Abstract
Background
The availability and affordability of germline genetic testing (GGT) has resulted in a broader utilization in daily clinical practice. However, adherence to testing guidelines is low, especially among older patients, where testing is often not offered.
Methods
In this study, consecutive, newly diagnosed patients with breast cancer (BC) aged ≥ 65 years and eligible for GGT, as per the National Comprehensive Cancer Network (NCCN) guidelines (version 1, 2021), were invited to participate, from March 2021 to December 2022. Patients were offered a restricted (two- or 20-gene panel), or an expanded 84-gene panel.
Results
During the study period, 204 patients were enrolled. The mean (standard deviation (SD)) age at BC diagnosis was 70.5 (5.13) years, ranging 65 - 81 years. All patients were Arab and the majority were Jordanian. The majority (n = 188, 92.2%) had early-stage (stages I and II) disease. One hundred three (50.5%) patients were tested with a restricted two-gene (n = 13) or 20-gene (n = 90) panel, while the remaining 101 (49.5%) patients had an expanded 84-gene panel. Family history of close blood relative(s) with BC was the most common indication for testing (n = 110, 53.9%). Among the entire study cohort, 22 (10.8%) had pathogenic/likely pathogenic germline variants (PGVs) and another 97 (47.5%) had ≥ 1 variants of uncertain significance (VUS). PGV rates were significantly higher with the expanded panel (14.9%) compared to restricted testing (6.8%) (P = 0.032). Similarly, VUS rates were significantly higher with the expanded panel (64.4%) compared to the restricted panel (31.1%) (P < 0.001). The most prevalent genes with PGVs were BRCA1/2 (31.3% of all PGV-positive patients), CHEK2 (23.1%) and ATM (19.2%).
Conclusion
GGT should not be overlooked in older BC patients, as this study demonstrates that > 10% of patients have PGVs, largely in potentially actionable genes.
Keywords: Germline genetic testing, Breast cancer, Predisposition genes, Older patients
Introduction
Breast cancer (BC) is the most common cancer among women worldwide, the incidence of which increases with age; almost a third of all cases are diagnosed in patients older than 70 years [1]. The median age at diagnosis is 62 years in Western societies [2, 3], but 10 years younger in developing countries [4].
Though most BCs are sporadic, up to 20% of cases are familial and some are associated with pathogenic germline variants (PGVs) in cancer-predisposing genes [5, 6]. Some of these genes are highly penetrant, such as BRCA1, BRCA2 and PALB2 [7, 8], while others like ATM and CHEK2 are associated with lower risks for cancer [9-11]. Hereditary BC is encountered more frequently in younger patients, those with positive family history, and those with triple-negative (TN) disease [12, 13].
Identifying carriers of such PGVs may help in cancer prevention through enhanced screening for breast and other cancers, and provide options for risk-reducing interventions like mastectomy and bilateral salpingo-oophorectomy (BSO) [14, 15]. However, the impact of PGVs in BRCA1 or BRCA2 has now moved beyond cancer prevention to involve treatment decisions of both early- and advanced-stage disease. The EMBRACA [16] and the OlympiAD [17] clinical trials have demonstrated that a poly (ADP-ribose) polymerase (PARP) inhibitor is superior to chemotherapy for BRCA1/2 PGV carriers with advanced-stage, human epidermal growth factor receptor 2 (HER2)-negative BC. More recently, the OlympiA study has demonstrated significant improvement in outcomes, including overall survival (OS), in patients with BRCA1/2 PGVs who received PARP inhibitors in the adjuvant treatment of high-risk early-stage BC [18].
Several studies had shown that more than half of eligible patients with BC, as recommended by the National Comprehensive Cancer Network (NCCN) guidelines, never undergo germline genetic testing (GGT). Such underutilization is highest among minorities, underserved populations and older patients [19-22]. A study from the United Kingdom reviewed genetic counseling, referral and genetic testing of over 47,000 women. History of BC was identified in 2.7% of the cohort and 35.6% met one or more eligibility criteria for genetic testing; of these, 29.0% had a discussion with their healthcare provider, 20.2% were advised to undergo, and only 15.3% underwent genetic testing [23]. Other studies utilizing the Surveillance, Epidemiology, and End Results (SEER)-based databases from Seattle [24], California and Georgia [25] reached similar conclusions.
Current NCCN guidelines rely heavily on the patients’ age at BC diagnosis with testing recommended for all BC patients diagnosed at age 50 or younger, regardless of their personal or family history of breast or other cancers, whereas older patients should have additional risk factors to be tested.
The aim of our study was to determine the prevalence of PGVs among Arab patients with BC diagnosed at age 65 years or older utilizing a multi-gene panel (MGP) of cancer-predisposing genes.
Materials and Methods
This was a prospective study of consecutive, newly diagnosed patients with BC aged 65 years or older and eligible for GGT, as per the NCCN guidelines. All patients were Arab, mostly Jordanians, diagnosed and treated at King Hussein Cancer Center, a comprehensive cancer center. Per patient choice, GGT was performed with a standard 20-gene guidelines-based or an expanded 84-gene panel testing. Prior to the availability of MGP testing, 13 patients were tested with BRCA1 and BRCA2 only. The 20-gene panel consists of ATM, BARD1, BRCA1, BRCA2, BRIP1, CDH1, CHEK2, EPCAM, MLH1, MSH2, MSH6, NBN, NF1, PALB2, PMS2, PTEN, RAD51C, RAD51D, STK11 and TP53. Genes included in the 84-gene panel are detailed in Supplementary Material 1 (www.wjon.org).
DNA sequencing and analysis
Genetic testing was performed using a peripheral blood sample at two reference laboratories: Leeds Cancer Centre, St James’s Institute of Oncology (Leeds, UK) for the BRCA1 and BRCA2-only testing [12], and Invitae Corp. (San Francisco, USA) for the 20- and the 84-gene MGP testing. Whole gene sequencing, deletion, and duplication analysis of all coding exons, +/- 20 base pairs of flanking introns, and other special targets, and variant interpretations were performed at Invitae as previously described [26]. Variants were classified as negative, pathogenic/likely pathogenic (positive) and variant of uncertain significance (VUS). Demographics and clinical history were collected and analyzed from patients’ electronic medical records.
Statistical analysis
Frequencies of pathogenic variants in each gene were assessed for patients with BC and stratified by gene panel used (restricted versus expanded), age group (≤ 70 versus > 70 years), presence of family history, multiple BCs, and TN disease. Proportions were compared using Fisher’s exact test and significance was set at P < 0.05. All analyses were performed utilizing version 9.4 of SAS software (SAS Institute Inc., Cary, NC).
This study was approved by the Institutional Review Board at King Hussein Cancer Center (protocol number 20 KHCC 202), and was conducted in compliance with the ethical standards of the institution on human subjects as well as with the Helsinki Declaration.
Results
Between March 2021 and December 2022, 204 eligible patients were enrolled. The mean (standard deviation (SD)) age at BC diagnosis was 70.5 (5.13) years, ranging 65 - 81 years; 45 (22.1%) were > 75 years. The majority (n = 188, 92.2%) had early-stage disease, 13 (6.4%) were male and 33 (16.2%) had TN disease. All patients were Arab and the majority were Jordanian. Family history of close blood relatives (first-, second- or third-degree) with BC was reported by 143 (70.1%); 110 (53.9%) had close relatives diagnosed at age 50 years or younger, which was the most common indication for GGT in our cohort (Table 1).
Table 1. Patients’ Characteristics (n = 204).
| Characteristics | Number | % |
|---|---|---|
| Age at diagnosis (years) | ||
| Median (range) | 70.5 | |
| 65 - 70 | 102 | 50.0% |
| 71 - 75 | 57 | 27.9% |
| > 75 | 45 | 22.1% |
| Stage | ||
| Early-stage | 188 | 92.2% |
| Advanced-stage | 16 | 7.8% |
| Hormonal status | ||
| ER-positive | 155 | 75.9% |
| PR-positive | 148 | 72.5% |
| HER2 status | ||
| HER2-positive | 31 | 15.2% |
| HER2-negative | 165 | 80.9% |
| Unknown | 8 | 3.9% |
| Triple-negative | ||
| Yes | 33 | 16.2% |
| No | 171 | 83.8% |
| Positive family historya | ||
| Breast | 143 | 70.1% |
| Ovarian, pancreas, high-grade prostate | 34 | 16.7% |
| Nationality | ||
| Jordanians | 192 | 94.1% |
| Non-Jordanians | 12 | 5.9% |
aFirst, second or third-degree. ER: estrogen receptor; PR: progesterone receptor; HER2: human epidermal growth factor receptor 2.
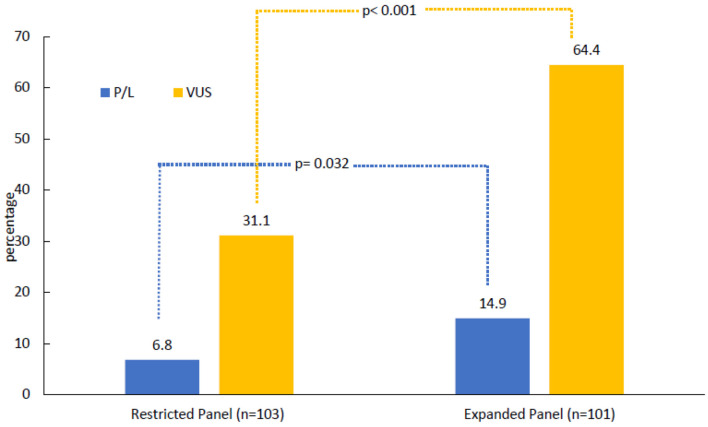
Patients were tested based on NCCN guidelines, 103 (50.5%) were tested with restricted panels, BRCA1 and BRCA2 only (n = 13, 6.4%) or a 20-gene panel (n = 90, 44.1%), while the remaining 101 (49.5%) patients were tested using an expanded 84-gene panel. Among the entire study cohort, 22 (10.8%) had PGVs and 97 (47.5%) had VUS results in the absence of a PGV finding. PGVs were detected among seven (6.8%) patients who were tested using the restricted panels and among 15 (14.9%) patients who underwent the expanded 84-gene panel (P = 0.032). VUS rate was significantly higher in the expanded 84-gene panel (n = 65, 64.4%) compared to those who had restricted panel testing (n = 32, 31.1%, P < 0.001) (Fig. 1). One patient in the expanded-panel testing had increased risk allele (APC), two patients were carriers of MUTYH gene associated with autosomal recessive cancer risk and were excluded from the analysis. Among the 13 male patients included in the study, three (23.1%) had PGVs, all in BRCA2.
Figure 1.
Results of germline genetic testing. One patient in the expanded-panel testing had increased risk allele (APC). Two carriers of autosomal recessive mutations were excluded. P/L: pathogenic/likely pathogenic; VUS: variants of uncertain significance.
PGV rates were similar in patients ≤ 70 years (8.8%) and in those > 70 (12.7%) (P = 0.18). Additionally, we did not observe a significant difference in the prevalence of PGVs in patients with TN disease (9.1%) compared to other subtypes (11.1%) (P = 0.37). Family history of breast, ovarian, pancreatic, or prostate cancers was not associated with higher rates of PGV as detailed in Table 2.
Table 2. Rates of Positive Pathogenic Variants by Indication.
| Variable | Total | Pathogenic/likely pathogenic mutations |
|||||
|---|---|---|---|---|---|---|---|
| All variants |
BRCA1/2
|
Non-BRCA1/2 |
|||||
| n (%) | P-value | n (%) | P-value | n (%) | P-value | ||
| Age at diagnosis (years) | 0.18 | 0.23 | 0.29 | ||||
| ≤ 70 | 102 | 9 (8.8%) | 3 (2.9%) | 6 (5.8%) | |||
| > 70 | 102 | 13 (12.7%) | 5 (4.9%) | 8 (7.8%) | |||
| Triple-negative disease | 0.37 | 0.24 | 0.33 | ||||
| Yes | 33 | 3 (9.1%) | 2 (6.1%) | 1 (3.0%) | |||
| No | 171 | 19 (11.1%) | 11 (6.4%) | 8 (4.7 %) | |||
| ≥ close blood relative with breast cancer diagnosed at age ≤ 50 years | 0.47 | 0.17 | 0.21 | ||||
| Yes | 110 | 12 (10.9%) | 3 (2.7%) | 9 (8.2%) | |||
| No | 94 | 10 (10.6%) | 5 (5.3%) | 5 (5.3%) | |||
| ≥ 1 close blood relative with: Epithelial ovarian cancer at any age; Exocrine pancreatic cancer at any age; Metastatic prostate cancer at any age | 0.18 | 0.24 | 0.29 | ||||
| Yes | 33 | 5 (15.2%) | 2 (6.1%) | 3 (9.1%) | |||
| No | 171 | 17 (9.9%) | 6 (3.5%) | 11 (6.4%) | |||
| ≥ 2 close relatives with breast cancer diagnosed at any age | 0.38 | 0.39 | 0.27 | ||||
| Yes | 86 | 10 (11.6%) | 3 (3.5%) | 7 (8.1%) | |||
| No | 118 | 12 (10.2%) | 5 (4.2%) | 7 (5.9%) | |||
| More than one primary breast cancers, first diagnosed before age of 65 years | 0.36 | 0.23 | 0.45 | ||||
| Yes | 13 | 1 (7.7%) | 0 | 1 (7.7%) | |||
| No | 191 | 21 (10.9%) | 8 (4.2%) | 13 (6.8%) | |||
| Diagnosed at any age with a close male relative with breast cancer at any age | 0.24 | 0.34 | 0.29 | ||||
| Yes | 4 | 0 | 0 | 0 | |||
| No | 200 | 22 (11%) | 8 (4%) | 14 (7%) | |||
| All patients | 204 | 22 (10.8%) | 8 (3.9%) | 14 (6.9%) | |||
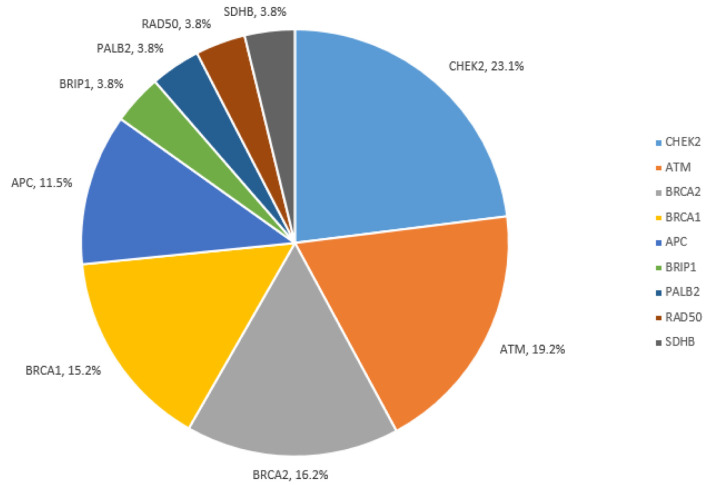
PGVs were most frequent in BRCA1/2 (31.3%), CHEK2 (23.1%), ATM (19.2%), and APC I1307K (11.5%) (Fig. 2). Detailed PGV findings are reported in Supplementary Material 2 (www.wjon.org). Notably, all identified PGVs were in genes included on the 20-gene panel.
Figure 2.
Pathogenic/likely pathogenic variants among all cohort.
Discussion
The emphasis in testing guidelines on age at diagnosis as a predictor for carrying a PGV in a cancer-predisposing gene, is a barrier to the identification of actionable PGVs [27]. Age was moved in the NCCN guidelines from 40, to 45, and lately to 50 years, as a cutoff for GGT. However, in its most recent guidelines update, the American Society of Clinical Oncology (ASCO) and the American Society of Surgical Oncology, both raised the age to commence testing to 65 years [28, 29]. Additionally, the most recently updated NCCN guidelines recommend germline testing of any patient with BC, regardless of their age, if such test results can aid in systemic treatment decisions using newly approved drugs like the PARP inhibitors in both early- and advanced-stage disease [16-18]. Additionally, age was disregarded in patients with TN disease, multiple BCs (synchronous or metachronous), and in those with lobular pathology with personal or family history of diffuse gastric cancer [30]. However, the decision to offer GGT for older women is often based on the coexistence of other risk factors for carrying cancer-predisposing genes including Ashkenazi Jewish ancestry, positive family history, TN disease and those with multiple primary BCs.
When testing older patients, the pre-test counseling should include a discussion of the relatively lower likelihood of positive results, compared to younger patients. Additionally, counseling should address, in addition to screening and preventive measures, the therapeutic implications of a positive test. However, given the age of patients under discussion, and the potential existing comorbidities, such patients might not be candidates for PARP inhibitors, enrollment in clinical trials, or other interventions [16-18]. Age, existing comorbidities and life expectancy should be the main factors on how extensive screening or risk-reducing interventions should be recommended for such patients. Nevertheless, identification of PGVs allows for cascade testing of at-risk family members and potentially life-saving early detection and risk-reduction opportunities.
Our study showed a relatively high rate of moderate-risk PGVs such as CHEK2 and ATM. Women with such PGVs can have lifetime risks of BC that exceed 20%, which justifies more careful surveillance with annual breast magnetic resonance imaging (MRI) [31]. Additionally, CHEK2 PGVs are associated with an increased risk of colorectal cancer, and ATM PGVs may increase the risk of pancreatic cancer. However, the clinical utility of increased surveillance in older patients with moderate-risk PGVs warrants additional studies.
Our PGV rate was slightly higher than what had been reported in Western literature. Kurian et al utilized data from the Women’s Health Initiative which enrolled 161,808 postmenopausal women aged 50 to 79 years at 40 US sites from 1993 through 1998 [32]. A nested case-control study of women who were diagnosed with invasive BC (cases, n = 2,195) or remained cancer-free (controls, n = 2,322) was performed. The median age at BC diagnosis was 73 years for case participants and 81 years for the control. GGT was performed using a 28-gene panel. PGVs were detected in 6.7% (95% confidence interval (CI): 5.7-7.9%) and in 4.0% (95% CI: 3.2-4.0%) (P < 0.001), among the case participants and the control group, respectively. Only 30.8% of case participants and 20% of controls with BRCA1/2 PGVs met the NCCN guidelines. Age, above or below 70 years, had no impact on the frequency of PGVs in BRCA1/2 (P = 0.34) or other BC-associated genes (P = 0.54) [32].
In another case-control study that aimed to address the effect of age on the frequency of PGVs in women with BC diagnosed at age over 65 years, 26,707 women from population-based studies (51.5% with BC and 48.5% unaffected) were enrolled. All women were tested utilizing a 12-gene panel of established BC risk genes (ATM, BARD1, BRCA1, BRCA2, CDH1, CHEK2, NF1, PALB2, PTEN, RAD51C, RAD51D, and TP53). The frequency of PGVs among women older than 65 years was 3.2% for those with BC and 1.5% for controls. PGVs were most frequent in BRCA2 and PALB2 for those with BC, while CHEK2 and ATM PGVs were common among those with and without cancer. Similar to our data, the frequency of pathogenic variants was not different across age groups; women diagnosed at age (75 - 85) years and (66 - 75) years had similar rates for BRCA2, CHEK2, and PALB2. However, BRCA1 was less common among the older subgroup [33].
Though the rate of pathogenic variants was higher in the group of patients tested with the expanded (84-gene) panel, most of the identified variants, except APC and RAD50, in the expanded panel were included within the restricted (20-gene) panel. As such, no real benefit of expanding the gene tested from 20- to 84-gene panel. On the other hand, such expansion may significantly increase the rate of VUS as illustrated in our study, from 31.1% to 64.4%. Such significant increase in the rate of VUS may increase patients’ anxiety and add to the evolving pressure on health care systems to come up with appropriate plans to follow up on potential reclassifications of such variants. Given the ease of tailoring individual panel of gene tested, both APC and RAD50 will be added to our BC gene panel for future testing.
Given the lack of accuracy of current testing guidelines, researchers are utilizing artificial intelligence and deep learning to better predict positive mutations based on distinct endophenotypes associated with different cancer predisposition genes in BC patients. Such a model achieved superior performance in identifying germline pathogenic variant carriers among Chinese BC patients in a recently published study [34]. More recently, our group reported our experience on universal testing of all cancer patients at diagnosis. Such argument is more convincing when considering BC [35].
Our study is not without limitations; the number of patients enrolled is relatively small, but that can be explained by demographics of Jordan population, with less than 5% of the population being > 65. Second, patients enrolled were from one center; however, given that the center treats over 50% of country’s load of cancer cases, we believe the enrolled cohort is representable.
Conclusions
This study demonstrates that the rate of potentially actionable PGVs among older patients with BC is probably higher than that of Western societies, and is high enough to justify screening of eligible patients. Identification of PGVs can confer eligibility for targeted therapies and clinical trials, enhanced screening and risk-reducing interventions, and cascade testing of at-risk relatives.
Supplementary Material
The 84-gene panel.
List of all detected pathogenic/likely pathogenic variants.
Acknowledgments
None to declare.
Funding Statement
This project was partially funded by intramural grant from King Hussein Cancer Center, Pfizer International, New York, NY and Invitae, San Francisco, CA.
Conflict of Interest
Sarah M. Nielsen, Brandie Heald, Kathryn E. Hatchell and Edward D. Esplin, are (or were) employees at Invitae corporation, San Francisco, CA, USA. All other authors declared no competing interests.
Informed Consent
Written informed consent has been obtained from the all enrolled patients.
Author Contributions
Conceptualization: HAR and SMN. Data curation: FT, BS, KA, HBH, RM, AA, and MA. Formal analysis: HAR, FT, and HBH. Investigation: FT, BS, KA, HBH, RM, AA, and MA. Project administration: HAR, BS, and HBH. Supervision: HAR and HBH. Writing-original draft: HAR, FT, BS, SMN, BH, KEH, EDE, and HBH. Writing-review and editing: HAR, SMN, BH, KEH, and EDE.
Data Availability
The data supporting the findings of this study are available through the corresponding author upon reasonable request. All variants, including those reported in this manuscript, are submitted regularly to ClinVar. Our most recent submission has been processed in March 2023, which covered variants and interpretations to the end of 2022. Details are available at: https://www.ncbi.nlm.nih.gov/clinvar/submitters/500031
References
- 1.Carleton N, Nasrazadani A, Gade K, Beriwal S, Barry PN, Brufsky AM, Bhargava R. et al. Personalising therapy for early-stage oestrogen receptor-positive breast cancer in older women. Lancet Healthy Longev. 2022;3(1):e54–e66. doi: 10.1016/s2666-7568(21)00280-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Risk Factors: Age - National Cancer Institute Available online: https://www.cancer.gov/about-cancer/causes-prevention/risk/age (Accessed on May 16, 2024)
- 3. Cancer of the Breast (Female) - Cancer Stat Facts. SEER Available online: https://seer.cancer.gov/statfacts/html/breast.html (Accessed on May 16, 2024)
- 4.Abdel-Razeq H, Mansour A, Jaddan D. Breast cancer care in Jordan. JCO Glob Oncol. 2020;6:260–268. doi: 10.1200/JGO.19.00279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Breast Cancer Association Consortium. Dorling L, Carvalho S, Allen J, Gonzalez-Neira A, Luccarini C, Wahlstrom C. et al. Breast cancer risk genes - association analysis in more than 113,000 women. N Engl J Med. 2021;384(5):428–439. doi: 10.1056/NEJMoa1913948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hu C, Hart SN, Gnanaolivu R, Huang H, Lee KY, Na J, Gao C. et al. A population-based study of genes previously implicated in breast cancer. N Engl J Med. 2021;384(5):440–451. doi: 10.1056/NEJMoa2005936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang X, Leslie G, Doroszuk A, Schneider S, Allen J, Decker B, Dunning AM. et al. Cancer risks associated with germline PALB2 pathogenic variants: an international study of 524 families. J Clin Oncol. 2020;38(7):674–685. doi: 10.1200/JCO.19.01907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Antoniou AC, Casadei S, Heikkinen T, Barrowdale D, Pylkas K, Roberts J, Lee A. et al. Breast-cancer risk in families with mutations in PALB2. N Engl J Med. 2014;371(6):497–506. doi: 10.1056/NEJMoa1400382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cybulski C, Wokolorczyk D, Jakubowska A, Huzarski T, Byrski T, Gronwald J, Masojc B. et al. Risk of breast cancer in women with a CHEK2 mutation with and without a family history of breast cancer. J Clin Oncol. 2011;29(28):3747–3752. doi: 10.1200/JCO.2010.34.0778. [DOI] [PubMed] [Google Scholar]
- 10.Lowry KP, Geuzinge HA, Stout NK, Alagoz O, Hampton J, Kerlikowske K, de Koning HJ. et al. Breast cancer screening strategies for women with ATM, CHEK2, and PALB2 pathogenic variants: a comparative modeling analysis. JAMA Oncol. 2022;8(4):587–596. doi: 10.1001/jamaoncol.2021.6204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marabelli M, Cheng SC, Parmigiani G. Penetrance of ATM gene mutations in breast cancer: a meta-analysis of different measures of risk. Genet Epidemiol. 2016;40(5):425–431. doi: 10.1002/gepi.21971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Abdel-Razeq H, Abujamous L, Abunasser M, Edaily S, Bater R. Prevalence and predictors of germline BRCA1 and BRCA2 mutations among young patients with breast cancer in Jordan. Sci Rep. 2021;11(1):14906. doi: 10.1038/s41598-021-94403-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Abdel-Razeq H, Tamimi F, Abujamous L, Edaily S, Abunasser M, Bater R, Salama O. Patterns and prevalence of BRCA1 and BRCA2 germline mutations among patients with triple-negative breast cancer: Regional Perspectives. Cancer Manag Res. 2021;13:4597–4604. doi: 10.2147/CMAR.S316470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Domchek SM, Friebel TM, Singer CF, Evans DG, Lynch HT, Isaacs C, Garber JE. et al. Association of risk-reducing surgery in BRCA1 or BRCA2 mutation carriers with cancer risk and mortality. JAMA. 2010;304(9):967–975. doi: 10.1001/jama.2010.1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Finch AP, Lubinski J, Moller P, Singer CF, Karlan B, Senter L, Rosen B. et al. Impact of oophorectomy on cancer incidence and mortality in women with a BRCA1 or BRCA2 mutation. J Clin Oncol. 2014;32(15):1547–1553. doi: 10.1200/JCO.2013.53.2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Litton JK, Rugo HS, Ettl J, Hurvitz SA, Goncalves A, Lee KH, Fehrenbacher L. et al. Talazoparib in patients with advanced breast cancer and a germline BRCA mutation. N Engl J Med. 2018;379(8):753–763. doi: 10.1056/NEJMoa1802905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Robson M, Im SA, Senkus E, Xu B, Domchek SM, Masuda N, Delaloge S. et al. Olaparib for metastatic breast cancer in patients with a germline BRCA mutation. N Engl J Med. 2017;377(6):523–533. doi: 10.1056/NEJMoa1706450. [DOI] [PubMed] [Google Scholar]
- 18.Tutt ANJ, Garber JE, Kaufman B, Viale G, Fumagalli D, Rastogi P, Gelber RD. et al. Adjuvant olaparib for patients with BRCA1- or BRCA2-mutated breast cancer. N Engl J Med. 2021;384(25):2394–2405. doi: 10.1056/NEJMoa2105215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McCarthy AM, Bristol M, Domchek SM, Groeneveld PW, Kim Y, Motanya UN, Shea JA. et al. Health care segregation, physician recommendation, and racial disparities in BRCA1/2 testing among women with breast cancer. J Clin Oncol. 2016;34(22):2610–2618. doi: 10.1200/JCO.2015.66.0019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Childers KK, Maggard-Gibbons M, Macinko J, Childers CP. National distribution of cancer genetic testing in the United States: evidence for a gender disparity in hereditary breast and ovarian cancer. JAMA Oncol. 2018;4(6):876–879. doi: 10.1001/jamaoncol.2018.0340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Knerr S, Bowles EJA, Leppig KA, Buist DSM, Gao H, Wernli KJ. Trends in BRCA test utilization in an integrated health system, 2005-2015. J Natl Cancer Inst. 2019;111(8):795–802. doi: 10.1093/jnci/djz008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fackenthal JD, Olopade OI. Breast cancer risk associated with BRCA1 and BRCA2 in diverse populations. Nat Rev Cancer. 2007;7(12):937–948. doi: 10.1038/nrc2054. [DOI] [PubMed] [Google Scholar]
- 23.Childers CP, Childers KK, Maggard-Gibbons M, Macinko J. National estimates of genetic testing in women with a history of breast or ovarian cancer. J Clin Oncol. 2017;35(34):3800–3806. doi: 10.1200/JCO.2017.73.6314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Clark NM, Roberts EA, Fedorenko C, Sun Q, Dubard-Gault M, Handford C, Yung R. et al. Genetic testing among patients with high-risk breast, ovarian, pancreatic, and prostate cancers. Ann Surg Oncol. 2023;30(3):1312–1326. doi: 10.1245/s10434-022-12755-y. [DOI] [PubMed] [Google Scholar]
- 25.Kurian AW, Ward KC, Howlader N, Deapen D, Hamilton AS, Mariotto A, Miller D. et al. Genetic testing and results in a population-based cohort of breast cancer patients and ovarian cancer patients. J Clin Oncol. 2019;37(15):1305–1315. doi: 10.1200/JCO.18.01854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nykamp K, Anderson M, Powers M, Garcia J, Herrera B, Ho YY, Kobayashi Y. et al. Sherloc: a comprehensive refinement of the ACMG-AMP variant classification criteria. Genet Med. 2017;19(10):1105–1117. doi: 10.1038/gim.2017.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tung N, Desai N. Germline genetic testing for women with breast cancer: shifting the paradigm from whom to test to whom not to test. J Clin Oncol. 2021;39(31):3415–3418. doi: 10.1200/JCO.21.01761. [DOI] [PubMed] [Google Scholar]
- 28.Bedrosian I, Somerfield MR, Achatz MI, Boughey JC, Curigliano G, Friedman S, Kohlmann WK. et al. Germline testing in patients with breast cancer: ASCO-society of surgical oncology guideline. J Clin Oncol. 2024;42(5):584–604. doi: 10.1200/JCO.23.02225. [DOI] [PubMed] [Google Scholar]
- 29.Kurian AW, Bedrosian I, Kohlmann WK, Somerfield MR, Robson ME. Germline testing in patients with breast cancer: ASCO-society of surgical oncology guideline Q and A. JCO Oncol Pract. 2024;20(4):466–471. doi: 10.1200/OP.23.00771. [DOI] [PubMed] [Google Scholar]
- 30. Genetic/Familial High-Risk Assessment: Breast, Ovarian, and Pancreatic. NCCN Available at: https://www.nccn.org/guidelines/guidelines-detail?category=2&id=1503 (Accessed on May 16, 2024)
- 31.Tung N, Domchek SM, Stadler Z, Nathanson KL, Couch F, Garber JE, Offit K. et al. Counselling framework for moderate-penetrance cancer-susceptibility mutations. Nat Rev Clin Oncol. 2016;13(9):581–588. doi: 10.1038/nrclinonc.2016.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kurian AW, Bernhisel R, Larson K, Caswell-Jin JL, Shadyab AH, Ochs-Balcom H, Stefanick ML. Prevalence of pathogenic variants in cancer susceptibility genes among women with postmenopausal breast cancer. JAMA. 2020;323(10):995–997. doi: 10.1001/jama.2020.0229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Boddicker NJ, Hu C, Weitzel JN, Kraft P, Nathanson KL, Goldgar DE, Na J. et al. Risk of late-onset breast cancer in genetically predisposed women. J Clin Oncol. 2021;39(31):3430–3440. doi: 10.1200/JCO.21.00531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu J, Zhao H, Zheng Y, Dong L, Zhao S, Huang Y, Huang S. et al. DrABC: deep learning accurately predicts germline pathogenic mutation status in breast cancer patients based on phenotype data. Genome Med. 2022;14(1):21. doi: 10.1186/s13073-022-01027-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Abdel-Razeq H, Sharaf B, Bani Hani H, Abu Hijlih R, Alkyam M, Al-Azzam K, Elemian S. et al. Implementation of Universal Pan-Cancer Germline Genetic Testing in an Arab population: the jordanian exploratory cancer genetics study. JCO Glob Oncol. 2024;10:e2400068. doi: 10.1200/GO.24.00068. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The 84-gene panel.
List of all detected pathogenic/likely pathogenic variants.
Data Availability Statement
The data supporting the findings of this study are available through the corresponding author upon reasonable request. All variants, including those reported in this manuscript, are submitted regularly to ClinVar. Our most recent submission has been processed in March 2023, which covered variants and interpretations to the end of 2022. Details are available at: https://www.ncbi.nlm.nih.gov/clinvar/submitters/500031