Abstract
Although γ-tubulin complexes (γ-TuCs) are known as microtubule (MT) nucleators, their function in vivo is still poorly defined. Mto1p (also known as mbo1p or mod20p) is a γ-TuC–associated protein that recruits γ-TuCs specifically to cytoplasmic MT organizing centers (MTOCs) and interphase MTs. Here, we investigated γ-TuC function by analyzing MT behavior in mto1Δ and alp4 (GCP2 homologue) mutants. These cells have free, extra-long interphase MTs that exhibit abnormal behaviors such as cycles of growth and breakage, MT sliding, treadmilling, and hyperstability. The plus ends of interphase and spindle MTs grow continuously, exhibiting catastrophe defects that are dependent on the CLIP170 tip1p. The minus ends of interphase MTs exhibit shrinkage and pauses. As mto1Δ mutants lack cytoplasmic MTOCs, cytoplasmic MTs arise from spindle or other intranuclear MTs that exit the nucleus. Our findings show that mto1p and γ-TuCs affect multiple properties of MTs including nucleation, nuclear attachment, plus-end catastrophe, and minus-end shrinkage.
INTRODUCTION
The proper organization and dynamics of microtubules (MTs) is essential for cellular processes such as cell migration, cell polarity, and cell division. The nucleation of microtubules at microtubule organizing centers (MTOCs) is thought to be mediated by the γ-tubulin complex (γ-TuC). In many organisms, γ-tubulin is found in large ring-shaped complexes (γ-TuRCs; Zheng et al., 1995; Oegema et al., 1999; Murphy et al., 2001), which consist of 5–6 small complexes (γ-TuSCs) and at least five other proteins. These complexes are embedded in the centrosomal matrix, where they anchor MT minus ends. In vitro, γ-TuRCs template new MT assembly and cap MT minus ends (Moritz and Agard, 2001; Job et al., 2003).
The in vivo functions of γ-TuRCs, however, remain largely unexplored. Conditional mutants in γ-TuC components in yeast and fungi have defects in mitosis, suggestive of roles in SPB function and in a mitotic spindle checkpoint (Sobel and Snyder, 1995; Marschall et al., 1996; Spang et al., 1996; Paluh et al., 2000; Vardy and Toda, 2000; Hendrickson et al., 2001; Jung et al., 2001; Prigozhina et al., 2001; Vardy et al., 2002; Prigozhina et al., 2004). RNAi depletion of γ-tubulin in Caenorhabditis elegans leads to defects in MT nucleation primarily during interphase (Strome et al., 2001; Hannak et al., 2002). Paradoxically, the most striking defects seen in yeast cells are in the regulation of MT length, suggestive of a function in modulating MT plus ends (Paluh et al., 2000; Vardy and Toda, 2000; Vogel et al., 2001).
The fission yeast Schizosaccharomyces pombe has at least three types of MTOCs throughout its cell cycle (Hagan, 1998). During mitosis, the spindle pole body (SPB) organizes intranuclear spindle MTs from its nuclear face and astral microtubules from its cytoplasmic face. During midanaphase, the SPB is extruded from the nuclear envelope and is established in the cytoplasm adjacent to the nucleus, where it remains through interphase (Ding et al., 1997). During interphase, 3–5 cytoplasmic MT bundles are organized from the outer nuclear envelope. One of these is associated with the SPB, whereas the others are organized from interphase MTOCs (iMTOCs; Tran et al., 2001). These MTs are arranged along the long axis of the cell and form antiparallel bundles in which MT minus ends are stable within an MT overlap zone near the nucleus, and dynamic MT plus ends grow toward the cell tips (Drummond and Cross, 2000; Tran et al., 2001). After contact with cell tips, MTs undergo catastrophe and shrink back to the nucleus. These MTs position the nucleus at the cell middle (Tran et al., 2001). The equatorial MTOC (eMTOC) forms at the contractile ring and nucleates MTs into both daughter cells during cytokinesis (Heitz et al., 2001). S. pombe γ-TuC components include gtb1p (γ-tubulin), alp4p (homologue of Saccharomyces cerevisiae spc97/human GCP2), and alp6p (homologue of S. cerevisiae spc98/human GCP3), which all localize to all three types of MTOCs and are essential for viability and mitosis (Horio et al., 1991; Paluh et al., 2000; Vardy and Toda, 2000; Hendrickson et al., 2001; Fujita et al., 2002; Zimmerman et al., 2004b). These γ-TuC components are also distributed in satellites that move bidirectionally along interphase microtubules and are even present at MT plus ends (Sawin et al., 2004; Zimmerman et al., 2004b). Mutants in these proteins have abnormally long MTs, but the role of γ-TuCs in regulating MT dynamics has not been well characterized.
Mto1p (also known as mbo1p or mod20p) is a γ-TuC–associated protein that mediates γ-TuC recruitment specifically to cytoplasmic MTOCs (Sawin et al., 2004; Venkatram et al., 2004). Mto1p is predicted to be composed predominantly of coiled-coils and shares significant homology along its length with the Aspergillus nidulans nuclear positioning gene ApsB (Suelmann et al., 1998) and with regions of S. pombe pcp1p and Drosophila centrosomin at its N-terminus (Sawin et al., 2004). mto1Δ mutants are unable to localize other γ-TuC components to cytoplasmic MTOCs and have severe defects in the organization of cytoplasmic interphase MTs (Sawin et al., 2004; Venkatram et al., 2004). However, they still target γ-TuCs to the SPBs and are able to form mitotic spindles and divide. The viability of mto1Δ mutants therefore makes them more amenable for phenotypic analyses of nonessential processes than conditional lethal γ-TuC mutants.
Here, we characterize highly unusual MT behaviors in mutants defective in γ-TuC function, mto1Δ and alp4. We find that cytoplasmic MTs in mto1Δ mutants do not originate from cytoplasmic MTOCs, but from intranuclear MTs that exit the nucleus through the nuclear envelope. Quantitative analysis of MT dynamics showed persistent growth without catastrophe at MT plus ends and MT treadmilling. Similar phenotypes are also seen in the γ-TuC mutant, alp4, indicating that these phenotypes reflect a loss of γ-TuC function. Thus, these studies provide new insights into multiple cellular functions of γ-TuCs.
MATERIALS AND METHODS
Yeast Strains, Media, and Genetic Methods
S. pombe strains used are listed in Table 1. Standard methods for media and genetic manipulations are described at http://www.sanger.ac.uk/Post-Genomics/S_pombe/links.shtml. mto1Δ, mto1-GFP, and mto1-CFP strains were constructed using a PCR-directed method for site-directed recombination (Bahler et al., 1998) in haploid strains and confirmed by PCR. For mto1-GFP and mto1-CFP, primers 5′-ACAGGAGTCTTTCAAGGCAAATAAAAACGGATAAAGAAAGCAATTCTAGGTCTCCTTCTATATCATCACA AGAACATAAACGGATCCCCGGGTTAATTAA-3′ and 5′-TGTCAATTAAATTCTCCAACTAAGTAGATATGTACTGGTGATTAAGAAAAGCTATATACTTCAACCGCAAACTATCCAGAGAATTCGAGCTCGTTTAAAC-3′ were used to amplify GFP-kanMX from pFA6a-GFP (S65T)-kanMX6 and CFP-KanMX from pFA6a-CFP (S65T)-kanMX6, respectively. For construction of mto1Δ, primers 5′-TCGCTCCTTTGAAATTGTTACAAGAGTCAATGCCTACTGCTTTGGATTTGCTTATGTCGAAGTTCGTTTTCTAATCAATTCGGATCCCCGGGTTAATTAA-3′ and 5′-TGTCAATTAAATTCTCCAACTAAGTAGATATGTACTGGTGATTAAGAAAAGCTATATACTTCAACCGCAAACTATCCAGAGAATTCGAGCTCGTTTAAAC-3′ were used to amplify kanMX6 from pFA6a-kanMX6. pDQ105 (Ding et al., 1998) was obtained from D. Q. Ding and Y. Hiraoka.
Table 1.
Strains used in this study
| Strain no. | Mating type | Description | Auxotrophies | Source | ||
|---|---|---|---|---|---|---|
| FC421 | h- | ura4-D18 | leu1-32 | ade6-216 | Chang lab | |
| FC1025 | h- | nup107:GFP [pDQ105; nmt-GFP-atb2: LEU2] | leu1 | Chang lab | ||
| FC1188 | h- | mto1:GFP-kanMX6 | ura4-D18 | leu1-32 | ade6-216 | This study |
| FC1189 | h+ | mto1::kanMX6 | ura4-D18 | leu1-32 | ade6 | This study |
| FC1190 | h- | mto1::kanMX6 alp4:GFP-kanMX6 | ura4 | leu1 | ade6 | This study |
| FC1191 | h+ | alp4:GFP-kanMX6 | ura4 | leu1 | ade6 | This study |
| FC1192 | h- | mto1::kanMX6 alp4:GFP-kanMX6 [pDQ105; nmt-GFP-atb2: LEU2] | ura4 | leu1 | ade6 | This study |
| FC1193 | h+ | alp4:GFP-kanMX6 [pDQ105; nmt-GFP-atb2: LEU2] | ura4 | leu1 | ade6 | This study |
| FC1194 | h- | mto1:CFP-kanMX6 alp4:YFP-kanMX6 | ura4-D18 | leu1-32 | ade6 | This study |
| FC1195 | h- | mto1::kanMX6 tea1:GFP-kanMX6 | ura4-D18 | leu1-32 | ade6 | This study |
| FC1196 | h+ | tea1:GFP-kanMX6 | ura4-D18 | leu1-32 | ade6 | This study |
| FC1197 | h+ | tea1:YFP-kanMX6 [pRL74; nmt-CFP-atb2: ura4+] | ura4-D18 | leu1-32 | ade6 | This study |
| FC1198 | h- | mto1::kanMX6 tea1:YFP-kanMX6 [pRL74; nmt-CFP-atb2: ura4+] | ura4-D18 | leu1-32 | ade6 | This study |
| FC1199 | h- | alp4-1891[pDQ105; nmt-GFP-atb2: LEU2] | leu1 | This study | ||
| FC1200 | h+ | mto1::kanMX6 cut11:GFP ura4+ | leu1-32 | ade6 | This study | |
| FC1201 | h+ | mto1::kanMX6 tip1:YFP-kanMX6 [pRL74; nmt-CFP-atb2: ura4+] | ura4-D18 | leu1-32 | ade6 | This study |
| FC1202 | h- | tip1:YFP-kanMX6 [pRL74; nmt-CFP-atb2: ura4+] | ura4-D18 | leu1-32 | ade6 | |
| FC1203 | h+ | mto1::kanMX6 cut11:GFP ura4+ [pDQ105; nmt-GFP-atb2: LEU2] | leu1-32 | ade6 | This study | |
| FC1204 | h- | mto1::kanMX6 [pDQ105; nmt-GFP-atb2: LEU2] | ura4-D18 | leu1-32 | ade6 | This study |
| FC1205 | h- | mto1::kanMX6 alp4:YFP-kanMX6 [pRL74; nmt-CFP-atb2: ura4+] | ura4-D18 | leu1-32 | ade6 | This study |
| FC1226 | h+ | mto1::kanMX6NLS-GFP cut11:GFP ura4+ | leu1-32 | ade6 | This study | |
| FC1227 | h+ | tip1:YFP-kanMX6 mto1::kanMX6 | ura4-D18 | leu1-32 | ade6 | This study |
| FC1228 | h- | tip1:YFP-kanMX6 | ura4-D18 | leu1-32 | ade6 | This study |
| FC1229 | h- | tip1::kanMX6 mto1::kanMX6 [pDQ105; nmt-GFP-atb2: LEU2] | leu1-32 | This study | ||
| FC1230 | h- | tip1::kanMX6 [pDQ105; nmt-GFP-atb2: LEU2] | leu1-32 | This study | ||
| FC1231 | h- | alp4-1891 tip1::kanMX6 [pDQ105; nmt-GFP-atb2: LEU2] | leu1-32 | This study | ||
| FC1232 | h+ | tip1::kanMX6 mto1::kanMX6 cut11:GFP ura4+[pDQ105; nmt-GFP-atb2: LEU2] | leu1-32 | This study | ||
Cytoskeletal Analysis
Rhodamine phalloidin staining was performed on formaldehyde-fixed cells as described (Pelham and Chang, 2001). Cells expressing integrated fusions from endogenous promoters, such as mto1p-GFP and tip1p-YFP, were grown in exponential phase overnight in 2–3-ml liquid cultures with shaking at 30°C. For imaging microtubules in living cells, cells carrying pDQ105 (Ding et al., 1998) were grown as described (Zimmerman and Chang, 2004), except those used for measurements, which were grown to exponential phase overnight in 2–3 ml liquid cultures with shaking at 30°C. mto1Δ tip1Δ and alp4-1891 tip1Δ double mutants were likewise grown overnight. For MT regrowth experiments, cells were kept on ice for 20 min and then either imaged immediately on a cold agar pad (Tran et al., 2001) or allowed to recover for incremental periods of time at 25°C before imaging. Cells were imaged on agar pads for time-lapse acquisition over long time periods or under a coverslip in 1 μl of liquid media and sealed with VALAP (Tran et al., 2004). Methyl-benzidazole-carbamate (MBC; Aldrich, Milwaukee, WI) was used at 25 μg/ml final concentration (Tran et al., 2001).
Generally, MT imaging was carried out in multiple focal planes 0.4–0.5 μm apart using a spinning disk confocal microscope. MT dynamics and mto1p-GFP movements were measured in cells grown overnight at 30°C (mto1Δ, tip1Δ, mto1Δ tip1Δ, mto1p-GFP, wild type) or in 5–8 h cultures at 30°C or 36°C (alp4-1891). Images were recorded in 3.5-s (to measure growth and shrinkage rates), 5.5-s (to measure dwell times), or 13-s (mto1p-GFP) intervals at 25°C (except alp4-1891 was imaged at 30 or 36°C). Kymographs were created using NIH ImageJ 1.30. Fluorescent speckle microscopy measurements were done using a single focal plane. Measurements of MT growth, shrinkage rates, and dwell times were carried out relative to MT speckles in kymographs. Speckles present in MTs were imaged in cells grown to exponential phase overnight in 2–3-ml liquid cultures with shaking at 30°C.
Fluorescence intensity of tip1p-YFP was measured by recording the maximum intensity of single dots in single focal planes using Openlab 3 (Improvision, Lexington, MA).
Spindle orientation and elongation were measured in mto1Δ and wild-type cells by spinning disk confocal microscopy at 29°C in 45-s intervals. Measurements of spindle length and angle were obtained using NIH ImageJ 1.30.
Microscopy
Microscopy was carried out using a wide-field microscope station (for DIC and mto1p-GFP imaging) or a spinning disk confocal imaging station (Perkin Elmer-Cetus Wallac, Norwalk, CT), using 100× NA 1.4 and 60× NA 1.4 objectives and with acquisition and analysis using Openlab 3 (Improvision) and NIH Image J software (Pelham and Chang, 2001). FRAP experiments were performed using a separate wide-field imaging system described by Khodjakov et al. (2004).
RESULTS
mto1Δ Mutants Have Defects in Microtubule Organization
We independently identified mto1p in a two-hybrid screen as a protein that interacts with rsp1p, an MTOC protein involved in disassembly of the eMTOC at cell division (Zimmerman et al., 2004b). The relationship between rsp1p and mto1p will be described in a subsequent article.
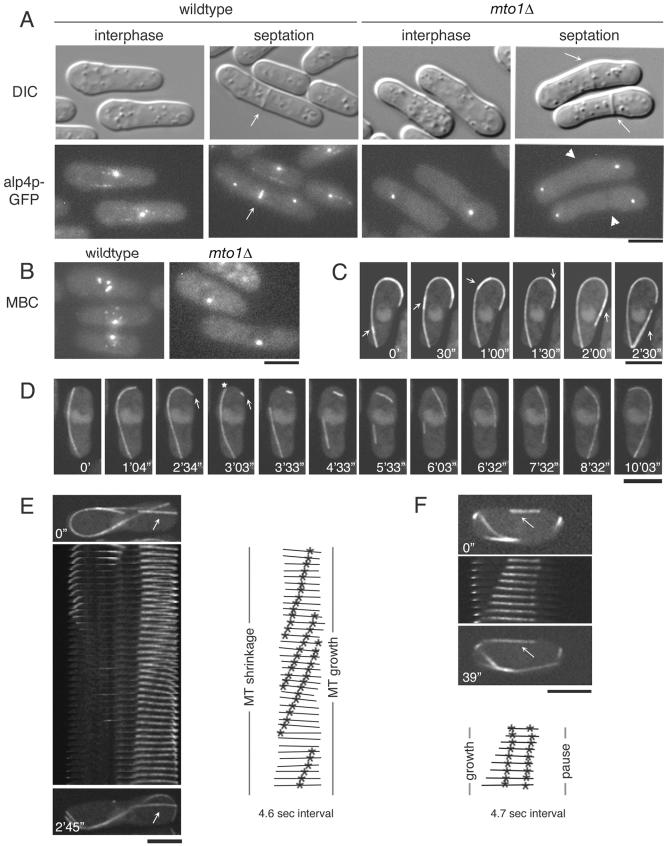
As shown previously (Sawin et al., 2004; Venkatram et al., 2004), mto1Δ cells were viable and exhibited near normal growth rates, but many cells were bent (31% showed >10° bend relative to the long axis of the cell vs. 2% wt, n = 200) and often divided asymmetrically (Figure 1), suggestive of microtubule defects (Toda et al., 1983). 3D time-lapse confocal microscopy and GFP markers revealed strikingly abnormal MT organization (Supplementary Movies 1–4). mto1Δ cells typically possess fewer interphase MT bundles than wild-type cells (Figure 1, B–F). Although most wild-type cells (76%; n = 319) have three or more MT bundles, 91% of mto1Δ cells had 2 or fewer MT bundles. MTs were absent in 20% of cells, although diffuse GFP-tubulin fluorescence was observed (Figure 1C; n = 467). The interphase MTs in mto1Δ cells were often abnormally long and continued to grow and curve upon reaching a cell tip, rather than undergoing catastrophe (Figure 1, B and F). Occasionally, these curved MTs broke, and the resulting fragments either continued to grow or shrank away (Figure 1F, Supplementary Movie 3). No clear instances of cytoplasmic MT nucleation were detected in 45 cells imaged >30 min each. Instead, MTs appeared to regenerate continually from preexisting MTs through multiple cycles of breakage and fragment elongation. We also noted that many MTs formed thick dynamic bundles, in which MTs often moved along each other and sometimes were observed to merge or slide apart (Figure 1E).
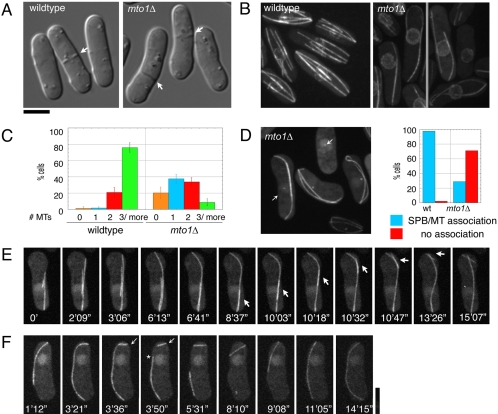
Figure 1.
mto1Δ cells have defects in MT organization. (A) Differential interference contrast (DIC) images of wild-type (FC421) and mto1Δ (FC1189) cells. Arrows indicate septum position. (B–F) Cells expressing GFP-α-tubulin (pDQ105; GFP-atb2p) were imaged by 3D confocal fluorescence microscopy in multiple focal planes. (B) Maximum projection images of nup107p-GFP (FC1025) and cut11p-GFPmto1Δ (FC1203). (C) Number of interphase MTs in wild-type (FC1193; n = 319) and mto1Δ (FC1192; n = 467). (D) mto1Δ alp4p-GFP (FC1192) cells expressing GFP-α-tubulin were imaged (left). Arrows point to SPBs. SPB-MT association was quantified in interphase wild-type (FC1193) and mto1Δ (FC1102) cells expressing GFP-α-tubulin and alp4p-GFP as a SPB marker (right). An association was counted if the SPB overlapped an MT for two time points 120 s apart (n > 160). (E) Time-lapse images of an mto1Δ cell (FC1204) in which an MT slides along another MT (arrow). (F) Time-lapse images of MT growth around the cell tip and subsequent breakage in a mto1Δ cell. Arrow marks MT growing tip, and asterisk refers to the MT break. Scale bars, 5 μm.
In addition, mto1Δ mutants had a strong defect in the attachment of MTs to the nuclear envelope. mto1Δ MTs were often dissociated from the nucleus and the SPB (Figure 1, D and F). Although the SPB was always associated with one MT bundle in wild-type cells (98%, n = 177), in mto1Δ cells, the SPB was often not associated with any cytoplasmic MTs (71% of cells with MTs, n = 161). In addition, mto1Δ MTs did not originate from or shrink back toward the nucleus (Figure 1F), showing that they were not organized from the nucleus. Consistent with a defect in MT attachment, the nucleus was mispositioned in most mto1Δ cells, (69% vs. 4% wt, n = 300; Figure 1, B, E, and F). Thus, mto1p is required for iMTOC function at the nuclear envelope.
mto1Δ Cells Show Abnormal Dynamics at Both MT Plus and Minus Ends
Next, we measured the dynamic parameters of these interphase MTs in mto1Δ mutants. Quantitative measurements of MT dynamics in γ-TuC mutants have not been reported previously, potentially because of problems of MT bundling and sliding. We sought to analyze individual MTs by using GFP-α-tubulin speckles as fiduciary marks (fluorescent speckle microscopy) to consider the movement of the MT lattice (Tran et al., 2001; Waterman-Storer and Danuser, 2002). In wild-type cells, we measured an MT plus-end growth rate of 3.6 ± 0.86 μm/min (n = 25) and an MT plus-end shrinkage rate of 12.2 ± 1.7 μm/min (n = 15), consistent with previous data (Drummond and Cross, 2000; Feierbach and Chang, 2001; Tran et al., 2001; Figure 2A). MT minus ends were generally nondynamic and were embedded in iMTOCs and the SPB (Tran et al., 2001). In mto1Δ cells, we analyzed MTs with clear speckle patterns, which were more likely to be individual MTs. In these MTs, we observed a strikingly consistent pattern: one end grew, whereas the other end did not (100% cells, n = 17).
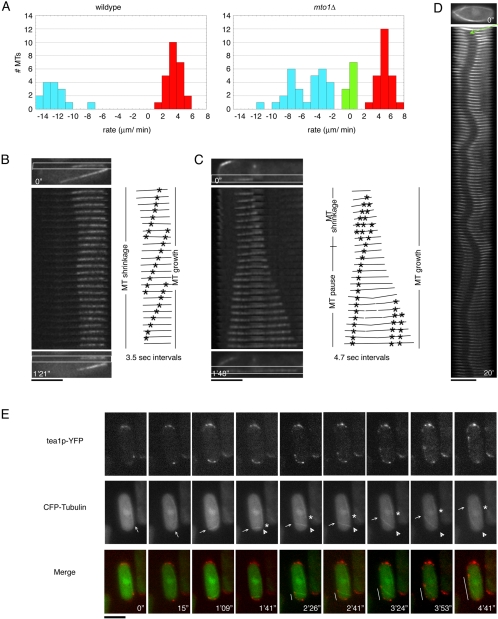
Figure 2.
Abnormal MT dynamics in mto1Δ mutant cells. (A) Interphase MT growth and shrinkage rates were measured in single focal planes by fluorescence speckle microscopy (see Materials and Methods) of wild-type (FC1025) and mto1Δ cells (FC1204, FC1193) expressing GFP-α-tubulin. Positive rates (red) represent MT growth, negative rates (blue) represent MT shrinkage, and zero rates (green) represent MT pauses. (B and C) Examples of MT treadmilling and pausing, as shown by location of MT ends relative to GFP-α-tubulin speckles in mto1Δ cells (FC1204, B; FC1193, C). Kymographs show confocal images in a single focal plane of an MT (box) over time. Line drawings illustrate the behavior of the MTs marked with representative speckles. (D) Kymograph showing the behavior of a stable photobleach mark (arrow) on an MT bundle in mto1Δ cell (FC1204). Images are taken 10 s apart. (E) Time-lapse of mto1Δ cells expressing tea1p-YFP and CFP-tubulin (FC1198). Arrows in CFP-tubulin refer to the MT plus end marked by tea1p and arrowheads denote the predicted MT minus end lacking tea1p. The asterisk marks the site of MT breakage and then the shrinking MT (the predicted plus end). Lines in Merge panels mark tea1p-YFP dots deposited along cell cortex. Scale bars, 5 μm.
Analysis of speckled MTs showed that MT growth rates in mto1Δ cells were slightly higher than those of wild-type MT plus ends (4.4 ± 0.85 μm/min; n = 24; Figure 2A). We confirmed that these were MT plus ends by localizing MT plus-end binding proteins (tea1p and the CLIP170 homologue tip1p) to the growing MT tip (see below).
Significantly, we found that the MT plus ends had a profound defect in MT catastrophe. In our analysis of speckled MTs, we noted no instances of MT plus-end shrinkage within the time frames of the movies (2–5 min). These ends did not shrink upon hitting the cell tip, but instead continued to grow and curved around the cell tips (n = 15). MT plus ends labeled with the MT plus-end protein tea1p-YFP showed a similar behavior (Figure 2E). A reduction in MT catastrophe events was apparent in other time lapse images of interphase MTs, but the polarity of the MTs in many of these cases was less clear.
Rates of MT shrinkage were more heterogeneous and could be sorted into at least two distinct groups (Figure 2A, right, blue). In one group, shrinkage rates of 3–5 μm/min were measured at predicted MT minus ends. Because this shrinkage rate was similar to the rate of MT plus-end growth, striking examples of treadmilling were observed, in which the length of the MT remained constant but the movement of GFP-tubulin fluorescent speckles showed continuous flux (Figure 2, A and B, Supplementary Movie 5). A second group of MT shrinkage rates (>8 μm/min) occurred at both predicted MT minus and plus ends. Plus end shrinkage often occurred at a newly generated plus-end after an MT break, indicating that these do not exhibit the same stability as an established growing plus end. This rapid shrinkage rate often led to the loss of the entire MT. The reason for these two classes of MT minus-end shrinkage rates is not clear.
We also detected many examples of paused MT ends. In most cases, pauses occurred at predicted MT minus ends and did not necessarily occur near the cell surface. Figure 2C shows an example of an MT minus end that transitions from a shrinking to a pausing state.
To analyze the behavior of MTs within bundles in mto1Δ cells, we made photobleach marks on GFP-tubulin–labeled MT bundles. These photobleach marks on interphase MT bundles could be maintained for at least 20 min (Figure 2D), whereas similar marks in wild-type MT bundles were maintained for less than 5 min (unpublished data). Thus, mto1Δ cells possess hyperstable MTs.
In summary, mto1Δ cells exhibit significant abnormalities at MT minus ends and plus ends. These properties lead to novel MT behaviors, such as treadmilling and hyperstability.
Mto1p Affects eMTOC and Astral MT Formation and Spindle Dynamics in Mitosis
Although mto1Δ cells were competent to assemble mitotic spindles, they did exhibit a number of defects during mitosis. Time-lapse measurements of individual spindles labeled with GFP-tubulin showed that all three phases of mitosis (Phase 1, Prophase and Spindle Assembly; Phase 2, Metaphase-Anaphase A; Phase 3, Anaphase B spindle elongation; Nabeshima et al., 1998; Mallavarapu et al., 1999) generally occurred with normal kinetics in mto1Δ cells (Figure 3). In contrast to Venkatram et al. (2004) but in agreement with the less direct measurements of Sawin et al. (2004), we observed no defects in the rate of spindle elongation in Anaphase B (0.92 ± 0.17 μm/min for mto1Δ;n = 13 vs. 0.97 ± 0.11 μm/min for wt; n = 15; Figure 3C, Supplementary Movie 6).
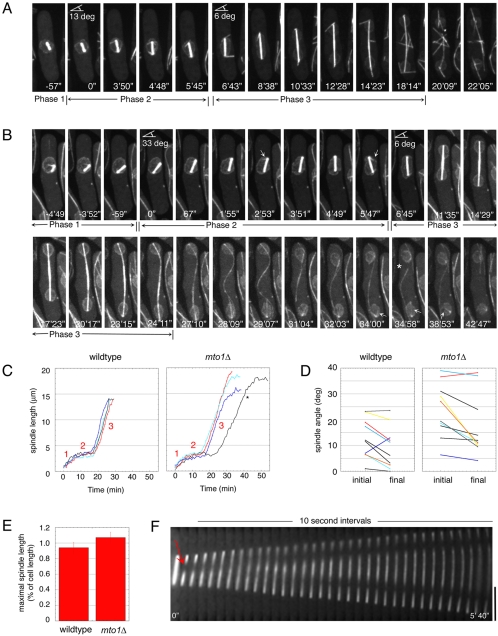
Figure 3.
Mto1p regulates the assembly of astral and eMTOC MTs and the disassembly of spindle MTs. Wild-type (FC1025) and mto1Δ (FC1203 or FC1204) cells expressing GFP-tubulin and either nup107p-GFP (wild-type) or cut11p-GFP (mto1Δ; FC1203) were imaged by 3D confocal microscopy at 29°C. (A) Normal spindle in wild-type cell (FC1025). (B) Spindle in mto1Δ cell. Note the absence of anaphase astral MTs and eMTOC MTs. The asterisk marks spindle breakage, and arrows refer to intranuclear MTs. (C) Spindle elongation in four representative wild-type and mto1Δ cells. The asterisk marks one of two spindles (n = 13) that elongated at a slower rate than wild type. mto1Δ spindles persist over time. (D) Changes in spindle orientation over Phase 2 (metaphase/anaphase A) in representative cells. (E) Quantitation of the spindle length at the time of spindle breakdown. (F) Kymograph of an mto1Δ spindle that was photobleached in the middle in early anaphase (arrow). Scale bars, 5 μm.
mto1Δ mitotic spindles lacked cytoplasmic astral MTs (Phase 3; 100% cells, n = 50; Figure 3, A and B). This may reflect a loss of γ-TuC activity from the cytoplasmic face of the SPB. The normal spindle elongation rate indicates that cytoplasmic astral MTs are not required for spindle elongation. This finding supports conclusions from recent laser ablation studies that physically cut astral MTs or SPBs (Khodjakov et al., 2004; Tolic-Norrelykke et al., 2004).
Intranuclear MTs (astral-like MTs in Phase 2) were present normally in mto1Δ cells (Figure 3B, Phase 2, see arrows; Sagolla et al., 2003; Zimmerman et al., 2004a). As also reported by Venkatram et al. (2004), mto1Δ cells possessed more misoriented spindles at the onset of metaphase than did wild-type spindles (33% of spindles >30° off, n = 18, vs. 8% of wild-type spindles, n = 25). However, mto1Δ Phase 2 spindles still rotated and corrected the spindle angle to a similar extent as wild-type cells (for spindles that were 15–30° offset, avg. rotation = 9.8° in mto1Δ, n = 7, vs. 9.2° in wt, n = 8; Figure 3D). Thus, mto1Δ cells did not have a defect in spindle rotation; rather the apparent spindle misorientation in Phase 2 in mto1Δ cells may arise from abnormal SPB positions before mitosis, which could be secondary to defects in interphase MT attachment.
mto1Δ cells also lacked eMTOC function. MTs did not grow out from the future cell division site during anaphase and after anaphase (Figure 3B). However, cells were able to septate and divide without obvious cytokinesis defects, suggesting that the eMTOC is not required for cell division and contractile ring function (our unpublished data). The eMTOC has been proposed to maintain the position of the contractile ring in a septation mutant (Pardo and Nurse, 2003), but we did not detect significant problems in ring stability in mto1Δ (our unpublished observations).
At the end of mitosis, mto1Δ cells exhibited a defect in spindle breakdown (Figure 3, C and E). Instead of disassembling when the nuclei reached the cell tips, spindles continued to grow (Figure 3, B, C, and E) and bent into S-shaped structures (Figure 3B, 28′09″-29′07″). To determine the cause of this overelongation, we generated photobleach marks in GFP-tubulin labeled spindles. In wild-type cells, photobleach marks made in the middle of the spindle during early anaphase initially disappeared, reflecting plus-end polymerization of spindle MTs. Later in anaphase, two photobleach marks reappeared near the ends of the spindle and then moved apart with the poles (Mallavarapu et al., 1999; Khodjakov et al., 2004). This behavior shows that MT minus ends at the SPB are nondynamic and do not undergo minus-end directed flux. Photobleach marks in mto1Δ cells behaved similarly (n = 13; Figure 3F). Notably, the distance between the poles and the photobleach marks did not change over time (final distance/initial distance = 0.97 ± 0.14; n = 13; Figure 3F), showing that in contrast to the minus ends of cytoplasmic MTs, MT minus ends were not dynamic in these spindles. The behavior of the photobleach marks also showed that no additional abnormal MTs were nucleated from the SPBs during anaphase. Therefore, spindle overelongation may result from a defect in stopping plus-end elongation of spindle MTs at the end of anaphase.
mto1Δ Cells Nucleate MTs Only from Inside the Nucleus
The apparent absence of cytoplasmic MTOCs poses the question: how do cytoplasmic MTs form in these cells? To further characterize MT nucleation activity, we examined MT regrowth after cytoplasmic MTs were depolymerized by cold shock for 20 min (Mata and Nurse, 1997; Chen et al., 1999; Tran et al., 2001; Figure 4, A, left, and B, left). Within 1 min of shifting to 25°C, wild-type cells began to renucleate MTs and restored a normal MT array by 5 min. In contrast, most mto1Δ cells either slowly formed short MTs or nucleated no MTs at all (as was also seen in Sawin et al., 2004). Short MTs were found in 61% of cells within 5 min (n = 50) and in 74% after a 15-min shift at 25°C (n = 76; Figure 4C). Z-section images of cells coexpressing a nuclear envelope marker confirmed that these MT bundles were intranuclear (Figure 4E). Generally, one end of these MTs was in close proximity with the SPB (unpublished data). Most of these MTs grew and shrank within the nuclear envelope, whereas some were more stable (Figure 4D, Supplementary Movie 7). Many MT bundles projected outside the body of the nucleus (Figure 4E). Consistent with this, we observed that the nuclear envelope formed thin protrusions that grew and shrank (Figure 4, G and H, Supplementary Movie 8). Such intranuclear MTs and nuclear envelope distortions were not observed in wild-type cells. Several criteria confirmed that these cells with intranuclear MTs were in interphase and not in mitosis: these cells did not exhibit cytokinetic actin rings or separated SPBs and were often shorter than mitotic cells (Figure 4, B and H; our unpublished observations).
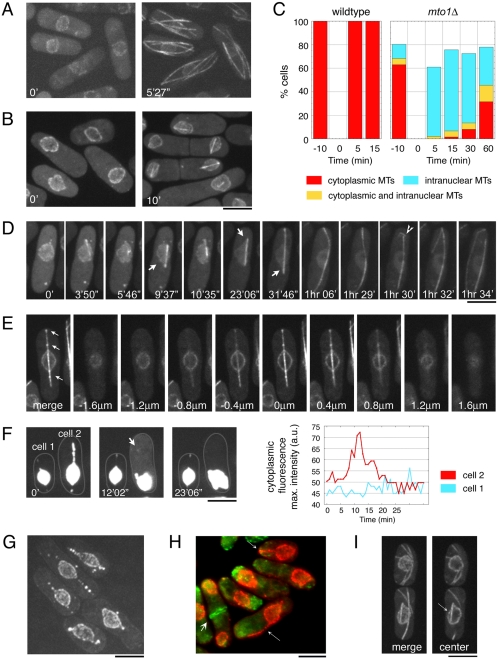
Figure 4.
Abnormal MT nucleation in the mto1Δ mutant. MTs in wild-type (FC1025) and mto1Δ (FC1203) cells expressing GFP-α-tubulin and nuclear pore markers were depolymerized by a 20-min treatment at 0°C and were monitored for MT regrowth upon shift to 25°C. (A) Images of wild-type cells at indicated times after cold shock. (B) Images of mto1Δ cells at indicated times after cold shock. (C) Characterization of MT types that develop after cold shock in nonseptated interphase cells (n > 50 cells at each time point). (D) Time-lapse images of an mto1Δ cell. Arrows mark stretching of the nuclear envelope during MT growth and arrowhead indicates the entry of an intranuclear MT into the cytoplasm. (E) Z-sections of an mto1Δ cell expressing GFP-α-tubulin and a nuclear pore marker cut11p-GFP 30 min after cold shock. Sections show that the MT bundle resides inside the nuclear envelope. Note the presence of nuclear pores (dots marked by cut11p-GFP) on the sides of the MT bundle (arrows). (F) Time-lapse images of two cells expressing cut11p-GFP and a soluble NLS-GFP-βgal marker (FC1226). In each cell, maximum cytoplasmic fluorescence intensity was measured at a point adjacent to the nucleus and plotted in the graph to the right. The increase, at 12′02″, in cytoplasmic fluorescence intensity in cell 2 (arrow) indicates a transient breakage of the nuclear envelope. (G) mto1Δ cells expressing the nuclear pore marker cut11p-GFP (FC1200) after 30 min of recovery after cold shock. (H) F-actin staining (green) of mto1Δ cells expressing cut11p-GFP (red; FC1200) after 30 min of recovery after cold shock. Small arrows show abnormal projections of the nuclear envelope in interphase cells without actin rings. Large arrow marks dividing cell with actin ring and no nuclear projections. (I) Intranuclear MT (arrow) in mto1Δ cells expressing GFP-α-tubulin and cut11p-GFP (FC1203) without cold shock. Maximum intensity projection (left) and a single focal plane (right) are shown. Scale bars, 5 μm.
Cytoplasmic MTs began to appear only 30–60 min after recovery from cold shock (Figure 4C, unpublished data). We found that the intranuclear MTs contributed to the formation of these cytoplasmic MTs. Although interphase intranuclear MTs remained relatively static for extended periods of time, we observed instances where they suddenly began to grow more rapidly, moved throughout the cytoplasm and propagated themselves by growing and breaking like cytoplasmic MTs, as seen in non-cold shocked mto1Δ cells (Figure 4D, 1 h 30′ to 1 h 34′, Supplementary Movie 7). We tested whether the abrupt change in MT behavior could represent the entry of the intranuclear MT into the cytoplasm by puncturing the nuclear envelope. We assayed for breaks in nuclear envelope integrity using a soluble nuclear GFP marker, NLS-GFP-βgal (Yoshida and Sazer, 2004). We found that after cold shock, some cells exhibited small, transient bursts of cytoplasmic NLS-GFP-βgal fluorescence, which were sometimes accompanied by abrupt retractions in the nuclear envelope (Figure 4F, Supplementary Movie 9). These breakthroughs were observed in 12% of cells imaged for 1 h after cold shock (as assayed by changes in MT dynamics; n = 48; Figure 4D, unpublished data). Thus, interphase intranuclear MTs that pierce the nuclear envelope contribute to the formation of a significant fraction of the cytoplasmic MTs, but there are probably additional sources as well.
Additional observations revealed that the majority of cytoplasmic MTs originated from mitotic spindles. After mitosis, some spindle MTs in mto1Δ cells recovering from cold shock were released into the cytoplasm (unpublished data). Consistent with this, 67 ± 8% of newly divided cells possessed cytoplasmic MTs, irrespective of the recovery time after cold shock (n ≥ 25 for samples treated without cold shock and 30′, 60′, 120′, and 180′ after cold shock). In addition, after 3 h (about a cell cycle period), cytoplasmic MTs were reestablished in 70% of cells (n = 102).
We next tested whether persistent spindle MTs also contribute to the establishment of interphase cytoplasmic MTs in non–cold-shocked mto1Δcells. Time-lapse images showed that after anaphase, persistent spindle MTs frequently grew past the nucleus, split from the main spindle, and curved around cell tips. These MTs then typically broke into fragments that continued to propagate within the cytoplasm (Figure 3B, 28′0″ to 42′47″; see also Sawin et al., 2004). Images of the nuclear envelope and the NLS-GFP-βgal marker suggested that these spindle MTs do not puncture the nuclear envelope as during interphase, but rather enter the cytoplasm when the nuclear envelope fissions at the spindle midzone at the end of anaphase (our unpublished data). Time-lapse images showed that most cytoplasmic MTs originated in this manner. Short intranuclear MTs were also present in postmitotic cells (Figure 3B, 31′03″ to 38′53″; see also Figure 4I, arrow) and occasionally seeded the formation of cytoplasmic MTs. However, most of these intranuclear MTs did not persist later in interphase (12% of cells exhibited both interphase intranuclear MTs and cytoplasmic MTs; n = 85). In summary, these findings in mto1Δ mutant cells document an unusual mechanism of generating cytoplasmic MTs from intranuclear MTs.
alp4 Mutants Have Similar Phenotypes
We next examined if mto1p functions primarily through its effects on γ-TuCs. Consistent with previous findings (Sawin et al., 2004; Venkatram et al., 2004), in mto1Δ cells, alp4p-GFP was present only at the SPB and was absent from MT satellites, iMTOCs, and the eMTOC (Figure 5, A and B). Thus, mto1p is required to localize γ-TuC components to cytoplasmic MTOCs.
Figure 5.
Localization and mutant phenotype of the γ-tubulin complex protein alp4p. (A and B) Cells expressing alp4p-GFP were imaged in multiple focal planes by wide-field microscopy in wild-type (FC1191) or mto1Δ (FC1190) cells. Alp4p-GFP localization was analyzed throughout the cell cycle (A) and after MBC treatment (B). Arrows refer to the septum or eMTOC, and arrowheads mark the position where eMTOCs should form. (C–F) alp4-1891 cells expressing GFP-Tubulin (FC1199) were grown at 30°C and imaged in multiple (C and D) or single (E and F) focal planes by confocal microscopy. (C) Time-lapse imaging of MT sliding in alp4-1891 cell. Arrows refer to the sliding MT bundle. (D) Time-lapse imaging of MT snapping and elongation in the alp4-1891 mutant. The arrow marks the tip of the original MT, and the asterisk (*) marks the site of MT breakage. (E and F) Kymographs demonstrating (E) MT treadmilling and (F) MT pause in the alp4-1891 mutant. Arrows highlights the MT analyzed. Models adjacent to the kymograph mark representative speckles (*). Scale bars, 5 μm.
If mto1p functions primarily with γ-TuCs, we predicted that other γ-TuC mutants would share common phenotypes. γ-TuC mutants (gtb1, alp4-1891, alp6-719, and alp16Δ) have abnormally long interphase and spindle MTs, similar to those seen in mto1Δ (Paluh et al., 2000; Vardy and Toda, 2000; Fujita et al., 2002; Tange et al., 2004). We found that alp4-1891 cells at a semipermissive temperature of 30°C exhibited many of the MT phenotypes seen in mto1Δ mutants, including MT growth around cell tips, MT breakage, MT disassociation from the nucleus, MT treadmilling, and pausing (Figure 5, C–F, Supplementary Movie 10). The eMTOC was either nonfunctional or very weak, nucleating just one or two MTs at a time (Supplementary Movie 11). Astral MTs (Phase 3) were also absent, weak, or unstable (Supplementary Movie 11). After anaphase, intranuclear MTs were occasionally present. Thus, the properties of the partial loss-of-function mutant alp4-1891 were similar to, but less severe than, the mto1Δ mutant, indicating that these mto1Δ phenotypes are likely to be caused by a loss of γ-TuC function.
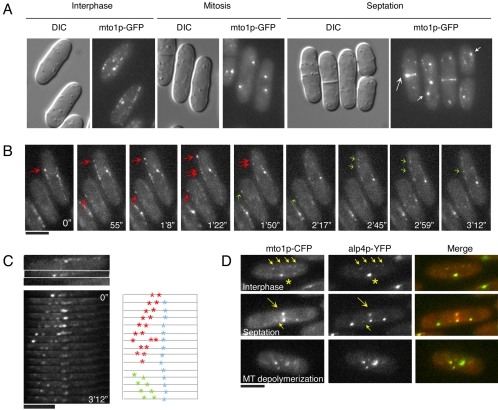
Mto1p Localizes to MTOCs and Motile MT Satellites
We reexamined the localization of mto1p, using live cell imaging of functional GFP and CFP fusions expressed under the endogenous mto1 promoter (see also Sawin et al., 2004; Venkatram et al., 2004). Mto1p-CFP colocalized with alp4p-YFP at all cellular MTOCs: the SPB throughout the cell cycle, the eMTOC during septation, iMTOCs and satellites during interphase (Figure 6, A and D). Mto1p colocalized precisely with alp4p in satellites (Figure 6D, top panel). In contrast to the perinuclear staining seen in fixed cells (Sawin et al., 2004), live cell imaging showed that mto1p-GFP localized, like alp4p and rsp1p, to 2–4 discrete perinuclear iMTOC dots in MBC-treated cells (Zimmerman et al., 2004b).
Figure 6.
Mto1p localization to MTOCs and satellites. (A) mto1p-GFP localization in different cell cycle stages. Maximum intensity projections of confocal images are shown. Large arrow marks the eMTOC, and smaller arrows point to SPBs. (B) Time-lapse images of mto1p-GFP satellites moving in the predicted MT plus direction toward the cell tips (red arrows) and in the predicted MT minus direction toward the nucleus (green arrows; FC1188). Images were taken in a single focal plane. (C) Kymograph showing mto1p-GFP satellite behavior on an MT bundle (box). Line drawing shows representative behaviors of individual satellites. Red asterisks mark satellites moving in the plus direction, green asterisks denote the minus direction, and blue asterisks refer to satellites that move little. (D) Colocalization of mto1p-CFP and alp4p-YFP (FC1194) at satellites (small yellow arrows), SPB (asterisks), eMTOC (large yellow arrows), and iMTOCs (MBC-treated cells) in single focal planes. Scale bars, 5 μm.
γ-TuC components also localize to satellites, which exhibit complex patterns of movement on MTs (Zimmerman et al., 2004b). As the mto1p-GFP fusion was brighter than the alp4p-GFP fusion, we were able to further analyze the movement of satellites on microtubules using kymographs, which revealed patterns of satellite movements not previously reported (Figure 6, B and C). Satellites were present on growing and shrinking MT plus ends, as well as possibly along MTs. Many satellites moved as groups toward the MT plus or minus ends. Interestingly, a subset of satellites near the medial regions of MT overlap, moved very little (Figure 6C, blue asterisks). Occasionally, dots apparently reversed directions before reaching the cell tip (Figure 6B, bottom cell). Although the function of these movements is not well understood, they may facilitate the distribution of γ-TuCs on microtubules.
Role of the CLIP170 tip1p in Regulating MT Catastrophe
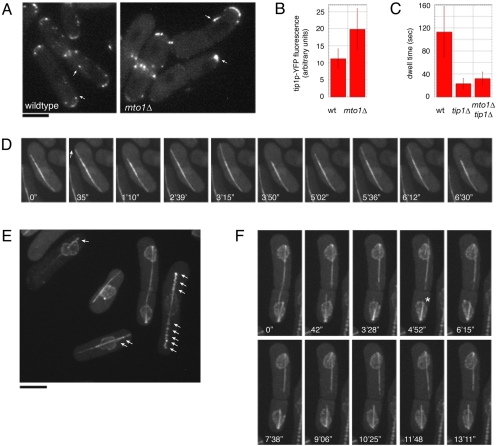
An apparent paradox is why γ-TuCs are needed for MT catastrophe at the plus end. We present a number of possible models in the discussion. One attractive possibility is that γ-TuCs affect the loading, function, or dynamics of MT plus-end factors that regulate MT catastrophe. The CLIP170-like protein tip1p is an MT plus-end protein that prevents premature MT catastrophe. In tip1Δ mutants, MTs grow at normal rates, but catastrophe upon hitting the side or tip of the cell and have a decreased dwell time at the cell surface before catastrophe (Brunner and Nurse, 2000).
We found that the strong MT catastrophe defect in γ-TuC mutants was dependent on tip1p. Instead of long, curved MTs, mto1Δtip1Δ double mutant cells exhibited a single straight MT bundle that did not curve around cell tips (Figure 7, D–F, Supplementary Movies 12 and 13). MTs within a bundle grew to the cell tips and shrank back repeatedly to the MT bundle. The dwell time in these cells was comparable to that of tip1Δ (32 ± 11 s for tip1Δmto1Δ vs. 23 ± 9 s for tip1Δ and 113 ± 44 s for wt; n = 16, 18, and 12, respectively; Figure 7C). In other MT bundles, the ends grew very slowly or not at all (Supplementary Movie 13: bottom right cell, top bundle, also top cell), similar to intranuclear MTs in mto1Δ mutants. Similar types of bundles were seen in alp4-1891 tip1Δ mutants (at 30°C; unpublished data). Thus, the ability to undergo catastrophe is restored in these γ-TuC mutants by deleting tip1.
Figure 7.
γ-TuCs affect the regulation of tip1p (CLIP170 homologue). (A) Tip1p-YFP localization in wild-type (FC1228) and in mto1Δ (FC1227) cells. Cells were imaged in multiple focal planes by wide-field microscopy. Arrows mark dots of tip1p that are likely to be at MT plus ends. (B) Quantification of fluorescence intensity of tip1p-YFP dots that are likely to be at MT plus ends. Measurements were obtained from single focal planes from images such as those in A. (C) Quantification of dwell time in wild-type (FC1025), tip1Δ (FC1230), and mto1Δtip1Δ (FC1229) double mutants. (D) 3D confocal time-lapse images of MT growth and catastrophe from a single MT bundle in mto1Δtip1Δ (FC1229). The top portion of the MT bundle (arrow) is dynamic, whereas the lower portion is not. (E and F) Maximum intensity projections of confocal images of mto1Δtip1Δ cells expressing cut11p-GFP (nuclear pore marker) and GFP-tubulin (FC1232). (E) Arrows mark examples of nuclear pores spread along projecting MT bundle (see Supplementary Movie 13). (F) Time-lapse images of the spindle and other intranuclear MTs during cell division. One of the half spindles (top cell) persists as a projecting, dynamic MT bundle. The other half spindle (in bottom cell) shrinks, and a nonspindle intranuclear MT (asterisk) projects out of the nucleus. Scale bars, 5 μm.
Consistent with this phenotype, tip1p-YFP often accumulated in large dots at the ends of MTs in mto1Δ mutants (Figure 7A, see arrows). Fluorescence intensity measurements of tip1p-YFP showed a significant increase in the amount of tip1p at MT plus ends (19.8 ± 6 arbitrary fluorescence units in mto1Δ, compared with 11.2 ± 2.9 in wild-type; n = 39 and 43, respectively; Figure 7B). Immunoblots showed no clear change (<2-fold) in tip1p protein levels in mto1 mutant extracts (our unpublished observations). Because excess tip1p could be imagined to stabilize MTs and prevent catastrophe, these accumulations of tip1p provide a potential explanation for MT plus-end stabilization.
Another difference between mto1Δtip1Δ and mto1Δ cells was that most of the MT bundles appeared to be attached to the nucleus. 3D confocal microscopy of cells expressing markers for the nuclear envelope and microtubules showed that a part of each MT bundle resided inside the nucleus in at least 65% of interphase cells (n = 335). Portions of the MT bundles projecting from the nuclear envelope were partially or entirely covered with nuclear pores, indicating that many of the projected MTs were encased within the nuclear envelope (Figure 7E, see arrows; Supplementary Movie 13). Images of nuclear pore-GFP alone confirmed that most cells contained projections of the nuclear envelope (unpublished observations). Dynamic portions of the MT bundles were free of nuclear pores, while nondynamic bundles were often decorated with them (Supplementary Movie 13). Thus a single MT bundle may have portions inside and outside the nucleus: the MTs outside the nuclear envelope were dynamic, whereas the MTs entirely inside were less dynamic.
Time-lapse images showed that many of these MT bundles were derived from persistent spindle MTs. In some cells, the spindle did not break down entirely after anaphase, but persisted through interphase as a single straight MT bundle (Figure 7F, top cell, Supplementary Movie 13). In other cells, the spindle shrank back to the newly divided nucleus after anaphase, and a new intranuclear MT grew into an MT bundle projecting from the nucleus (Figure 7F, bottom cell). There were only rare free MT bundles in mto1Δ tip1Δ cells. These free MTs, which were apparently derived from these intranuclear MTs, formed antiparallel bundles and exhibited catastrophe at cell tips like the nuclear-attached bundles. Thus, many of the unusual interphase MT bundles in mto1Δtip1Δ cells represent persistent mitotic spindle MTs or other intranuclear MTs that are unable to dissociate from the nucleus.
In summary, the CLIP170-like protein tip1p modulates the MT plus-end behavior of mto1 and alp4 mutants. The catastrophe defects were dependent on tip1p, and tip1p itself accumulates at higher levels on the MT plus ends. One possibility is that γ-TuC directly or indirectly regulates the dissociation of tip1p from MT plus ends, a step that may contribute to MT catastrophe.
Localization of Other MT Plus-end Factors
We also examined two other factors that affect MT catastrophe: the kelch repeat protein tea1p and the kinesin klp5p (Mata and Nurse, 1997; West et al., 2001; Behrens and Nurse, 2002; Garcia et al., 2002). Both tea1p and klp5p were localized properly on MTs in mto1Δ mutants and unlike tip1p, did not accumulate at higher levels (Figure 2E, Supplementary Figure 1A, unpublished data).
However, we noted some abnormal distributions of tea1p at other locations that may explain other aspects of mto1 mutants. Tea1p regulates cell polarity and is deposited by MTs at cell tips (Mata and Nurse, 1997; Behrens and Nurse, 2002; Feierbach et al., 2004). In mto1Δ cells, tea1p was often present on the sides of cells and was either not concentrated at the cell tips or was unevenly distributed between them (Supplementary Figure 1B; unpublished data). We observed that a growing MT plus end appeared to deposit tea1p dots along the side of the cell (Figure 2E, arrows in merge), suggesting that MT catastrophe is not required for tea1p deposition. The abnormal distribution of tea1p at the cortex may explain the altered cell shapes seen in mto1Δ mutants. In addition, we also observed tea1p-YFP on the mitotic spindle in mto1Δ cells, which was never seen in wild-type cells (Supplementary Figure 1, C and D). Tea1p localized along the length of short spindles (47% of short spindles; n = 18), and localized either diffusely or on the poles of long spindles (Supplementary Figure 1D, unpublished data). In contrast, tip1p was not detected on the spindle (unpublished data). This is the first observed example of tea1p in the nucleus and provides a potential explanation for the altered spindle dynamics and mild chromosomal segregation defects seen in mto1Δ cells (Sawin et al., 2004).
DISCUSSION
Mto1p and the γ-Tubulin Complex
Here, in order to understand the in vivo function of γ-TuCs, we have studied a number of novel abnormal MT behaviors in cells defective for γ-TuC function. We used mto1Δ mutants as a way to study the effects of γ-TuCs on cytoplasmic MTs in otherwise viable cells. As mto1p is required to anchor γ-TuCs specifically at cytoplasmic MTOCs and MTs, the mto1Δ phenotype may be equivalent to a loss-of-function of γ-TuCs at these sites. Similar MT behaviors were also found in an alp4 mutant. Our results largely agree with the previous reports on this gene product (also known as mbo1p or mod20p; Sawin et al., 2004; Venkatram et al., 2004), with several noted exceptions, and are consistent with previous studies of other S. pombe γ-TuC proteins (Vardy and Toda, 2000; Hendrickson et al., 2001; Fujita et al., 2002; Horio and Oakley, 2003). However, our study extends these previous ones in several different ways. First, we provide one of the first quantitative measurements of MT dynamics in loss-of-function γ-TuC mutants in any organism, which reveal a striking defect on MT plus catastrophe. Second, we demonstrate several novel MT behaviors such as treadmilling, pausing, and exit of MTs from the nucleus. Third, we find a possible role for the CLIP170 homologue, tip1p, in the regulation of MT catastrophe by γ-TuCs. Thus, our studies provide new insights into the in vivo functions of γ-TuCs in MT dynamics and organization at both ends of the microtubule.
Nuclear Origin of Cytoplasmic Microtubules
Two clear functions of mto1p and γ-TuCs are in MT nucleation and MT attachment to the nucleus. Mto1p is required for MT nucleation from all cytoplasmic MTOCs (the cytoplasmic face of the SPB, iMTOCs, and the eMTOC). During interphase, mto1Δ cells lack iMTOCs and possess either no MTs or small numbers of free, abnormal MTs. Notably, mto1Δ mutants are among the first mutants identified with a strong defect in MT-nuclear attachment and support a model that iMTOCs and γ-TuCs are required for MT attachment.
In trying to understand the origin of cytoplasmic MTs in mto1Δ cells lacking cytoplasmic nucleating sites, we found that most, if not all, cytoplasmic MTs are originally nucleated inside the nucleus, either as spindle MTs or as interphase intranuclear MTs. The interphase intranuclear MTs, which are not normally seen in wild-type cells, may arise from γ-TuC activity inside the nucleus, or possibly even from γ-TuC-independent nucleation activity, for instance from the kinetochores (Khodjakov et al., 2003). Spindle MTs exit the nucleus at the end of anaphase, as the nuclear envelope is broken. Intranuclear MTs can also exit the nucleus by directly puncturing the nuclear envelope during interphase and producing a break in the nuclear envelope that is then rapidly repaired. Once in the cytoplasm, these MTs grow into the cytoplasm and become free MTs that regenerate via repeated cycles of MT fragment breakage and growth or stabilization. The mto1Δtip1Δ double mutant gives strong support to the intranuclear origin of these MTs. In this double mutant, the spindle or intranuclear MTs also project out of the nucleus, but remain bundled with the nuclear MTs, creating an unusual situation where the MT bundle skewers the nuclear envelope. The difference between mto1Δ and mto1Δtip1Δ mutants suggests that continued growth of the projected MTs is required for generation of free cytoplasmic MTs. These behaviors document unusual and novel mechanisms of cytoplasmic MT formation and propagation.
Regulation of MT Plus-end Catastrophe
Our quantitative analysis of MT dynamics reveals effects of γ-TuCs at both MT plus and minus ends. We found strong defects in MT plus-end catastrophe: MT plus ends continued to grow even after hitting cell ends, leading to overly long, curled MTs. We also found that free MT minus ends exhibited shrinkage (in at least two classes of rates) and also pauses, but no growth, consistent with MTs in other cell types (Dammermann et al., 2003). The phases of pause and shrinkage may reflect inherent properties of the MT minus end, as such behaviors are observed in vitro (Desai and Mitchison, 1997). Although treadmilling MTs and hyperstable MTs have been noted previously in other cell types (Rodionov and Borisy, 1997; Margolis and Wilson, 1998; Rodionov et al., 1999; Dammermann et al., 2003; Shaw et al., 2003; Gundersen et al., 2004), this is the first report (to our knowledge) of such behaviors in yeast cells.
mto1Δ mitotic spindle MTs have normal nondynamic MT minus ends and abnormal MT plus ends that continue to grow at the end of anaphase. Although this may reflect a possible checkpoint defect at the end of mitosis (Prigozhina et al., 2004), we suggest that this spindle defect is caused by the same MT plus-end catastrophe defect seen in interphase. Importantly, this spindle phenotype suggests that the defects at MT plus ends are separable from nucleation defects at MT minus ends.
We can imagine several models to explain how γ-TuCs may influence MT plus ends: First, mto1Δ mutants generally have fewer MTs than wild-type cells and therefore probably have an increased concentration of tubulin dimers in the soluble pool. This difference in dimer concentration could explain the slight increase in MT plus-end polymerization rate and perhaps some of the variability in MT minus-end depolymerization rates. An increased dimer pool could also potentially inhibit MT catastrophe. However, we found that another mutant, rsp1Δ, also has reduced number of MT bundles, but still shows normal MT catastrophe (Zimmerman et al., 2004b; our unpublished data). Preliminary measurements of GFP-tubulin fluorescence levels suggest that at least some of these rsp1Δ cells have less total MT polymer than some mto1Δ cells. Therefore, an increase in soluble tubulin levels alone may not be sufficient to account for the MT catastrophe defect in γ-TuC mutant cells.
Second, γ-TuCs could affect the loading or unloading of MT regulatory factors. We provide evidence that γ-TuCs affect the CLIP170 homologue tip1p, as mto1Δ cells have increased amounts of tip1p at MT plus ends, which may contribute to MT stabilization. The MT catastrophe defect in mto1Δ cells is dependent on tip1p, as mto1Δ tip1Δ double mutants exhibited MT catastrophe. Thus, one function of γ-TuCs may be to remove tip1p from the MT plus end for MT catastrophe at the cell end. Alternatively, the increased amounts of tip1p at MT ends may reflect the prolonged growth phase of the MT plus end. Whether the effects of γ-TuCs on tip1p are direct or indirect remains to be tested. γ-TuC satellites localize to MT plus ends and thus could potentially regulate tip1p at MT plus ends in a direct manner. However, one hint that satellites may not be strictly necessary for regulating MT plus ends lies in the fact that rsp1 mutants, which have no or reduced satellites, exhibit normal MT plus-end dynamics (Zimmerman et al., 2004b). γ-TuCs could also affect the loading of MT regulatory factors from MTOCs. For instance, in budding yeast, properties at the SPB regulate the specific loading of factors onto astral MTs (Liakopoulos et al., 2003; Maekawa et al., 2003; Usui et al., 2003). However, we note that tip1p, as well other MT regulatory proteins tea1p and klp5p, are all loaded properly on mto1Δ MTs.
Third, γ-TuCs affect MT anchorage to the nucleus, which may be necessary to generate MT compression forces that can contribute to MT catastrophe. Elegant in vitro studies show that isolated MTs attached to a substrate will catastrophe after hitting a barrier because of MT compression forces (Janson et al., 2003). In mto1Δ mutants, MT minus ends are not anchored, and thus there may be less compressive force on plus ends when they contact the cell tip. However, we have observed that in alp4 mutant cells, occasional MTs that are still attached to the nucleus can also exhibit catastrophe defects (unpublished data). Further, the free MTs that are sometimes seen in normal cells, exhibit normal catastrophe patterns.
Fourth, γ-TuCs could affect the MT lattice structure. γ -TuCs may template the normal 13 protofilaments of MTs, and MTs formed in the absence of γ-TuCs generally have abnormal numbers of protofilaments (often 14; Chretien et al., 1992; Chretien and Fuller, 2000). Although the effects of abnormal protofilaments have not been characterized in vivo, these differences could possibly alter MT flexibility or interactions with MT associated proteins, such as the CLIP170 homologue, tip1p. However, this template model does not account for the catastrophe defects in mto1Δ spindle MTs, which are presumably still nucleated from γ-TuCs on the nuclear face of the SPB.
In summary, the reasons for MT catastrophe defects in mto1Δ are still not clear, and multiple mechanisms may be contributory. Further testing of these models will provide general insights into the molecular mechanism of MT catastrophe. These studies illustrate that there is still much to learn about the functions of γ-TuCs, and their continued study promises to illuminate molecular and functional aspects of regulation on both ends of the microtubule.
Acknowledgments
We are grateful for T. Toda for providing valuable reagents. We thank R. Vallee, A. Khodjokov, J. Glynn, and members of the Chang lab for valuable discussion and comments on the manuscript. We thank A. Khodjokov, S. La Terra, and Nikon for technical assistance and use of the imaging system for FRAP studies at the Marine Biological Laboratories, Woods Hole, MA. This work was supported by March of Dimes research grant and National Institutes of Health GM56836 to F.C.
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E04–08–0676) on March 16, 2005.
Abbreviations used: γ-TuCs, gamma tubulin complexes; γ-TuRCs, gamma ring complexes; SPB, spindle pole body; MT, microtubule; MTOC, microtubule organizing centers; iMTOC, interphase MTOC; eMTOC, equatorial MTOC.
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
References
- Bahler, J., Wu, J. Q., Longtine, M. S., Shah, N. G., McKenzie, A., 3rd, Steever, A. B., Wach, A., Philippsen, P., and Pringle, J. R. (1998). Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14, 943–951. [DOI] [PubMed] [Google Scholar]
- Behrens, R., and Nurse, P. (2002). Roles of fission yeast tea1p in the localization of polarity factors and in organizing the microtubular cytoskeleton. J. Cell Biol. 157, 783–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunner, D., and Nurse, P. (2000). CLIP170-like tip1p spatially organizes microtubular dynamics in fission yeast. Cell 102, 695–704. [DOI] [PubMed] [Google Scholar]
- Chen, C. R., Li, Y. C., Chen, J., Hou, M. C., Papadaki, P., and Chang, E. C. (1999). Moe1, a conserved protein in Schizosaccharomyces pombe, interacts with a Ras effector, Scd1, to affect proper spindle formation. Proc. Natl. Acad. Sci. USA 96, 517–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chretien, D., and Fuller, S. D. (2000). Microtubules switch occasionally into unfavorable configurations during elongation. J. Mol. Biol. 298, 663–676. [DOI] [PubMed] [Google Scholar]
- Chretien, D., Metoz, F., Verde, F., Karsenti, E., and Wade, R. H. (1992). Lattice defects in microtubules: protofilament numbers vary within individual microtubules. J. Cell Biol. 117, 1031–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dammermann, A., Desai, A., and Oegema, K. (2003). The minus end in sight. Curr. Biol. 13, R614–R624. [DOI] [PubMed] [Google Scholar]
- Desai, A., and Mitchison, T.J. (1997). Microtubule polymerization dynamics. Annu. Rev. Cell Dev. Biol. 13, 83–117. [DOI] [PubMed] [Google Scholar]
- Ding, D. Q., Chikashige, Y., Haraguchi, T., and Hiraoka, Y. (1998). Oscillatory nuclear movement in fission yeast meiotic prophase is driven by astral microtubules, as revealed by continuous observation of chromosomes and microtubules in living cells. J. Cell Sci. 111 (Pt 6), 701–712. [DOI] [PubMed] [Google Scholar]
- Ding, R., West, R. R., Morphew, D. M., Oakley, B. R., and McIntosh, J. R. (1997). The spindle pole body of Schizosaccharomyces pombe enters and leaves the nuclear envelope as the cell cycle proceeds. Mol. Biol. Cell 8, 1461–1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond, D. R., and Cross, R. A. (2000). Dynamics of interphase microtubules in Schizosaccharomyces pombe. Curr. Biol. 10, 766–775. [DOI] [PubMed] [Google Scholar]
- Feierbach, B., and Chang, F. (2001). Roles of the fission yeast formin for 3 p in cell polarity, actin cable formation and symmetric cell division. Curr. Biol. 11, 1656–1665. [DOI] [PubMed] [Google Scholar]
- Feierbach, B., Verde, F., and Chang, F. (2004). Regulation of a formin complex by the microtubule plus end protein tea1p. J. Cell Biol. 165, 697–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita, A., Vardy, L., Garcia, M. A., and Toda, T. (2002). A fourth component of the fission yeast gamma-tubulin complex, Alp16, is required for cytoplasmic microtubule integrity and becomes indispensable when gamma-tubulin function is compromised. Mol. Biol. Cell 13, 2360–2373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia, M. A., Koonrugsa, N., and Toda, T. (2002). Two kinesin-like Kin I family proteins in fission yeast regulate the establishment of metaphase and the onset of anaphase A. Curr. Biol. 12, 610–621. [DOI] [PubMed] [Google Scholar]
- Gundersen, G. G., Gomes, E. R., and Wen, Y. (2004). Cortical control of microtubule stability and polarization. Curr. Opin. Cell Biol. 16, 106–112. [DOI] [PubMed] [Google Scholar]
- Hagan, I. M. (1998). The fission yeast microtubule cytoskeleton. J. Cell Sci. 111(Pt 12), 1603–1612. [DOI] [PubMed] [Google Scholar]
- Hannak, E., Oegema, K., Kirkham, M., Gonczy, P., Habermann, B., and Hyman, A. A. (2002). The kinetically dominant assembly pathway for centrosomal asters in Caenorhabditis elegans is gamma-tubulin dependent. J. Cell Biol. 157, 591–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heitz, M. J., Petersen, J., Valovin, S., and Hagan, I. M. (2001). MTOC formation during mitotic exit in fission yeast. J. Cell Sci. 114, 4521–4532. [DOI] [PubMed] [Google Scholar]
- Hendrickson, T. W., Yao, J., Bhadury, S., Corbett, A. H., and Joshi, H. C. (2001). Conditional mutations in γ-tubulin reveal its involvement in chromosome segregation and cytokinesis. Mol. Biol. Cell 12, 2469–2481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horio, T., and Oakley, B. R. (2003). Expression of Arabidopsis gamma-tubulin in fission yeast reveals conserved and novel functions of gamma-tubulin. Plant Physiol. 133, 1926–1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horio, T., Uzawa, S., Jung, M. K., Oakley, B. R., Tanaka, K., and Yanagida, M. (1991). The fission yeast gamma-tubulin is essential for mitosis and is localized at microtubule organizing centers. J. Cell Sci. 99(Pt 4), 693–700. [DOI] [PubMed] [Google Scholar]
- Janson, M. E., de Dood, M. E., and Dogterom, M. (2003). Dynamic instability of microtubules is regulated by force. J. Cell Biol. 161, 1029–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Job, D., Valiron, O., and Oakley, B. (2003). Microtubule nucleation. Curr. Opin. Cell Biol. 15, 111–117. [DOI] [PubMed] [Google Scholar]
- Jung, M. K., Prigozhina, N., Oakley, C. E., Nogales, E., and Oakley, B. R. (2001). Alanine-scanning mutagenesis of Aspergillus gamma-tubulin yields diverse and novel phenotypes. Mol. Biol. Cell 12, 2119–2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodjakov, A., Copenagle, L., Gordon, M. B., Compton, D. A., and Kapoor, T. M. (2003). Minus-end capture of preformed kinetochore fibers contributes to spindle morphogenesis. J. Cell Biol. 160, 671–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodjakov, A., La Terra, S., and Chang, F. (2004). Laser microsurgery in fission yeast; role of the mitotic spindle midzone in anaphase B. Curr. Biol. 14, 1330–1340. [DOI] [PubMed] [Google Scholar]
- Liakopoulos, D., Kusch, J., Grava, S., Vogel, J., and Barral, Y. (2003). Asymmetric loading of Kar9 onto spindle poles and microtubules ensures proper spindle alignment. Cell 112, 561–574. [DOI] [PubMed] [Google Scholar]
- Maekawa, H., Usui, T., Knop, M., and Schiebel, E. (2003). Yeast Cdk1 translocates to the plus end of cytoplasmic microtubules to regulate bud cortex interactions. EMBO J. 22, 438–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallavarapu, A., Sawin, K., and Mitchison, T. (1999). A switch in microtubule dynamics at the onset of anaphase B in the mitotic spindle of Schizosaccharomyces pombe. Curr. Biol. 9, 1423–1426. [DOI] [PubMed] [Google Scholar]
- Margolis, R. L., and Wilson, L. (1998). Microtubule treadmilling: what goes around comes around. Bioessays 20, 830–836. [DOI] [PubMed] [Google Scholar]
- Marschall, L. G., Jeng, R. L., Mulholland, J., and Stearns, T. (1996). Analysis of Tub4p, a yeast gamma-tubulin-like protein: implications for microtubule-organizing center function. J. Cell Biol. 134, 443–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mata, J., and Nurse, P. (1997). tea1 and the microtubular cytoskeleton are important for generating global spatial order within the fission yeast cell. Cell 89, 939–949. [DOI] [PubMed] [Google Scholar]
- Moritz, M., and Agard, D. A. (2001). Gamma-tubulin complexes and microtubule nucleation. Curr. Opin. Struct. Biol. 11, 174–181. [DOI] [PubMed] [Google Scholar]
- Murphy, S. M., Preble, A. M., Patel, U. K., O'Connell, K. L., Dias, D. P., Moritz, M., Agard, D., Stults, J. T., and Stearns, T. (2001). GCP5 and GCP 6, two new members of the human gamma-tubulin complex. Mol. Biol. Cell 12, 3340–3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabeshima, K., Nakagawa, T., Straight, A. F., Murray, A., Chikashige, Y., Yamashita, Y. M., Hiraoka, Y., and Yanagida, M. (1998). Dynamics of centromeres during metaphase-anaphase transition in fission yeast: Dis1 is implicated in force balance in metaphase bipolar spindle. Mol. Biol. Cell 9, 3211–3225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oegema, K., Wiese, C., Martin, O. C., Milligan, R. A., Iwamatsu, A., Mitchison, T. J., and Zheng, Y. (1999). Characterization of two related Drosophila gamma-tubulin complexes that differ in their ability to nucleate microtubules. J. Cell Biol. 144, 721–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paluh, J. L., Nogales, E., Oakley, B. R., McDonald, K., Pidoux, A. L., and Cande, W. Z. (2000). A mutation in gamma-tubulin alters microtubule dynamics and organization and is synthetically lethal with the kinesin-like protein pkl1p. Mol. Biol. Cell 11, 1225–1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardo, M., and Nurse, P. (2003). Equatorial retention of the contractile actin ring by microtubules during cytokinesis. Science 300, 1569–1574. [DOI] [PubMed] [Google Scholar]
- Pelham, R. J., Jr., and Chang, F. (2001). Role of actin polymerization and actin cables in actin-patch movement in Schizosaccharomyces pombe. Nat. Cell Biol. 3, 235–244. [DOI] [PubMed] [Google Scholar]
- Prigozhina, N. L., Oakley, C. E., Lewis, A. M., Nayak, T., Osmani, S. A., and Oakley, B. R. (2004). gamma-tubulin plays an essential role in the coordination of mitotic events. Mol. Biol. Cell 15, 1374–1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prigozhina, N. L., Walker, R. A., Oakley, C. E., and Oakley, B. R. (2001). Gamma-tubulin and the C-terminal motor domain kinesin-like protein, KLPA, function in the establishment of spindle bipolarity in Aspergillus nidulans. Mol. Biol. Cell 12, 3161–3174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodionov, V., Nadezhdina, E., and Borisy, G. (1999). Centrosomal control of microtubule dynamics. Proc. Natl. Acad. Sci. USA 96, 115–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodionov, V. I., and Borisy, G. G. (1997). Microtubule treadmilling in vivo. Science 275, 215–218. [DOI] [PubMed] [Google Scholar]
- Sagolla, M. J., Uzawa, S., and Cande, W. Z. (2003). Individual microtubule dynamics contribute to the function of mitotic and cytoplasmic arrays in fission yeast. J. Cell Sci. 116, 4891–4903. [DOI] [PubMed] [Google Scholar]
- Sawin, K. E., Lourenco, P. C., and Snaith, H. A. (2004). Microtubule nucleation at non-spindle pole body microtubule-organizing centers requires fission yeast centrosomin-related protein mod20p. Curr. Biol. 14, 763–775. [DOI] [PubMed] [Google Scholar]
- Shaw, S. L., Kamyar, R., and Ehrhardt, D. W. (2003). Sustained microtubule treadmilling in Arabidopsis cortical arrays. Science 300, 1715–1718. [DOI] [PubMed] [Google Scholar]
- Sobel, S. G., and Snyder, M. (1995). A highly divergent gamma-tubulin gene is essential for cell growth and proper microtubule organization in Saccharomyces cerevisiae. J. Cell Biol. 131, 1775–1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spang, A., Geissler, S., Grein, K., and Schiebel, E. (1996). gamma-Tubulin-like Tub4p of Saccharomyces cerevisiae is associated with the spindle pole body substructures that organize microtubules and is required for mitotic spindle formation. J. Cell Biol. 134, 429–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strome, S., Powers, J., Dunn, M., Reese, K., Malone, C. J., White, J., Seydoux, G., and Saxton, W. (2001). Spindle dynamics and the role of gamma-tubulin in early Caenorhabditis elegans embryos. Mol. Biol. Cell 12, 1751–1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suelmann, R., Sievers, N., Galetzka, D., Robertson, L., Timberlake, W. E., and Fischer, R. (1998). Increased nuclear traffic chaos in hyphae of Aspergillus nidulans: molecular characterization of apsB and in vivo observation of nuclear behaviour. Mol. Microbiol. 30, 831–842. [DOI] [PubMed] [Google Scholar]
- Tange, Y., Fujita, A., Toda, T., and Niwa, O. (2004). Functional dissection of the gamma-tubulin complex by suppressor analysis of gtb1 and alp4 mutations in Schizosaccharomyces pombe. Genetics 167, 1095–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toda, T., Umesono, K., Hirata, A., and Yanagida, M. (1983). Cold-sensitive nuclear division arrest mutants of the fission yeast Schizosaccharomyces pombe. J. Mol. Biol. 168, 251–270. [DOI] [PubMed] [Google Scholar]
- Tolic-Norrelykke, I. M., Sacconi, L., Thon, G., and Pavone, F. S. (2004). Positioning and elongation of the fission yeast spindle by microtubule-based pushing. Curr. Biol. 14, 1181–1186. [DOI] [PubMed] [Google Scholar]
- Tran, P. T., Marsh, L., Doye, V., Inoue, S., and Chang, F. (2001). A mechanism for nuclear positioning in fission yeast based on microtubule pushing. J. Cell Biol. 153, 397–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran, P. T., Paoletti, A., and Chang, F. (2004). Imaging green fluorescent protein fusions in living fission yeast cells. Methods 33, 220–225. [DOI] [PubMed] [Google Scholar]
- Usui, T., Maekawa, H., Pereira, G., and Schiebel, E. (2003). The XMAP215 homologue Stu2 at yeast spindle pole bodies regulates microtubule dynamics and anchorage. EMBO J. 22, 4779–4793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vardy, L., Fujita, A., and Toda, T. (2002). The gamma-tubulin complex protein Alp4 provides a link between the metaphase checkpoint and cytokinesis in fission yeast. Genes Cells 7, 365–373. [DOI] [PubMed] [Google Scholar]
- Vardy, L., and Toda, T. (2000). The fission yeast gamma-tubulin complex is required in G(1) phase and is a component of the spindle assembly checkpoint. EMBO J. 19, 6098–6111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatram, S., Tasto, J. J., Feoktistova, A., Jennings, J. L., Link, A. J., and Gould, K. L. (2004). Identification and characterization of two novel proteins affecting fission yeast γ-tubulin complex function. Mol. Biol. Cell 15, 2287–2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel, J., Drapkin, B., Oomen, J., Beach, D., Bloom, K., and Snyder, M. (2001). Phosphorylation of gamma-tubulin regulates microtubule organization in budding yeast. Dev. Cell 1, 621–631. [DOI] [PubMed] [Google Scholar]
- Waterman-Storer, C. M., and Danuser, G. (2002). New directions for fluorescent speckle microscopy. Curr. Biol. 12, R633–640. [DOI] [PubMed] [Google Scholar]
- West, R. R., Malmstrom, T., Troxell, C. L., and McIntosh, J. R. (2001). Two related kinesins, klp5+ and klp6+, foster microtubule disassembly and are required for meiosis in fission yeast. Mol. Biol. Cell 12, 3919–3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida, M., and Sazer, S. (2004). Nucleocytoplasmic transport and nuclear envelope integrity in the fission yeast Schizosaccharomyces pombe. Methods 33, 226–238. [DOI] [PubMed] [Google Scholar]
- Zheng, Y., Wong, M. L., Alberts, B., and Mitchison, T. (1995). Nucleation of microtubule assembly by a gamma-tubulin-containing ring complex. Nature 378, 578–583. [DOI] [PubMed] [Google Scholar]
- Zimmerman, S., Daga, R. R., and Chang, F. (2004a). Intra-nuclear microtubules and a mitotic spindle orientation checkpoint. Nat. Cell Biol. 6, 1245–1246. [DOI] [PubMed] [Google Scholar]
- Zimmerman, S., Tran, P. T., Daga, R. R., Niwa, O., and Chang, F. (2004b). Rsp1p, a J domain protein required for disassembly and assembly of microtubule organizing centers during the fission yeast cell cycle. Dev. Cell 6, 497–509. [DOI] [PubMed] [Google Scholar]