Abstract
Coronin 1 is a member of the coronin protein family specifically expressed in leukocytes and accumulates at sites of rearrangements of the F-actin cytoskeleton. Here, we describe that coronin 1 molecules are coiled coil-mediated homotrimeric complexes, which associate with the plasma membrane and with the cytoskeleton via two distinct domains. Association with the cytoskeleton was mediated by trimerization of a stretch of positively charged residues within a linker region between the N-terminal, WD repeat-containing domain and the C-terminal coiled coil. In contrast, neither the coiled coil nor the positively charged residues within the linker domain were required for plasma membrane binding, suggesting that the N-terminal, WD repeat-containing domain mediates membrane interaction. The capacity of coronin 1 to link the leukocyte cytoskeleton to the plasma membrane may serve to integrate outside-inside signaling with modulation of the cytoskeleton.
INTRODUCTION
Coronin 1 is expressed exclusively by leukocytes (Suzuki et al., 1995; Ferrari et al., 1999) and is a member of the WD repeat protein family termed coronins, which are collectively defined as F-actin–associated proteins widely expressed in the eukaryotic kingdom (de Hostos, 1999). In Dictyostelium discoideum, coronin colocalizes with F-actin filaments at crown-shaped phagocytic cups and macropinosomes (de Hostos et al., 1991, 1993; Maniak et al., 1995; Fukui et al., 1999). Dictyostelium deleted for coronin displays strong reduction in cell locomotion, phagocytosis, macropinocytosis, and cytokinesis, indicating that in this slime mold coronin is functionally involved in F-actin–based motility-related processes (de Hostos et al., 1993). In Saccharomyces cerevisiae, the single coronin isoform Crn1p was found to localize to cortical F-actin patches in an actin-dependent manner (Heil-Chapdelaine et al., 1998). In vitro, Crn1p can nucleate and cross-link F-actin filaments and bind to microtubules (Goode et al., 1999). Recently, yeast Crn1 was proposed to promote the formation of actin filament networks based on its interaction with the Arp2/3 complex (Humphries et al., 2002). Interestingly, unlike the Dictyostelium coronin-null mutant, an S. cerevisiae Crn1p-null-mutant does not show any phenotype in actin-dependent processes (Heil-Chapdelaine et al., 1998), suggesting that in this organism coronin does not perform an essential function in regulating the actin cytoskeleton. Although lower eukaryotes have one coronin gene, database searches have revealed the existence of several coronins in humans and mice (denoted coronins 1–7) (Okumura et al., 1998; de Hostos, 1999; Rybakin et al., 2004).
In macrophages and lymphocytes, coronin 1 concentrates at sites of rearrangement of the cytoskeleton. In lymphocytes, coronin 1 assembles at the immunological synapse formed during activation of T-cells (Nal et al., 2004). In mouse macrophages, coronin 1 accumulates during phagocytosis at the cytosolic face of phagosomes and is actively retained by pathogenic mycobacteria, preventing lysosomal delivery and allowing these bacteria to survive intracellularly (Ferrari et al., 1999; Gatfield and Pieters, 2000). Similarly, in human dendritic cells coronin 1 accumulates at the mycobacterial phagosome, whereas in human macrophages coronin 1 retention seems to be dependent upon the activation state of the macrophages (Schuller et al., 2001). Together, these studies suggest that coronin 1 may have a function in the modulation of cytoskeletal rearrangements during leukocyte-specific processes.
Based on sequence comparison among different species of the coronin family members, three conserved domain structures were identified (de Hostos, 1999). The ∼400-residue long N-terminal domain contains five highly conserved WD (tryptophan-aspartate) repeats reminiscent of the ones found in the β-subunits of G proteins (Neer et al., 1994; Lambright et al., 1996; Sondek et al., 1996). Based on this finding, it has been hypothesized that the N-terminal domain of the coronins folds into a five-bladed β-propeller structure (de Hostos, 1999). The most C-terminally located 30–40 residues are strongly predicted to fold into a α-helical coiled coil structure. For the Xenopus coronin homologues (Xcoronins), as well as coronin 3, the coiled coil has been shown to mediate the formation of higher molecular weight complexes (de Hostos, 1999; Asano et al., 2001; Spoerl et al., 2002) whose exact oligomerization states are not fully clear. Finally, a unique region located between the propeller and coiled coil domains was identified that varies greatly in length (50–200 amino acids) and sequence among coronin homologues (de Hostos, 1999). Besides this sequence information and their classification as actin-associated proteins, little is known about the structure–function relationship of the different coronins in mammals (Suzuki et al., 1995; Spoerl et al., 2002; Oku et al., 2003).
In this article, we describe that coronin 1 is organized into three domains: 1) an N-terminal, WD repeat-containing β-propeller connected by 2) a linker region to 3) a coiled coil. Whereas the N-terminal, WD repeat-containing β-propeller domain was found to mediate plasma membrane binding, a stretch of positively charged residues within the linker region was responsible for interaction with the F-actin cytoskeleton. Cytoskeletal association occurred exclusively upon trimerization that was mediated via the coiled coil domain. As such, coronin 1 may function as a bridging protein between plasma membrane domains and the dynamic actin cytoskeleton of leukocytes and may allow for the highly efficient remodeling of the cytoskeleton in response to the versatile outside signals transmitted into leukocytes.
MATERIALS AND METHODS
DNA Constructs
The C-terminally hemagglutinin (HA)-tagged version of coronin 1 (Cor1-HA) was generated by PCR, by using as template the coronin 1 cDNA cloned into pBluescript via the EcoRI site (Veithen et al., 1996; Ferrari et al., 1999). The forward primer was 5′-ACAAGCAGCGGAGTCCTGCTACC-3′ (coronin 1 nucleotides 796–818) and the EcoRI site-containing reverse primer was 5′-CGCGAATTCCTATGCGTAGTCTGGTACGTCGTATGGGTACTTGGCCTGAACAGTCTC3-′ encoding the HA tag (underlined) and the EcoRI site (in italics). The product was digested with HindIII and EcoRI, and the HindIII-EcoRI fragment was used for exchanging the corresponding wild-type fragment in the pBluescript construct.
The coiled coil deletion mutant Cor1-ΔCC-HA encoding amino acids 1–432 fused to the HA-tag was generated from the pBluescript-Cor1-HA template by site-directed mutagenesis using the QuikChange kit (Stratagene) and the primers 5′-GATACCGTGTCAAGG//TACCCATACGACGTACCAGAC-3′ (forward) and 5′-TACGTCGTATGGGTA//CCTTGACACGGTATCCGAGCT-3′ (reverse) where //indicates the deletion position. For expression in mammalian cells, the constructs were cloned via the EcoRI site into the cytomegalovirus promoter-driven pCB6 expression vector.
The deletion mutant Cor1-Δ400-416-HA was generated as follows. In a first PCR, the coding sequence for the N-terminal region, including the deletion (residue 400–416) was generated using the primer pair 5′-GTGAATTCCATGAGCCGGCAGGTGGTTCG-3′ (EcoR1 site in italics) and 5′-GGCTACGTGCCCCCA//GCTACACCAGAGCCC AGCGGCA-3′ (//indicates the deletion position), resulting in the EcoR1-Cor1nt1–1197//1249-1270 fragment. The coding sequence for the C-terminal region, including the deletion (residue 400–416), was generated using the primer pair 5′-GGGCTCTGGTGTAGC//TGGGGGCACGTAGCCATCCTTG-3′ (//indicates the deletion position) and 5′-GAGGATCCCTAAGCGTAATCTGGAACATCGTATGGGTACTTGGCCTGAACAGTCTCC3′ (BamH1 site in italics, HA-tag underlined), resulting in the Cor1nt1176-1197/1249-1383-HA-BamH1 DNA fragment. In a subsequent PCR, EcoR1-Cor1nt1–1197//1249–1270 together with Cor1nt1176–1197/1249–1383-HA-BamH1 were used as templates and with primers 5′GTGAATTCCATGAGCCGGCAGGTGGTTCG-3′ and 5′-GAGGATCCCTAAGCGTAATCTGGAACATCGTATGGGTACTTGGCCTGAACAGTCTCC-3′ the EcoR1-Cor1nt1–1197//1249-1383-HA-BamH1fragment was amplified and subcloned into the pGEM-T-Vector (Promega, Madison, WI). For expression in mammalian cells, the pGEM-Cor1-Δ400-416-HA construct was digested with HindIII and BamH1, and the HindIII-BamHI fragment containing the deletion was used for exchanging the corresponding wild-type fragment in the pCB6-Cor1-HA construct.
The pEGFP-Cor1429-461 (pEGFP-CC) fusion protein expression construct was generated by PCR-based amplification of the sequences Cor1nt1285-1386 by using the primer pair 5′-CGCCTCGAGTGTCAAGGCTGGAGGAGGACG-3′ (XhoI site in italics) and 5′GCGGGATCCTACTTGGCCTGAACAGTCTCC-3′ (BamHI site in italics). The amplified DNA was digested with XhoI and BamHI and ligated via these sites into the pEGFP-C1 vector (Stratagene, La Jolla, CA). All sequences were verified by DNA sequencing by using the BigDye reagent (PerkinElmer Life and Analytical Sciences, Boston, MA) on an ABI 310 Sequencer (Applied Biosystems, Foster City, CA).
Cell Lines, Antibodies, and Reagents
J774A.1 (ATCC TIB-67) mouse macrophage cells, RAW 264.7 (ATCC TIB-71) mouse macrophage cells, human embryonic kidney (HEK)293 (ATCC CRL-1573) cells were grown in DMEM (Invitrogen, Carlsbad, CA) supplemented with 10% fetal calf serum (FCS) (Invitrogen). Jurkat cells were grown in RPMI 1640 medium supplemented with 6% FCS. For the generation of anti-coronin 1 antiserum, the coding sequence of coronin 1 was fused to the glutathione S-transferase (GST) sequence in the pGEX-4T-1 expression vector, the fusion protein was expressed in Escherichia coli, purified, and used for immunization of rabbits to obtain an anti-coronin 1 antiserum that was used for immunofluorescence staining and immunoaffinity purification of coronin 1. For immunoblotting, anti-peptide antibodies were generated in rabbits against coronin 1 peptide sequence Cor15–20 (anti-N-terminal coronin 1 antiserum) (Ferrari et al., 1999). A rabbit anti-vimentin antiserum was raised against the mouse vimentin peptide sequence vimentin1-20. The monoclonal anti-tubulin antibody (clone E7) was obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the National Institute of Child Health and Human Development and maintained by Department of Biological Sciences (University of Iowa, Iowa City, IA). For immunodetection of actin, HA-tag, CD14, and enhanced green fluorescent protein (EGFP) following Western blotting, anti-actin ascites (mouse monoclonal antibody [mAb] clone C4; Chemicon International, Temecula, CA), anti-HA-tag (mouse mAb clone 16B12; Covance Research Products, Princeton, NJ), anti-CD14 rat monoclonal antibodies (rmC5-3; BD Biosciences PharMingen, San Diego, CA) and anti-green fluorescent protein (GFP) mouse monoclonal antibodies (clone 7.1 and 13.1; Roche Diagnostics, Mannheim, Germany) were used. For depolymerization of F-actin, latrunculin B (Sigma-Aldrich, St. Louis, MO) was used at the concentrations indicated.
Subcellular Fractionation and Size Exclusion Chromatography
Subcellular fractionation was performed according to standard protocols (Tulp et al., 1994; Ferrari et al., 1997). Briefly, confluent monolayers were washed three times with cold homogenization buffer (10 mM triethanolamine, 10 mM acetic acid, 1 mM EDTA, 250 mM sucrose, pH 7.4), and cells were harvested by scraping and homogenized with a syringe and a 22-gauge 1/4 needle. The postnuclear supernatant was obtained by centrifugation at 240 × g (15 min). Membranes were separated from cytosol by ultracentrifugation of the postnuclear supernatant at 100,000 × g (30 min).
Isolation of the cytoskeleton-containing detergent-insoluble fraction was performed by carefully resuspending the harvested cell pellets in 10 volumes of ice-cold cytoskeletal isolation buffer (1% Triton X-100 in 80 mM PIPES, pH 6.8, 5 mM EGTA, 1 mM MgCl2) and immediate centrifugation at 3000 × g (2 min) resulting in a Triton X-100–insoluble pellet. For analysis, cell equivalents rather than protein equivalents were loaded on SDS-PAGE.
For size exclusion chromatography, cells were homogenized in homogenization buffer, and the cytosol and membrane fractions were obtained as described above. Membranes and cytosol were supplemented with 2% (wt/vol final concentration) octylglucopyranoside (Sigma-Aldrich) followed by an incubation on ice for 30 min. Subsequently the samples were filtered (0.2 μm), the protein concentrations were adjusted to 1 mg/ml, and 50 μl were separated on a Superdex 200 gel filtration column using the SMART system (Amersham Biosciences, Piscataway, NJ), and 75-μl fractions were collected. For calibration, 180 μg of gel filtration standard (151-1901; Bio-Rad, Hercules, CA) were used.
Protein Analysis
After standard SDS-PAGE separation of proteins (10 or 15% acrylamide), proteins were blotted onto Hybond-C Super nitrocellulose membrane (Amersham Biosciences) by using the semidry transfer cell (Bio-Rad). Detection of coronin 1, actin, hemagglutinin-tag, vimentin, and tubulin was performed using anti-N-terminal coronin 1 peptide antiserum (1:1000), anti-actin mAb (1:2000), anti-HA mAb (1:1000; Covance Research Products), anti-vimentin antiserum (1:500), and anti-tubulin ascites (1:5000), respectively, followed by goat anti-rabbit or goat anti-mouse antisera coupled to horseradish peroxidase (Southern Biotechnology Associates (Birmingham, AL). Blots were exposed to x-ray films (Hyperfilm ECL; Amersham Biosciences) after enhanced chemiluminescence reaction (ECL; Amersham Bioscience). Biochemical experiments were performed at least twice, and representative results are shown. Signal intensities were quantified by densitometric analysis.
Transfection of Mammalian Cell Lines
Transient transfection of HEK293 cells was performed using Lipofectin (Invitrogen) or Polyfectin (QIAGEN, Valencia, CA) following the manufacturer's protocol. RAW 264.7 macrophages were transfected by electroporation with 40 μg of circular DNA in a volume of 800 μl in 4-mm cuvettes (975 μF, 300 V; Bio-Rad gene pulser II).
Immunostaining and Imaging
For staining of J774, RAW 264.7, and HEK293 cells, cells were grown on 10-well Teflon-coated glass slides (Polysciences, Warrington, PA). After fixation (10 min, 3% formaldehyde in phosphate-buffered saline [PBS], 37°C), cells were permeabilized in 0.1% saponin/2% bovine serum albumin in PBS and incubated for 30 min with primary antibodies (anti-coronin 1 antiserum, 1:4000; anti-HA, clone 12CA5, mouse IgG2b, 2 μg/ml; Roche Diagnostics) (Cella et al., 1997; Ferrari et al., 1999). After washing (3× saponin/bovine serum albumin), phalloidin-Texas Red or phalloidin-488 (Molecular Probes, Eugene, OR) and the secondary antibodies (goat anti-mouse Alexa Fluor 633, goat anti-rabbit Alexa Fluor 488 or 568; Molecular Probes) were applied for 30 min at 1:200 dilutions, the slides were washed three times with saponin/bovine serum albumin and three times with PBS, and mounted using Fluoroguard antifade mounting medium (Bio-Rad). Specimen observation and image acquisition was performed using the confocal laser scanning module LSM510 Meta (Carl Zeiss, Jena, Germany) connected to an Axiovert 200M (Carl Zeiss) by using the software supplied.
Purification of Coronin 1 Complexes
Coronin 1 complexes were purified from cytosol of J774 cells harvested from 15 confluent 15 φ cm tissue culture plates. Using the Amersham Bioscience fast-performance liquid chromatography system, the filtered (0.2 μm) cytosol was passed over 1 ml of HiTrap NHS-Sepharose column (Amersham Bioscience) coupled with protein A-Sepharose-purified rabbit anti-coronin 1 antiserum. After extensive washing with 20 mM Tris, pH 8.0, 150 mM NaCl, the bound protein was eluted with 100 mM glycine-HCl, pH 2.5, and the resulting fractions were immediately neutralized using 1 M Tris, pH 9.3. The purity of the eluted material was analyzed by SDS-PAGE and silver staining.
Dephosphorylation of Coronin 1 Complexes
Dephosphorylation was carried out with coronin 1 complexes purified from metabolically labeled J774 cells by immunoprecipitation. Briefly, J774 cells were incubated with 0.2 mCi/ml [35S]methionine for 16 h, washed, and lysed in 0.5% SDS in 50 mM HEPES, pH 7.5, supplemented with protease inhibitors (1 mM phenylmethylsulfonyl fluoride, 100 μg/ml chymostatin/leupeptin/aprotinin/antipain/pepstatin) and phosphatase inhibitors (2 mM orthovanadate, 10 mM NaF, 0.5 mM EDTA). After heating for 5 min at 95°C, the samples were diluted 10-fold in Triton X100 lysis buffer (Pieters et al., 1991). Immunoprecipitation of coronin 1 was performed as described previously (Tulp et al., 1994), and immune complexes were incubated for 2 h at 37°C in the absence or presence of 10 U of alkaline phosphatase (ALP) from calf intestine (Roche Diagnostics). Analysis of dephosphorylation was performed subsequently by two-dimensional gel electrophoresis (Engering et al., 2003) and autoradiography.
Silver Staining of SDS-PAGE Gels
Gels were fixed for 30 min in 50% ethanol, 10% acetic acid. Then, they were incubated in 0.5 M sodium acetate, 0.2% sodium thiosulfate, 0.5% glutaraldehyde in 30% ethanol for 60 min, followed by four 5-min washes in distilled water. Gels were then incubated in 0.1% silver nitrate, 0.01% formaldehyde for 30 min, washed briefly in distilled water, and developed in 2.5% sodium carbonate, 0.01% formaldehyde with agitation. The reaction was stopped in 50 mM EDTA.
Two-dimensional Gel Electrophoresis
Proteins were analyzed by two-dimensional isoelectric focusing (IEF)/SDS-PAGE as described previously (Lefkovits et al., 1985; Engering et al., 1997) (first dimension long Iso Resolyte, pH 4–8 (Merck-BDH, Poole, United Kingdom); second-dimension acrylamide gradient 11–18%). For mass spectrometry, two-dimensional gels were fixed and stained using colloidal Coomassie (Novex, San Diego, CA). For autoradiography, gels were fixed in 20% ethanol/10% acetic acid and exposed to Hyperfilm MP (Amersham Biosciences).
Sequence Analysis
The Jpred consensus method for protein secondary structure prediction (Cuff et al., 1998) was run on the server available at http://www.compbio.dundee.ac.uk/~www-jpred. The 3D-pssm method for protein fold recognition by using one- and 3D sequence profiles coupled with secondary structure and solvation potential information (Kelley et al., 2000) was run on the server available at http://www.sbg.bio.ic.ac.uk/~3dpssm. The Coils method (Lupas, 1996) is available at http://www.ch.embnet.org/software/COILS_form.html. The MTIDK matrix, a window width of 21, and no weights were used. The exon-intron organization of the mouse Cor1 gene was taken from the GeneLynx mouse genome portal at http://www.genelynx.org/MOUSE/.
Circular Dichroism (CD) Spectroscopy
Far-UV CD spectra and thermal unfolding profiles were recorded on a Jasco J 720 spectropolarimeter equipped with a temperature-controlled quartz cell of 0.1-cm path length. The spectrum shown is the average of five accumulations. A ramping rate of 50°C h–1 was used to record the thermal unfolding profile. The midpoint of the transition, Tm, was taken as the maximum of the derivative d[Θ]222/dT.
Analytical Ultracentrifugation
Analytical ultracentrifugation was performed on an Optima XL-A analytical ultracentrifuge (Beckman Coulter, Fullerton, CA) equipped with an An-60ti rotor. The ccCor1 peptide was analyzed in 5 mM sodium phosphate buffer, pH 7.4, containing 150 mM NaCl, and peptide concentrations were adjusted to 0.1–0.5 mg/ml. Sedimentation velocity experiments were performed at 56,000 rpm in a 12-mm Epon double-sector cell. Sedimentation coefficients were corrected to water by the standard procedure (Eason, 1986). Sedimentation equilibrium runs were performed at 29.000, 38.000, and 48.000 rpm. The partial specific volume of the synthetic peptide was calculated from its amino acid sequence
Specimen Preparation and Transmission Electron Microscopy (TEM)
A 5-μl aliquot of affinity-purified coronin 1 complexes (70 μg/ml) was applied to a weakly glow-discharged carbon coated 400-mesh/inch copper grid. The sample was allowed to adsorb for 30 s, washed twice with distilled water, and negatively stained with 0.75% (wt/vol) uranyl formiate, pH 4.25, as described previously (Steinmetz et al., 1997). Specimens were examined in a Phillips EM400 TEM operated at an accelerating voltage of 80 kV and at a nominal magnification of 55,000×. Micrographs were recorded on Kodak SO-163 electron image film that was developed for 6 min at room temperature in KODAK D-19 developer diluted threefold with water. Magnification was calibrated according to Wrigley (1968).
Tryptic Digest
The postnuclear supernatant from a J774 macrophage homogenate was digested by the addition of 25 μg of trypsin (Roche Diagnostics, Basel, Switzerland) per mg of protein and incubation for 5 min at 37°C. The digestion was quenched by addition of 25 μg of soybean trypsin inhibitor (Calbiochem) per μg of trypsin and the cytosolic fraction was obtained by ultracentrifugation at 100.000 × g (30 min).
RESULTS
Subcellular Localization of Coronin 1
Coronin 1, also known as P57 or TACO, is exclusively expressed in leukocytes, suggesting a cell type-specific function. To determine the precise subcellular localization of coronin 1 in leukocytes, macrophages (J774), or lymphocytes (Jurkat human T-cell lymphoma) were fixed, permeabilized, and stained with anti-coronin 1 antiserum as well as Texas Red-conjugated phalloidin to visualize the F-actin cytoskeleton. Immunofluorescence microscopy revealed colocalization of coronin 1 with the cortical F-actin cytoskeleton at the plasma membrane of both cell types (Figure 1A, left and middle) and no coronin 1 staining of other subcellular organelles. Also, the nonleukocyte human embryo kidney cell line HEK293 transfected with the HA-tagged coronin 1 molecule displayed a similar distribution of coronin 1 at the plasma membrane and colocalization with F-actin (Figure 1A, right).
Figure 1.
Subcellular localization of coronin 1. (A) Macrophages (J774), lymphocytes (Jurkat T-cell lymphoma), or coronin 1-transfected HEK293 cells were grown on microscopy slides, fixed, and permeabilized followed by immunolocalization of coronin 1 and F-actin localization by using anti-coronin 1 antiserum (middle) and Texas Red-labeled phalloidin (bottom). Top, corresponding Nomarski images. Bar, 10 μm. (B) Biochemical analysis of coronin 1 interaction with macrophage membranes. The postnuclear supernatant of a J774 cell homogenate was separated into membrane fraction and cytosol (100,000 × g, 30 min) and analyzed for the presence of coronin 1 by SDS-PAGE and immunoblotting. (C) Biochemical analysis of coronin 1 localization in the detergent-insoluble fraction of macrophages. J774 macrophages were lysed in cytoskeleton isolation buffer containing 1% Triton X-100 and directly subjected to low-speed centrifugation (3000 × g, 2 min; see Materials and Methods). Subsequently, the detergent-insoluble pellets and the supernatants were subjected to SDS-PAGE and immunoblotting for detection of coronin 1, actin, tubulin, vimentin, and the raft marker protein CD14. (D) J774 macrophages were treated for 30 min with 5 μM LatB, processed as in C, and analyzed for the presence of coronin 1, actin, vimentin, and CD14 in pellets and supernatants.
To further analyze the subcellular distribution of coronin 1 between plasma membrane and the cytoskeleton of J774 macrophages, cell fractionation was used. Macrophages were homogenized in sucrose-containing buffer, and membrane and cytosolic fractions were prepared by ultracentrifugation, as described in Materials and Methods. SDS-PAGE and immunoblotting for coronin 1 with an N-terminal–specific antiserum revealed that ∼20% of the molecules were associated with the membrane fraction, whereas ∼80% were soluble (Figure 1B). To analyze the cytoskeletal association of coronin 1, the cytoskeleton from J774 cells was isolated by cell lysis in 1% Triton X-100/80 mM PIPES and low-speed centrifugation (3000 × g, 2 min). Immunoblotting of pellet and supernatant revealed that virtually all coronin 1 molecules were found in the sedimented detergent-insoluble fraction (Figure 1C, top), suggesting that in resting, non-phagocytosing macrophages coronin 1 is associated with the cytoskeleton. To characterize further the detergent-insoluble fraction, the distribution of the three major macrophage cytoskeletal filament systems was determined by immunoblotting by using anti-actin, anti-tubulin, and anti-vimentin antibodies. Whereas actin was equally distributed between the insoluble and the soluble fraction, tubulin was exclusively detected in the soluble fraction, whereas vimentin was recovered in the insoluble fraction (Figure 1C). To analyze the distribution of cholesterol-rich microdomains, which are known to be Triton X-100 insoluble, the presence of the glycosylphosphatidyl inositol-linked raft constituent CD14 in pellet and supernatant was determined. Immunoblotting using anti-CD14 antibodies revealed that rafts were coisolated with the cytoskeletal fraction (Figure 1C).
Because coronins are described as F-actin–associated proteins, we tested whether the presence of coronin 1 in the Triton X-100–insoluble pellet was due to F-actin association. Thus far, coronin 1 binding to F-actin filaments has been analyzed in vitro by a two-component cosedimentation by using recombinant coronin 1 and F-actin and putative direct interaction sites have been mapped to the N-terminal globular domain of coronin 1 (Suzuki et al., 1995; Oku et al., 2003). To analyze whether the observed Triton X-100 insolubility of coronin 1 was due to an in vivo association of coronin 1 with the actin cytoskeleton, the actin cytoskeleton was disrupted by treating J774 cells for 30 min with the F-actin–depolymerizing drug latrunculin B (LatB; Figure 1D). Isolation of the detergent-insoluble fraction followed by separation of proteins by SDS-PAGE and immunoblotting with anti-actin antibodies revealed fully detergent-soluble actin in the latrunculin B-treated macrophages, indicating efficient depolymerization of the actin cytoskeleton (Figure 1D). Blotting with anti-coronin 1 antibodies demonstrated a simultaneous release of coronin 1 into the supernatant (Figure 1D). In contrast, vimentin distribution as well as the CD14/raft distribution remained essentially unaffected by the latrunculin B treatment. Thus, detergent-insolubility of coronin 1 occurs as the consequence of association with the actin cytoskeleton.
Together, in a J774 macrophage homogenate ∼20% of coronin 1 molecules were present in the membrane fraction, whereas all molecules cofractionated with the detergent-insoluble cytoskeleton. This suggests that coronin 1 associates with the cytoskeleton in an F-actin–dependent manner as well as with the plasma membrane, possibly via two distinct binding sites.
Sequence Analysis of Coronin 1
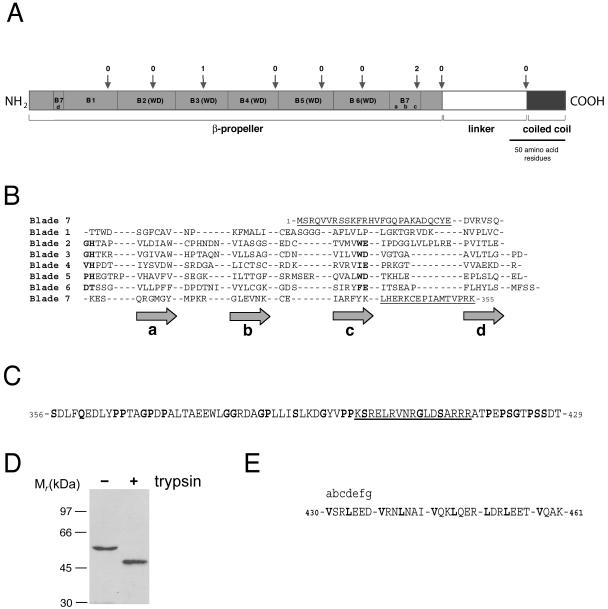
To map both plasma membrane association and actin cytoskeleton association to individual domains of the coronin 1 molecule, a detailed sequence analysis was performed. Secondary structure prediction by using Jpred (Cuff et al., 1998) confirmed the presence of three structural domains (Spoerl et al., 2002), however, with significantly different domain boundaries (Figure 2A): An N-terminal domain rich in β-sheet (residues 1–355; referred to as β-propeller), a region with little regular secondary structure (residues 356–429; referred to as linker domain), and a C-terminal domain rich in α-helix (residues 430–461; referred to as coiled coil).
Figure 2.
Sequence analysis of coronin 1. (A) Schematic representation of the proposed domain organization. The N-terminal domain containing the seven predicted propeller blades is shown in light gray. The linker region is shown in white, and the coiled coil domain is shown in dark gray. The intron positions are marked with arrows with the phases of the introns (Patthy, 1987) on top. The domains, repeats and intron positions are shown to scale. (B) Secondary structure prediction by Jpred and fold recognition by 3D-pssm suggests a seven-bladed β-propeller fold reminiscent of the ones of the yeast transcriptional repressor Tup1 and β-subunit of the G protein. Coronin 1 was aligned with Tup1 and the G protein β-subunit according to the positions of the predicted and known β-strands. Sequence similarities were then noted that confirmed or fine-tuned the alignment. The GlyHis and TrpAsp dipeptides of the five WD repeats are highlighted in bold. Predicted blade numbers and corresponding β-strands (gray arrows) are shown on the left and on the bottom of the alignment, respectively. The N- and C-terminal domain extensions are underlined. C, linker domain is predicted to contain little regular secondary structure. Flexible residues (Gly, Pro, Ser, and Gln) are highlighted in bold. The stretch of positively charged residues is underlined. D, limited trypsin digestion of coronin 1. Western blot detection was performed using an antibody directed toward the N-terminal portion of coronin 1. E, C-terminal domain is predicted by Coils to fold into a α-helical coiled coil structure. The assignment of heptad repeats (abcdefg) is indicated and hydrophobic residues at the a and d core positions are highlighted in bold.
In the N-terminal domain, 30 short β-strands each separated by loops consisting of 7 ± 5 amino acid residues were predicted. As previously documented, five consecutive WD repeats (residues 65–296) of 46 ± 3 residues in length form the core of this domain (Figure 2, A and B; van der Voorn and Ploegh, 1992; Neer et al., 1994).
WD repeat proteins, although frequently sharing only limited sequence similarity, were shown to adopt a highly symmetrical β-propeller fold (Smith et al., 1999) containing four-stranded antiparallel β-sheets as individual propeller blades. These four β-strands, denoted consecutively d, a, b, and c according to the definition of Smith et al. (1999), are predicted for each of the five WD repeats of coronin 1 (Figure 2B). However, two additional sequence stretches of 46 and 44 residues, respectively, that flank the WD repeat-containing core sequence are predicted to form four short β-strands and align with the corresponding β-strands of the five WD repeats (Figure 2B). Because WD repeats are not strictly necessary to assert a propeller fold (Fulop and Jones, 1999; Smith et al., 1999; Jawad and Paoli, 2002), the prediction suggests that the coronin 1 propeller domain is, in fact, made up of at least seven blades instead of the previously proposed five blades (Fulop and Jones, 1999; Smith et al., 1999; Jawad and Paoli, 2002). Although the determination of the precise β-propeller fold of coronin 1 awaits solution of the structure of the N-terminal domain, the existence of two additional propeller blades is further supported by sequence threading by using the protein fold recognition program 3D-pssm (Kelley et al., 2000), which predicted a similarity between the coronin 1 N-terminal domain and the yeast transcriptional repressor Tup1 (Sprague et al., 2000) and the G protein β-subunit (Lambright et al., 1996), both WD repeat containing seven-bladed β-propeller proteins.
Finally, the exon–intron organization of the coronin 1 gene was analyzed because a concordance of exon structure with domain structure is frequently found for domains composed of multiple sequence repeats, such as β-propellers (van der Voorn et al., 1990; Springer, 1998). As shown in Figure 2A, both the five WD repeats as well as the two flanking sequence stretches in N-terminal domain do not correlate with intron positions. In fact, the exon boundaries and phases strongly support the prediction that the modular unit comprises the sequence stretch Met1-Lys355, which frames the seven predicted propeller blades. In summary, this analysis indicates that coronin 1 does not exist as a five-bladed, but probably as a seven-bladed β-propeller molecule. High resolution structural analysis is needed to define the exact topology of N-terminal mammalian coronin domains.
After the β-propeller lies the 74-residue-long linker domain, which harbors the unique region of coronin homologues (de Hostos, 1999). Less than 15% regular secondary structure (i.e., α-helix or β-strand) is predicted by Jpred for the linker domain, suggesting that it might be at least partially disordered (Figure 2C; Vihinen et al., 1994). Notably, an enrichment of the flexible amino acids Gly, Pro, Ser, or Gln (Vihinen et al., 1994) to 34% is observed throughout the linker domain (Figure 2C). Moreover, the C-terminal part of this domain contains a stretch of positively charged residues (underlined in Figure 2C). Consistent with a partially flexible domain, limited tryptic digestion of coronin 1 produced an N-terminal fragment of 46 kDa. This cleavage was not observed to occur in a mutant lacking residues 400–416 (cfr. later), suggesting that cleavage occurred within this region (Figure 2D).
The C terminal domain has previously been predicted to adopt an α-helical coiled coil conformation (de Hostos, 1999). As assigned by the Coils method (Lupas, 1996), the a and d core positions of the heptad repeat are occupied by hydrophobic Val and Leu residues (Figure 2E). The exon boundaries and phases suggest that both the linker domain and the coiled coil correspond to modular units (Figure 2A).
Molecular Organization of Coronin 1
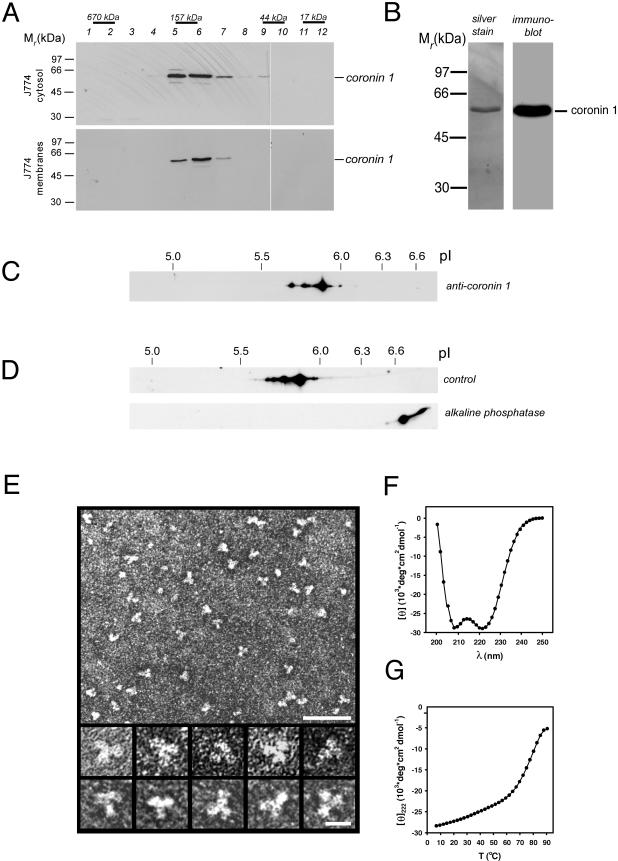
Coronin 1 monomers (Mr of 51 kDa) are predicted to assemble into oligomers via the C-terminal coiled coil (see above). To determine the oligomerization state of coronin 1, membranes and cytosol were prepared from J774 macrophages and analyzed by analytical size exclusion chromatography followed by SDS-PAGE of the eluted fractions and immunoblotting by using anti-coronin 1 antibody. Both the cytosolic as well as the membrane-bound form of coronin 1 eluted at a position corresponding to a molecular mass of ∼160 kDa (Figure 3A), suggesting the presence of a complex.
Figure 3.
Molecular organization of the coronin 1 complex. (A) Isolated cytosol (top) and membranes (bottom) from mouse macrophages were solubilized in 2% octyl glucopyranoside and subjected to size exclusion chromatography on a Superdex 200 column. The eluted fractions were assayed for the presence of coronin 1 by SDS-PAGE and immunoblotting. The positions of proteins of known Mr are indicated. (B) Silver-stained SDS-PAGE (left) and immunoblot (right) of affinity-purified coronin 1 molecules from macrophage cytosol by using anti-coronin antiserum. (C) Detection of different coronin 1 isoforms in macrophage lysates by two-dimensional IEF/SDS-PAGE and immunoblotting by using anti-coronin 1 antibodies. (D) Analysis of control and ALP-treated affinity-purified coronin 1. J774 cells were metabolically labeled for 16 h with [35S]methionine, lysed in Triton X-100 lysis buffer, and coronin 1 complexes were purified from the cell lysate by immunoprecipitation by using an anti-coronin 1 antibody. The purified coronin 1 complexes were incubated at 37°C for 2 h with ALP or buffer alone, before analysis by two-dimensional IEF/SDS-PAGE and fluorography. (E) Transmission electron micrographs of affinity-purified and negatively stained coronin 1 complexes. Top, low-magnification overview. Bar, 50 nm. Bottom, high-magnification gallery. Bar, 10 nm. (F and G) CD analysis of ccCor1. Far-UV CD spectra (F) and thermal unfolding recorded by CD at 222 nm (G) were performed in 5 mM sodium phosphate, pH 7.4, containing 150 mM NaCl and at a peptide concentration of 0.15 mg/ml.
To analyze coronin 1 complexes biochemically and morphologically, they were isolated from J774 macrophage cytosol by immunoaffinity chromatography. SDS-PAGE and silver staining of the eluted fractions revealed the presence of a single protein band with an apparent molecular mass of ∼51 kDa (Figure 3B, left) that is recognized by an independent coronin 1-specific anti-peptide antiserum (Figure 3B, right). Mass spectrometry did not detect coronin isoforms other than coronin 1. Two-dimensional IEF/SDS-PAGE and immunoblotting of a macrophage lysate by using an independent antibody against coronin 1 that does not cross react with any other isoform in these cells identified a series of spots with the same molecular weight but differing in pI (Figure 3C). The difference in pI suggests posttranslational modification by multiple phosphorylation. To directly analyze phosphorylation, coronin 1 immunoprecipitated from metabolically labeled macrophages was dephosphorylated and analyzed by two-dimensional IEF/SDS-PAGE. As shown in Figure 3D, after phosphatase treatment all spots shift to the basic end of the first dimension. Together, these data indicate that in macrophages coronin 1 forms homooligomers, which are differentially phosphorylated.
The morphology of affinity-purified coronin 1 was assessed by transmission electron microscopy. Low-magnification overviews of negatively stained specimens revealed uniformly distributed particles with an apparent threefold symmetry (Figure 3E, top). The images shown in the high magnification gallery of Figure 3E, suggest that three globular domains, each 5–6 nm in diameter, are joined together to form a trefoil-like flexible structure with an overall diameter of 12–15 nm. The globular domains most likely correspond to individual β-propeller domains.
To analyze whether the C-terminal residues of coronin 1 formed a α-helical coiled coil structure as predicted, a peptide comprising residues Val430-Lys461 (referred to as cc-Cor1) was synthesized and characterized by circular dichroism spectroscopy and analytical ultracentrifugation. The far-UV circular dichroism spectrum recorded at 5°C from ccCor1 exhibited well defined minima centered at 208 and 222 nm, which is characteristic of proteins with a helical conformation (Figure 3F). At a concentration of 0.15 mg/ml, ccCor1 revealed a fully reversible sigmoidal thermal unfolding profile with a single midpoint of the transition centered at Tm = 79°C (Figure 3G). The oligomeric state of the peptide (3.8 kDa) was assessed by analytical ultracentrifugation. At 20°C, sedimentation velocity and equilibrium runs of 0.1–0.5 mg/ml ccCor1 solutions yielded a sedimentation coefficient sw,20 and an average molecular mass of 1.2 S and 11.8 kDa, respectively, consistent with an elongated trimeric structure. In agreement with the circular dichroism and analytical ultracentrifugation results, the x-ray crystal structure of cc-Cor1 revealed a three-stranded, in-register, parallel α-helical coiled coil (Kammerer, Kostrewa, Progias, Honnappa, Avila, Lustig, Winkler, Pieters, and Steinmetz, unpublished data). Recently, Oku et al. (2004) found that coronin 1 migrates as a dimer as determined by gel filtration and sucrose density analysis. Using these methods, however, molecular mass can only be estimated because they are not shape independent. Our results, based on analytical ultracentrifugation and electron microscopy, clearly demonstrate that coronin 1 is a trimer. Together, these results suggest that in vivo three N-terminal coronin 1 β-propeller domains are joined in parallel by their C-terminal coiled coil domains.
Functional Analysis of Coronin 1 Domains
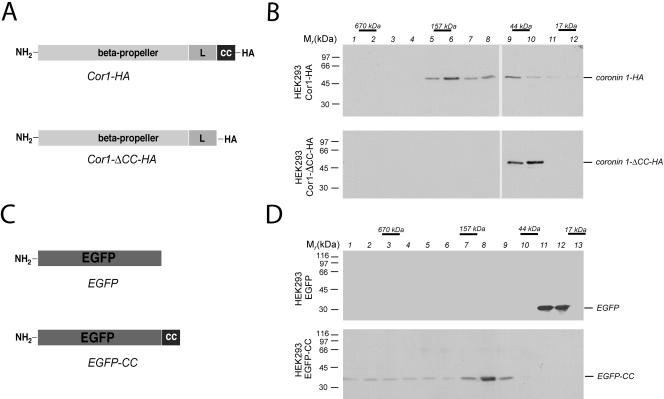
To analyze the role of the C-terminal coiled coil domain of coronin in vivo, a coronin 1 mutant lacking the C-terminal 29 residues encompassing the coiled coil domain was constructed (referred to as Cor1-ΔCC-HA; Figure 4A). Cor1-ΔCC-HA or wild-type coronin 1 (referred to as Cor1-HA; Figure 4A) protein, both hemagglutinin-tagged, were transiently expressed in the coronin 1-negative cell line HEK293 cells, and the cytosol was subjected to analytical size exclusion chromatography. SDS-PAGE and immunoblotting of the eluted fractions with anti-coronin 1 antibodies revealed that Cor1-HA existed as a 160-kDa complex whereas Cor1-ΔCC-HA migrated at a position corresponding to the monomeric subunit (Figure 4B). These findings define the coiled coil as necessary for coronin 1 complex formation.
Figure 4.
Functional analysis of the coiled coil domain of coronin 1. (A) Schematic representation of coronin 1 deletion mutant used in this study. (B) HEK293 cells were transiently transfected with the indicated constructs, and after 24 h the cytosol was analyzed by size exclusion chromatography. The presence of the coronin 1 constructs was analyzed using immunoblotting following SDS-PAGE of the fractions indicated. (C) Schematic representation of EGFP fusion protein. (D) Size exclusion chromatography analysis of cytosol from EGFP or EGFP-CC–expressing HEK293 cells. Size exclusion chromatography was performed as described in the legend to Figure 3A and Materials and Methods.
Whether the coiled coil domain itself was sufficient for trimerization was analyzed by adopting the following strategy. The coiled coil sequence (coronin 1429–461) was fused to the C terminus of the enhanced green fluorescent protein (termed EGFP-CC; Figure 4C) and transfected transiently into HEK293 cells. To analyze the oligomeric state of EGFP and EGFP-CC, HEK293 transfectants were homogenized and their cytosol was subjected to analytical size exclusion chromatography followed by SDS-PAGE and immunoblotting by using anti-GFP antibodies. Figure 4D demonstrates that EGFP migrated at the expected molecular mass of ∼30 kDa (fraction 11/12, top), whereas EGFP-CC migrated at a molecular mass of ∼110 kDa (fraction 8, bottom), indicating a coiled coil-mediated trimerization of EGFP in the absence of any detectable monomers.
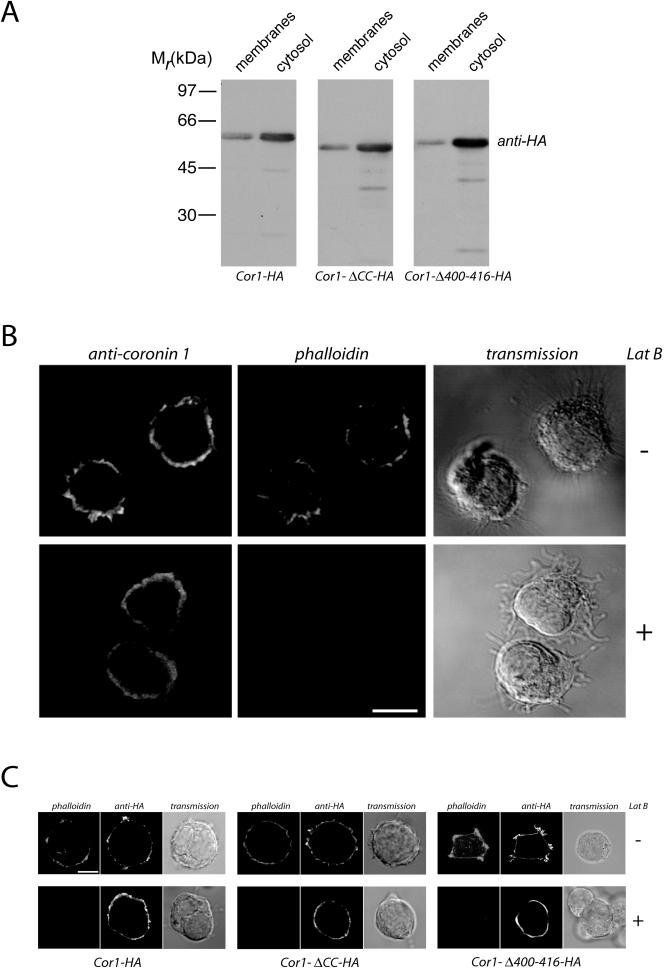
Next, the contribution of the coiled coil to cytoskeletal interaction of coronin 1 was analyzed. To that end, the cytoskeleton from HEK293 cells expressing either wild-type or the coronin 1 mutant lacking the coiled coil (Cor1-ΔCC-HA) was isolated. Although a significant portion of wild-type coronin (Cor1-HA) was recovered in the detergent-insoluble pellet, the coronin 1 mutant lacking the coiled coil (Cor1-ΔCC-HA) was fully detergent soluble (Figure 5A, left and middle). These data demonstrate that the coiled coil domain is necessary for mediating the in vivo association of coronin 1 with the cytoskeleton.
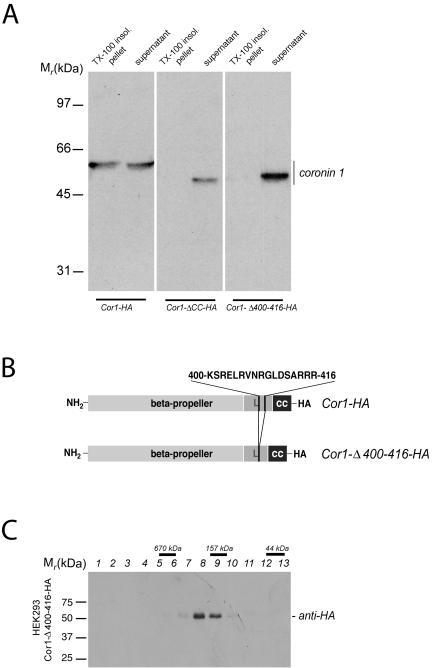
Figure 5.
Association of coronin 1 with the cytoskeleton. (A) Biochemical analysis of F-actin association of coronin 1. HEK293 cells transiently transfected with Cor1-HA, Cor1-Δ CC-HA, or Cor1-Δ400-416-HA were lysed in cytoskeleton isolation buffer containing 1% Triton X-100, and a detergent-insoluble fraction was obtained by low-speed centrifugation (see Materials and Methods). Subsequently, detergent-insoluble pellets and supernatants were analyzed by SDS-PAGE and immunoblotting by using an anti-coronin 1 antibody. (B) Schematic representation of coronin 1 deletion mutant Cor1-Δ400-416-HA. (C) Size exclusion chromatography analysis of cytosol from Cor1-Δ400-416-HA expressing HEK293 cells. Size exclusion chromatography was performed as described in Figure 3A and Materials and Methods.
To analyze a function for the linker region in cytoskeletal binding, a mutant coronin 1 was constructed, (Cor1-Δ361-422), lacking the complete linker domain. However, expression of this construct in HEK293 cells lead to coronin 1 aggregation, as revealed by analytical gel filtration (our unpublished data). Because this linker region contains a stretch of positively charged amino acid residues that may be responsible for such cytoskeletal interaction (Tang et al., 1996; Wohnsland et al., 2000), a mutant Cor1-Δ400-416-HA was constructed in which these residues are deleted (Figure 5B). This construct was correctly oligomerized upon expression in HEK293 cells (Figure 5C). When the cytoskeletal association was analyzed by isolation of Triton X-100–insoluble cytoskeleton, this mutant was retrieved in the soluble fraction (Figure 5A, right). Together, these results indicate that the stretch of positively charged residues within the linker domain mediate cytoskeletal association. Furthermore, this confirms that the coronin 1 coiled coil functions to trimerize the linker region, which is necessary to generate the cytoskeleton binding site.
In macrophages, coronin 1 molecules interact with the plasma membrane (Figure 1, A and B). To analyze the region of coronin 1 required for plasma membrane binding, HEK293 cells were transfected with cDNA constructs encoding Cor1-HA, Cor1-ΔCC-HA, or Cor1-Δ400-416-HA, homogenized in sucrose-containing buffer, and membrane and cytosolic fractions were prepared. SDS-PAGE of membranes and cytosol followed by immunoblotting using anti-HA antibodies revealed that the wild-type coronin 1 molecule distributed between membrane and cytosolic fraction in a 20:80 ratio (Figure 6A), as was observed in J774 cells. Interestingly, both the coronin 1 molecule lacking the coiled coil as well the mutant lacking the cytoskeleton binding site were distributed in a similar proportion between membranes and cytosol (Figure 6A) suggesting that these domains are not required for plasma membrane association.
Figure 6.
Association of coronin 1 with the plasma membrane. (A) The postnuclear supernatants of HEK293 cells expressing Cor1-HA, Cor1-ΔCC-HA, or Cor1-Δ400-416-HA expression constructs were subjected to subcellular fractionation (100,000 × g, 30 min). Membrane and cytosolic fractions were analyzed for the presence of coronin 1 by SDS-PAGE and immunoblotting with an anti-HA antibody. (B) Immunofluorescence analysis of coronin 1 and F-actin localization in control and LatB-treated macrophages. J774 cells were left untreated or treated with 5 μM latrunculin B for 30 min before formaldehyde fixation and saponin permeabilization. Cells were stained for coronin 1 and F-actin by using an anti-coronin 1 antiserum (secondary reagent goat anti-rabbit-Alexa Fluor 488) and phalloidin-Texas Red. Bar, 10 μm. (C) Immunofluorescence analysis of coronin 1 and F-actin localization in latrunculin B-treated HEK293 cells. Cells were transfected with the indicated constructs and after 24 h treated with 20 μM latrunculin B (30 min) or left untreated. Cells were fixed with formaldehyde followed by saponin permeabilization and stained for coronin 1 and F-actin using anti-HA (secondary reagent goat anti-mouse Alexa Fluor 488) and phalloidin-Texas Red. Bar, 10 μm.
To directly analyze whether coronin 1 membrane localization was independent of its F-actin association, J774 macrophages were treated with the F-actin–depolymerizing drug latrunculin B for 30 min, fixed and stained for coronin 1 and F-actin. As shown in Figure 6B, the cortical coronin 1 staining remained essentially unaffected, whereas the actin cytoskeleton was efficiently depolymerized as indicated by a lack of phalloidin fluorescence. Additionally, when HEK293 cells expressing Cor1-HA, Cor1-ΔCC-HA, or Cor1-Δ400-416-HA were treated with latrunculin B, F-actin was fully depolymerized, whereas the cortical localization of Cor1-HA as well as Cor1-ΔCC-HA and Cor1-Δ400-416-HA remained unchanged (Figure 6C). These observations demonstrate that coronin 1 association with the plasma membrane is independent of F-actin.
DISCUSSION
Leukocytes possess a dynamic actin cytoskeleton to fulfill their immunological functions such as phagocytosis, cell-cell contact, cell migration, and immunoreceptor signaling in response to outside signals (Allen and Aderem, 1996; Fischer et al., 1998; Fuller et al., 2003; Gruenheid and Finlay, 2003). The coronin family of actin-associated proteins have been shown to be implicated in actin dynamics (de Hostos et al., 1991; Suzuki et al., 1995). All coronins have a similar architecture, in that they posses a large, WD40 repeat containing region at the N terminus (β-propeller), followed by a flexible, unstructured domain (linker domain) and a C-terminal coiled coil region. The mammalian isoform coronin 1 (also known as P57 or TACO) is exclusively expressed in leukocytes and therefore likely to possess immune cell-specific functions compared with other family members that also are expressed in other cell types (Okumura et al., 1998; de Hostos, 1999). Here, we describe that coronin 1 molecules are homotrimers, which are assembled through their C termini via coiled coil interactions (Figure 7). The reported molecular masses of coronin 3 and Xcoronin complexes (Asano et al., 2001; Spoerl et al., 2002) would in fact be consistent with trimeric complex formation. Furthermore, we found that coronin 1 molecules are associated constitutively with the cytoskeleton in an F-actin–dependent manner through a region of positively charged residues within the linker domain connecting the actin cytoskeleton via their N-terminal, WD40-containing domain with the leukocyte plasma membrane. Trimerization of the linker region through the coiled coil region is essential for the generation of the cytoskeletal binding site. As such, coronin 1 may integrate outside-inside signaling at the plasma membrane with the actin cytoskeleton in leukocytes.
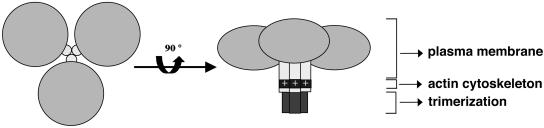
Figure 7.
Coronin 1 serves as a linker between the plasma membrane and the actin cytoskeleton in leukocytes. Coronin 1 is a parallel homotrimeric protein consisting of three globular N-terminal β-propellers (light gray) assembled via the C-terminal coiled coil (dark gray). Association of coronin 1 with the cytoskeleton occurs via a stretch of positively charged residues in the linker region (light gray) and is dependent on trimerization. The F-actin–independent binding to the plasma membrane is mediated via the N-terminal globular β-propeller domain.
The primary sequences of all coronin family members have suggested the presence of an N-terminal WD repeat-containing domain and a C-terminal coiled coil domain (Suzuki et al., 1995; de Hostos, 1999). Notably, rather than forming a five-bladed propeller, the N-terminal part of coronin 1 is predicted to fold at least into a seven-bladed β-propeller (Figure 2). Because the primary sequences of the N-terminal domains are highly conserved throughout the coronin family members, it is likely that all these homologous domains adopt a similar β-propeller topology.
Biophysical analyses of the coronin 1 C terminal domain demonstrates that this portion of the coronin 1 molecule folds into a parallel three-stranded coiled coil that mediates trimerization of coronin 1 monomers. In coronin 1 molecules, we found that coiled coil mediated trimerization generates a cytoskeletal binding site comprising positively charged residues located within the linker domain, whereas plasma membrane binding is mediated by the globular β-propeller (Figure 7). Indeed, basic peptides have been shown to be important for interaction with actin (Tang et al., 1996, Wohnsland et al., 2000). Many plasma membrane-cytoskeletal linker molecules contain two isolated binding sites such as filamin in platelets, myosin1 in nonmuscle cells, ezrin in epithelial cells, and dystrophin in muscle cells (Rafael et al., 1996; Jontes and Milligan, 1997; Bretscher, 1999; Stossel et al., 2001; Schafer, 2002). However, in contrast to coronin 1, these linker molecules bind F-actin via globular domains and the plasma membrane via α-helical domains.
In contrast to a previous report, which mapped the F-actin binding sites in recombinant coronin 1 to the N-terminal propeller by using an in vitro two-component cosedimentation assay (Oku et al., 2003), our results indicate that in vivo, association to the cytoskeleton is mediated by the unique linker domain. Interestingly, in S. cerevisiae the C terminal part including the linker region and the coiled coil domain of Crn1p can bind and modulate Arp2/3 activity in vitro (Humphries et al., 2002; Rodal et al., 2004). Whether coronin 1 in mammalian cells also interacts with the Arp 2/3 complex remains to be established.
In the absence of the coiled coil or the cytoskeleton interaction site within the linker region, plasma membrane binding still occurred, suggesting that the N-terminal, β-propeller-containing domain is responsible for plasma membrane association.
The binding partners of the N terminal, β-propeller containing domain of coronin 1 at the plasma membrane remain unknown, and might include integral or peripheral proteins as well as lipid moieties (Gatfield and Pieters, 2000). Also, how coronin 1 localization is controlled is currently unknown. Regulation of membrane and cytoskeletal association via a switch in the coronin 1 oligomerization state is unlikely because trimers are the only detectable species in macrophages (Figure 3A), but, as is the case, e.g., for ezrin (Gautreau et al., 2000), regulation might involve phosphorylation.
The finding that the coronin 1 coiled coil domain is not involved in plasma membrane targeting is in contrast to observations on human coronin 3 (Spoerl et al., 2002). Deletion of the coronin 3 coiled coil abolishes plasma membrane binding and reduces cell spreading similar to the transfection of the Xenopus coronin mutant (Asano et al., 2001; Spoerl et al., 2002). However, we found that cell spreading of macrophages devoid of coronin 1 was unaltered, suggesting alternative roles for the different coronin family members in different cell types.
Many receptors in immune cells trigger rapid rearrangements of the actin cytoskeleton. Among these are lymphocyte antigen receptors, phagocytic receptors as well as cell adhesion molecules (Allen and Aderem, 1996; Fischer et al., 1998; Fuller et al., 2003; Gruenheid and Finlay, 2003). Several of these receptors such as T-cell receptors and B-cell receptors are located in or recruited to cholesterol-rich microdomains during ligand mediated triggering (Moran and Miceli, 1998; Cheng et al., 1999; van't Hof and Resh, 1999; Cherukuri et al., 2001). Interestingly, recruitment into lipid rafts also has been shown for the complement receptor involved in mycobacterial uptake by macrophages where cholesterol-rich microdomains and coronin 1 molecules accumulate at the site of entry (Gatfield and Pieters, 2000; Peyron et al., 2000). Triggering of phagocytic receptors is associated with extensive remodeling of the underlying actin cytoskeleton. By linking the plasma membrane to the underlying actin cytoskeleton in immune cells, coronin 1, either via direct or indirect binding to transmembrane receptors, may facilitate the integration of extracellular signals with F-actin remodeling. A role for coronins as integrators of cellular structural components also had been suggested previously for S. cerevisiae Crn1p (Heil-Chapdelaine et al., 1998), which has binding sites for the actin cytoskeleton and microtubules.
Besides F-actin interaction, little is known about the different functions of the coronin family members. It is possible that coronins as multimeric or even exclusively trimeric WD repeat proteins, such as the coronin 1 isoform, serve as specific integrators of the actin cytoskeleton with major structural or signaling complexes in the cell.
Acknowledgments
We thank L. Kuhn for performing the two-dimensional IEF/SDS-PAGE. We are indebted to A. Lustig for performing the analytical ultracentrifugation experiments and D. Avila for preparing the synthetic ccCor1 peptide. We acknowledge the Interdisciplinary Center for Microscopy (IZM) and the Department of Biophysical Chemistry of the University of Basel for access to transmission electron microscopy and circular dichroism facilities, respectively. We thank U. Aebi for critical reading of the manuscript and F. Winkler for support. This work was supported by F. Hoffmann-La Roche, which founded the Basel Institute for Immunology where part of this work was performed. This work was further supported by grants from the Swiss National Science Foundation, the World Health Organization, and the Olga Maeyenfish Stiftung.
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E05–01–0042) on March 30, 2005.
References
- Allen, L. A., and Aderem, A. (1996). Mechanisms of phagocytosis. Curr. Opin. Immunol. 8, 36–40. [DOI] [PubMed] [Google Scholar]
- Asano, S., Mishima, M., and Nishida, E. (2001). Coronin forms a stable dimer through its C-terminal coiled coil region: an implicated role in its localization to cell periphery. Genes Cells 6, 225–235. [DOI] [PubMed] [Google Scholar]
- Bretscher, A. (1999). Regulation of cortical structure by the ezrin-radixinmoesin protein family. Curr. Opin. Cell Biol. 11, 109–116. [DOI] [PubMed] [Google Scholar]
- Cella, M., Engering, A., Pinet, V., Pieters, J., and Lanzavecchia, A. (1997). Inflammatory stimuli induce accumulation of MHC class II complexes on dendritic cells. Nature 388, 782–787. [DOI] [PubMed] [Google Scholar]
- Cheng, P. C., Dykstra, M. L., Mitchell, R. N., and Pierce, S. K. (1999). A role for lipid rafts in B cell antigen receptor signaling and antigen targeting. J. Exp. Med. 190, 1549–1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherukuri, A., Dykstra, M., and Pierce, S. K. (2001). Floating the raft hypothesis: lipid rafts play a role in immune cell activation. Immunity 14, 657–660. [DOI] [PubMed] [Google Scholar]
- Cuff, J. A., Clamp, M. E., Siddiqui, A. S., Finlay, M., and Barton, G. J. (1998). JPred: a consensus secondary structure prediction server. Bioinformatics 14, 892–893. [DOI] [PubMed] [Google Scholar]
- de Hostos, E. L. (1999). The coronin family of actin-associated proteins. Trends. Cell Biol. 9, 345–350. [DOI] [PubMed] [Google Scholar]
- de Hostos, E. L., Bradtke, B., Lottspeich, F., Guggenheim, R., and Gerisch, G. (1991). Coronin, an actin binding protein of Dictyostelium discoideum localized to cell surface projections, has sequence similarities to G protein beta subunits. EMBO J. 10, 4097–4104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Hostos, E. L., Rehfuess, C., Bradtke, B., Waddell, D. R., Albrecht, R., Murphy, J., and Gerisch, G. (1993). Dictyostelium mutants lacking the cytoskeletal protein coronin are defective in cytokinesis and cell motility. J. Cell Biol. 120, 163–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eason, R. (1986). Analytical ultracentrifugation. In: Centrifugation, A Practical Approach., ed. D. Rickwood, Oxford: IRL Press, 251–286.
- Engering, A., Kuhn, L., Fluitsma, D., Hoefsmit, E., and Pieters, J. (2003). Differential post-translational modification of CD63 molecules during maturation of human dendritic cells. Eur. J. Biochem. 270, 2412–2420. [DOI] [PubMed] [Google Scholar]
- Engering, A., Lefkovits, I., and Pieters, J. (1997). Analysis of subcellular organelles involved in major histocompatibility complex (MHC) class II-restricted antigen presentation by electrophoresis. Electrophoresis 18, 2523–2530. [DOI] [PubMed] [Google Scholar]
- Ferrari, G., Knight, A. M., Watts, C., and Pieters, J. (1997). Distinct intracellular compartments involved in invariant chain degradation and antigenic peptide loading of major histocompatibility complex (MHC) class II molecules. J. Cell Biol. 139, 1433–1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari, G., Naito, M., Langen, H., and Pieters, J. (1999). A coat protein on phagosomes involved in the intracellular survival of mycobacteria. Cell 97, 435–447. [DOI] [PubMed] [Google Scholar]
- Fischer, K. D., Tedford, K., and Penninger, J. M. (1998). Vav links antigen-receptor signaling to the actin cytoskeleton. Semin. Immunol. 10, 317–327. [DOI] [PubMed] [Google Scholar]
- Fukui, Y., Engler, S., Inoue, S., and de Hostos, E. L. (1999). Architectural dynamics and gene replacement of coronin suggest its role in cytokinesis. Cell Motil. Cytoskeleton 42, 204–217. [DOI] [PubMed] [Google Scholar]
- Fuller, C. L., Braciale, V. L., and Samelson, L. E. (2003). All roads lead to actin: the intimate relationship between TCR signaling and the cytoskeleton. Immunol. Rev. 191, 220–236. [DOI] [PubMed] [Google Scholar]
- Fulop, V., and Jones, D. T. (1999). Beta propellers: structural rigidity and functional diversity. Curr. Opin. Struct. Biol. 9, 715–721. [DOI] [PubMed] [Google Scholar]
- Gatfield, J., and Pieters, J. (2000). Essential role for cholesterol in entry of mycobacteria in macrophages. Science 288, 1647–1650. [DOI] [PubMed] [Google Scholar]
- Gautreau, A., Louvard, D., and Arpin, M. (2000). Morphogenic effects of ezrin require a phosphorylation-induced transition from oligomers to monomers at the plasma membrane. J. Cell Biol. 150, 193–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goode, B. L., Wong, J. J., Butty, A. C., Peter, M., McCormack, A. L., Yates, J. R., Drubin, D. G., and Barnes, G. (1999). Coronin promotes the rapid assembly and cross-linking of actin filaments and may link the actin and microtubule cytoskeletons in yeast. J. Cell Biol. 144, 83–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruenheid, S., and Finlay, B. B. (2003). Microbial pathogenesis and cytoskeletal function. Nature 422, 775–781. [DOI] [PubMed] [Google Scholar]
- Heil-Chapdelaine, R. A., Tran, N. K., and Cooper, J. A. (1998). The role of Saccharomyces cerevisiae coronin in the actin and microtubule cytoskeletons. Curr. Biol. 8, 1281–1284. [DOI] [PubMed] [Google Scholar]
- Humphries, C. L., Balcer, H. I., D'Agostino, J. L., Winsor, B., Drubin, D. G., Barnes, G., Andrews, B. J., and Goode, B. L. (2002). Direct regulation of Arp2/3 complex activity and function by the actin binding protein coronin. J. Cell Biol. 159, 993–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jawad, Z., and Paoli, M. (2002). Novel sequences propel familiar folds. Structure 10, 447–454. [DOI] [PubMed] [Google Scholar]
- Jontes, J. D., and Milligan, R. A. (1997). Brush border myosin-I structure and ADP-dependent conformational changes revealed by cryoelectron microscopy and image analysis. J. Cell Biol. 139, 683–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley, L. A., MacCallum, R. M., and Sternberg, M. J. (2000). Enhanced genome annotation using structural profiles in the program 3D-PSSM. J. Mol. Biol. 299, 499–520. [DOI] [PubMed] [Google Scholar]
- Lambright, D. G., Sondek, J., Bohm, A., Skiba, N. P., Hamm, H. E., and Sigler, P. B. (1996). The 2.0 A crystal structure of a heterotrimeric G protein. Nature 379, 311–319. [DOI] [PubMed] [Google Scholar]
- Lefkovits, I., Young, P., Kuhn, L., Kettman, J., Gemmell, A., Tollaksen, S., Anderson, L., and Anderson, N. [eds.] (1985). Use of large-scale two-dimensional ISODALT gel electrophoresis systems in immunology. In: Immunological Methods III. Orlando, FL: Academic Press, 163–185.
- Lupas, A. (1996). Coiled coils: new structures and new functions. Trends. Biochem. Sci. 21, 375–382. [PubMed] [Google Scholar]
- Maniak, M., Rauchenberger, R., Albrecht, R., Murphy, J., and Gerisch, G. (1995). Coronin involved in phagocytosis: dynamics of particle-induced relocalization visualized by a green fluorescent protein Tag. Cell 83, 915–924. [DOI] [PubMed] [Google Scholar]
- Moran, M., and Miceli, M. C. (1998). Engagement of GPI-linked CD48 contributes to TCR signals and cytoskeletal reorganization: a role for lipid rafts in T cell activation. Immunity 9, 787–796. [DOI] [PubMed] [Google Scholar]
- Nal, B., Carroll, P., Mohr, E., Verthuy, C., Da Silva, M. I., Gayet, O., Guo, X. J., He, H. T., Alcover, A., and Ferrier, P. (2004). Coronin-1 expression in T lymphocytes: insights into protein function during T cell development and activation. Int. Immunol. 16, 231–240. [DOI] [PubMed] [Google Scholar]
- Neer, E. J., Schmidt, C. J., Nambudripad, R., and Smith, T. F. (1994). The ancient regulatory-protein family of WD-repeat proteins. Nature 371, 297–300. [DOI] [PubMed] [Google Scholar]
- Oku, T., Itoh, S., Ishii, R., Suzuki, K., Nauseef, W. M., Toyoshima, S., and Tsuji, T. (2005). Homotypic dimerisation of the actin-binding protein p57/coronin-1 mediated by a leucine zipper motif in the C-terminal region. Biochem. J. 387, 325–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oku, T., Itoh, S., Okano, M., Suzuki, A., Suzuki, K., Nakajin, S., Tsuji, T., Nauseef, W. M., and Toyoshima, S. (2003). Two regions responsible for the actin binding of p57, a mammalian coronin family actin-binding protein. Biol. Pharm. Bull. 26, 409–416. [DOI] [PubMed] [Google Scholar]
- Okumura, M., Kung, C., Wong, S., Rodgers, M., and Thomas, M. L. (1998). Definition of family of coronin-related proteins conserved between humans and mice: close genetic linkage between coronin-2 and CD45-associated protein. DNA Cell. Biol. 17, 779–787. [DOI] [PubMed] [Google Scholar]
- Patthy, L. (1987). Intron-dependent evolution: preferred types of exons and introns. FEBS Lett. 214, 1–7. [DOI] [PubMed] [Google Scholar]
- Peyron, P., Bordier, C., N′Diaye, E. N., and Maridonneau-Parini, I. (2000). Nonopsonic phagocytosis of Mycobacterium kansasii by human neutrophils depends on cholesterol and is mediated by CR3 associated with glycosylphosphatidylinositol-anchored proteins. J. Immunol. 165, 5186–5191. [DOI] [PubMed] [Google Scholar]
- Pieters, J., Horstmann, H., Bakke, O., Griffiths, G., and Lipp, J. (1991). Intracellular transport and localization of major histocompatibility complex class II molecules and associated invariant chain. J. Cell Biol. 115, 1213–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rafael, J. A., Cox, G. A., Corrado, K., Jung, D., Campbell, K. P., and Chamberlain, J. S. (1996). Forced expression of dystrophin deletion constructs reveals structure-function correlations. J. Cell Biol. 134, 93–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodal, A. A., Sokolova, O., Robins, D. B., Daugherty, K. M., Hippenmeyer, S., Riezman, H., Grigorieff, N., and Goode, B. L. (2004). Conformational changes in the Arp2/3 complex leading to actin nucleation. Nat. Struct. Mol. Biol. 12, 26–31. [DOI] [PubMed] [Google Scholar]
- Rybakin, V., Stumpf, M., Schulze, A., Majoul, I. V., Noegel, A. A., and Hasse, A. (2004). Coronin 7, the mammalian POD-1 homologue, localizes to the Golgi apparatus. FEBS Lett. 573, 161–167. [DOI] [PubMed] [Google Scholar]
- Schafer, D. A. (2002). Coupling actin dynamics and membrane dynamics during endocytosis. Curr. Opin. Cell Biol. 14, 76–81. [DOI] [PubMed] [Google Scholar]
- Schuller, S., Neefjes, J., Ottenhoff, T., Thole, J., and Young, D. (2001). Coronin is involved in uptake of Mycobacterium bovis BCG in human macrophages but not in phagosome maintenance. Microbiol. 3, 785–793. [DOI] [PubMed] [Google Scholar]
- Smith, T. F., Gaitatzes, C., Saxena, K., and Neer, E. J. (1999). The WD repeat: a common architecture for diverse functions. Trends Biochem. Sci. 24, 181–185. [DOI] [PubMed] [Google Scholar]
- Sondek, J., Bohm, A., Lambright, D. G., Hamm, H. E., and Sigler, P. B. (1996). Crystal structure of a G-protein beta gamma dimer at 2.1A resolution. Nature 379, 369–374. [DOI] [PubMed] [Google Scholar]
- Spoerl, Z., Stumpf, M., Noegel, A. A., and Hasse, A. (2002). Oligomerization, F-actin interaction, and membrane association of the ubiquitous mammalian coronin 3 are mediated by its carboxyl terminus. J. Biol. Chem. 277, 48858–48867. [DOI] [PubMed] [Google Scholar]
- Sprague, E. R., Redd, M. J., Johnson, A. D., and Wolberger, C. (2000). Structure of the C-terminal domain of Tup1, a corepressor of transcription in yeast. EMBO J. 19, 3016–3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer, T. A. (1998). An extracellular beta-propeller module predicted in lipoprotein and scavenger receptors, tyrosine kinases, epidermal growth factor precursor, and extracellular matrix components. J. Mol. Biol. 283, 837–862. [DOI] [PubMed] [Google Scholar]
- Steinmetz, M. O., Goldie, K. N., and Aebi, U. (1997). A correlative analysis of actin filament assembly, structure, and dynamics. J. Cell Biol. 138, 559–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stossel, T. P., Condeelis, J., Cooley, L., Hartwig, J. H., Noegel, A., Schleicher, M., and Shapiro, S. S. (2001). Filamins as integrators of cell mechanics and signalling. Nat. Rev. Mol. Cell. Biol. 2, 138–145. [DOI] [PubMed] [Google Scholar]
- Suzuki, K., Nishihata, J., Arai, Y., Honma, N., Yamamoto, K., Irimura, T., and Toyoshima, S. (1995). Molecular cloning of a novel actin-binding protein, p57, with a WD repeat and a leucine zipper motif. FEBS. Lett. 364, 283–288. [DOI] [PubMed] [Google Scholar]
- Tang, J. X., and Janmey, P. A. (1996). The polyelectrolyte nature of F-actin and the mechanism of actin bundle formation. J. Biol. Chem. 271, 8556–8563. [DOI] [PubMed] [Google Scholar]
- Tulp, A., Verwoerd, D., Dobberstein, B., Ploegh, H. L., and Pieters, J. (1994). Isolation and characterization of the intracellular MHC class II compartment. Nature 369, 120–126. [DOI] [PubMed] [Google Scholar]
- van der Voorn, L., Gebbink, M., Plasterk, R. H., and Ploegh, H. L. (1990). Characterization of a G-protein beta-subunit gene from the nematode Caenorhabditis elegans. J. Mol. Biol. 213, 17–26. [DOI] [PubMed] [Google Scholar]
- van der Voorn, L., and Ploegh, H. L. (1992). The WD-40 repeat. FEBS Lett. 307, 131–134. [DOI] [PubMed] [Google Scholar]
- van't Hof, W., and Resh, M. D. (1999). Dual fatty acylation of p59(Fyn) is required for association with the T cell receptor zeta chain through phosphotyrosine-Src homology domain-2 interactions. J. Cell Biol. 145, 377–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veithen, A., Cupers, P., Baudhuin, P., and Courtoy, P. J. (1996). v-Src induces constitutive macropinocytosis in rat fibroblasts. J. Cell Sci. 109, 2005–2012. [DOI] [PubMed] [Google Scholar]
- Vihinen, M., Torkkila, E., and Riikonen, P. (1994). Accuracy of protein flexibility predictions. Proteins 19, 141–149. [DOI] [PubMed] [Google Scholar]
- Wohnsland, F., Schmitz, A. A., Steinmetz, M. O., Aebi, U., and Vergeres, G. (2000). Interaction between actin and the effector peptide of MARCKS-related protein. Identification of functional amino acid segments. J. Biol. Chem. 275, 20873–20879. [DOI] [PubMed] [Google Scholar]
- Wrigley, N. G. (1968). The lattice spacing of crystalline catalase as an internal standard of length in electron microscopy. J. Ultrastruct. Res. 24, 454–464. [DOI] [PubMed] [Google Scholar]