Abstract
Reorganization of the nuclear machinery after mitosis is a fundamental but poorly understood process. Here, we investigate the recruitment of the nucleolar processing proteins in the nucleolus of living cells at the time of nucleus formation. We question the role of the prenucleolar bodies (PNBs), during migration of the processing proteins from the chromosome periphery to sites of rDNA transcription. Surprisingly, early and late processing proteins pass through the same PNBs as demonstrated by rapid two-color four-dimensional imaging and quantification, whereas a different order of processing protein recruitment into nucleoli is supported by differential sorting. Protein interactions along the recruitment pathway were investigated using a promising time-lapse analysis of fluorescence resonance energy transfer. For the first time, it was possible to detect in living cells the interactions between proteins of the same rRNA processing machinery in nucleoli. Interestingly interactions between such proteins also occur in PNBs but not at the chromosome periphery. The dynamics of these interactions suggests that PNBs are preassembly platforms for rRNA processing complexes.
INTRODUCTION
In higher eukaryotes, the nucleus is disassembled when chromosomes condense at the beginning of mitosis and reassemble at the end of mitosis. During mitosis, there is redistribution and/or inactivation of the nuclear machineries that will be further involved in rebuilding nuclear functions. One of the fundamental features of nuclear organization is that many components of the RNA synthesis and processing machineries are grouped into compartments (Spector, 1993; Lamond and Earnshaw, 1998; Matera, 1999; Gall, 2000; Misteli, 2000; Spector, 2001; Hernandez-Verdun et al., 2002; Huang, 2002; Lamond and Spector, 2003). This implies that the recruitment of dedicated machineries and formation of discrete nuclear domains are crucial events at the beginning of interphase. In addition, recent data indicate that the entry as well as correct relocation of the nuclear machineries into newly forming nuclei is a step-dependent and ordered process (Savino et al., 2001; Prasanth et al., 2003; Bubulya et al., 2004; Leung et al., 2004). However, recruitment of the nucleolar processing proteins on transcription sites is still a poorly understood process. To better understand the rebuilding of nucleolar functions after mitosis, we chose to investigate the assembly of the nucleolar processing machinery in real time in living cells.
The nucleolus is a functional compartment of the nucleus generated by ribosome biogenesis (Hadjiolov, 1985; Mélèse and Xue, 1995; Leung et al., 2003). It is also a plurifunctional domain involved in the three-dimensional (3D) organization of chromatin in the nucleus (Chubb et al., 2002), in the assembly of several ribonucleoprotein complexes (Pederson, 1998; Olson et al., 2000), and in the formation of nuclear speckles (Bubulya et al., 2004). The nucleolus therefore seems to be a key player of the nuclear functional architecture. Its functions depend on the activation and recruitment of the nucleolar machineries involved in transcription of the ribosomal genes (rDNA) and processing of ribosomal RNAs (rRNA). These machineries are inherited through mitosis from the previous interphase (Hernandez-Verdun et al., 2002). During mitosis, the nucleolar machineries that participate in rRNA processing within the nucleolus are distributed around all the chromosomes and therefore at some distance from the rDNA sequences. When rDNA transcription is activated during telophase, the processing machineries are targeted to the sites of rRNA synthesis. Along the translocation pathway between chromosome periphery and sites of transcription, prenucleolar bodies (PNBs) are formed (Ochs et al., 1985; Azum-Gélade et al., 1994; Jiménez-Garcia et al., 1994; Dundr et al., 2000; Savino et al., 2001). The PNBs are discrete fibrogranular structures characterized by electron microscopy in animal and plant cells (Stevens, 1965; Ochs et al., 1985; Savino et al., 2001). PNB formation occurs on the chromosome surface during telophase and is under the control of cyclin-dependent kinases (Sirri et al., 2002). The PNBs also are formed during Xenopus development before complete nucleolar assembly, and they can be assembled in vitro in Xenopus egg extracts (Bell et al., 1992; Verheggen et al., 1998, 2001). Thus, PNB formation is a general phenomenon occurring during cell cycle and development. However, the role of this steady state along the recruitment pathway of the nucleolar processing complexes is presently unknown.
To investigate the role of PNBs in the establishment of nucleolar functions, we analyzed the dynamics and the possible interactions between processing proteins along the assembly pathway in living cells. The early rRNA processing machinery is cotranscriptionally associated with rRNAs, whereas the late rRNA processing machinery is involved in rRNA processing after termination of transcription. Here, we chose fibrillarin as representative protein of 90S pre-rRNA particles (early processing) and Nop52, Bop1, and B23 as representative proteins of 60s pre-rRNA particles (late processing) (Fatica and Tollervey, 2002; Fromont-Racine et al., 2003). The behavior of early and late rRNA processing proteins at the exit of mitosis was analyzed by 1) immunocytochemistry; 2) fast two-color four-dimensional (4D) imaging in living cells; and 3) time-lapse fluorescence resonance energy transfer (FRET) monitored by time domain fluorescence lifetime imaging microscopy (tdFLIM), a new, direct, and noninvasive method to follow protein–protein interactions in living cells. Based on data presented here, we propose that PNBs are preassembly platforms of rRNA processing complexes.
MATERIALS AND METHODS
Cells
HeLa cells were grown on glass coverslips in modified medium (minimal essential medium; Invitrogen, Carlsbad, CA) supplemented with 10% (vol/vol) fetal calf serum (FCS), 10 μg ml–1 antibiotics (penicillin and streptomycin), and 2 mM l-glutamine (Invitrogen) at 37°C in 5% CO2.
They were transfected at 60% confluence by using the Superfect reagent (QIAGEN, Valencia, CA). The following constructs were used: green fluorescent protein (GFP)-Nop52 and GFP-fibrillarin (Savino et al., 2001), GFP-B23 provided by Dr. S. Huang (Chen and Huang, 2001), GFP-Bop1 provided by Dr. D. Pestov (Pestov et al., 2001b), and Discosoma red fluorescence protein (DsRed)-Nop52 and DsRed-B23 inserted into pDsRed2-C1 (BD Biosciences Clontech, Palo Alto, CA). In all cases, GFP and DsRed were fused to the NH2 terminus of the proteins.
Several stably transformed cell lines were used: GFP-Nop52, GFP-fibrillarin, GFP-B23, GFP-Bop1, DsRed-B23, and DsRed-Nop52. Selection of the GFP-B23, GFP-Bop1, DsRed-B23, and DsRed-Nop52 stably transformed cells was carried out as described previously (Savino et al., 2001). Doubly transfected cells GFP-Nop52/DsRed-B23, GFP-fibrillarin/DsRed-B23, GFP-B23/DsRed-Nop52, GFP-Bop1/DsRed-B23, and DsRed-B23/GFP-Nop52 were generated from stably transformed cells transiently transfected with DsRed-B23, DsRed-Nop52, or GFP-Nop52. For FRET analysis, living adhesive cell lines permanently expressing the donor (GFP-Nop52, GFP-Bop1, or GFP-fibrillarin) were cultured on glass coverslips in Dulbecco's modified Eagle's medium, supplemented with 0.5% FCS, at 37°C in 5% CO2. The cells were transfected with DsRed-B23 1 d before measurements. The coverslips were mounted in a special holder allowing reconstruction of a Petri dish and placed on an inverted microscope. Measurements were carried out in culture medium without phenol red at 37°C in 5% CO2. A paraffin oil (Merck, Darmstadt, Germany) layer was deposited on top of the medium to avoid evaporation and to allow long-term observations at 37°C.
Antibodies
The following antibodies were used: a mouse monoclonal anti-fibrillarin, 72B9 (Reimer et al., 1987); a human serum directed against Nop52 (Savino et al., 1999); and a goat polyclonal anti-B23 (C19; Santa Cruz Biotechnology, Santa Cruz, CA). Anti-mouse secondary antibodies conjugated to fluorescein isothiocyanate (FITC) or to Cy5, anti-human antibodies conjugated to Cy5, and goat antibodies conjugated to Texas Red were from Jackson ImmunoResearch Laboratories (West Grove, PA). For immunolabeling, cells were treated as described previously (Savino et al., 2001).
Time-Lapse Microscopy
The cells were grown on glass coverslips, mounted in a Ludin observation chamber filled with complete medium supplemented with 10 mM HEPES, pH 7.4. The microscope and the chamber were kept at 37°C. A Leica DM IRB microscope, equipped with a piezoelectric translator (PIFOC; Physik Instrumente, Karlsruhe, Germany) placed at the base of a 100× PlanApo numerical aperture (NA) 1.4 objective and a 5-MHz Micromax 782Y interline charge-coupled device (CCD) camera (Roper Scientific, Evry Cedex, France) were used. To visualize two differently tagged proteins, rapid wavelength selection was achieved by a Shutter DG4 illuminator and wavelength changer. For imaging of GFP and DsRed, a dual narrow pass band FITC/tetramethylrhodamine B isothiocyanate (TRITC) filter block was used. In the DG-4 illuminator, short pass KP 500 and long pass LP 515 filters were mounted in positions 1 and 2, respectively. The acquisition software (MetaMorph; Universal Imaging, Downingtown, PA) was set to trigger rapid wavelength changes to acquire two images at each Z-step (0.3 μm). Imaging at full overlapped speed of the CCD device assured that the two fluorescent tags were recorded sequentially at maximum speed, without movement of the filters. The stacks were assembled after image deconvolution as described previously (Savino et al., 2001).
Fluorescence intensities in PNBs were quantified on the sum of three consecutive nondeconvoluted slices. The measurement on three slices was found necessary to include PNB movement. Measurements on regions of interest (ROI) were carried out in different subnuclear domains corresponding to diffuse or foci areas from telophase to early G1. Foci are defined as regions of local intensity greater than three times that of diffuse areas. The same ROI was used for GFP and DsRed fluorescence. The signals were quantified using the ImageJ software. The mean gray value, area, SD, minimum, and maximum gray value were recorded.
FRET Determination by tdFLIM Measurements
The apparatus used for FRET determination performs tdFLIM by the time- and space-correlated single-photon counting method and has been described previously (Emiliani et al., 2003). This technique directly gives the picosecond time-resolved fluorescence decay for every pixel by counting and sampling single emitted photons according to 1) the time delay between photon arrival and laser pulse (picosecond time scale, 4096 channels); 2) their x–y coordinate (256 × 256 pixel image), and 3) their absolute time. A titanium sapphire laser (Millennia 5W/Tsunami 3960-M3BB-UPG kit; Spectra-Physics, France) that delivers picosecond pulses was tuned at 960 nm to obtain a 480 nm excitation wavelength after frequency doubling. The repetition rate was 4 MHz after pulse-picker (3980-35; Spectra-Physics, Paris, France). The laser beam was expanded and inserted into an inverted epifluorescence microscope (Leica DMIRBE, Rueil-Malmaison, France) for wide field illumination. Green fluorescence decay images were taken using a Leica Plan-APOCHROMAT 100×1.3 NA oil objective, a dichroic beam splitter (505DRLP, Omega; Optophotonics, Eaubonne, France), an emission filter (535AF45, Omega; Optophotonics), and the quadrant-anode TSCSPC detector (QA; Europhoton, Berlin, Germany). The count rate was up to 50 kHz. The microscope stage was equipped with an incubator system for temperature and CO2 regulation (37°C, 5% CO2). TdFLIM images were obtained by mapping pixel by pixel the mean lifetime determined from a single exponential fit. For qualitative determination of the occurrence of FRET, the fluorescence decays corresponding to a nucleolus or PNB area were extracted from the acquisition matrix, and the decays of donor-tagged proteins in the presence of the acceptor-tagged B23 were compared with the control decays of donor-tagged proteins measured in the absence of acceptor. The experimental curves were further fitted with a Marquardt nonlinear least-square algorithm (Globals Unlimited software, University of Illinois, Urbana-Champaign, IL) by using a Gaussian distribution of lifetimes as theoretical model. Time-lapse FRET images were obtained from the Gaussian distribution analysis of the fluorescence decays associated with each region of interest and at different elapsed times of acquisition. A reduction in the center of the fluorescence lifetime distribution of GFP-tagged protein superior or equal to 0.200 ns was considered as occurrence of FRET.
RESULTS
Early and Late rRNA Processing Proteins Colocalize in Anaphase and Form PNBs in Telophase
In anaphase, both types of rRNA processing proteins formed a faint and continuous sheath around chromosomes visualized by 4,6-diamidino-2-phenylindole (DAPI). Never was a detectable difference observed between the distributions of early (fibrillarin) and late (B23, Nop52, and Bop1) proteins (Figure 1 and Video 1) in the entire cell volume. These observations provide evidence for the colocalization of the proteins during anaphase and hence demonstrate that during their migration toward the poles the proteins of these machineries do so in association with chromosomes. This hypothesis of chromosome association also is supported by DNase solubilization of B23 during mitosis (Zatsepina et al., 1997), by the presence of the proteins at the surface of isolated chromosomes (Gautier et al., 1992b), and by localization of the proteins in electron microscopy (Gautier et al., 1992a). During telophase and early G1, the rRNA processing proteins form PNBs at the chromosome periphery and are progressively recruited to the site of transcription. As already described, early processing proteins (fibrillarin) are recruited first, whereas the majority of late processing proteins are in PNBs (Savino et al., 1999). It is noticeable that Bop1 and B23 are in the same PNBs (Figure 2), as well as Nop52 (our unpublished data). Fibrillarin is also in PNBs before recruitment to the nucleolus during a short period (Savino et al., 2001). We cannot exclude the possibility that early and late processing proteins are in the same PNBs during this short period (∼10 min) of telophase as visible in Figure 2.
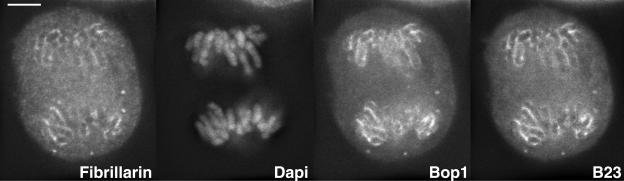
Figure 1.
Relative distribution of early (fibrillarin) and late (Bop1 and B23) rRNA processing proteins during anaphase. Immunolocalization of fibrillarin and B23 in a permanently transfected GFP-Bop1 cell. The chromosomes are visualized by DAPI. The same focal plane is shown after deconvolution. Fibrillarin, GFP-Bop1, and protein B23 show the same distribution, mainly at the chromosome periphery and some dispersed in the cytoplasm. On chromosomes, the distribution is homogeneous and the few foci visible correspond to bending of chromosomes. A 3D reconstruction comprising all the focal planes of the same cell is presented in Video 1. Bar, 10 μm.
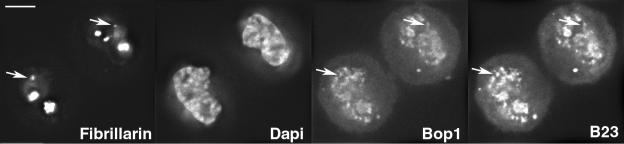
Figure 2.
Relative distribution of early (fibrillarin) and late (Bop1 and B23) rRNA processing proteins during early G1. Immunolocalization of fibrillarin and B23 in a permanently transfected GFP-Bop1 cell. The chromosomes are visualized by DAPI. The same focal plane is shown after deconvolution. Fibrillarin is recruited in the nucleoli and few foci are visible outside the nucleoli (arrows). Protein GFP-Bop1 and B23 show the same distribution in PNBs at the chromosome periphery and weak accumulation in nucleoli. The fibrillarin foci colocalize with Bop1 and B23 PNBs (arrows). Bar, 10 μm.
PNBs Contain Both Early and Late rRNA Processing Proteins in Telophase
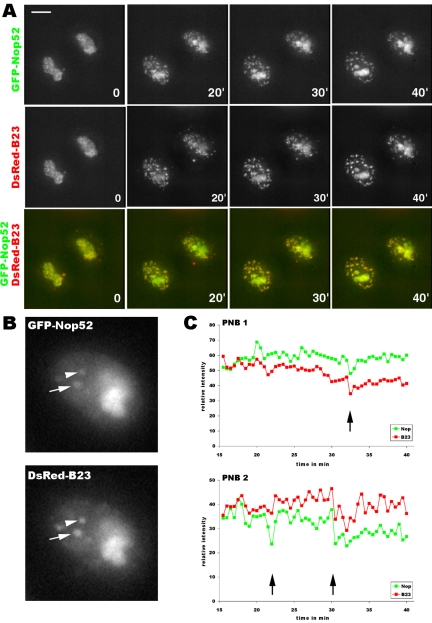
We assessed the relative dynamics of early and late rRNA processing proteins at the time of PNB formation. Using cells coexpressing GFP-fibrillarin and DsRed-B23, we analyzed in the same living cell the kinetics of translocation of the two proteins by rapid two-color 4D imaging (volume + time for both proteins). Mitotic cells expressing a similar and low level of both tagged proteins were selected, eliminating cells with high expression. We assume that GFP-fibrillarin is functional because the permanent cell line was selected on the basis of identical distribution for endogenous and GFP-fibrillarin, of normal cell cycle, and of the level of GFP-fibrillarin (Savino et al., 2001). In all the cells (n = 24) studied, once near the poles, one or two min after the onset of telophase, numerous bright fluorescent foci containing both GFP-fibrillarin and DsRed-B23 became visible almost simultaneously (Figure 3 and Video 2). In particular, in corresponding focal planes, the same foci contained GFP-fibrillarin and DsRed-B23 and looked orange in the color merge (Figure 3, arrow, and Video 2). To confirm this observation, the dynamics of this colocalization was measured. GFP and DsRed were quantified in the same foci and compared with dispersed proteins in the same nucleus (as described in Materials and Methods). The identification of foci corresponding to PNBs versus incipient nucleoli was based on the gradual growth of the structures with time; incipient nucleoli increased in size and intensity contrary to PNBs (see GFP-fibrillarin, Figure 3). Also, several tens of PNBs are formed, but only a maximum of six incipient nucleoli in this cell line (Roussel et al., 1996). Starting in telophase, 12 PNBs were analyzed in four different cells for periods lasting 20 min (Figure 4). The relative amount of B23 in PNBs was 5 to 6 times that of the dispersed proteins (Figure 4, compare continuous and dotted red curves). Interestingly, the amount of fibrillarin in these PNBs was 3 to 4 times that of dispersed proteins for ∼10 min (Figure 4, compare continuous and dotted green curves). After this time, fibrillarin was released, whereas B23 was still present in PNBs, either decreasing or remaining at high levels (Figure 4A, 19′, and graphs B and C). This clearly illustrates the presence of the two types of nucleolar processing proteins in the same PNBs and suggests differential sorting of these proteins.
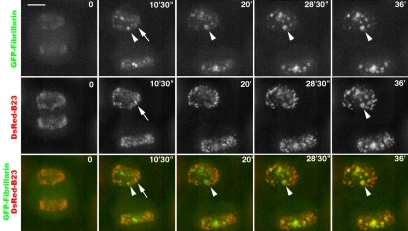
Figure 3.
Different dynamics of fibrillarin and B23 in the same living cell during nucleolar assembly. Two-color 4D imaging from telophase to early G1. At time 0 (i.e., 9′ 30″ after the onset of anaphase) both GFP-fibrillarin and DsRed-B23 are in PNBs; in comparison, the same focal planes with the merged picture indicates that the same PNBs contain both GFP-fibrillarin and DsRed-B23 (arrow at 10′ 30″). At 10′ 30″, fibrillarin is already delivered to incipient nucleoli (arrowhead), whereas DsRed-B23 is recruited in nucleoli later, at 28′ 30″. Differential sorting of fibrillarin and B23 from PNBs is visualized in merged focal planes by a progression with time from orange to red of PNBs that have lost fibrillarin and by progression of nascent nucleoli from green to yellow. See Video 2 for kinetics starting in metaphase. Bar, 10 μm.
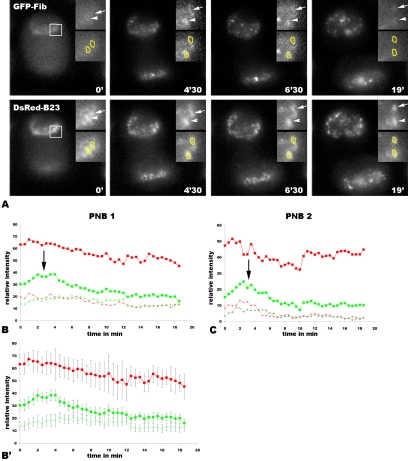
Figure 4.
Differential sorting of GFP-fibrillarin/DsRed-B23 from the same PNBs. (A) Time-lapse sequence from three consecutive focal planes containing two PNBs (white square) visible in Figure 3 and Video 2. Time 0 correspond to 9′ 30″ after the onset of anaphase visualized by chromosome segregation. Enlarged PNBs are shown in the inserts (PNB1, arrow, and PNB2, arrowhead). PNBs containing DsRed-B23 are visible during the time-lapse sequence. GFP-fibrillarin rapidly accumulates in the same PNBs during short time. (B and C) Graphs represent relative fluorescence of GFP and DsRed in each PNB during 19 min. The curves with large red squares represent DsRed-B23 in PNB, and the green squares represent GFP-fibrillarin. The dotted red and green curves represent DsRed and GFP in an area without PNBs, i.e., defined as diffuse proteins. The black arrows in B and C indicate, in B23-containing PNBs, 4 to 5 times higher fibrillarin concentrations compared with dispersed proteins. (B′) The same DsRed-B23 and GFP-fibrillarin curves with standard deviations (vertical bars), respectively, indicate that the relative intensity of GFP-fibrillarin is significantly higher in PNB during 5 min compared with diffuse proteins.
Accordingly, in all the cells studied, the timing of recruitment of these two proteins into incipient nucleoli was different (Figures 2 and 3 and Video 2). In the example shown, 10 min after the onset of telophase, GFP-fibrillarin was seen in incipient nucleoli, whereas in the same nucleoli, DsRed-B23 was detected only 20 min later (Figure 3 and Video 2).
Similar Dynamics of Nop52 and B23
The dynamics of GFP-Nop52 and DsRed-B23 were analyzed simultaneously in the same living cells. Both proteins participate in processing of the internal transcribed spacer 2 of rRNAs (Savkur and Olson, 1998; Savino et al., 1999). We found similar dynamics and flows of both proteins within the same PNBs when simultaneously observed by two-color 4D microscopy (Figure 5A and Video 3). The dynamics of this colocalization was quantified in the same PNBs as described for fibrillarin and B23. Starting in telophase, eight PNBs were analyzed in four different cells for periods lasting 25 min. The relative amounts of Nop52 and B23 in PNBs were very similar (Figure 5C, compare red and green curves). Interestingly, the amounts of both proteins varied simultaneously in particular when decreasing (Figure 5C, arrows). This could indicate that these late processing proteins are released from PNBs as complexes and could interact along the pathway and be recruited together into nucleoli (see Figure 5 legend for details). This prompted us to investigate the interactions of these proteins from the chromosome periphery to PNBs and nucleoli.
Figure 5.
Similar kinetics of GFP-Nop52/DsRed-B23. (A) Time-lapse sequence from telophase to early G1. At time 0 (i.e., 20′ after the beginning of anaphase), GFP-Nop52 and DsRed-B23 show the same dynamics during the formation of PNBs as well as during recruitment of these proteins and accumulation in nucleoli. In the same focal planes, images of GFP and DsRed are exactly superimposable. The bright dots outside the nucleus correspond to the nucleolar derived foci (NDF). These bodies moved within the cytoplasm (as described by Dundr et al., 2000 and Video 3). In NDF, the concentration of B23 is higher than of Nop52, red foci are visible in merge images (Figure 5A, 0 and 20′). The time-lapse images were recorded in auto-scale that consequently decreased the general intensity level when bright foci are included in the images. This is the case of B23 during the first 20 min and because there is no Nop52 foci, the signals are brighter for Nop52 in the nucleus. See Video 3 for kinetics. Bar, 10 μm. (B) Enlargement of a nucleus to show the PNBs in three consecutive merged optical sections that were analyzed (PNB1, arrow, and PNB2, arrowhead). (C) Relative fluorescence intensity from 15 to 40 min. The green curves correspond to GFP-Nop52 and red curves to Dsred-B23. The black arrows indicate similar fluctuations for both proteins, suggesting simultaneous release.
Interaction of Nop52 and B23 in Nucleoli
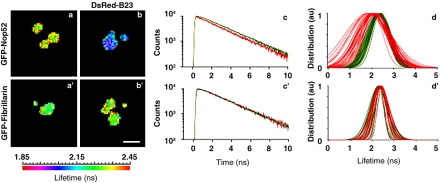
To determine whether Nop52 associates with B23 along the PNB pathway, tdFLIM was used to monitor FRET between GFP-Nop52 (donor) and DsRed-B23 (acceptor). If donor and acceptor are in proximity (typically <7 nm for the GFP/DsRed couple), FRET reduces the fluorescence lifetime of GFP. First, FRET was determined between GFP-Nop52 and DsRed-B23 in nucleoli. A significant reduction in the fluorescence lifetime of GFP-Nop52 in the presence of DsRed-B23, compared with the fluorescence lifetime of the donor in the absence of the acceptor, was detected as seen in the fluorescence lifetime pseudocolored maps (Figure 6, a and b). In contrast, there was no reduction in the fluorescence lifetime of the donor when GFP-fibrillarin was coexpressed with DsRed-B23 (Figure 6b′), compared with its fluorescence lifetime measured in the absence of the acceptor (Figure 6a′). The absence of FRET between GFP-fibrillarin and DsRed-B23 was expected because fibrillarin is concentrated in the dense fibrillar component of nucleoli, whereas B23 is localized in the granular component, preventing direct interaction between fibrillarin and B23. Importantly, the fact that no reduction of fluorescence lifetime could be detected in nucleoli coexpressing GFP-fibrillarin and DsRed-B23, compared with nucleoli expressing only GFP-fibrillarin, means that the green fluorescence coming from either the immature DsRed or from intramolecular FRET between immature (green fluorescence) and mature (red fluorescence) DsRed (Cotlet et al., 2001; Tramier et al., 2002) was not detected in our conditions. Consequently, the reduction of the fluorescence lifetime of GFP-Nop52 measured in the presence of DsRed-B23 does not arise from a DsRed-associated artifact. In addition, it was not possible to detect FRET between two late processing proteins, GFP-Bop1 and DsRed-B23 (our unpublished data), indicating that colocalization in the nucleolar granular component is not sufficient to record a positive FRET signal. Together, these results indicate that in nucleoli, FRET occurs between GFP-Nop52 and DsRed-B23.
Figure 6.
During interphase, Nop52 and protein B23 interact in the nucleolus of living cells. tdFLIM-FRET measurements were carried out by acquiring fluorescence decay images of the GFP donor (515 nm < λ < 560 nm) in permanent cell lines expressing GFP-Nop52 (a–d) or GFP-fibrillarin (a′–d′), alone (a, a′, and green curves) or in the presence of the DsRed acceptor after transfection with DsRed-B23 (b, b′, and red curves). The tdFLIM images were obtained by analyzing pixel by pixel the fluorescence decays with a single lifetime and are displayed as fluorescence lifetime pseudocolored maps (a, a′, b, and b′). The lifetime between 2.45 and 1.85 ns is indicated by colors presented in the scale. The nucleolus-associated fluorescence decay of GFP-tagged proteins is visible and compared in c and c′ to donor alone (green curves, GFP-Nop52 and GFP-fibrillarin, respectively) and to donor with acceptor (red curves, DsRed-B23). Fits of these fluorescence decays were carried out using a Gaussian distribution lifetime model, and the complete results are plotted in d for Nop52GFP alone (green curves, n = 53) and in the presence of B23DsRed (red curves, n = 35), and in d′ for GFP-fibrillarin alone (green curves, n = 22) and in the presence of DsRed-B23 (red curves, n = 21). Bar, 10 μm.
The kinetics of decay of nucleolus-associated fluorescence in the presence or absence of DsRed-B23, of, respectively, GFP-Nop52 (Figure 6c) and GFP-fibrillarin (Figure 6c′) was analyzed by Gaussian distributions of the fluorescence lifetime (Figure 6, d and d′, respectively). Each bell-shaped curve represents the lifetime distributions of GFP-Nop52 fluorescence (Figure 6d) and of GFP-fibrillarin fluorescence (Figure 6d′) within a given single nucleolus, measured in the absence (green curves) and in the presence (red curves) of the acceptor (DsRed-B23). The entire fluorescence lifetime distribution of GFP-Nop52 measured in the presence of DsRed-B23 in several nucleoli (n = 35) was shifted toward short fluorescence lifetime values compared with the distribution measured in the absence of DsRed-B23 (n = 53) (Figure 6d). This was accompanied by a concomitant broadening of the fluorescence lifetime distribution (Figure 6d). In contrast, the fluorescence lifetime distributions of GFP-fibrillarin measured in the absence (n = 22) and presence (n = 21) of DsRed-B23 display complete overlap with each other (Figure 6d′). The shift of the center of the fluorescence lifetime distribution in nucleoli of GFP-Nop52/DsRed-B23 cells (from 2.33 ± 0.05 ns in control GFP-Nop52 cells to 2.04 ± 0.06 ns) makes it possible to calculate a mean FRET efficiency of 12.4% between GFP-Nop52 and DsRed-B23 in nucleoli of living cells (from the expression: E = 1 – τDA/τD, with E being the FRET efficiency, τDA the fluorescence life-time of the donor in the presence of the acceptor, and τD the fluorescence lifetime of the donor alone). This is to be compared with the absence of shift between the center of the fluorescence lifetime distribution in nucleoli of GFP-fibrillarin/DsRed-B23 cells (2.36 ± 0.04 ns) and that in nucleoli of control GFP-fibrillarin cells (2.34 ± 0.04 ns). The broadening of the lifetime distribution in nucleoli of GFP-Nop52/DsRed-B23 cells provides evidence of an increasing heterogeneity of GFP lifetime due to variable situations of GFP-Nop52/DsRed-B23 proximity/orientation in the same nucleolus.
Late rRNA Processing Proteins Already Interact in PNBs
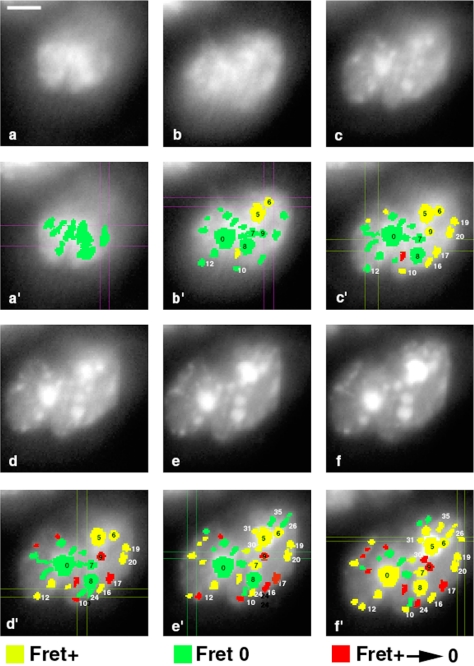
Because it is possible to detect FRET between B23 and Nop52 in nucleoli, we decided to track FRET during the recruitment of these proteins into nucleoli from anaphase to early G1. The principles of the analysis are presented in Figure 7, i.e., acquisition of GFP fluorescence decay images in living cells, then manual drawing of the regions of interest around the GFP signals and analysis of the FRET in these regions throughout recording. Time-lapse tdFLIM-FRET measurements were carried out by acquiring fluorescence decay images of the GFP donor in GFP-Nop52/DsRed-B23 along the PNB pathway from late anaphase to the early G1 (Figure 8, a–f); only one of the two daughter cells is presented in Figure 8. We carried out FRET analysis for 35 PNBs and nucleoli in the same living cell. Analysis of the experimental data showed that FRET was never detected during anaphase at the periphery of the chromosomes (Figure 8a′), whereas FRET was registered in 20% PNBs at the beginning of telophase (Figure 8b′), in ∼40% at the end of telophase (Figure 8, c′ and d′), and in 55% in early G1 (Figure 8, e′ and f′). Thus, interaction between GFP-Nop52 and DsRed-B23 was established progressively in PNBs, as the number of PNBs exhibiting FRET increased. Such data indicate that Nop52 and B23 did not interact until they were recruited in PNBs. It is noteworthy that a given PNB can alternatively present FRET or not present FRET (for example, see PNB 16, Figure 8).
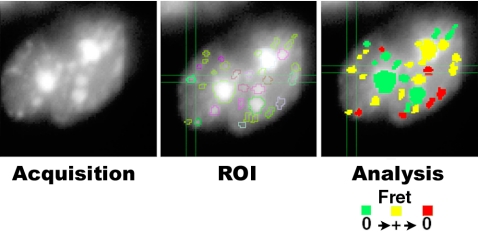
Figure 7.
Detection of FRET in regions of interest. Global acquisition of tdFLIM data is displayed as total fluorescence intensity image to manually draw the different ROI for the analysis of FRET. Fluorescence decays coming from these regions were then individually fitted with a Marquardt nonlinear least-square algorithm by using a Gaussian distribution of lifetimes. The region is green for negative FRET (no variation in the center of the fluorescence lifetime distribution of the GFP-tagged protein in the presence of the DsRed-tagged protein compared with the control experiment of GFP-tagged protein alone); it is yellow when FRET occurs (reduction in the position of the center of the fluorescence lifetime distribution of the GFP-tagged protein superior or equal to 200 ps); and it is red if a region was considered negative after having been positive.
Figure 8.
Time-lapse of the Nop52/B23 interactions studied during nucleolus assembly by tdFLIM-FRET. Time-lapse tdFLIM-FRET measurements were carried out by acquiring fluorescence decay images of the GFP donor (515 nm < λ < 560 nm) in permanent cell lines expressing GFP-Nop52 in the presence of the acceptor (DsRed) of protein DsRed-B23. The measurement was carried out continuously for the 120-min acquisition time from late anaphase to early G1. The data were separated in six parts of 20 min each to build up time-lapse tdFLIM data. Time-lapse total fluorescence intensity images (equivalent to steady state fluorescence intensity images) are displayed in a (t = 0–20 min), b (t = 20–40 min), c (t = 40–60 min), d (t = 60–80 min), e (t = 80–100 min), and f (t = 100–120 min). The fluorescence decays of the regions of interest (numbered from 0 to 35) corresponding to PNBs or incipient nucleoli were obtained by extraction from each set of data. Using a Gaussian distribution of lifetime as model fit, decays were analyzed and sorted into three FRET groups: negative (green), positive (yellow), and negative after having been positive (red). Visualization of time-lapse tdFLIM-FRET are done in a′ (t = 0–20 min), b′ (t = 20–40 min), c′ (t = 40–60 min), d′ (t = 60–80 min), e′ (t = 80–100 min), and f′ (t = 100–120 min). Bar, 10 μm.
DISCUSSION
The rRNA processing proteins are maintained at the periphery of the chromosomes during mitosis. At the mitosis/interphase transition, recruitment of the rRNA processing machinery to transcription sites is not direct but involves the formation of large complexes, the PNBs (Stevens, 1965; Ochs et al., 1985; Jiménez-Garcia et al., 1994; Savino et al., 1999, 2001; Dundr et al., 2000). This is a general phenomenon that occurs in most cells, but the role of these large complexes in the delivery of the rRNA processing machinery is presently unclear.
The formation of PNBs depends on the inactivation of the cyclin-dependent kinase (CDK) 1-cyclin B, and recruitment in the nucleoli of early and late processing proteins is differentially regulated (Sirri et al., 2002). The processing proteins cotranscriptionally associated with rRNA such as fibrillarin are recruited first, followed by proteins involved in late steps of processing such as B23 and Nop52 (Savino et al., 2001; Leung et al., 2004). Accordingly, in telophase no granular component is visible in incipient nucleoli (Hernandez-Verdun et al., 1980). PNB formation could contribute to the temporal order of recruitment of the processing complexes. This temporal order could be established either by different PNBs with different lifetimes or by differential sorting from the same PNBs. The former possibility has been the preferred one so far (Ochs et al., 1985; Jiménez-Garcia et al., 1994; Savino et al., 2001). In the present study, comparing the kinetics of early and late processing proteins in the same cells, we obtained evidence that initially both early and late processing proteins are concentrated in the same PNBs from which they are differentially sorted. In retrospect, given the short time window of the events described here (∼10 min), it was important to compare the kinetics in the same cells by using the rapid two-color 4D microscopy approach, and this explains why this mechanism was not reported previously.
An important question concerns the timing of the formation of the rRNA processing protein complexes. Here, we investigated during nucleolar assembly, the interaction of Nop52 and B23, proteins involved in the processing of 60S ribosome particles. The choice of these proteins was dictated by their function in the internal transcribed spacer 2 and because immunoprecipitation indicated that they are in the same complexes (our unpublished data). In yeast, the recent advent of proteomic analyses demonstrates that rRNA processing implies large protein complexes associated sequentially to nascent ribosomal particles. These complexes are specific of each processing step and are different for 40 and 60S ribosomal subunits (Dragon et al., 2002; Fatica and Tollervey, 2002; Fromont-Racine et al., 2003; Horsey et al., 2004). In mammals, the characterization of these rRNA processing complexes lags far behind that in yeast. However, it is possible to take advantage of the high conservation of some rRNA processing proteins between yeast and humans to determine the roles of the human proteins. Rrp1p is the yeast homologue of the human Nop52 (Savino et al., 1999; Horsey et al., 2004). A proteomic analysis of the Rrp1p partners has demonstrated that the preribosome particles contain at least 28 nonribosomal proteins necessary for the production of 60S (Horsey et al., 2004). Interestingly, Erb1p, the yeast homologue of the human Bop1 (Pestov et al., 2001a), was detected in the complex. No yeast homologue of B23 has been identified, but this protein is known to be involved in the processing of the ITS2 (Savkur and Olson, 1998) as Nop52 and Bop1.
To analyze protein interactions in living cells, energy transfer and consequently the distance between GFP and DsRed of tagged processing proteins was determined using tdFLIM. Monitoring FRET by tdFLIM has the advantage of being independent of chromophore concentration and lightpath length (Bastiaens and Squire, 1999; Selvin, 2000), as opposed to steady-state fluorescence intensity-based measurements. The use of a time-correlated single-photon counting method to perform tdFLIM (Emiliani et al., 2003; Tramier et al., 2003) provides the additional advantage of allowing precise determination of short fluorescence lifetimes and of detecting small variations in these lifetimes due to the excellent time resolution (∼100 ps) of the fluorescence decay kinetics. Moreover, this technique is minimally invasive because it requires only a very low level of laser excitation intensity (Gautier et al., 2001; Tramier et al., 2002). It is therefore very well suited to carry out time-lapse FRET imaging microscopy in living cells during extended periods.
In contrast to other GFP-tagged proteins for which fluorescence lifetimes have already been analyzed in living cells (Gautier et al., 2001; Tramier et al., 2002, 2003), the fluorescence decays of nucleolar GFP-tagged proteins GFP-fibrillarin, GFP-Nop52, GFP-Bop1, and GFP-B23 could not be fitted with a single fluorescence lifetime model in the absence of the acceptor (our unpublished data). This particularity is not understood and might be due to various conformational constraints on GFP when embedded in huge macromolecular complexes such as nucleoli and PNBs. The fact that the fluorescence decay of the donor (GFP-Nop52) was not monoexponential prevented a quantitative analysis of FRET (determination of the intrinsic FRET efficiency and of the ratio of bound and unbound donor). Such an analysis indeed requires the existence of a single fluorescence lifetime of GFP in the absence of acceptor that would allow the determination of both FRET parameters (Tramier et al., 2002; Emiliani et al., 2003). Here, the presence of FRET between GFP-Nop52 and DsRed-B23 was deduced from the shift between the center of the GFP-Nop52 lifetime distribution in the nucleoli and PNBs of GFP-Nop52/DsRed-B23 cells and of GFP-Nop52–containing cells. The associated broadening of the lifetime distribution suggests the existence of a distribution of the true FRET efficiency between GFP-Nop52 and DsRed-B23 (conformational changes within the nucleus, i.e., distance/orientation changes between donor and acceptor), and/or the existence of various relative amounts of bound and unbound GFP-Nop52 within a single nucleolus or PNB. Time-lapse tdFLIM experiments showed that the FRET signal is transient within a single PNB. This could arise from either transient interactions between GFP-Nop52 and DsRed-B23 within a single PNB or stable interactions but release of the GFP-Nop52/DsRed-B23 complexes toward the nucleoli. Both explanations reveal unanticipated dynamics within the PNB compartment. However, the fact that the steady-state intensity of GFP within a single PNB also decreased during nucleoli reconstruction suggests that the release of GFP-Nop52 and DsRed-B23 toward the nucleoli most likely occurs in the form of stable complexes. This hypothesis also supported by the simultaneous periodic decrease of both proteins from PNBs observed in two-color 4D imaging (Figure 5C).
The formation of PNBs occurs during telophase. Given the dynamic nature of many of the nucleolar proteins composing these bodies (Phair and Misteli, 2000; Huang, 2002), it is surprising that these structures exist at precisely this period of the cell cycle. As proposed for other nuclear bodies (Janicki and Spector, 2003), high-affinity binding sites could induce the formation of the PNBs. This is exactly what was found in the present study. During translocation of the rRNA processing proteins, interactions between GFP-Nop52 and DsRed-B23 were detected in PNBs and not at the chromosome periphery where these two proteins colocalize. Hence, the capacity of these two proteins to interact is modified at the telophase/early G1 transition and is initiated in PNBs. Such interactions between Nop52 and B23 occur in PNBs even before their recruitment in the incipient nucleoli (Figure 9). The presence of rRNAs as well as of small nucleolar RNAs has been demonstrated in PNBs (Jiménez-Garcia et al., 1994; Verheggen et al., 1998; Dousset et al., 2000). The interaction of B23 with rRNAs depends on phosphorylation of the protein by CDK (Okuwaki et al., 2002). In particular, the RNA binding activity of B23 is disrupted by CDK1-cyclin B during mitosis. Thus, the presence of rRNAs could contribute to the assembly of the rRNA processing complexes in PNBs when CDK1-cyclin B is inactivated, an event that normally occurs at the end of mitosis. The release of late processing proteins (B23 and Nop52) from PNBs (not the early ones) seems to be regulated by a presently uncharacterized CDK (Sirri et al., 2002). However, it is impossible to predict what inhibits this PNB release because the interactions depending on phosphorylation of this CDK are not characterized. One possible explanation could be that the late processing machinery assembled with rRNAs in PNBs is activated by this unknown CDK, thus becoming able to process rRNA and consequently be released from PNBs. This model could explain why formation of PNBs occurs only during telophase/early G1 transition and why the recruitment of late processing proteins takes longer than that of early ones. However, uncovering the mechanisms that govern the recruitment of the nucleolar processing machinery will require further investigation of the interactions occurring in PNB complexes and of the transit between PNBs and the nucleolus.
Figure 9.
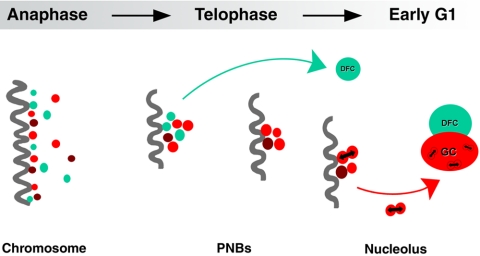
Kinetics of rRNA processing proteins during nucleolar reconstruction. During anaphase, proteins of early (fibrillarin, green spots) and late (Nop52 and B23, red spots) rRNA processing machineries colocalize at the periphery of chromosomes (dark fiber). At the beginning of telophase, these proteins regroup in PNBs; early and late rRNA processing proteins pass through the same PNBs. Recruitment of rRNA processing proteins in the nucleolus then occurs according to differential sorting from the same PNBs of early (fibrillarin into dense fibrillar component; DFC) and late (Nop52 and B23 into granular component; GC) processing proteins (see Results). As a consequence, at the beginning of telophase, PNBs contain all the rRNA processing proteins. In contrast, after recruitment of the early rRNA processing proteins by the incipient nucleolus, PNBs only retain the late rRNA processing proteins. Interactions detected between proteins of the same rRNA processing machinery (visualized by bidirectional arrows between Nop52 and B23) in both PNBs and nucleolus suggest that PNBs are preassembly platforms for rRNA processing complexes.
In conclusion, we observed that early and late rRNA processing proteins pass through the same PNBs, that differential sorting from the same PNBs ensures differential nucleolar recruitment, and that interactions between protein partners occur in PNBs. We therefore propose that PNBs function as assembly platforms of rRNA processing complexes.
Supplementary Material
Acknowledgments
We are grateful to Drs. S. Huang (Northwestern University Medical School, Chicago, IL) and D. G. Pestov (University of Illinois at Chicago College of Medicine, Chicago, IL) for gifts of probes and to S. Chamot for help with cell cultures. We thank A.-L. Haenni for critical reading of the manuscript. This work was supported in part by the Centre National de la Recherche Scientifique, the Association pour la Recherche sur le Cancer (contracts 4290 to D.H.-V. and 4421 to M.C.-M.), and the Fondation pour la Recherche Médicale (ICP1.36 to M.C.-M.).
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E05–01–0041) on April 6, 2005.
Abbreviations used: DsRed, Discosoma red fluorescence protein; FRET, fluorescence resonance energy transfer; GFP, green fluorescence protein; PNB, prenucleolar body; ROI, regions of interest; tdFLIM, time domain fluorescence lifetime imaging microscopy.
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
References
- Azum-Gélade, M.-C., Noaillac-Depeyre, J., Caizergues-Ferrer, M., and Gas, N. (1994). Cell cycle redistribution of U3 snRNA and fibrillarin. Presence in the cytoplasmic nucleolus remnant and in the prenucleolar bodies at telophase. J. Cell Sci. 107, 463–475. [DOI] [PubMed] [Google Scholar]
- Bastiaens, P. I., and Squire, A. (1999). Fluorescence lifetime imaging microscopy: spatial resolution of biochemical processes in the cell. Trends Cell Biol. 9, 48–52. [DOI] [PubMed] [Google Scholar]
- Bell, P., Dabauvalle, M. C., and Scheer, U. (1992). In vitro assembly of prenucleolar bodies in Xenopus egg extract. J. Cell Biol. 118, 1297–1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bubulya, P. A., Prasanth, K. V., Deerinck, T. J., Gerlich, D., Beaudouin, J., Ellisman, M. H., Ellenberg, J., and Spector, D. L. (2004). Hypophosphorylated SR splicing factors transiently localize around active nucleolar organizing regions in telophase daughter nuclei. J. Cell Biol. 167, 51–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, D., and Huang, S. (2001). Nucleolar components involved in ribosome biogenesis cycle between the nucleolus and nucleoplasm in interphase cells. J. Cell Biol. 153, 169–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chubb, J. R., Boyle, S., Perry, P., and Bickmore, W. A. (2002). Chromatin motion is constrained by association with nuclear compartments in human cells. Curr. Biol. 12, 439–445. [DOI] [PubMed] [Google Scholar]
- Cotlet, M., Hofkens, J., Habuchi, S., Dirix, G., Van Guyse, M., Michiels, J., Vanderleyden, J., and De Schryver, F. C. (2001). Identification of different emitting species in the red fluorescent protein DsRed by means of ensemble and single-molecule spectroscopy. Proc. Natl. Acad. Sci. USA 98, 14398–14403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dousset, T., Wang, C., Verheggen, C., Chen, D., Hernandez-Verdun, D., and Huang, S. (2000). Initiation of nucleolar assembly is independent of RNA polymerase I transcription. Mol. Biol. Cell 11, 2705–2717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dragon, F., et al. (2002). A large nucleolar U3 ribonucleoprotein required for 18S ribosomal RNA biogenesis. Nature 417, 967–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dundr, M., Misteli, T., and Olson, M.O.J. (2000). The dynamics of postmitotic reassembly of the nucleolus. J. Cell Biol. 150, 433–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emiliani, V., Sanvitto, D., Tramier, M., Piolot, T., Petrasek, Z., Kemnitz, K., Durieux, C., and Coppey-Moisan, M. (2003). Low-intensity two-dimensional imaging of fluorescence lifetimes in living cells. Appl. Phys. Lett. 83, 2471–2473. [Google Scholar]
- Fatica, A., and Tollervey, D. (2002). Making ribosomes. Curr. Opin. Cell Biol. 14, 313–318. [DOI] [PubMed] [Google Scholar]
- Fromont-Racine, M., Senger, B., Saveanu, C., and Fasiolo, F. (2003). Ribosome assembly in eukaryotes. Gene 313, 17–42. [DOI] [PubMed] [Google Scholar]
- Gall, J. G. (2000). Cajal bodies: the first 100 years. Annu. Rev. Cell Dev. Biol. 16, 273–300. [DOI] [PubMed] [Google Scholar]
- Gautier, I., Tramier, M., Durieux, C., Coppey, J., Pansu, R. B., Nicolas, J. C., Kemnitz, K., and Coppey-Moisan, M. (2001). Homo-FRET microscopy in living cells to measure monomer-dimer transition of GFT-tagged proteins. Biophys. J. 80, 3000–3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautier, T., Dauphin-Villemant, C., André, C., Masson, C., Arnoult, J., and Hernandez-Verdun, D. (1992a). Identification and characterization of a new set of nucleolar ribonucleoproteins which line the chromosomes during mitosis. Exp. Cell Res. 200, 5–15. [DOI] [PubMed] [Google Scholar]
- Gautier, T., Robert-Nicoud, M., Guilly, M.-N., and Hernandez-Verdun, D. (1992b). Relocation of nucleolar proteins around chromosomes at mitosis - a study by confocal laser scanning microscopy. J. Cell Sci. 102, 729–737. [DOI] [PubMed] [Google Scholar]
- Hadjiolov, A. A. (1985). The Nucleolus and Ribosome Biogenesis, Vienna, Springer.
- Hernandez-Verdun, D., Bourgeois, C. A., and Bouteille, M. (1980). Simultaneous nucleologenesis in daughter cells during late telophase. Biol. Cell 37, 1–4. [Google Scholar]
- Hernandez-Verdun, D., Roussel, P., and Gébranne-Younès, J. (2002). Emerging concepts of nucleolar assembly. J. Cell Sci. 115, 2265–2270. [DOI] [PubMed] [Google Scholar]
- Horsey, E. W., Jakovljevic, J., Miles, T. D., Harnpicharnchai, P., and Woolford, J. L., Jr. (2004). Role of the yeast Rrp1 protein in the dynamics of pre-ribosome maturation. RNA 10, 813–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, S. (2002). Building an efficient factory: where is pre-rRNA synthesized in the nucleolus? J. Cell Biol. 157, 739–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janicki, S. M., and Spector, D. L. (2003). Nuclear choreography: interpretations from living cells. Curr. Opin. Cell Biol. 15, 149–157. [DOI] [PubMed] [Google Scholar]
- Jiménez-Garcia, L. F., Segura-Valdez, M. L., Ochs, R. L., Rothblum, L. I., Hannan, R., and Spector, D. L. (1994). Nucleologenesis: U3 snRNA-containing prenucleolar bodies move to sites of active pre-rRNA transcription after mitosis. Mol. Biol. Cell 5, 955–966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamond, A. I., and Earnshaw, W. C. (1998). Structure and function in the nucleus. Science 280, 547–553. [DOI] [PubMed] [Google Scholar]
- Lamond, A. I., and Spector, D. L. (2003). Nuclear speckles: a model for nuclear organelles. Nat. Rev. Mol. Cell. Biol. 4, 605–612. [DOI] [PubMed] [Google Scholar]
- Leung, A. K., Andersen, J. S., Mann, M., and Lamond, A. I. (2003). Bioinformatic analysis of the nucleolus. Biochem. J. 376, 553–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung, A. K., Gerlich, D., Miller, G., Lyon, C., Lam, Y. W., Lleres, D., Daigle, N., Zomerdijk, J., Ellenberg, J., and Lamond, A. I. (2004). Quantitative kinetic analysis of nucleolar breakdown and reassembly during mitosis in live human cells. J. Cell Biol. 166, 787–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matera, A. G. (1999). Nuclear bodies: multifaceted subdomains of the interchromatin space. Trends Cell Biol. 9, 302–309. [DOI] [PubMed] [Google Scholar]
- Mélèse, T., and Xue, Z. (1995). The nucleolus: an organelle formed by the act of building a ribosome. Curr. Opin. Cell Biol. 7, 319–324. [DOI] [PubMed] [Google Scholar]
- Misteli, T. (2000). Cell biology of transcription and pre-mRNA splicing: nuclear architecture meets nuclear function. J. Cell Sci. 113, 1841–1849. [DOI] [PubMed] [Google Scholar]
- Ochs, R. L., Lischwe, M. A., Shen, E., Caroll, R. E., and Busch, H. (1985). Nucleologenesis: composition and fate of prenucleolar bodies. Chromosoma 92, 330–336. [DOI] [PubMed] [Google Scholar]
- Okuwaki, M., Tsujimoto, M., and Nagata, K. (2002). The RNA binding activity of a ribosome biogenesis factor, nucleophosmin/B23, is modulated by phosphorylation with a cell cycle-dependent kinase and by association with its subtype. Mol. Biol. Cell 13, 2016–2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson, M.O.J., Dundr, M., and Szebeni, A. (2000). The nucleolus: an old factory with unexpected capabilities. Trends Cell Biol. 10, 189–196. [DOI] [PubMed] [Google Scholar]
- Pederson, T. (1998). The plurifunctional nucleolus. Nucleic Acids Res. 26, 3871–3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestov, D. G., Stockelman, M. G., Strezoska, Z., and Lau, L. F. (2001a). ERB1, the yeast homolog of mammalian Bop1, is an essential gene required for maturation of the 25S and 5.8S ribosomal RNAs. Nucleic Acids Res. 29, 3621–3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestov, D. G., Strezoska, Z., and Lau, L. F. (2001b). Evidence of p53-dependent cross-talk between ribosome biogenesis and cell cycle: effects of nucleolar protein Bop1 on G1/S transition. Mol. Cell. Biol. 21, 4246–4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phair, R. D., and Misteli, T. (2000). High mobility of proteins in the mammalian cell nucleus. Nature 404, 604–609. [DOI] [PubMed] [Google Scholar]
- Prasanth, K. V., Sacco-Bubulya, P. A., Prasanth, S. G., and Spector, D. L. (2003). Sequential entry of components of gene expression machinery into daughter nuclei. Mol. Biol. Cell 14, 1043–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimer, G., Pollard, K. M., Pennig, C. A., Ochs, R. L., Lischwe, M. A., Busch, H., and Tan, E. M. (1987). Monoclonal autoantibody from NZB/NZW F1 mouse and some human scleroderma sera target a Mr 34000 nucleolar protein of the U3-ribonucleoprotein particle. Arthritis Rheum. 30, 793–800. [DOI] [PubMed] [Google Scholar]
- Roussel, P., André, C., Comai, L., and Hernandez-Verdun, D. (1996). The rDNA transcription machinery is assembled during mitosis in active NORs and absent in inactive NORs. J. Cell Biol. 133, 235–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savino, T. M., Bastos, R., Jansen, E., and Hernandez-Verdun, D. (1999). The nucleolar antigen Nop52, the human homologue of the yeast ribosomal RNA processing RRP1, is recruited at late stages of nucleologenesis. J. Cell Sci. 112, 1889–1900. [DOI] [PubMed] [Google Scholar]
- Savino, T. M., Gébrane-Younès, J., De Mey, J., Sibarita, J.-B., and Hernandez-Verdun, D. (2001). Nucleolar assembly of the rRNA processing machinery in living cells. J. Cell Biol. 153, 1097–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savkur, R. S., and Olson, M.O.J. (1998). Preferential cleavage in pre-ribosomal RNA by protein B23 endoribonuclease. Nucleic Acids Res. 26, 4508–4515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selvin, P. R. (2000). The renaissance of fluorescence resonance energy transfer. Nat. Struct. Biol. 7, 730–734. [DOI] [PubMed] [Google Scholar]
- Sirri, V., Hernandez-Verdun, D., and Roussel, P. (2002). Cyclin-dependent kinases govern formation and maintenance of the nucleolus. J. Cell Biol. 156, 969–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spector, D. L. (1993). Macromolecular domains within the cell nucleus. Annu. Rev. Cell Biol. 9, 265–315. [DOI] [PubMed] [Google Scholar]
- Spector, D. L. (2001). Nuclear domain. J. Cell Sci. 114, 2891–2893. [DOI] [PubMed] [Google Scholar]
- Stevens, B. (1965). The fine structure of the nucleolus during mitosis in the grasshopper neuroblast cell. J. Cell Biol. 24, 349–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tramier, M., Gautier, I., Piolot, T., Ravalet, S., Kemnitz, K., Coppey, J., Durieux, C., Mignotte, V., and Coppey-Moisan, M. (2002). Picosecond-hetero-FRET microscopy to probe protein-protein interactions in live cells. Biophys. J. 83, 3570–3577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tramier, M., Piolot, T., Gautier, I., Mignotte, V., Coppey, J., Kemnitz, K., Durieux, C., and Coppey-Moisan, M. (2003). Homo-FRET versus hetero-FRET to probe homodimers in living cells. Methods Enzymol. 360, 580–597. [DOI] [PubMed] [Google Scholar]
- Verheggen, C., Le Panse, S., Almouzni, G., and Hernandez-Verdun, D. (1998). Presence of pre-rRNAs before activation of polymerase I transcription in the building process of nucleoli during early development of Xenopus laevis. J. Cell Biol. 142, 1167–1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verheggen, C., Le Panse, S., Almouzni, G., and Hernandez-Verdun, D. (2001). Maintenance of nucleolar machineries and pre-rRNAs in remnant nucleolus of erythrocyte nuclei and remodeling in Xenopus egg extracts. Exp. Cell Res. 269, 23–34. [DOI] [PubMed] [Google Scholar]
- Zatsepina, O. V., Todorov, I. T., Philipova, R. N., Krachmarov, C. P., Trendelenburg, M. F., and Jordan, E. G. (1997). Cell cycle-dependent translocations of a major nucleolar phosphoprotein, B23, and some characteristics of its variants. Eur. J. Cell Biol. 73, 58–70. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.