Abstract
Mono-, di-, and trimethylation of specific histone residues adds an additional level of complexity to the range of histone modifications that may contribute to a histone code. However, it has not been clear whether different methylated states reside stably at different chromatin sites or whether they represent dynamic intermediates at the same chromatin sites. Here, we have used recently developed antibodies that are highly specific for mono-, di-, and trimethylated lysine 9 of histone H3 (MeK9H3) to examine the subnuclear localization and replication timing of chromatin containing these epigenetic marks in mammalian cells. Me1K9H3 was largely restricted to early replicating, small punctate domains in the nuclear interior. Me2K9H3 was the predominant MeK9 epitope at the nuclear and nucleolar periphery and colocalized with sites of DNA synthesis primarily in mid-S phase. Me3K9H3 decorated late-replicating pericentric heterochromatin in mouse cells and sites of DAPI-dense intranuclear heterochromatin in human and hamster cells that replicated throughout S phase. Disruption of the Suv39h1,2 or G9a methyltransferases in murine embryonic stem cells resulted in a redistribution of methyl epitopes, but did not alter the overall spatiotemporal replication program. These results demonstrate that mono-, di-, and trimethylated states of K9H3 largely occupy distinct chromosome domains.
INTRODUCTION
Histone methylation has emerged as a primary epigenetic mark, central to the regulation of local and global chromatin structure. In particular, lysine 9 methylation of histone H3 (MeK9H3) has been correlated with both local and global repression of transcription and the formation of large constitutive heterochromatin domains. The complexity that can be achieved with this one modification alone is quite remarkable. K9H3 residues can be either mono-, di- or trimethylated (Waterborg, 1993; Santos-Rosa et al., 2002; Peters et al., 2003; Rice et al., 2003; Wang et al., 2003; Zhang et al., 2003). At least five methyltransferases have been shown to methylate K9H3: Suv39h1 and Suv39h2 (Rea et al., 2000; Peters et al., 2001), G9a (Tachibana et al., 2001), ESET/SETDB1 (Schultz et al., 2002), and EuHMTase1 (Ogawa et al., 2002). These enzymes have different affinities for the un-, mono- or dimethylated states and produce different methylation states; some may act only on previously methylated lysines, whereas others may carry out de novo methylation. In vivo, these enzymes differentially methylate histones in euchromatin versus heterochromatin, and knockout mice for Suv39h1,2 and G9a have very different phenotypes, suggesting that methyltransferases may be differentially targeted to specific chromatin contexts for regulatory purposes.
Attempts to determine whether these different methylation states reside stably at different chromatin sites have not produced a clear picture. Imunolocalization experiments in mouse cells have revealed that the majority of Me3K9H3 is localized to the prominent clusters of pericentromeric heterochromatin (chromocenters) that are observed in this species, whereas the mono- and dimethylation states could not be resolved (Peters et al., 2003; Rice et al., 2003). Here, we have localized the epitopes for each of these antibodies relative to sites of coordinated DNA synthesis (replicon clusters) at different times during S phase. These ∼1-Mb segments of coordinately replicated DNA constitute one of the most recognizable subnuclear units of chromosome organization (Jackson and Pombo, 1998; Ma et al., 1998; Dimitrova and Gilbert, 1999; Zink et al., 1999; Leonhardt et al., 2000; Sadoni et al., 2004). Because specific classes of chromosomal domains replicate at particular times, the combination of spatial and temporal separation provides an added dimension of resolution to the subnuclear localization of specific epitopes (Wu et al., 2004). Furthermore, because cells exploit the spatio-temporal separation of replication to assemble different types of chromatin at different times during S phase (Bozhenok et al., 2002; Gilbert, 2002; Zhang et al., 2002; McNairn and Gilbert, 2003), one can infer a relationship between spatiotemporal localization and function. We find that mono-, di-, and trimethylated K9H3 can be distinguished by the spatiotemporal properties of the domains in which they reside, indicating that they are largely targeted to different chromosomal domains. These results strongly suggest that each of the three states of MeK9H3 serve distinct functions within the nucleus.
MATERIALS AND METHODS
Cell Culture
C127 (mouse mammary tumor), CHOC400, HeLa S3 were all grown in DMEM (Life Technologies, Rockville, MD) with appropriate supplements at 37°C in 5% CO2. Medium for C127 was supplemented with 10% fetal calf serum (FCS; Hyclone, Logan, UT). Medium for CHOC 400 was supplemented with 5% FCS and 1% nonessential amino acids (Life Technologies). Medium for HeLa S3 was supplemented with 10% cosmic calf serum (Hyclone). ES cells were grown on GMEM (Life Technologies) supplemented with 5% newborn calf serum (NCS; Hyclone), 5% FCS, 1% nonessential amino acid, 2 mM l-glutamine, 0.1 mM β-mercaptoethanol, and 500 U/ml leukemia inhibitor factor (LIF; Chemicon, Temecula, CA). ES cells were subcultured every day and fresh medium was always changed on the same day as the experiment. Suv39h1/h2 dn (double null) ES cells were constructed by first targeting the single, X-linked Suv39h1 allele, followed by sequential targeting of the two Suv39h2 alleles in XY HM1 ES cells. A complete description of the Suv39h1/h2dn null ES cell line will be given in Kourmouli et al. (unpublished results).
Labeling of Nascent DNA with Nucleotide Analogs in Cultured Cells
To follow the spatiotemporal patterns of DNA replication in cells, exponentially growing cells on 12-mm-diameter round coverslips were pulse-labeled with 10 μg ml–1 CldU (5-chloro-2′-deoxyuridine; Sigma-Aldrich, St. Louis, MO) for 10 min and, after different chase times, pulse-labeled with 10 μgml–1 IdU (5-iodo-2′-deoxyuridine; Sigma-Aldrich) for 10 min. Cells were then fixed and stored in ice-cold ethanol. To examine colocalization between K9 methyl groups and replication foci, cells were pulse-labeled with 30 μg ml–1 BrdU (5-bromo-2′-deoxyuridine; Sigma-Aldrich) for 15 min before fixing in 2% paraformaldehylde in phosphate-buffered saline (PBS) for 10 min at room temperature.
Antibodies and Detection of Labeled DNA and K9 Methyl Groups
Secondary antibodies in this study are Alexa-Fluor 488– or Alexa-Fluor 594– conjugated (Molecular Probes, Eugene, OR) and dilution ranges from 1:200 to 1:800 dilutions for general use. Primary antibodies for detection of CldU and IdU substituted DNAs were rat monoclonal anti-BrdU antibody (Accurate Chemicals, Westbury, NY; OBT0030) and mouse monoclonal antibody (Becton-Dickinson, Mountain View, CA; 347580), respectively. The staining was performed as described (Dimitrova and Gilbert, 1999). To examine colocalization between K9 methyl groups and BrdU, paraformaldehydefixed cells were permeabilized by incubating with 0.2% Triton X-100 in PBS for 5 min. Then cells were incubated in blocking buffer (3% bovine serum albumin and 0.5% Tween 20 in PBS) for 20 min before a 45-min to 1-h incubation with K9H3 mono-, di-, or trimethylation antibodies (raised in rabbit and 1:1000 diluted with blocking buffer; Peters et al., 2003). After incubation with secondary antibody and washing, the cells were refixed with 4% paraformaldehyde in PBS for 15 min. Cells were then incubated with 0.3 M glycine in PBS for 5 min, 0.5% NP40 in PBS for 15 min, and 1.5 N HCl for 30 min, washing with PBS between each step, before the incubation with mouse monoclonal anti-BrdU antibody for 1 h. After application of the secondary antibody for BrdU detection, all coverslips were incubated with 0.02 μgml–1 4′,6-diamidino-2-phenyl-indole (DAPI) in PBS/T (0.5% Tween 20 in PBS) for 5 min followed by three washes with PBS/T. All coverslips were mounted on slides with Vectashield (Vector Laboratories, Burlingame, CA).
Microscopy
Stained specimens were observed with a Nikon Labophot-2 microscope (Melville, NY) equipped with a 100× 1.4 NA oil immersion Nikon PlanApo objective and epifluorescence images were collected with a CCD camera (SPOT RT Slider, Diagnostic Instruments, Sterling Heights, MI). For deconvolution microscopy, images were collected with 0.5-μm Z-series, using the same apparatus, and then deconvolved using Autodeblur software version 9.1 (AutoQuant Imaging, Watervliet, NY). To examine colocalization between K9H3 methyl groups and BrdU, a single optical section through the middle of each nucleus was sequentially scanned with a Bio-Rad MRC-1024 confocal microscope (Richmond, CA) that is mounted on a Nikon Eclipse 600 microscope. To avoid picking specific targets, nuclei from consecutive fields are scanned. Each nucleus was designated a replication pattern according to the BrdU confocal image. The merged confocal images were then subjected to colocalization analysis with LaserPix software (Bio-Rad) to determine the “coefficient of colocalization.” The line intensity analysis of MeK9H3 across the nucleus was analyzed using LaserPix software.
RESULTS
Different Methylated States of K9H3 Localize to Spatiotemporally Distinct Chromosome Domains
The spatial distribution and replication timing of chromatin containing specific proteins can be determined by pulse-labeling cells with BrdU and coimmunostaining with antibodies against BrdU and the protein of interest (Wu et al., 2004). Because the spatial patterns of DNA synthesis are unique to specific times during S phase, this analysis can be carried out without the need for cumbersome cell synchronization methods, whose efficacy can vary significantly between cell types and can often perturb or distort cell cycle progression (Wu et al., 2004). Therefore, this approach allows one to compare different cell lines using the same methodology. However, the distribution of replication sites and the temporal order in which they appear during S phase can differ significantly in different cell lines, so it is important to carefully characterize these patterns in each cell line before such analysis.
We have previously described a pulse-chase-pulse method to accurately define the temporal order and length of replication patterns in any given cell line (Dimitrova and Gilbert, 1999). Figure 1 shows an example of such an analysis in mouse C127 cells. Asynchronously growing cells were pulse-labeled with CldU, chased for various lengths of time, pulse-labeled with IdU, and then stained with red (CldU) and green (IdU) fluorochrome-conjugated antibodies. Six distinct patterns of replication were discerned in C127, termed I-VI (Figure 1A). When the IdU and CldU images were merged (Figure 1B), each pattern was reproducibly followed by the subsequent pattern in a logical progression. Because the length of the chase period sufficient for nuclei to switch from one spatial pattern to the next is proportional to the duration of each pattern, a temporal map of these patterns could be constructed (Figure 1, C and D).
Figure 1.
Characterization of the spatial patterns of replication in mouse C127 Cells. (A) Asynchronous cultures were pulse-labeled with BrdU and stained with anti-BrdU antibodies. Shown are deconvolution images of each labeling pattern (I–VI). DAPI shows intense staining at the positions of mouse chromocenters, as well as visible staining at the nuclear periphery. Chromocenters are generally found to associate with nucleoli, visualized as DNA-free areas that are devoid of DAPI or BrdU staining. (B) Asynchronous cultures were pulse-labeled with CldU and chased for the indicated times before they were pulse-labeled with IdU. Sites of CldU (red) and IdU (green) incorporation were visualized using CldU- and IdU-specific antibodies. Representative confocal images from this “pulse-chase-pulse” experiment are shown. Similar to other mammalian cell lines (O'Keefe et al., 1992; Dimitrova and Gilbert, 1999; Dimitrova and Berezney, 2002), DNA synthesis begins at many small, discrete foci in the internal, euchromatic region of the nucleus, excluding the nucleoli (and associated chromocenters) and nuclear periphery region (pattern I). In pattern II, replication continues throughout the euchromatic region, but also is observed in the perinucleolar and nuclear periphery regions. Pattern III is characterized by decreased euchromatic foci in the interior and increased replication foci at the nuclear (and nucleolar) periphery. By the middle of S phase, most euchromatic foci have finished replication and DNA synthesis begins within the pericentric heterochromatin (pattern IV). Thereafter, replication at the nuclear periphery becomes more predominant, coinciding with the replication of a few internal but non-pericentric domains (pattern V). Finally, small numbers of large speckles are observed in both the interior and periphery of the nuclei (Pattern VI). In B, examples of nuclei chased for the indicated time periods are shown. Because each replicon cluster takes 45–60 min to complete DNA synthesis (Ma et al., 1998; Dimitrova and Gilbert, 1999; Leonhardt et al., 2000), there is substantial colocalization of IdU and CldU after 15 min, much less colocalization at 45 min, and no detectable colocalization after 1 h. After longer pulses, some nuclei can be seen to exit S phase into G2 phase and no longer incorporate IdU (e.g., right-most panel; 3 Hrs.). (C) The percentage of each type of CldU-labeled nuclei (I–VII) that had proceeded to subsequent stages of S phase (or to G2 phase) during the chase was scored. Results shown were derived from an experiment in which more than 200 S phase nuclei were scored for each chase period. (D) Schematic diagram approximating the duration of each replication pattern, derived by doubling the time for 50% of nuclei to move from one pattern to the subsequent pattern and taking into account the total length of the C127 S phase (12 h).
To determine the spatial distribution and replication timing of chromatin containing different methyl K9H3 epitopes, C127 cells were pulse-labeled with BrdU and then immunostained with anti-BrdU antibodies and one of three recently developed anti-methyl K9H3 antibodies generated against branched mono-, di-, and trimethylated peptides. Antibodies previously generated against different types of methyl K9H3 epitopes have been found to display different degrees of specificity for mono-, di-, and trimethylated K9, as well as different degrees of cross-reactivity with other methylated lysine epitopes or other proteins (Perez-Burgos et al., 2004). The antibodies used here have been extensively evaluated for cross-reactivity by ELISA, peptide arrays, and Western blotting, as well as peptide competition in Westerns and immunofluorescence (Peters et al., 2003; Perez-Burgos et al., 2004) and are highly specific for their respective antigen.
Confocal microscopy revealed significant differences in spatial distribution and replication timing of chromatin enriched for mono-, di-, and trimethyl K9H3 (Figure 2). The monomethylated antibody decorated many small punctate foci within the interior of the nucleus, excluding the nuclear periphery, nucleoli, and associated chromocenters. Nucleoli and chromocenters were identified by their well-defined spatial replication patterns and by staining with DAPI (Figure 1A). Colocalization of Me1K9H3 with sites of DNA synthesis was found almost exclusively during the first half of S phase. In contrast, foci containing Me2K9H3 were generally larger than those harboring Me1K9H3, and the overall pattern of their distribution was quite different. Me2K9H3 was more concentrated at the nuclear periphery and also decorated perinucleolar regions (more easily seen in Figure 4), in addition to a subset of internal foci. Colocalization of Me2K9H3 with BrdU was detected throughout most of S phase, unlike for Me1K9H3. Me3K9H3 was very strongly enriched in the pericentric heterochromatin, with only a minority of small punctate foci found at other sites throughout the nucleus, consistent with previous reports (Cowell et al., 2002; Peters et al., 2003; Rice et al., 2003). Pericentric chromatin containing this epitope replicated almost exclusively during the mid-late period of S phase.
Figure 2.
Replication timing of MeK9H3 chromatin in C127 cells. Asynchronous C127 cells were pulse-labeled with BrdU and then stained with antibodies to BrdU (red) and either mono-, di-, or trimethylated K9H3 (green). Shown are single optical sections through the center of each nucleus obtained by dual-color confocal laser scanning microscopy as described in Materials and Methods. Bar, 10 μm.
Figure 4.
Spatial distribution of MeK9H3 epitopes in mouse, hamster, and human cells. C127, CHO, and HeLa cells were stained with anti-MeK9H3 antibodies as in Figure 2 and counterstained with DAPI. To avoid comparisons of cells with different relative amounts of chromatin replicated at different times during S phase, cells in G1 phase (small, BrdU negative nuclei) were compared. Images were subjected to deconvolution analysis and pseudocolored before merging. Hamster and human cells do not contain the intensely DAPI-dense chromocenters that are characteristic of mouse cells. Nonetheless, nucleoli can be easily identified as DAPI-negative regions surrounded by more DAPI-dense DNA. Colocalization of DAPI-dense regions (red) with MeK9H3 (green) is found almost exclusively with Me2K9H3 and Me3K9H3, and each modification displays a unique spatial distribution in the nucleus. Bar, 10 μm.
To quantitatively assess the replication timing of chromatin containing these epitopes, we determined the “coefficient of colocalization” for each of 60 confocal images from two independent staining experiments (Figure 3). This analysis evaluates the amount of yellow coloration relative to the total amount of one of the fluorochromes. Colocalization relative to the anti-methylated epitope (Figure 3, A and C) is proportional to the fraction of each methylated epitope that is engaged in replication at each brief pulse-labeling time during S phase. From these results, we conclude that Me1K9H3 is replicated primarily in the first half of S phase, Me2K9H3 is replicated throughout most of S phase, and Me3K9H3 is replicated primarily in mid- to late-S phase. On the other hand, colocalization relative to anti-BrdU is proportional to the fraction of synchronously replicated domains that are decorated with each epitope. This analysis reveals that most of the DNA synthesized in the first half of S phase is packaged into chromatin containing Me1K9H3, and most of the DNA synthesized in the middle of S phase is the Me3K9H3-containing chromocenters, confirming the results in Figure 3, A and B. However, this analysis emphasizes that, although roughly equal proportions of the Me2K9H3 replicate at most times during S phase, most of the DNA synthesized in the last 3 h of S phase contains Me2K9H3. Taken together, these differences in morphology, location and replication timing strongly suggest that the majority of these K9H3 methylation states are segregated into separate chromosomal domains.
Figure 3.
Analysis of the degree of colocalization of MeK9H3 epitopes with DNA synthesized at each of three stages of S phase. Confocal images (from the experiment shown in Figure 2) of a single optical section through the middle of each nucleus were subjected to colocalization analysis with LaserPix software to determine the “coefficient of colocalization” (the amount of yellow coloration relative to either red or green). This was normalized to the highest value within each antibody group and displayed as either a line graph (A and B) versus the time during S phase of each spatial pattern (as determined in Figure 1C) or as a bar graph (C and D), dividing S phase into three spatiotemporal groups: early (E), middle (M), and late (L). In A and C the coefficient of colocalization relative to the total amount of signal for each MeK9H3 epitope was evaluated. In B and D, the coefficient of colocalization relative to the total amount of BrdU was evaluated. For each methylation state, at least 60 S phase nuclei were analyzed. Error bars show the SE of the mean.
Spatiotemporal Organization of MeK9H3 States in Different Cell Lines
Our results suggest that the Me1K9H3 marks early replicating euchromatin, Me2K9H3 marks perinuclear and perinucleolar heterochromatin, whereas Me3K9H3 is largely restricted to pericentric heterochromatin. Because the presence of such prominent pericentric compartments in mouse cells is not observed in most other mammalian species, we wanted to determine whether the same properties of these epitopes would be found in cell lines from other species. Two cell lines have been extensively characterized for the temporal order of spatial patterns, Chinese hamster ovary (CHO) cells (O'Keefe et al., 1992; Dimitrova and Gilbert, 1999) and human HeLa cells (O'Keefe et al., 1992). Figure 4 shows a comparison of the spatial distribution of the three states of MeK9H3 in C127, CHO, and HeLa, relative to DAPI staining. In all three cell lines, Me1K9H3 was localized to a large number of small foci distributed throughout the interior of the nucleus, mostly or completely excluded from DAPI-dense areas. In contrast, Me2K9H3 was much more prominent than Me1K9H3 at the nuclear and nucleolar periphery, as revealed by the more extensive colocalization with these DAPI-stained regions. Me3K9H3 localized to more clustered foci that were not enriched at the nuclear and nucleolar periphery but which colocalized extensively with DAPI-dense regions. These results demonstrate that Me1K9H3 resides within intranuclear regions that stain poorly with DAPI, Me2K9H3 stains DAPI-dense DNA at the nuclear and nucleolar periphery, and Me3K9H3 stains predominantly intranuclear DAPI-dense DNA.
Figures 5 and 6 illustrate the colocalization of MeK9H3 states with sites of DNA synthesis in CHO and HeLa. Only five distinguishable spatial patterns are observed in these cell lines. The six patterns observed in mouse cells are likely due to the unique organization of chromocenters that creates an additional distinguishable pattern. In both lines, Me1K9H3 replication was largely restricted to the first half of S phase (% of methyl epitope; Figures 5B and 6B), and the percentage of simultaneously replicating foci colocalizing with Me1K9H3 is highest early in S phase (% of BrdU label; Figures 5C and 6C). In contrast, most Me2K9H3-rich domains were replicated in early-middle S phase, just at the onset of the transition to late replication, whereas a high percentage of DNA replicated at this time, as well as several hours later in S, was associated with Me2K9H3. For Me3K9H3, some cell-type differences were observed. Me3K9H3-containing domains in HeLa cells colocalized best with domains replicated in the middle of S phase, whereas a similar percentage of DNA replicated at all times was associated with Me3K9H3. However, CHO cells replicated a similar fraction of Me3K9H3 at all times during S phase, whereas virtually all the DNA replicated late in S phase was associated with Me3K9H3. A comparison to Figure 3 also reveals subtle differences in these replication profiles of Me2K9H3 and Me3K9H3, but not Me1K9H3, with C127 cells. These subtle differences may reflect differences in the relative amounts of DNA replicated at different times during S phase in different cell lines, or they may reflect true differences in the proportion of Me2K9H3 and Me3K9H3 replicated at different times during S phase. Regardless, these results demonstrate that the three different states of MeK9H3 are differentially enriched in separate chromatin domains in cell lines derived from different mammalian species.
Figure 5.
Replication timing of MeK9H3 chromatin in HeLa cells. (A) Asynchronous HeLa cells were pulse-labeled with BrdU and stained as in Figure 2. Replication patterns were identified as per the previously described patterns for this cell line (O'Keefe et al., 1992). (B and C) At least 60 nuclei displaying each pattern of replication were subjected to colocalization analysis as shown in Figure 3. Bar in A, 10 μm.
Figure 6.
Replication timing of MeK9H3 chromatin in CHO cells. (A) Asynchronous CHO cells were pulse-labeled with BrdU and stained as in Figure 2. Replication patterns were identified as per the previously described patterns for this cell line (O'Keefe et al., 1992; Dimitrova and Gilbert, 1999). (B and C) At least 60 nuclei displaying each pattern of replication were subjected to colocalization analysis as shown in Figure 3. Bar in A, 10 μm.
Redistribution of Methylation States Does Not Affect the Overall Spatiotemporal Replication Program
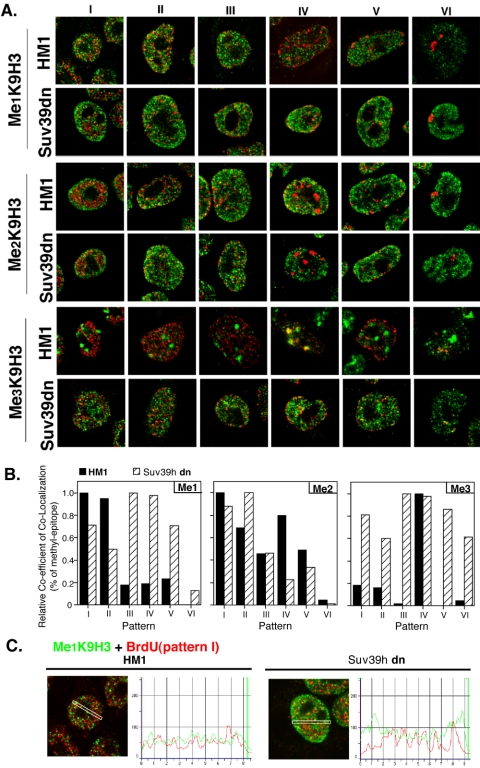
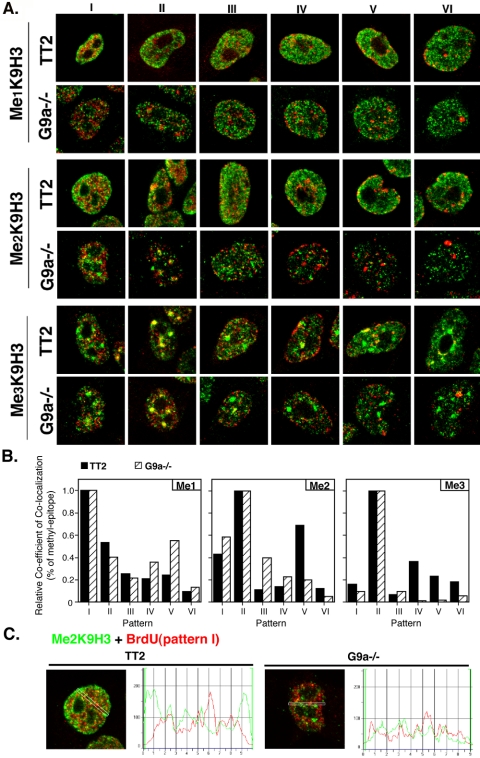
The results described above indicate that the three states of MeK9H3 reside in domains that are distinguishable by their replication time. Because replication timing can be influenced by chromatin modifying enzymes in yeast (Vogelauer et al., 2002; Zappulla et al., 2002; Aparicio et al., 2004), a question arises as to whether the different MeK9H3 states are involved in the determination of the temporal replication program. Recently, mouse embryonic stem (ES) cell lines containing targeted gene disruptions of either the Suv39h1,2 (Kourmouli et al., unpublished results) or G9a methyltransferases (Tachibana et al., 2002) have become available. These disruptions result in significant changes in the amount and localization of the affected modifications (Peters et al., 2003; Rice et al., 2003). We reasoned that these cell lines might reveal effects of histone methylation on replication timing. For these experiments, the spatiotemporal order of replication was first characterized in both the mutant cell types and the wild-type ES cell lines from which they were derived. As for mouse C127 cells, six distinct replication patterns in each line were identified (Figures 7 and 8). Interestingly, the two wild-type ES cell lines (HM1 and TT2) replicated their pericentric heterochromatin at measurably different times during S phase (HM1 in pattern IV, Figure 7 vs. TT2 in pattern II, Figure 8). Importantly, however, we did not detect differences in the overall spatiotemporal order of these patterns between mutant cell lines and their corresponding wild-type counterparts.
Figure 7.
Analysis of MeK9H3 localization and replication timing in HM1 versus Suv39h1,2 double null (Suv39h dn) mutants. The overall spatiotemporal sequence of replication patterns in HM1 and derivative Suv39hdn ES cells was first determined as shown in Figure 1 and was found to be similar to C127 (unpublished data). Cells were then pulse-labeled with BrdU and stained as in Figure 2. For Me3K9H3 staining of Suv39hdn, the confocal gain was increased relative to wild-type cells because of the low intensity of signal with this antibody. Shown in A are the merged confocal images and in B the colocalization analysis performed as in Figure 3. In C, a line intensity analysis for the region indicated with a rectangle is shown, comparing Me1K9H3 (green) with early replication patterns (red) to illustrate the enrichment of Me1K9H3 staining at the nuclear periphery in mutant relative to wild-type cells. Bar in A, 10 μm.
Figure 8.
Analysis of MeK9H3 localization and replication timing in TT2 versus G9a–/– mutants. TT2 and G9a–/– cells were analyzed as in Figure 7. For Me1K9H3 and Me2K9H3 staining of G9a–/–, the confocal gain was increased relative to wild-type cells because of the low intensity of signal with this antibody. Note that the spatiotemporal replication patterns are similar to those in HM1 and Suv39h dn, except that pericentric heterochromatin replicates earlier in these cell lines than in the other mouse lines in this report. However, this overall temporal order is not affected by the G9a disruption. Bar in A, 10 μm.
Figure 7 illustrates the results from the Suv39h1,2 double null (Suv39h dn) and corresponding HM1 wild-type line. Consistent with previously published results (Peters et al., 2003), Me1K9H3 in Suv39h dn ES cells was detected within pericentric heterochromatin at an equal, or occasionally greater intensity than the rest of the nucleus (unpublished data). However, our confocal analysis also detected Me1K9H3 at the nuclear periphery (Figure 7C). Importantly, while replication of chromatin containing Me1K9H3 primarily took place during the first two spatiotemporal periods in HM1, Me1K9H3 could be found in chromatin replicating at all times during S phase in the Suv39hdn. In contrast, we could find no significant differences in replication timing or distribution of Me2K9H3 between Suv39hdn and HM1. Finally, because the Suv39h1,2 knockout removes all detectable pericentric Me3K9H3 staining, which constitutes ∼75% of all Me3K9H3 in the cell (Peters et al., 2003; Rice et al., 2003), it provided an opportunity to evaluate the replication timing of non-pericentric Me3K9H3-containing chromatin that results from the activity of other methyltransferases. The overall Me3K9H3 staining was much reduced, as expected, but measurable (see legend to Figure 7). These non-pericentric, Me3K9H3-containing chromatin domains were found to replicate throughout S phase, suggesting that a class of Me3K9H3 domains, methylated by other methyltransferases, are replicated at various times during S phase, as was seen for CHO and HeLa cells (Figures 5 and 6).
Figure 8 illustrates the results from the G9a null and corresponding TT2 wild-type line. We found the overall signal for Me1K9H3 and Me2K9H3 to be significantly reduced but detectable, consistent with a 50–60% reduction in the total amount of these methylation states in these cell lines, as determined by mass spectrometry (Peters et al., 2003). The replication timing of chromatin containing the remaining Me1K9H3 and Me2K9H3 did not differ significantly from that of TT2. However, Me2K9H3 staining was no longer predominant at the nuclear periphery (Figure 8C) and was clearly visible within the early-middle S phase replicating pericentric heterochromatin. The amount, localization, and replication timing of Me3K9H3 domains was unchanged by the G9a mutation. Hence, the redistribution of methyl-epitopes resulting from disruptions in either G9a or Suv39h1,2 does not affect the overall spatiotemporal program for replication.
DISCUSSION
We conclude that mono-, di-, and trimethylation states of K9H3 largely reside in separate chromosome domains that can be distinguished by their distinct subnuclear localization and replication timing. Prior immunolocalization of these epitopes by conventional microscopy suggested that mono- and dimethyl epitopes coexist within euchromatin (Peters et al., 2003; Rice et al., 2003). By combining the spatial resolution of confocal and deconvolution analysis, with the temporal resolution achieved by replication timing analysis, we could resolve different spatiotemporal patterns for each of these three states. These patterns were generally preserved among different mammalian cell lines and species, suggesting that different states of K9H3 methylation may represent a well-conserved regulatory network in mammals. Altogether, our results suggest that Me1K9H3 primarily resides in early replicating euchromatin, whereas Me2K9H3 and Me3K9H3 are found within different types of heterochromatin, possibly distinguishing facultative and constitutive heterochromatin, respectively.
The differential enrichment of MeK9H3 epitopes in chromatin domains and the conservation of this overall spatiotemporal organization suggest that they each play unique roles in the structural and functional organization of chromosomes. This may involve differences in the interaction of each of these methylation states with specific cohorts of chromatin proteins, creating higher order levels of chromatin organization. However, our results do not imply that these different states are mutually exclusive; the resolution of our analysis does not rule out the existence of smaller domains or individual histone tails harboring one or more of the alternate methylation states. Instead, they reveal domain-wide differences in the overall density of these modifications that provide a global signature for each domain within which more localized levels of functional control must operate. Individual domains of chromatin enriched for specific histone modifications may help to separate independently regulated parts of the genome, possibly punctuated by boundary elements that prevent the spreading of one modification into neighboring domains (Litt et al., 2001a; Noma et al., 2001). The formation of such higher order domains has been shown to be essential for imprinting, dosage compensation, chromosome condensation and segregation (Bernard et al., 2001; Litt et al., 2001b; Noma et al., 2001; Delaval and Feil, 2004; Heard, 2004; Taddei et al., 2004).
Our results also demonstrate that the K9H3 methylation state alone is not sufficient to dictate spatial localization or replication timing of chromosome domains. First, mutations in two different methyltransferases resulted in a redistribution of methylation states, but did not change in the overall spatial organization and temporal order of replication of these domains. Second, the loss of Suv39h1,2 eliminates 75% of cellular Me3K9H3 (Peters et al., 2003), revealing that the remaining Me3K9H3-containing chromatin domains, apparently methylated by as yet unidentified enzymes, replicate at different times throughout S phase. Third, in Suv39h dn mutant cells Me1K9H3 replicated throughout S phase, demonstrating that this modification is not sufficient to dictate early replication. Finally, the subnuclear localization and temporal order of replication of different types of heterochromatin showed subtle variations in different cell lines and species.
Given the intimate link that has been shown between the structure and function of chromatin domains, their spatial organization within the nucleus (Williams and Fisher, 2003; Misteli, 2004; Taddei et al., 2004) and their organization into temporally distinct replication domains (Gilbert, 2002), it is somewhat surprising that a dramatic change in methyl epitope distribution had no detectable effect on replication timing and subnuclear position. However, these results do not rule out a role for these modifications in regulating replication timing. It is possible that multiple modifications work together to establish spatial and temporal control so that no single change can disrupt this control or that other modifications can compensate for the change in MeK9H3 state. Finally, our experiments cannot rule out the possibility that small or selective (localized) changes in replication timing are not detected in these experiments.
Our results also suggest insights into the activities and localization of some of the methyltransferases that mediate these modifications, which unfortunately have not been directly localized because of the lack of antibodies sufficient for immunolocalization of these enzymes (T. Jenuwein, personal communication). In the G9a knockout ES cells, mass spectrometry analysis demonstrated that total cellular Me1K9H3 and Me2K9H3 are reduced by 50–60% relative to wild-type, whereas Me3K9H3 levels are unchanged (Peters et al., 2003), consistent with the weaker but still detectable staining of Me1K9H3 and other Me2K9H3 observed here. This implies that enzymes maintain Me1K9H3 and Me2K9H3 at some domains in the absence of G9a. We did not see a change in the spatiotemporal distribution of these remaining Me1K9H3 domains, demonstrating that the monomethylated state largely resides in the early replicating euchromatic domains, regardless of which enzyme carries out the methylation reaction. Me2K9H3 staining, however, was no longer predominant at the nuclear periphery, suggesting that G9a is responsible for the Me2K9H3 in these peripheral, late-replicating domains. In addition, we detected some Me2K9H3 within the early-middle S phase replicating pericentric heterochromatin in G9a mutants. This pericentric Me2K9H3 localization appeared to be cell cycle–regulated, because it was found in early S phase but not late S phase cells. However, Rice et al. (2003) did not detect pericentric Me2K9H3 in these same cell lines, so the source of this discrepancy needs to be resolved before the significance of Me2K9H3 localization to chromocenters in G9a mutants can be appreciated.
In the Suv39hdn ES cells, we observed a low to moderate pericentric localization of Me1K9H3, consistent with previous results (Peters et al., 2003). Because our Suv39hdn ES cells were derived independently from those used by Peters et al. (2003), this confirms that redistribution of this epitope is not cell-line–specific but reflects the loss of Suv39h1,2 enzymes. Because mass spectrometry analysis detected a moderate increase in the total amount of Me1K9H3 (∼20%) in cells lacking Suv39h1,2 (Peters et al., 2003), this altered localization is consistent with an accumulation of Me1K9H3 at sites that are normally trimethylated by the Suv39h1,2 enzymes, which prefer monomethyl as a substrate (Rice et al., 2003). However, in contrast to ES cells, we and others (Rice et al., 2003) did not find pericentric localization of Me1K9H3 in Suv39h dn murine embryonic fibroblasts (MEFs; R. Wu, unpublished results). This observation suggests that the distribution of one or more methyltransferases may differ between ES cells and MEFs.
Intriguingly, our confocal analysis also revealed Me1K9H3 at the nuclear periphery in the Suv39hdn ES cells (as well as Suv39hdn MEFs; R. Wu, unpublished results), which was not detected in the corresponding wild-type cell line. This redistribution of Me1K9H3 was not detected by prior conventional microscopy (Peters et al., 2003; Rice et al., 2003; Perez-Burgos et al., 2004). Although we cannot rule out indirect consequences of Suv39h1,2 loss, it is tempting to consider the possibility that Suv39h1,2 plays an active role at the nuclear periphery. One possibility is that Suv39h1,2 predominantly dimethylates K9H3 at the periphery. This seems unlikely though, because we could find no differences in replication timing or loss of peripheral localization for Me2K9H3 between HM1 and Suv39h dn cells, and previous mass spectrometry did not detect a significant change in the total amount of Me2K9H3 (Peters et al., 2003).
An intriguing possibility is that there is a form of Suv39h1,2-mediated trimethylation at the periphery that goes undetected in the conditions used here. This would be consistent with the localization of HP1 proteins, which are known to recruit Suv39h1,2, at the nuclear periphery (Kourmouli et al., 2000, 2001; Polioudaki et al., 2001; Singh and Georgatos, 2002). Moreover, we recently reported that a different anti-Me3K9H3 antibody (Cowell antibody) strongly paints the nuclear periphery in C127 (Cowell et al., 2002), HM1, and murine embryonic fibroblasts cells (R. Wu, unpublished results). This antibody is also highly specific for the trimethylated state, and although some studies have detected cross-reactivity with Me3K27H3 (Tamaru et al., 2003; Perez-Burgos et al., 2004) and Me3K20H4 (Perez-Burgos et al., 2004), the independent localization of Me3K27H3 (Peters et al., 2003; R. Wu, unpublished results) and Me3K20H4 (Kourmouli et al., 2004; Schotta et al., 2004; R. Wu, unpublished results) do not suggest enrichment for these modifications at the periphery. Because the Cowell antibody was generated against a 20 amino acid linear trimethylated antigen (aa 1–20), whereas the Me3K9H3 antibody used in this report was generated against an 11 amino acid di-branched antigen (aa 5–15), it is tempting to speculate that the Cowell antibody is detecting Suv39h1,2-mediated Me3K9H3 at the nuclear periphery that does not present itself to the antibody used in this report. Resolution of this dilemma will require methods that do not rely on antibody specificity, such as the biochemical purification of different subnuclear compartments (Makatsori et al., 2004) and the evaluation of their constitution by mass spectrometry.
Acknowledgments
We thank T. Jenuwein and A. Peters for kind gifts of the antibodies used in this study before their publication. We also thank Y. Shinkai for providing G9a knockout ES cells; B. Aucott and A. Leskovar for help with growing ES cells; M. C. Cardoso, T. Yokochi, and T. Jenuwein for helpful comments on the manuscript; and G. Ring for assistance with the confocal facility. This work was supported by National Science Foundation grant MCB-0077507 and National Institutes of Health grant GM-57233 to D.M.G.
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E04–11–0997) on March 23, 2005.
References
- Aparicio, J. G., Viggiani, C. J., Gibson, D. G., and Aparicio, O. M. (2004). The Rpd3-Sin3 histone deacetylase regulates replication timing and enables intra-S origin control in Saccharomyces cerevisiae. Mol. Cell. Biol. 24, 4769–4780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard, P., Maure, J. F., Partridge, J. F., Genier, S., Javerzat, J. P., and Allshire, R. C. (2001). Requirement of heterochromatin for cohesion at centromeres. Science 2539–2541. [DOI] [PubMed]
- Bozhenok, L., Wade, P. A., and Varga-Weisz, P. (2002). WSTF-ISWI chromatin remodeling complex targets heterochromatic replication foci. EMBO J. 21, 2231–2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowell, I. G. et al. (2002). Heterochromatin, HP1 and methylation at lysine 9 of histone H3 in animals. Chromosoma 111, 22–36. [DOI] [PubMed] [Google Scholar]
- Delaval, K., and Feil, R. (2004). Epigenetic regulation of mammalian genomic imprinting. Curr. Opin. Genet. Dev. 14, 188–195. [DOI] [PubMed] [Google Scholar]
- Dimitrova, D. S., and Berezney, R. (2002). The spatio-temporal organization of DNA replication sites is identical in primary, immortalized and transformed mammalian cells. J. Cell Sci. 115, 4037–4051. [DOI] [PubMed] [Google Scholar]
- Dimitrova, D. S., and Gilbert, D. M. (1999). The spatial position and replication timing of chromosomal domains are both established in early G1-phase. Mol. Cell 4, 983–993. [DOI] [PubMed] [Google Scholar]
- Gilbert, D. M. (2002). Replication timing and transcriptional control: beyond cause and effect. Curr. Opin. Cell Biol. 14, 377–383. [DOI] [PubMed] [Google Scholar]
- Heard, E. (2004). Recent advances in X-chromosome inactivation. Curr. Opin. Cell Biol. 16, 247–255. [DOI] [PubMed] [Google Scholar]
- Jackson, D. A., and Pombo, A. (1998). Replicon clusters are stable units of chromosome structure: evidence that nuclear organization contributes to the efficient activation and propagation of S phase in human cells. J. Cell Biol. 140, 1285–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kourmouli, N., Dialynas, G., Petraki, C., Pyrpasopoulou, A., Singh, P. B., Georgatos, S. D., and Theodoropoulos, P. A. (2001). Binding of heterochromatin protein 1 to the nuclear envelope is regulated by a soluble form of tubulin. J. Biol. Chem. 276, 13007–13014. [DOI] [PubMed] [Google Scholar]
- Kourmouli, N. et al. (2004). Heterochromatin and tri-methylated lysine 20 of histone H4 in animals. J. Cell Sci. 117, 2491–2501. [DOI] [PubMed] [Google Scholar]
- Kourmouli, N., Theodoropoulos, P. A., Dialynas, G., Bakou, A., Politou, A. S., Cowell, I. G., Singh, P. B., and Georgatos, S. D. (2000). Dynamic associations of heterochromatin protein 1 with the nuclear envelope. EMBO J. 19, 6558–6568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonhardt, H., Rahn, H. P., Weinzierl, P., Sporbert, A., Cremer, T., Zink, D., and Cardoso, M. C. (2000). Dynamics of DNA replication factories in living cells. J. Cell Biol. 149, 271–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litt, M. D., Simpson, M., Gaszner, M., Allis, C. D., and Felsenfeld, G. (2001a). Correlation between histone lysine methylation and developmental changes at the chicken beta-globin locus. Science 293, 2453–2455. [DOI] [PubMed] [Google Scholar]
- Litt, M. D., Simpson, M., Recillas-Targa, F., Prioleau, M. N., and Felsenfeld, G. (2001b). Transitions in histone acetylation reveal boundaries of three separately regulated neighboring loci. EMBO J. 20, 2224–2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, H., Samarabandu, J., Devdhar, R. S., Acharya, R., Cheng, P., Meng, C., and Berezney, R. (1998). Spatial and temporal dynamics of DNA replication sites in mammalian cells. J. Cell Biol. 143, 1415–1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makatsori, D., Kourmouli, N., Polioudaki, H., Shultz, L. D., McLean, K., Theodoropoulos, P. A., Singh, P. B., and Georgatos, S. D. (2004). The inner nuclear membrane protein lamin B receptor forms distinct microdomains and links epigenetically marked chromatin to the nuclear envelope. J. Biol. Chem. 279, 25567–25573. [DOI] [PubMed] [Google Scholar]
- McNairn, A. J., and Gilbert, D. M. (2003). Epigenomic replication: linking epigenetics to DNA replication. Bioessays 25, 647–656. [DOI] [PubMed] [Google Scholar]
- Misteli, T. (2004). Spatial positioning; a new dimension in genome function. Cell 119, 153–156. [DOI] [PubMed] [Google Scholar]
- Noma, K., Allis, C. D., and Grewal, S. I. (2001). Transitions in distinct histone H3 methylation patterns at the heterochromatin domain boundaries. Science 293, 1150–1155. [DOI] [PubMed] [Google Scholar]
- O'Keefe, R. T., Henderson, S. C., and Spector, D. L. (1992). Dynamic organization of DNA replication in mammalian cell nuclei—spatially and temporally defined replication of chromosome-specific alpha-satellite DNA sequences. J. Cell Biol. 116, 1095–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa, H., Ishiguro, K., Gaubatz, S., Livingston, D. M., and Nakatani, Y. (2002). A complex with chromatin modifiers that occupies E2F- and Myc-responsive genes in G0 cells. Science 296, 1132–1136. [DOI] [PubMed] [Google Scholar]
- Perez-Burgos, L., Peters, A. H., Opravil, S., Kauer, M., Mechtler, K., and Jenuwein, T. (2004). Generation and characterization of methyl-lysine histone antibodies. Methods Enzymol. 376, 234–254. [DOI] [PubMed] [Google Scholar]
- Peters, A. H. et al. (2003). Partitioning and plasticity of repressive histone methylation states in mammalian chromatin. Mol. Cell 12, 1577–1589. [DOI] [PubMed] [Google Scholar]
- Peters, A. H. et al. (2001). Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell 107, 323–337. [DOI] [PubMed] [Google Scholar]
- Polioudaki, H., Kourmouli, N., Drosou, V., Bakou, A., Theodoropoulos, P. A., Singh, P. B., Giannakouros, T., and Georgatos, S. D. (2001). Histones H3/H4 form a tight complex with the inner nuclear membrane protein LBR and heterochromatin protein 1. EMBO Rep. 2, 920–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rea, S. et al. (2000). Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature 406, 593–599. [DOI] [PubMed] [Google Scholar]
- Rice, J. C., Briggs, S. D., Ueberheide, B., Barber, C. M., Shabanowitz, J., Hunt, D. F., Shinkai, Y., and Allis, C. D. (2003). Histone methyltransferases direct different degrees of methylation to define distinct chromatin domains. Mol. Cell 12, 1591–1598. [DOI] [PubMed] [Google Scholar]
- Sadoni, N., Cardoso, M. C., Stelzer, E. H., Leonhardt, H., and Zink, D. (2004). Stable chromosomal units determine the spatial and temporal organization of DNA replication. J. Cell Sci. 117, 5353–5365. [DOI] [PubMed] [Google Scholar]
- Santos-Rosa, H., Schneider, R., Bannister, A. J., Sherriff, J., Bernstein, B. E., Emre, N. C., Schreiber, S. L., Mellor, J., and Kouzarides, T. (2002). Active genes are tri-methylated at K4 of histone H3. Nature 419, 407–411. [DOI] [PubMed] [Google Scholar]
- Schotta, G., Lachner, M., Sarma, K., Ebert, A., Sengupta, R., Reuter, G., Reinberg, D., and Jenuwein, T. (2004). A silencing pathway to induce H3–K9 and H4–K20 trimethylation at constitutive heterochromatin. Genes Dev. 18, 1251–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz, D. C., Ayyanathan, K., Negorev, D., Maul, G. G., and Rauscher, F. J., 3rd. (2002). SETDB1, a novel KAP-1-associated histone H3, lysine 9-specific methyltransferase that contributes to HP1-mediated silencing of euchromatic genes by KRAB zinc-finger proteins. Genes Dev. 16, 919–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, P. B., and Georgatos, S. D. (2002). HP 1, facts, open questions, and speculation. J. Struct. Biol. 140, 10–16. [DOI] [PubMed] [Google Scholar]
- Tachibana, M., Sugimoto, K., Fukushima, T., and Shinkai, Y. (2001). Set domain-containing protein, G9a, is a novel lysine-preferring mammalian histone methyltransferase with hyperactivity and specific selectivity to lysines 9 and 27 of histone H3. J. Biol. Chem. 276, 25309–25317. [DOI] [PubMed] [Google Scholar]
- Tachibana, M. et al. (2002). G9a histone methyltransferase plays a dominant role in euchromatic histone H3 lysine 9 methylation and is essential for early embryogenesis. Genes Dev. 16, 1779–1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taddei, A., Hediger, F., Neumann, F. R., and Gasser, S. M. (2004). The function of nuclear architecture: a genetic approach. Annu. Rev. Genet. 38, 305–345. [DOI] [PubMed] [Google Scholar]
- Tamaru, H., Zhang, X., McMillen, D., Singh, P. B., Nakayama, J., Grewal, S. I., Allis, C. D., Cheng, X., and Selker, E. U. (2003). Trimethylated lysine 9 of histone H3 is a mark for DNA methylation in Neurospora crassa. Nat. Genet. 34, 75–79. [DOI] [PubMed] [Google Scholar]
- Vogelauer, M., Rubbi, L., Lucas, I., Brewer, B. J., and Grunstein, M. (2002). Histone acetylation regulates the time of replication origin firing. Mol. Cell 10, 1223–1233. [DOI] [PubMed] [Google Scholar]
- Wang, H., An, W., Cao, R., Xia, L., Erdjument-Bromage, H., Chatton, B., Tempst, P., Roeder, R. G., and Zhang, Y. (2003). mAM facilitates conversion by ESET of dimethyl to trimethyl lysine 9 of histone H3 to cause transcriptional repression. Mol. Cell 12, 475–487. [DOI] [PubMed] [Google Scholar]
- Waterborg, J. H. (1993). Dynamic methylation of alfalfa histone H3. J. Biol. Chem. 268, 4918–4921. [PubMed] [Google Scholar]
- Williams, R. R., and Fisher, A. G. (2003). Chromosomes, positions please! Nat. Cell Biol. 5, 388–3890. [DOI] [PubMed] [Google Scholar]
- Wu, R., Terry, A. V., and Gilbert, D. M. (2004). Observing nuclear structure/chromatin changes with fluorescently-labelled antibodies. In: Methods in Molecular Biology: Nuclear Reprogramming, vol., ed. S. Pells, Totowa, NJ: Humana Press (in press).
- Zappulla, D. C., Sternglanz, R., and Leatherwood, J. (2002). Control of replication timing by a transcriptional silencer. Curr. Biol. 12, 869–875. [DOI] [PubMed] [Google Scholar]
- Zhang, J., Xu, F., Hashimshony, T., Keshet, I., and Cedar, H. (2002). Establishment of transcriptional competence in early and late S phase. Nature 420, 198–202. [DOI] [PubMed] [Google Scholar]
- Zhang, X., Yang, Z., Khan, S. I., Horton, J. R., Tamaru, H., Selker, E. U., and Cheng, X. (2003). Structural basis for the product specificity of histone lysine methyltransferases. Mol. Cell 12, 177–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zink, D., Bornfleth, H., Visser, A., Cremer, C., and Cremer, T. (1999). Organization of early and late replicating DNA in human chromosome territories. Exp. Cell Res. 247, 176–188. [DOI] [PubMed] [Google Scholar]