display additional defects later in the cell cycle. Macronuclear division and cytokinesis are significantly delayed, often producing a “cut phenotype” in which nuclear division and cell division are concurrent. Several nonexclusive scenarios can account for the effect of TIF1 depletion on macronuclear division and cytokinesis.
In the first model, TIF1 functions during and after S phase. Although the temporal peak in TIF1 rDNA binding activity and mRNA abundance suggest that TIF1's primary role occurs during S phase, TIF1 might associate with other nuclear or cytoplasmic targets at later stages in the cell cycle. By analogy, the Drosophila and human Orc6 subunit regulates critical cellular processes during mitosis. DmOrc6 associates with peanut, a septin family protein involved in cytokinesis (Chesnokov et al., 2003). Mutations that disrupt this interaction lead to the formation of multinucleate cells, by disrupting cytokinesis or nuclear positioning at the cleavage furrow. Similarly, silencing of the human Orc6 gene generates an array of M phase defects, including multipolar spindles, aberrant mitosis, and the formation of multinucleate cells (Prasanth et al., 2002). TIF1, by contrast, appears to have a minor role in cytoplasmic events associated with cell division. Although more frequent than in wild-type cells, defects in macronuclear migration and/or cytokinesis were observed in a small fraction of aberrant cell divisions. Moreover, in contrast to the paclitaxel-hypersensitive β-tubulin mutant, btu1-1 (Smith et al., 2004), the vast majority of aberrant nuclear divisions involved macronuclei that were properly localized to the cleavage furrow. Amacronucleate cells or cells with two macronuclei were rarely observed in the TIF1 mutant background. Furthermore, TIF1 mutants did not exhibit wide fluctuations in DNA content that are characteristic of btu1-1 cells.
In the second model, the role of TIF1 is restricted to S phase. We propose that the macronuclear and micronuclear phenotypes documented here (diminished rate of macronuclear DNA replication, aberrant macronuclear division, micronuclear chromosome loss) arise from a common defect: the inability to activate the S phase checkpoint or repair DNA damage at stalled replication forks. Preliminary experiments suggest a role for TIF1 at the replication fork in both the micro- and macronucleus, as TIF1 mutants are hypersensitive to DNA damaging agents and activate repair pathways in the absence of exogenous mutagens (Morrison and Kapler, unpublished results). By analogy, the metazoan checkpoint proteins, ATR and ATM, which arrest replication forks in response to DNA damage, were recently shown to regulate replication initiation and elongation in unperturbed cell cycles (Schechter et al., 2004). Like the TIF1 deficiency, inactivation of ATM and ATR results in precocious (global) origin firing. Additional Tetrahymena origins are needed to determine if TIF1 is a global regulator of origin activation. Because TIF1 bears no obvious sequence similarity to ATM or ATR and depletion of TIF1 has an opposite effect on the rate of DNA synthesis than ATM/ATR, we predict that TIF1 would act through different regulatory targets.
The differential sensitivity of the micro- and macronucleus to TIF1 depletion may reflect fundamental differences in how chromosomes are segregated during nuclear division. Micronuclei undergo conventional mitosis, whereas macronuclear chromosomes segregate by a poorly understood amitotic mechanism. Tetrahymena has evolved several ways to compensate for genic imbalances associated with amitotic macronuclear division. Excess macronuclear DNA is eliminated through the formation of chromatin extrusion bodies, whereas endo-replication occurs when macronuclear DNA content falls below a minimal threshold (Cleffmann, 1968). Because the DNA content of the macronucleus was maintained in the TIF1 mutant, these compensatory mechanisms appear intact. By contrast, the diploid mitotic micronucleus lacks these pathways. The diminished micronuclear DNA content and sterility observed in the TIF1 mutant indicates that TIF1 is essential for the long-term propagation of micronuclear chromosomes. Although a TIF1-like deficiency would be lethal in organisms that contain a single diploid nucleus, the ability of the “somatic” macronucleus to remain functional allows for the propagation of cells with a severely compromised micronuclear genome. Consequently, we are positioned to study chromosomal events that go awry in a dispensable, but otherwise conventional mitotic nucleus.
Acknowledgments
We acknowledge the contributions of Yeni Letti Trejo and Kenneth Carson to the cell cycle studies on DNA replication and TIF1 DNA binding. We also thank Dorothy Shippen and Mohammad Mohammad for their advice and thoughtful comments on the manuscript. This work was supported by National Institutes of Health grant GM53572 to G.M.K.
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E05–02–0107) on March 16, 2005.
References
- Abdurashidova, G., Danailov, M., Ochem, A., Triolo, G., Djeliova, V., Radulescu, S., Vindigni, A., Riva, S., and Falashi, A. (2004). Localization of proteins bound to a replication origin of human DNA along the cell cycle. EMBO J. 22, 4294–4303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aggarwal, B. D., and Calvi, B. R. (2004). Chromatin regulates origin activity in Drosophila follicle cells. Nature 430, 372–376. [DOI] [PubMed] [Google Scholar]
- Allen, S. L. (1967). Cytogenetids of genomic exclusion in Tetrahymena thermophila. Genetics 55, 797–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aparicio, J. G., Viggiani, C. J., Gibson, D. G., and Aparicio, O. M. (2004). The Rpd3-Sin3 histone deacetylase regulates replication timing and enable intra-S origin control in Saccharomyces cerevisiae. Mol. Cell. Biol. 24, 4769–4780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austin, R. J., Orr-Weaver, T. L., and Bell, S. P. (1999). Drosophila ORC specifically binds ACE3, an origin of DNA replication control element. Genes Dev. 13, 2639–2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beall, E. L., Bell, M., Georlette, D., and Botchan, M. R. (2004). Dm-myb mutant lethality in Drosophila is dependent upon mip130: positive and negative regulation of DNA replication. Genes Dev. 18, 1667–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beall, E. L., Manak, J. R., Zhou, S., Bell, M., Lipsick, J. S., and Botchan, M. R. (2002). Role for a Drosophila Myb-containing protein complex in site-specific DNA replication. Nature 420, 833–837. [DOI] [PubMed] [Google Scholar]
- Bell, S. P., and Stillman, B. (1992). ATP-dependent recognition of eukaryotic origins of DNA replication by a multiprotein complex. Nature 357, 128–134. [DOI] [PubMed] [Google Scholar]
- Blomberg, P., Randolph, C., Yao, C.-H., and Yao, M.-C. (1997). Regulatory sequences for amplification and replication of the ribosomal DNA minichromosome in Tetrahymena thermophila. Mol. Cell. Biol. 17, 7237–7247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodenbender, J., Prohaska, A., Jauker, F., Hipke, H., and Cleffmann, G. (1992). DNA elimination and its relation to quantities in the macronucleus of Tetrahymena. Dev. Genet. 13, 103–110. [DOI] [PubMed] [Google Scholar]
- Cassidy-Hanley, D., Bowen, J., Lee, J. H., Cole, e., VerPlank, L. A., Gaertig, J., Gorovsy, M. A., and Bruns, P. J. (1997). Germline and somatic transformation of mating Tetrahymena thermophila by particle bombardment. Genetics 146, 135–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesnokov, I., Chesnokova, O., and Botchan, M. (2003). A cytokinetic function of Drosophila ORC6 protein resides in a domain distinct from its replication activity. Proc. Natl. Acad. Sci. USA 100, 9150–9155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesnokov, I., Remus, D., and Botchan, M. (2001). Functional analysis of mutant and wild-type Drosophila origin recognition complex. Proc. Natl. Acad. Sci. USA 98, 11997–12002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang, R.-Y., and Kelly, T. J. (1999). The fission yeast homolog of Orc4p binds the replication origin via multiple AT-hooks. Proc. Natl. Acad. Sci. USA 96, 2656–2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleffmann, G. (1968). Regulierung der DNS-Menge im macronucleus von Tetrahymena. Exp. Cell Res. 50, 309–314. [DOI] [PubMed] [Google Scholar]
- Darzynkiewicz, Z., Juan, G., and Elzbie Bedner, E. (2003). In: Current Protocols in Cell Biology: http://www.mrw2.interscience.wiley.com/cponline/. Chapter 8, Section 8.4. [DOI] [PubMed]
- Desveaux, D., Allard, J., Brisson, N., and Sygusch, J. (2002). A new family of plant transcription factors displays a novel ss-DNA binding surface. Nat. Struct. Biol. 9, 512–517. [DOI] [PubMed] [Google Scholar]
- Desveaux, D., Despres, C., Joyeaux, A., Subramaniam, R., and Brisson, N. (2000). PBF-2 is a novel single-stranded DNA binding factor implicated in PR-10a gene activation in potato. Plant Cell 12, 1477–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doerder, F. P. (1979). Regulation of macronuclear DNA content in Tetrahymena thermophila. J. Protozool. 26, 28–35. [DOI] [PubMed] [Google Scholar]
- Fugita, M., Ishimi, Y., Nakamura, H., Kiyono, T., and Tsurmi, T. (2002). Nuclear organization of DNA replication initiation proteins in mammalian cells. J. Biol. Chem. 277, 10354–10361. [DOI] [PubMed] [Google Scholar]
- Gallagher, R. C., and Blackburn, E. H. (1998). A promoter mutation affecting replication of the Tetrahymena ribosomal DNA minichromosome. Mol. Cell. Biol. 18, 3021–3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishimi, Y. (1997). A DNA helicase activity is associated with an MCM4,-6,-7 protein complex. J. Biol. Chem. 272, 24508–24513. [DOI] [PubMed] [Google Scholar]
- Kapler, G. M., and Blackburn, E. H. (1994). A weak germline excision mutation blocks developmentally controlled amplification of the rDNA minichromosome of Tetrahymena thermophila. Genes Dev. 8, 84–95. [DOI] [PubMed] [Google Scholar]
- Kapler, G. M., Dobbs, D. L., and Blackburn, E. H. (1996). DNA replication in Tetrahymena. In: DNA Replication in Eukaryotic Cells, ed. M. L. DePamphilis, Cold Spring Harbor, NY: Cold Spring Harbor Press, 915–932.
- Karrer, K. (2000). Tetrahymena genetics: two nuclei are better than one. In: Methods in Cell Biology, vol. 62, eds. D. Asai and J. Forney, San Diego: Academic Press, 128–189. [DOI] [PubMed] [Google Scholar]
- Kemp, M. G., Ghosh, M., Liu, G., and Leffak, M. (2005). The histone deacetylase inhibitor trichostatin A alters the pattern of DNA replication origin activity in human cells. Nucleic Acids Res. 33, 325–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong, D., Coleman, T. R., and DePamphilis, M. L. (2003). Xenopus origin recognition complex (ORC) initiates DNA replication preferentially at sequences targeted by Schizosaccharomyces pombe ORC. EMBO J. 22, 3441–3450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong, D., and DePamphilis, M. (2001). Site-specific DNA binding of Schizosaccharomyces pombe origin recognition complex is determined by the ORC4 subunit. Mol. Cell. Biol. 21, 8095–8103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreitz, S., Ritzi, M., Baack, M., and Knippers, R. (2001). The human origin recognition complex protein 1 dissociates from chromatin during S phase in HeLa cells. J. Biol. Chem. 276, 6337–6342. [DOI] [PubMed] [Google Scholar]
- Labib, K., Tecero, J., and Diffley, J. (2000). Uninterrupted MCM2–7 function required for DNA replication fork progression. Science 288, 1643–1647. [DOI] [PubMed] [Google Scholar]
- Larson, D. D., Blackburn, E. H., Yaeger, P. C., and Orias, E. (1986). Control of rDNA replication in Tetrahymena involves a cis-acting upstream repeat of a promoter element. Cell 47, 229–240. [DOI] [PubMed] [Google Scholar]
- Li, C.-J., and DePamphilis, M. (2002). Mammalian Orc1 protein is selectively released from chromatin and ubiquitinated during the S-to-M transition in the cell division cycle. Mol. Cell. Biol. 22, 105–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipford, J. R., and Bell, S. P. (2001). Nucleosomes positioned by ORC facilitate the initiation of DNA replication. Mol. Cell 7, 21–30. [DOI] [PubMed] [Google Scholar]
- MacAlpine, D. M., Zhang, Z., and Kapler, G. M. (1997). Type I elements mediate replication fork pausing at conserved upstream sites in the Tetrahymena thermophila rDNA minichromsome. Mol. Cell. Biol. 17, 4517–4525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marahrens, Y., and Stillman, B. (1992). A yeast chromosomal origin of DNA replication defined by multiple functional elements. Science 255, 817–823. [DOI] [PubMed] [Google Scholar]
- Marsh, T. C., Cole, E. S., Stuart, K. R., Campbell, C., and Romero, D. P. (2000). RAD51 is required for propagation of the germinal nucleus in Tetrahymena thermophila. Genetics 154, 1587–1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNairn, A. J., and Gilbert, D. M. (2003). Epigenomic replication: linking epigenetics to DNA replication. BioEssays 25, 647–656. [DOI] [PubMed] [Google Scholar]
- Mohammad, M., Saha, S., and Kapler, G. M. (2000). Three different proteins recognize a multifunctional determinant that controls replication initiation, fork arrest and transcription in Tetrahymena. Nucleic Acids Res. 28, 843–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammad, M., York, R., Hommel, J., and Kapler, G. (2003). Characterization of a novel origin recognition complex-like complex: implications for DNA recognition, cell cycle control and locus-specific gene amplification. Mol. Cell. Biol. 23, 5005–5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishitani, H., Lygerou, Z., Nishimoto, T., and Nurse, P. (2000). The Cdt1 protein is required to license DNA replication in fission yeast. Nature 404, 625–628. [DOI] [PubMed] [Google Scholar]
- Oehlmann, M., Score, A., and Blow, J. (2004). The role of Cdc6 is ensuring complete genome licensing and S phase checkpoint activation. J. Cell Biol. 165, 181–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan, W.-J., and Blackburn, E. H. (1995). Tandem repeats of the 5′ nontranscribed spacer of Tetrahymena rDNA function as high copy number autonomous replicons in the macronucleus, but do not prevent rRNA gene dosage regulation. Nucleic Acids Res. 23, 1561–1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasero, P., Bensimon, A., and Schwob, E. (2002). Single-molecule analysis reveals clustering and epigenetic regulation of replication origins at the yeast rDNA locus. Genes Dev. 16, 2479–2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasanth, S., Prasanth, K., and Stillmna, B. (2002). Orc6 involved in DNA replication, chromosome segregation and cytokinesis. Science 297, 1026–1031. [DOI] [PubMed] [Google Scholar]
- Preer, J. R., and Preer, L. B. (1979). The size of macronuclear DNA and its relationship to models for maintaining genic balance. J. Protozool. 26, 14–18. [Google Scholar]
- Reischmann, K. P., Zhang, Z., and Kapler, G. M. (1999). Long range cooperative interactions regulate the initiation of replication in the Tetrahymena thermophila rDNA minichromosome. Nucleic Acids Res. 27, 3079–3089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha, S., and Kapler, G. M. (2000). Allele-specific protein-DNA interactions between the single-stranded DNA binding protein, ssA-TIBF, and DNA replication determinants in Tetrahymena. J. Mol. Biol. 295, 423–439. [DOI] [PubMed] [Google Scholar]
- Saha, S., Nicholson, A., and Kapler, G. M. (2001). Cloning and biochemical analysis of the Tetrahymena origin binding protein TIF 1, in vitro and in vivo binding to critical rDNA replication determinants. J. Biol. Chem. 276, 45417–45426. [DOI] [PubMed] [Google Scholar]
- Schechter, D., Costanzo, V., and Gauthier, J. (2004). ATR and ATM regulate the timing of DNA replication origin firing. Nat. Cell Biol. 6, 648–655. [DOI] [PubMed] [Google Scholar]
- Smith, J., Yakisich, J., Kapler, G., Cole, E., and Romero, D. (2004). A β-tubulin mutation selectively uncouples nuclear division and cytokinesis in Tetrahymena. Eukaryotic Cell 3, 1217–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theis, J., and Newlon, C. (1993). The structure and function of yeast ARS elements. Curr. Opin. Genet. Dev. 3, 752–758. [DOI] [PubMed] [Google Scholar]
- Turkewitz, A., Orias, E., and Kapler, G. (2002). Functional genomics: the coming of age for Tetrahymena thermophila. Trends Genet. 18, 35–40. [DOI] [PubMed] [Google Scholar]
- Umthun, A. R., Hou, Z., Sibenaller, Z. A., Shiau, W.-L., and Dobbs, D. L. (1994). Identification of DNA-binding proteins that recognize a conserved Type I repeat sequence in the replication origin region of Tetrahymena rDNA. Nucleic Acids Res. 22, 4432–4440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vashee, S., Cvetic, C., Lu, W., Simancek, P., Kelly, T. J., and Walters, J. C. (2003). Sequence-independent DNA binding and replication initiation by the human origin recognition complex. Genes Dev. 17, 1894–1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vashee, S., Simancek, P., Challberg, M., and Kelly, T. (2001). Assembly of the human origin recognition complex. J. Biol. Chem. 276, 26666–26673. [DOI] [PubMed] [Google Scholar]
- Venditti, P., Costanzo, G., Negri, R., and Camilloni, G. (1994). ABF1 contributes to the chromatin organization of the Saccharomyces cerevisiae ARS1 B-domain. Biochim. Biophys. Acta 1219, 677–689. [DOI] [PubMed] [Google Scholar]
- Wyrick, J., Aparicio, J., Chen, T., Barnett, J., Jennings, E., Young, R., Bell, S., and Aparicio, O. (2001). Genome-wide distribution of ORC and MCM proteins in S. cerevisiae: high resolution mapping of replication origins. Science 294, 2357–2360. [DOI] [PubMed] [Google Scholar]
- Yao, M.-C., Kimmel, A. R., and Gorovsky, M. A. (1974). A small number of cistrons for ribosomal RNA in the germline nucleus of a eukaryote, Tetrahymena pyriformis. Proc. Nat. Acad. Sci. USA 71, 3082–3086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Z., MacAlpine, D. M., and Kapler, G. M. (1997). Developmental regulation of DNA replication: replication fork barriers and programmed gene amplification in Tetrahymena. Mol. Cell. Biol. 17, 6147–6156. [DOI] [PMC free article] [PubMed] [Google Scholar]