Abstract
The Woronin body (WB) is a peroxisome-related organelle that is centered on a crystalline core of the HEX-1 protein, which functions to seal septal pores of filamentous ascomycetes in response to cellular damage. Here, we investigate the cellular and genetic control of WB-formation and show that polarized hex-1 gene expression determines WB-biogenesis at the growing hyphal apex. We find that intron splicing is coupled to efficient hex-1 gene expression and strikingly, when the yellow fluorescent protein was expressed from hex-1 regulatory sequences, we observed a fluorescent gradient that was maximal in apical cells. Moreover, endogenous hex-1 transcripts were specifically enriched at the leading edge of the fungal colony, whereas other transcripts accumulated in basal regions. Time-lapse confocal microscopy showed that HEX-1 crystals normally formed in the vicinity of the hyphal apex in large peroxisomes, which matured and were immobilized at the cell periphery as cells underwent septation. When the hex-1 structural gene was expressed from regulatory sequences of an abundant, basally localized transcript, WB-core formation was redetermined to basal regions of the colony, and these strains displayed loss-of-function phenotypes specifically in apical hyphal compartments. These results show that apically localized gene expression is a key determinant of spatially restricted WB-assembly. We suggest that this type of regulation may be widely used to determine cellular activity in apical regions of the fungal hypha.
INTRODUCTION
Hyphae are the predominant mode of vegetative cellular organization in the fungal kingdom (Alexopolous et al., 1996). Yet, relatively little is known about the molecular basis of hyphal organization and differentiation within the fungal colony. Filamentous ascomycetes (Euascomycetes) are a monophyletic group of filamentous fungi that grow through polarized tip extension, resulting in the formation of a tubular cell (hypha), which is subsequently compartmentalized by the formation of perforate septa. Individual hyphae branch and fuse, eventually forming a multicellular syncytium where septal pores allow the trafficking of organelles and solutes within the colony (Buller, 1933). This syncytial lifestyle is probably important for rapid invasive growth of saprobes and pathogens and the elaboration of multicellular reproductive structures.
The Euascomycetes possess a septal pore-associated organelle known as the Woronin body (WB) (Buller, 1933; Markham and Collinge, 1987). The WB is centered on an oval or hexagonal core, which is surrounded by a closely associated unit membrane and these organelles have been observed by electron microscopy in many Euascomycetes, including plant and human pathogens (Markham and Collinge, 1987). The WB was first purified from Neurospora crassa, and this allowed the identification of a key WB-associated protein, HEX-1 (Jedd and Chua, 2000; Tenney et al., 2000), which uses a peroxisome-targeting signal-1 (PTS1) for vesicular localization (Jedd and Chua, 2000). hex-1 expression in yeast produces intraperoxisomal protein assemblies and recombinant HEX-1 spontaneously crystallizes in vitro, suggesting that HEX-1 is a self-assembling structural protein responsible for formation of the crystalline core of the WB (Jedd and Chua, 2000). Consistently, a Neurospora hex-1 mutant is devoid of the refractive core of the WB and mutant hyphae bleed protoplasm through septal pores after cellular damage, defining a function in septal pore sealing (Jedd and Chua, 2000; Tenney et al., 2000). In the plant pathogen Magnaporthe grisea,a hex-1 mutant is also defective in cell damage-induced septal pore sealing (Soundararajan et al., 2004). The mutant is compromised in pathogenesis-related development and displays cell death in response to nitrogen starvation, suggesting that the WB may function in response to diverse environmental stress (Soundararajan et al., 2004).
The crystal structure of HEX-1 reveals a protein lattice composed of three conserved intermolecular interfaces that mediate HEX-1 self-assembly (Yuan et al., 2003). Mutation of crystal-contact residues results in the formation of spherical noncrystalline WB-cores. These achieve normal dimensions, but the core is physically unstable, and these vesicles fail to complement WB loss-of-function phenotypes (Yuan et al., 2003). Thus, the WB requires a solid core for function, and this probably reflects the high intracellular turgor pressure that must be resisted during septal-pore patching. Interestingly, hex-1 shares significant sequence and structural homology with the ancient eIF-5a proteins, suggesting that hex-1 evolved after eIF-5a gene duplication in the ancestral Euascomycete (Yuan et al., 2003). Thus, the WB is a specialized peroxisome, which performs an adaptive function in the maintenance of organismal integrity (Jedd and Chua, 2000; Tenney et al., 2000; Yuan et al., 2003; Soundararajan et al., 2004).
WBs were first observed in the apical hyphal compartment (defined by the hyphal tip and first septum) by light microscopy in 1900 (Ternetz, 1900; reviewed in Buller, 1933) and later by electron microscopy in a variety of Euascomycetes, where they occupy a region of cytoplasm that underlies a cluster of tip-associated vesicles known as the spitzenkörper (Brenner and Carrol, 1968; McClure et al., 1968; Collinge and Markham, 1982; Momany et al., 2002). Apical WBs move within the cytoplasm (Ternetz, 1900), and in some cases, they have been observed in association with membranous sacs that suggest budding intermediates in the process of extrusion from the peroxisome lumen (Brenner and Carrol, 1968; Wergin, 1973; Camp, 1977). By contrast, in subapical cellular compartments (defined by two adjacent septa), the WB is immobile and localized near the septal pore or docked at the cell periphery (Markham and Collinge, 1987) where it exhibits a tightly appressed unit membrane. Woronin body position also has been examined in Aspergillus germlings. Interestingly, in germlings that have undergone septation, fewer Woronin bodies are observed at the hyphal apex, suggesting a process of retrograde transport (Momany et al., 2002).
We have examined and manipulated the spatial localization of hex-1 expression and directly observed WB-formation by using time-lapse confocal microscopy. We find that WB-formation is localized to apical hyphal compartments and determined in part by polarized hex-1 gene expression.
MATERIALS AND METHODS
Strains and General Techniques
The Neurospora hex-1 mutant and transformation with the genomic hex-1 construct POKE103-derived plasmids were described previously (Yuan et al., 2003). For the analysis of yellow fluorescent protein (YFP) and green fluorescent protein (GFP) fusion proteins a wild-type strain (FGSC#465) was used. Strains were grown in synthetic Vogel's N medium (Davis and de Serres, 1970). Neurospora conidia were transformed by electroporation as described previously (Yuan et al., 2003). To determine the effects of intron position and structure on gene expression, pools of primary transformants were analyzed. For each construct being examined, 30–50 transformants were excised from the primary transformation plate and grown overnight in liquid medium. Hyphae were harvested by filtration through a 70-μm cell strainer (BD Biosciences, Franklin Lakes, NJ), excess medium was removed by blotting with paper towels, and hyphae were frozen in liquid nitrogen before storage at –80°C. Hyphae were subsequently ground to a fine powder under liquid nitrogen using a mortar and pestle and total RNA was prepared using the RNeasy plant mini kit (QIAGEN, Valencia, CA). This pooling technique was found to be highly reproducible with similar results being obtained in several independent experiments.
To determine the cellular frequency of WBs (Figure 4A), the zone 1 to zone 5 slab of agar was transferred to a microscope slide and covered with a coverslip. The largest hyphae present in zones 1 and 5 were selected and refractile HEX-1 crystals were counted (n = 20 hyphal compartments) by using compound microscopy and a 100× objective. Hyphal compartments were defined by adjacent septa. Abnormally long and short hyphal compartments were avoided.
Figure 4.
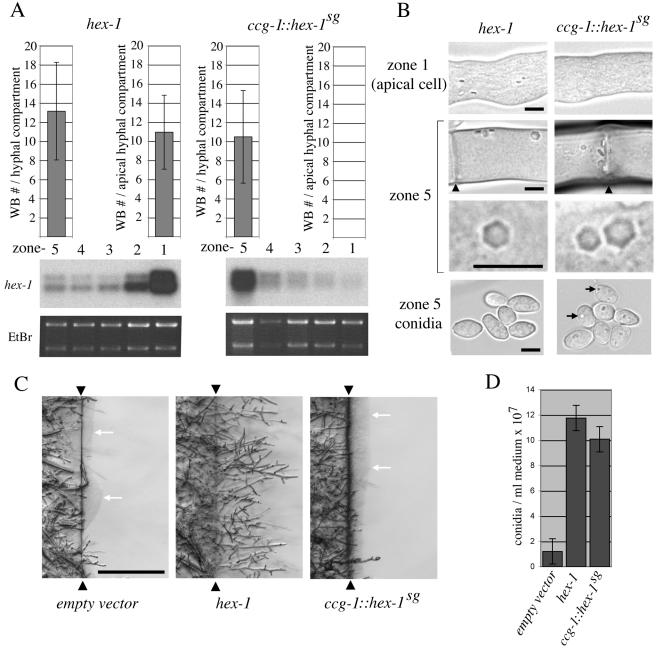
Woronin body-core formation can be redetermined to basal hyphal compartments producing a partial loss of function phenotype. (A) Gene expression and Woronin body number in hex-1–and ccg-1::hex-1sg–expressing strains. Transformants were grown as in Figure 2C, and total RNA from the fractionated colony was probed with the 3′ hex-1 probe (Figure 1). Ethidium bromide (EtBr) staining serves as a loading control. Top, Woronin body numbers in the zone 1 apical cells and in cells from zone 5. SD is indicated. (B) Woronin body appearance and distribution in hex-1- and ccg-1::hex-1sg-expressing strains. Micrographs from the indicated cell types and zones reveal the morphology of refractive HEX-1 crystals. Arrowheads point to septa and arrows indicate refractive HEX-1 assemblies that develop in ccg-1::hex-1sg-expressing strains. Bars, 5 μm. (C) ccg-1::hex-1sg apical compartments are defective in a hyphal regeneration after cell lysis. Hyphae from the indicated strains were severed at the interface between zone 1 and zone 2 (arrowheads), and the basal region of the colony was allowed to regenerate hyphae into a fresh block of agar for 2 h. White arrows indicate puddles of extruded protoplasm. Bar, 0.5 mm. (D) ccg-1::hex-1sg can largely complement conidiation defects. ccg-1::hex-1sg, hex-1, and empty vector transformants were grown on slants, and conidial production was determined. SD is indicated.
For the extraction of total RNA from the fungal colony (Figures 2C and 4A), the indicated strains were grown from conidia for 2 d in total darkness (all subsequent manipulation was performed using red safety lights). A rectangular fragment was excised from behind the growth front with a razor blade, and this fragment was used to inoculate a 2-cm-wide slab of fresh solid medium at one end. After 24 h of growth in darkness, the regions indicated in Figure 2C were excised with a razor blade, and total RNA was prepared as described above.
Figure 2.
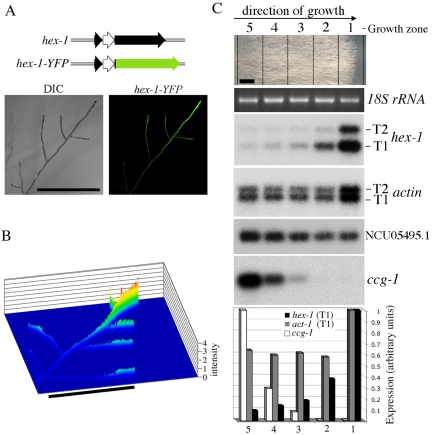
hex-1 gene expression is polarized. (A) Localization of YFP expressed within the context of the hex-1 promoter and intron. Structure of the hex-1-YFP fusion construct is shown at the top. Left, control bright-field image (DIC). Right, YFP fluorescence (hex-1-YFP). Bar, 500 μm. (B) Quantitation of the tip-localized fluorescence in a living hypha expressing hex-1-YFP. The y-axis reveals fluorescent intensity. Bar, 500 μm. (C) hex-1 transcripts are highly enriched at the leading edge of the fungal colony. Neurospora hyphae were grown for 24 h along a slab of solid medium (top, growth from left to right; bar, 0.5 cm), five zones were excised as indicated, and levels of hex-1,NCU05495.1, actin, and ccg-1 transcripts were examined by Northern blotting. Ethidium bromide (EtBr) staining serves as a loading control. Bottom, quantitation of transcript abundance in different regions of the fungal colony as determined by phosphorimaging.
For the determination of conidial production, the indicated strains were grown from conidia. Slants consisting of 2 ml of synthetic medium were incubated for 0–3 d in total darkness, followed by growth under conditions of alternating 12-h dark and 12-h light for a total of 9 d. All growth was conducted at 25°C. Conidia were harvested by the addition of 4 ml of water followed by vigorous shaking and filtration through a 40-μm mesh to remove large debris. Cells were then counted using a hemacytometer. Six independent measurements were made for each strain indicated in Figure 4D.
Mutants and Constructs
Mutants were constructed using the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA). In all cases where mutagenic oligonucleotide sequences are indicated, their complement also was used in the mutagenesis. To manipulate hex-1 gene structure, the POKE103::hex-1 plasmid was mutated to introduce NotI and PacI restriction sites at the 5′ and 3′ ends of the second hex-1 exon, respectively. The NotI site was introduced immediately down-stream of the 3′ splice site of the small hex-1 intron (Figure 1A) by using the oligonucleotide (oligo) NotI-1 (5′-CTT CCT CCA TTA CAG CTG CGG CCG CCG TTG AGG CTG ATG C-3′). This mutates amino acids 9–11 of HEX-1 to alanine. The PacI site was next introduced downstream of the hex-1 stop codon by using the oligo PacI-1 (5′-CCA CGG TTC CCG CCT TTA ATT AAA GGG TTC CGT CAT-3′). Transformation of the hex-1 mutant showed that this construct (GJP 595) was functional. To delete the small hex-1 intron, a second NotI site was introduced just downstream of the 5′ splice-site of GFP 595 by using the oligo NotI-2 (5′-CTA CGA CGA CGA CGC CGC GGC CGC ATC ATC CAT CAT CC-3′). This construct was digested with NotI to release the intron and then religated, producing the construct Δi (GJP 664), which has removed the small intron, leaving sequences equivalent to those produced by splicing of the parent construct (GJP 595).
Figure 1.
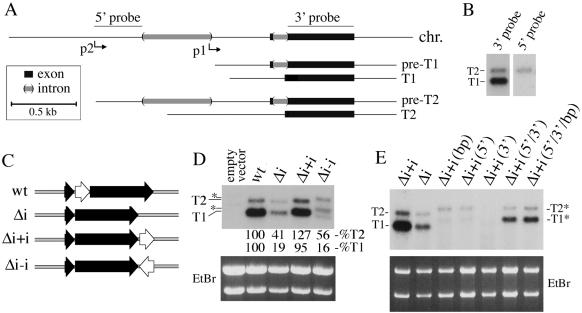
Intron splicing promotes hex-1 gene expression. (A) Organization of the genomic DNA fragment used to study hex-1 gene expression. Thick black lines indicate exons and thick gray lines indicate introns. Two transcripts, T1 and T2, are shown as both primary transcripts (preT1 and preT2) and mature transcripts (T1 and T2). The position of two probes used in Northern blotting also is shown. Scale bar is boxed. (B) Northern blot analysis reveals two hex-1 transcripts, T1 and T2. Total RNA from vegetative hyphae was probed with either the 5′ or 3′ probe as indicated. (C) Diagram of hex-1 constructs used to determine the function of the small hex-1 intron. Thick black arrows are exons and white arrow corresponds to the hex-1 intron. (D) Northern blot analysis of hex-1 gene expression from constructs shown in C. The indicated gene constructs were introduced into a hex-1 mutant background and RNA was isolated from pooled transformants. T1 and T2 are indicated. Asterisk denotes slightly longer T1 and T2 transcripts resulting from the antisense orientation intron. Relative levels of transcripts are indicated as a percentage of expression from the wild-type construct. (E) The intronic enhancement of gene expression requires splicing. The indicated constructs were introduced into the hex-1 mutant and analyzed as described in D. bp, branchpoint mutant; 5′,5′ splice site mutant; 3′, 3′ splice site mutant. Unspliced T1 and T2 transcripts are labeled as T1* and T2*.
To reintroduce introns into the hex-1 3′-untranslated region (UTR), we used the PCR to produce intron-cassettes with PacI ends. The small hex-1 intron was produced using the oligos hex-1int5′ (5′-CGC TTA ATT AAG GCT ACT ACG ACG ACG AC-3′) and hex-1int3′ (5′-GCC TTA ATT AAA GCC trichloroacetic acid (TCA) ACG TGG CCG TG-3′) and the third intron from the actin gene (NCU78026.1) was amplified using the oligos act-5′ (5′-GCC TTA ATT AAT CTA GAG CCC CCA GAG CTG TTT TCC-3′) and act-3′ (5′-GCC TTA ATT AAG GAC GAC CAA CGA TGG AAG-3′). These fragments were subsequently introduced into the PacI site of construct Δi, producing constructs Δi+i (GJP 613), Δi–i (GJP 614), and Δi+iact (GJP 641). For the mutation of splice site consensus sequences, the Δi+i construct was mutated using the following oligonucleotides: branch point mutant (5′-CTT CTC AAA ACA ATA AGC CGC GGT TGT CTT CCT CCA TTA C-3′), 5′ splice site mutant (5′-CTA CGA CGA CGA CGC AAA GCA CCC ATC ATC-3′), and 3′ splice site mutant (5′-GTC TTC CTC CAT TAC TCC TCA CGG CCA CGT TG-3′). For the combination of these mutations, mutants were either constructed simultaneously or sequentially. All constructs were verified by sequencing using the oligonucleotides hex-1.seq1 (5′-CCG TCC GGC TCA TAA GA-3′) and hex-1.seq2 (5′-TGT GTC CGA TTC CAG GGA-3′).
To replace the second exon of hex-1 with YFP and eGFP-PTS1, these were amplified using the oligos eG/C/YFP-NotI-5′ (5′-CGC GCG GCC GCG GTG AGC AAG GGC GAG-3′) and eG/C/YFP-PacI-3′ (5′-CGG TTA ATT AAC GGC CGC TTT ACT TGT AC-3′). These fragments were cloned into the NotI and PacI sites of GJP 595 producing the plasmids hex-1-YFP (GJP 607) and hex-1-GFP-PTS1 (GJP 602).
The ccg-1::hex-1sg plasmid was constructed in the following manner. A genomic ccg-1 fragment was produced using the oligonucleotides ccg.5′.sphI (5′-CCG GCA TGC GGC ATA ATG ACG TAG CTG CA-3′) and ccg.3′EcoRI (5′-CCG GAA TTC AAG TTC ATC GCG AGG GCT GA-3′), and this was cloned into the plasmid POKE 103. The ccg-1 fragment was further modified by site-directed mutagenesis to create NotI and PacI restriction sites directly upstream and downstream of the ccg-1 structural gene by using the oligonucleotides ccg.NotI (5′-GTG CTA CCC ACT AAG CGG CCG CAC CAA CAG TCG TTT TC-3′) and ccg.PacI (5′-CAA CCC CTC ACA TTT AAT TAA AAT GGA TAC CCT C-3′). The ccg-1 structural gene was subsequently replaced with the hex-1 structural gene (containing the intron) that was produced with the primers hex-1.5′ (5′-GCG TTA ATT AAC AAC ACA TCT TCC AAA-3′) and hex-1.3′ (5′-CCG GCG GCC GCG ATG ACG GAA CCC TCG AT-3′).
Northern Blotting
Total RNA was separated using denaturing agarose gel electrophoresis and transferred to Duralon-UV membranes (Stratagene) by using standard methods (Sambrook et al., 1989). PCR was used to create fragments from which probes were generated using random priming with the Megaprime DNA labeling system (Amersham Biosciences, Freiburg, Germany). The probes identified in Figure 1A were produced using the following oligos: 5′probe, 5′p-1 (5′-CCA CCT CAC TGA CAA GCT CC-3′) and 5′p-2 (5′-GAA CAA TCA ACT CAC CTG GT-3′); 3′ probe, 3′p-1 (5′-CTC ACG GCC ACG TTG AGG-3′) and 3′p-2 (5′-TAG AGG CGG GAA CCG TGG A-3′). The 3′ probe also was used to generate the hex-1 Northern blot shown in Figure 2C. The ccg-1 probe was generated using the oligos ccg-1.5′ (5′-TGG ATA CCC TCA AGA ACG CT-3′) and ccg-1.3′ (5′-GGT CAA TTC CTA GCT AGA AC-3′). The actin probe was generated using the oligos described below (5′ACT2 and 3′ACT2). The NCU05495.1 probe was made from a cloned genomic fragment that was digested with BglII and StuI, producing a fragment that contains the structural gene. Quantitation was achieved using phosphor imaging (Amersham Biosciences, Sunnyvale, CA).
Isolation of Cell Nuclei and Run-On Transcription
Nuclei were isolated essentially as described previously (Schuren et al., 1993), except that nuclei were separated through 0.75 and 1.0 M sucrose in a buffer A (10 mM PIPES, pH 6.9, 5 mM CaCl2, 5 mM MgSO4, and 1 mM phenylmethylsulfonyl fluoride) gradient. Nuclear suspensions (50 μl, ∼4.5 × 108 nuclei) were thawed and used in a 100-μl reaction mix containing 10 mM Tris-Cl, pH 7.6, 100 mM (NH4)2SO4, 10 mM MgCl2, 1 mM dithiothreitol, 0.5 mM each ATP, GTP, and CTP, and 100 μCi of [α-32P]UTP and incubated at 30°C for 30 min. The reaction was terminated by addition of 2.5 U of DNase I and incubated for 5 min at room temperature. Proteinase K (20 μg) was added, and samples were incubated for 30 min at 37°C. RNA was phenol-extracted, and ethanol precipitated. Before hybridization, RNA was separated from unincorporated [32P]UTP with a Sephadex G50 spin column (Amersham Biosciences, Freiburg, Germany). Linearized DNA containing a fragment of hex-1second exon or a linearized plasmid containing a fragment of the first exon of the actin gene or the empty pCR2.1 vector (Invitrogen, Carlsbad, CA) were resolved in a 1.2% agarose/1× Tris borate-EDTA gel and transferred to a nylon membrane by using standard methods (Sambrook et al., 1989). The hex-1 probe fragment was cloned using the oligos hex-1 internal1 (5′-CCG GCT CTG CTT CCC AGA-3′) and hex-1 3′ (5′-TGT CGT GTG TCC GAT TTC-3′), and the actin probe was cloned using the oligos 5′ACT2 (5′-CTC GTC ATC GAC AAT GGG TAA GC-3′) and 3′ACT2 (5′-CAT GAC AAT GTT GCC ATA GAG ATC C-3′).
Prehybridization and hybridization were carried out at 42°C in 50% formamide, 50 mM sodium phosphate, pH 6.5, 1% SDS, 4× Denhardt's, 5× SSC, and 100 μg/ml sperm DNA, 50 μg/ml E. coli tRNA, followed by three washes in 0.1× SSC, 0.1% SDS for 10 min at 55, 60, and 65°C successively. Blotted membranes were analyzed using a PhosphorImager (Amersham Biosciences, Sunnyvale, CA). Results of three independent experiments were normalized to the value of the actin signal set at 1.0 for reference.
Microscopy
Confocal imaging was performed on fluorescent protein-expressing Neurospora transformants grown for 12 h on solid Vogel's medium lacking supplemental pantothenate. Live cells were imaged using the “inverted agar block method” (Hickey et al., 2002). Briefly, a region ∼2 cm in length by 1 cm in width was cut out, inverted onto a coverglass, and imaged using an inverted Zeiss LSM 510 confocal microscope fitted with Plan Neofluar 10×/0.3 numerical aperture (NA), Plan Neofluar 25×/0.8 NA multi-immersion differential interference contrast (DIC), or C-Apochromat 63×/1.2 NA objective lenses. Simultaneous fluorescent and DIC images were acquired. Enhanced green fluorescent protein (eGFP) was imaged using 488-nm excitation and a 505- to 550-nm band-pass emission filter, whereas enhanced yellow fluorescent protein (eYFP) was imaged using 514-nm excitation and a 530-nm long-pass emission filter. To collect signal from a thicker optical slice of the hypha, the pinhole diameter was set at 2 Airy units (with the exception of the images shown in Figure 4B, where the pinhole was set to 1 Airy unit). For time-lapse movies, simultaneous GFP and DIC images were collected at intervals ranging between 1 and 5 s. To increase the visibility of HEX-1 crystals, the DIC alignment was sometimes adjusted to reduce the contrast of the background granular cytoplasm.
RESULTS
Structure and Expression of the Neurospora crassa hex-1 Gene
We examined hex-1 gene expression by using Northern blotting and RNA derived from vegetative hyphae (Figure 1B). A probe corresponding to the second exon of hex-1 (Figure 1A, 3′ probe) reveals two transcripts (T1 and T2), where T1 is the major transcript and T2 the minor transcript (Figure 1B). Examination of the 5′-ends of hex-1 expressed sequence tags (ESTs) confirmed that T1 is the major transcript and defined its 5′ end (Figure 1; Tenney et al., 2000). An additional EST (a3f07np) mapped upstream of the T1 start site, suggesting that it might be derived from the longer T2 transcript. Using this information, a T2 specific probe was designed (Figure 1A, 5′-probe) and used in Northern blotting (Figure 1B). Although both T1 and T2 hybridized to the common 3′ probe, only T2 hybridized with the 5′ probe. Thus, these two transcripts differ in their 5′-untranslated region (5′-UTR) and are expressed from distinct upstream activation sequence (UAS) elements (Figure 1A). The T2 EST also lacked a 537-base pair sequence present in the genomic DNA, suggesting the presence of a large T2-specific intron, and sequence analysis revealed consensus-5′, -3′, and -branch point sequences (Edelmann and Staben, 1994). We confirmed the splicing of this intron by using reverse transcription-PCR and oligos that span the putative T2-intron. Sequenced PCR fragments all (3 of 3) possessed a splice junction identical to the T2 EST (our unpublished data). Together, these data support the model of hex-1 gene structure shown in Figure 1A.
Intron Splicing Is Necessary for Efficient hex-1 mRNA Accumulation
We next sought to examine the spatial localization of hex-1 gene activity by expression of YFP from the hex-1 promoter. We developed a system that allows the reintroduction of a modified hex-1 genomic fragment into the hex-1 deletion strain. In the course of previous work, we failed to derive a hex-1 promoter suitable for hex-1 cDNA expression. However, a genomic fragment of hex-1 was well expressed and capable of complementing a hex-1 deletion (Yuan et al., 2003). This difference might be due to the small intron at the 5′ end of the hex-1 coding sequence (Figure 1C). To investigate this possibility, we first removed the small hex-1 intron (Figure 1C, Δi) and compared this with the wild-type (wt) hex-1 gene. Northern blotting revealed that the steady-state level of both hex-1 transcripts was significantly reduced in the intron-lacking construct (Figure 1D). To determine whether the intron's effect is position dependent, the hex-1 intron was introduced downstream of the intron lacking gene in both sense (Δi+i) and antisense (Δi–i) orientations (Figure 1C). The sense intron restored normal levels of hex-1 expression, whereas the antisense intron failed to do so (Figure 1D). Replacement of the hex-1 intron with the third intron from the actin gene (Tinsley et al., 1998) also could support wild-type levels of hex-1 expression (our unpublished data). Thus, the effect mediated by the hex-1 intron is independent of position, can be replaced by another intron, and probably requires pre-mRNA splicing.
We next constructed mutants in the 5′-splice sequence (5′), 3′-splice sequence (3′) and branch point (bp) consensus sequences in the context of the hex-1 intron engineered downstream of the hex-1 gene (Figure 1C, Δi+i). Mutants in any of the three consensus sequences reduced the accumulation of hex-1 transcripts to levels below those observed in the intron-lacking construct (Figure 1E), suggesting that defects in intron processing result in transcript degradation. By contrast, the combination of mutations in the 5′ and 3′ or 5′, 3′ and bp sequences resulted in transcript levels comparable with those observed in the intron-lacking construct (Δi). Together, these data show that functional splice sites are required for intron-dependent gene expression.
Finally, using nuclear run-on assays, we found that levels of nascent hex-1 transcripts are similar in intron-containing and intron-deleted transformants (Supplemental Figure 1). We conclude that intron splicing enhances hex-1 mRNA accumulation through a posttranscriptional mechanism, at the level of either mRNA transport or stability.
hex-1 Gene Expression Is Polarized
To examine the spatial regulation of hex-1 gene expression, the second exon of hex-1 was replaced with the gene encoding YFP, producing a hybrid transcript (hex1-YFP; Figure 2A). Translation of the spliced transcript produces a fusion protein containing the first seven amino acids of HEX-1 (derived from the first hex-1 exon) followed by full-length YFP. Living hyphae expressing YFP in this context were examined by confocal microscopy. As seen in Figure 2, A and B, vegetative hyphae display a gradient of fluorescent intensity that is maximal in apical cells and diminishes with distance from the tip.
To exclude the possible role of differences in protein translation and transport associated with the visualization of YFP, we localized endogenous hex-1 transcripts. Neurospora hyphae were grown on a slab of solid medium, and five zones were excised from the plate, with zone 1 corresponding to the growth front (Figure 2C; see Materials and Methods). Northern blotting of RNA derived from these zones shows that both T1 and T2 transcripts are highly enriched at the periphery of the fungal colony and their abundance dramatically drops with distance from the growth front (Figure 2C). These data demonstrate that hex-1 mRNA is enriched in apical cells and suggest that the gradient of YFP generated from the hex-1 promoter/intron construct is dependent at least in part on tip-cell localized gene expression.
To show that not all transcripts accumulate in an apically biased manner, we sought to identify transcripts that accumulate in basal regions of the colony. We examined the ccg-1 gene (clock controlled gene; Loros et al., 1989, also known as grg-1, glucose regulated gene; McNally and Free, 1988), which encodes an abundant transcript (McNally and Free, 1988) that is subject to complex physiological regulation (McNally and Free, 1988; Loros et al., 1989; Arpaia et al., 1995; Kimpel and Osiewacz, 1999). In our system, ccg-1 is enriched 200-fold in basal regions of the colony, whereas other transcripts are expressed throughout the colony (Figure 2C). These results define apical and basal patterns of gene expression within vegetative hyphae and suggest that Woronin body-core formation may be regulated in part at the level of hex-1 gene expression.
An Ordered Pathway of WB-Maturation at the Hyphal Apex
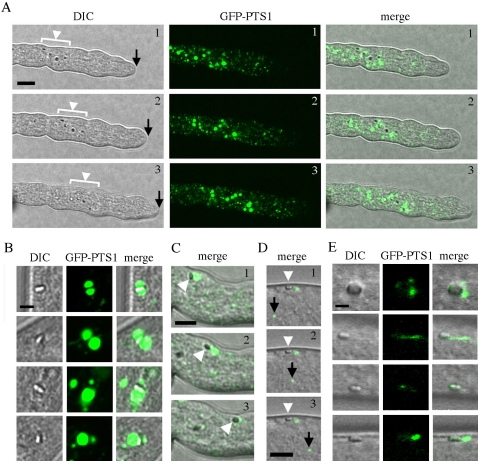
To observe the process of WB-maturation, we used GFP modified by a PTS1 identical to that used by HEX-1 (Ser-Arg-Leu). In this case, GFP-PTS1 and HEX-1 follow the same peroxisome import pathway, and GFP fluorescence defines the soluble matrix that surrounds crystalline HEX-1, which is observed as a distinct refractive structure (Jedd and Chua, 2000). In apical cells, GFP-PTS1 localized to large and small peroxisomes, which were mostly excluded from the extreme hyphal apex (Figure 3A). Small peroxisomes were found throughout the hypha, whereas large peroxisomes were generally associated with apical HEX-1 crystals (Figure 3, A and B), and these moved within the cytoplasm (Figure 3A). In apical WBs, HEX-1 crystals either bisect the large peroxisomes or are associated with its periphery (Figure 3B), suggesting that these structures correspond to intermediates in WB-formation. Consistently, time-lapse sequences also revealed the apparent fission of these peripheral GFP-PTS1–marked vesicular elements (Figure 3C and Video 1), resulting in HEX-1 cores that are associated with traces of peripheral GFP-PTS1.
Figure 3.
Woronin body maturation is a multistep, apically localized process. (A) HEX-1 crystals and peroxisomes at the hyphal apex. DIC microscopy (left) reveals a group of apical HEX-1 crystals (white arrowhead) that move at a rate similar to that of tip growth (black arrow) in a time sequence consisting of three frames taken at 50-s intervals. GFP-PTS1 fluorescence (middle) reveals large peroxisomes that are associated with apical HEX-1 assemblies. Merged image (right). Bar, 10 μm. (B) Details of relationship between large apical peroxisomes and HEX-1 crystals. Large peroxisomes are associated with the periphery of HEX-1 crystals and in some cases, the HEX-1 crystal bisects the peroxisome, suggesting that it forms contacts with the luminal face of the peroxisome membrane. Four apical crystals are shown. Bar, 2 μm. (C) Time sequence showing the apparent fission of a large peroxisome associated with a HEX-1 crystal (white arrowhead). This entire sequence can be viewed in Video 1. Bar, 5 μm. (D) In subapical cells, the WB (white arrowhead) is docked at the cell periphery and excluded from protoplasmic flow, which is revealed here by the movement of another peroxisome (black arrow) over three frames of a time-lapse sequence. Note that the docked WB has associated GFP-PTS1 signal that is also immobile over the time sequence. Also see Video 2. Bar, 5 μm. (E) In subapical cells, the WB remains associated with GFP-PTS1–positive tubules and vesicles, which seem diminished compared with those associated with the apical WB. Four subapical HEX-1 crystals are shown. Bar, 2 μm.
In subapical hyphal compartments, HEX-1 crystals retain associated GFP-PTS1 fluorescent vesicles and tubules, but these are generally diminished in fluorescence intensity and size (Figure 3E). Here, WBs were found exclusively at the cell periphery and were excluded from tip-directed proto-plasmic flow (Figure 3, D and E, and Video 2). Twenty-four of 25 subapical HEX-1 crystals observed were associated with peripheral GFP-PTS1, suggesting that mature WBs retains some peroxisome matrix proteins.
These observations suggested that apical WBs mature into subapical WBs. We performed direct observations of this process by acquiring long time-lapse sequences that encompass tip-growth and septation (Video 1). At early time points, apical WBs are associated with large peroxisomes, and these vesicles generally move forward in a tip-directed manner. However, other WBs slow (Video 1, blue circle) and begin to tumble at the cell periphery. These were observed undergoing fission events (Video 1, red circle) that diminished the GFP-PTS1 signal, and they later achieved a stationary position at the cell periphery approximately at septation (Video 1, arrows). Thus, the process of WB-maturation is largely tip cell localized.
Redirection of Woronin Body-Core Formation to Basal Regions of the Colony
If the apical localization of hex-1 transcripts is an essential determinant of localized WB-formation, conversion of hex-1 transcripts to a basal localization pattern should redirect WB-core formation to this region of the colony. We cloned a genomic fragment of the ccg-1 gene and replaced the structural gene with the corresponding region of the hex-1 (hex-1sg) producing a hybrid gene (ccg-1::hex-1sg), where ccg-1 regulatory sequences control expression of the hex-1 structural gene.
hex-1 deletion strains transformed with wild-type hex-1 accumulated transcripts in a polarized manner and produced Woronin body-cores in the apical hyphal compartment (Figure 4, A and B). In this context, the apical hyphal compartment contains an average of 11 refractive hex-1 crystals and this number increased to 13 in zone 5 of the colony. This distribution is consistent with apical crystal nucleation and growth, followed by slow subapical growth that could be mediated by low levels of subapical hex-1 transcripts (Figure 4, A and B). By contrast, ccg-1::hex-1sg-encoded transcripts were found at low levels in zone 1 and gradually accumulated to maximum levels in zone 5 hyphal compartments. In these transformants, WB-core structures were not observed in apical hyphal compartments but were instead found in zone 5 at almost wild-type levels (Figure 4A). These structures are excluded from protoplasmic flow and could reach dimensions similar to those encoded by the wild-type hex-1 gene (Figure 4B). Finally, ccg-1::hex-1sg expressing strains also produced refractive structures in asexually derived spores known as conidia (Figure 4B), suggesting that the ccg-1 promoter is active in all zone 5 cell types.
These data show that hex-1 transcript localization determines the site of WB-core formation and further predict that ccg-1::hex-1sg strains should behave like the hex-1 deletion mutant in apical hyphal compartments while displaying some degree of WB-function in basal regions of the colony. Consistently, like the deletion strain, ccg-1::hex-1sg strains were defective in hyphal regeneration from severed apical hyphae and extruded puddles of protoplasm from wounded hyphae (Figure 4C). To assess function in basal regions of the colony, we examined the production of conidia (Figure 4D). These are generally produced in later stages of colonial development, and their number is significantly reduced in the hex-1 deletion strain due to protoplasmic bleeding from aerial hyphae (Yuan et al., 2003). ccg-1::hex-1sg expressing strains were able to produce conidia nearly as well as wild-type hex-1 transformed strains, suggesting that ccg-1::hex-1sg encoded WB-core structures are largely functional (Figure 4D). Together, these results show that the apical localization of hex-1 expression determines apical WB-formation and that the normal execution of this process is required for full WB-functionality.
DISCUSSION
Here, we show that WB-formation is under both genetic and cell biological control and demonstrate the physiological importance of localizing this process to apical hyphal compartments. We propose a model in which the localization and level of hex-1 expression is a key determinants of WB-core formation. Normally, high levels of apically localized hex-1 transcript (Figure 2) determine the local production of HEX-1 protein, which is imported and concentrated in large apical peroxisomes. Under these conditions, HEX-1 crystals are nucleated, grow, and associate with the lumenal face of the peroxisome membrane, producing intermediate structures (Figure 3, B and C), which presumably correspond to peroxisome-associated WB-cores seen by electron microscopy (Brenner and Carrol, 1968; Wergin, 1973; Camp, 1977). These intermediates undergo maturation that may involve membrane fission (Figure 3C and Video 1), resulting in WBs that contains a large HEX-1 crystal and traces of matrix proteins (Figure 3E). Finally, the WB associates with the cell periphery roughly coinciding with septum formation (Video 1), and these vesicles are poised to execute their function in cell damage-induced septal pore-sealing (Jedd and Chua, 2000).
ccg-1::hex-1sg expressing strains demonstrate the physiological importance of apical WBs and further confirm that high levels of hex-1 transcripts are required for HEX-1 crystal nucleation and growth (Figure 4, A and B). In addition, the ability of ccg-1 regulatory sequences to localize WB-core formation within basal vegetative hyphae (Figure 4B) shows that ccg-1 and hex-1 transcripts are localized to different regions of the same cell type. This type of intrahyphal genetic differentiation may be widespread in hyphae of filamentous fungi. The zonal system of fungal growth defined here (Figure 2C) may be further exploited to define groups of genes, which show similar spatial regulation, and this may eventually further our understanding of how cellular and physiological processes are partitioned and coordinated within the fungal colony.
The ability of the hybrid ccg-1::hex-1sg transcript to direct the formation of functional WBs to basal hyphae (Figure 4), shows that these cells retain a high capacity for protein synthesis and PTS1-dependent import. In addition, these data suggest that the membranous component of the WB forms independently of HEX-1 dependent core formation. Consistently, GFP-PTS1 expression in the hex-1 deletion strain reveals large GFP-PTS1–containing vesicles that are tethered at the cell periphery (our unpublished observation). Thus, in basal hyphal compartments, HEX-1 protein produced from the hybrid ccg-1::hex-1sg transcript is probably imported into preexistent WB-ghosts.
Tip-localization of hex-1 transcripts (Figure 2) may be regulated at the level of transcription and/or through mRNA transport or stability. A local signal may activate hex-1 transcription in tip cell-localized nuclei. Alternatively or in addition, hex-l transcripts may be localized through actin microfilament or microtubule-dependent mechanisms analogous to those determining mRNA localization in other eukaryotic systems (Palacios and St. Johnson, 2001). The molecular dissection of hex-1 UAS sequences and mRNA noncoding regions and comparisons with basally localized transcripts such as ccg-1 should help resolve the mechanism of hex-1 mRNA localization.
We also demonstrate an important role for intron splicing in Neurospora, and similar findings in the filamentous basidiomycete Schizophyllum commune (Lugones et al., 1999) suggest that coupling between mRNA splicing and gene expression may be common in filamentous fungi. Intron splicing is required for the accumulation of hex-1 transcripts, and this effect is probably achieved at the posttranscriptional level (Supplemental Figure 1), suggesting that, as in metazoan systems (Maniatis and Reed, 2002), intron splicing is coupled to mRNA transport or stability. Transformants expressing hex-1 from the intron-lacking gene do not produce visible WB cores (our unpublished observation), suggesting that under these circumstances, HEX-1 protein is not produced at levels required for crystal nucleation. Determining the possible relationship between intron splicing and apical transcript localization will require additional work.
Interestingly, mutations in 5′,3′ and branch point-consensus sequences result in transcript levels that are significantly lower than those observed in the absence of an intron. This effect is suppressed by the combination of these mutations (Figure 1E), suggesting the presence of a spliceosome-associated surveillance-mechanism that recognizes and degrades transcripts containing aberrant introns. In the future, Neurospora should provide a good microbial system in which to study mRNA quality control and coupling within the gene expression pathway.
In summary, we show that WB-core formation occurs at the hyphal apex and that this process is determined in part by apically biased gene expression. These data show that fungi regulate some tip cell-associated processes by the spatial control of gene expression and coordinate organelle assembly with the production of constituent proteins. The continued study of localized gene expression and WB assembly should help determine the extent of this regulation and provide further insight into fundamental mechanisms of eukaryotic cell polarity.
Supplementary Material
Acknowledgments
G. J. thanks Nam-Hai Chua for support and encouragement and for providing laboratory space during initial stages of this work. We thank Dan Elreda for help with image processing and QuickTime movies, and Snezhana Oliferenko and Virginia Garreton for comments on the manuscript. This work was supported by Temasek Life Sciences Laboratory, Singapore, and in part by Grant 0090908 from the National Science Foundation.
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E04–10–0937) on March 30, 2005.
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
References
- Alexopolous, C. J., Mims, C. W., and Blackwell, M. (1996). Introductory Mycology, New York: John Wiley & Sons.
- Arpaia, G., Loros, J., Dunlap, J., Morelli, G., and Macino, G. (1995). Light induction of the clock-controlled gene ccg-1 is not transduced through the circadian clock in Neurospora crassa. Mol. Gen. Genet. 247, 157–163. [DOI] [PubMed] [Google Scholar]
- Brenner, D. M., and Carrol, G. C. (1968). Fine structural correlates of growth in hyphae of Ascodesmis sphaerospora. J. Bacteriol. 95, 658–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buller, A.H.R. (1933). The translocation of protoplasm through septate mycelium of certain Pyrenomycetes, Discomycetes and Hymenomycetes. In: Researches on Fungi, Vol. 5, London: Longmans, Green & Co., 75–167. [Google Scholar]
- Camp, R. R. (1977). Association of microbodies, Woronin bodies, and septa in intercellular hyphae of Cymadothea trifolii. Can. J. Bot.-Revue Canadienne De Botanique 55, 1856–1859. [Google Scholar]
- Collinge, A. J., and Markham, P. (1982). Hyphal tip ultrastructure of Aspergillus nidulans and Aspergillus giganteus and possible implications of Woronin bodies close to the hyphal apex of the latter species. Protoplasma 113, 209–213. [Google Scholar]
- Davis, R. H., and de Serres, F. J. (1970). Genetic and microbiological research techniques for Neurosopora crassa. Methods Enzymol. 27, 79–143. [Google Scholar]
- Edelmann, S. E., and Staben, C. (1994). A statistical-analysis of sequence features within genes from Neurospora crassa. Exp. Mycol. 18, 70–81. [Google Scholar]
- Hickey, P. C., Jacobsen, D., Read, N. D., and Glass, N. L. (2002). Live-cell imaging of vegetative hyphal fusion in Neurospora crassa. Fungal Genet. Biol. 37, 109–119. [DOI] [PubMed] [Google Scholar]
- Jedd, G., and Chua, N. H. (2000). A new self-assembled peroxisomal vesicle required for efficient resealing of the plasma membrane. Nat. Cell Biol. 2, 226–231. [DOI] [PubMed] [Google Scholar]
- Kimpel, E., and Osiewacz, H. D. (1999). PaGrg1, a glucose-repressible gene of Podospora anserina that is differentially expressed during lifespan. Curr. Genet. 35, 557–563. [DOI] [PubMed] [Google Scholar]
- Loros, J. J., Denome, S. A., and Dunlap, J. C. (1989). Molecular cloning of genes under control of the circadian clock in Neurospora. Science 243, 385–388. [DOI] [PubMed] [Google Scholar]
- Lugones, L. G., Scholtmeijer, K., Klootwijk, R., and Wessels, J.G.H. (1999). Introns are necessary for mRNA accumulation in Schizophyllum commune. Mol. Microbiol. 32, 681–689. [DOI] [PubMed] [Google Scholar]
- Maniatis, T., and Reed, R. (2002). An extensive network of coupling among gene expression machines. Nature 416, 499–506. [DOI] [PubMed] [Google Scholar]
- Markham, P., and Collinge, A. J. (1987). Woronin bodies of filamentous fungi. FEMS Microbiol. Rev. 46, 1–11. [Google Scholar]
- McClure, W. K., Park, D., and Robinson, P. M. (1968). Apical organization in the somatic hyphae of fungi. J. Gen. Microbiol. 50, 177–182. [DOI] [PubMed] [Google Scholar]
- McNally, M. T., and Free, S. J. (1988). Isolation and characterization of a Neurospora glucose-repressible gene. Curr. Genet. 14, 545–551. [DOI] [PubMed] [Google Scholar]
- Momany, M., Richardson, E. A., Van Sickle, C., and Jedd, G. (2002). Mapping Woronin body position in Aspergillus nidulans. Mycologia 94, 260–266. [PubMed] [Google Scholar]
- Palacios, I. M., and St. Johnson, D. (2001). Getting the message across: the intracellular localization of mRNAs in higher eukaryotes. Annu. Rev. Cell Dev. Biol. 17, 569–614. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E. F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual, Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
- Schuren, F.H.J., Vanderlende, T. R., and Wessels, J.G.H. (1993). Fruiting genes of Schizophyllum commune are transcriptionally regulated. Mycol. Res. 97, 538–542. [Google Scholar]
- Soundararajan, S., Jedd, G., Li, X., Ramos-Pamplona, M., Chua, N. H., and Naqvi, N. I. (2004). Woronin body function in Magnaporthe grisea is essential for efficient pathogenesis and for survival during nitrogen starvation stress. Plant Cell 16, 1564–1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenney, K., Hunt, I., Sweigard, J., Pounder, J. I., McClain, C., Bowman, E. J., and Bowman, B. J. (2000). hex-1, a gene unique to filamentous fungi, encodes the major protein of the Woronin body and functions as a plug for septal pores. Fungal Genet. Biol. 31, 205–217. [DOI] [PubMed] [Google Scholar]
- Ternetz, C. (1900). Protoplasmabewegung und Fruchtkoperbildung bei Ascophanus carneus. Jahrb. f. Wiss. Bot., Bd. XXXV, 273–312. [Google Scholar]
- Tinsley, J. H., Lee, I. H., Minke, P. F., and Plamann, M. (1998). Analysis of actin and actin-related protein 3 (ARP3) gene expression following induction of hyphal tip formation and apolar growth in Neurospora. Mol. Gen. Genet. 259, 601–609. [DOI] [PubMed] [Google Scholar]
- Wergin, W. P. (1973). Development of Woronin bodies from microbodies in Fusarium oxysporum f. sp. lycopersici. Protoplasma 76, 249–260. [DOI] [PubMed] [Google Scholar]
- Yuan, P., Jedd, G., Kumaran, D., Swaminathan, S., Shio, H., Hewitt, D., Chua, N. H., and Swaminathan, K. (2003). A HEX-1 crystal lattice required for Woronin body function in Neurospora crassa. Nat. Struct. Biol. 10, 264–270. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.