Abstract
DIABLO/Smac is a mitochondrial protein that can promote apoptosis by promoting the release and activation of caspases. To do so, DIABLO/Smac must first be processed by a mitochondrial protease and then released into the cytosol, and we show this in an intact cellular system. We propose that the precursor form of DIABLO/Smac enters the mitochondria through a stop-transfer pathway and is processed to its active form by the inner membrane peptidase (IMP) complex. Catalytic subunits of the mammalian IMP complex were identified based on sequence conservation and functional complementation, and the novel sequence motif RX5P in Imp1 and NX5S in Imp2 distinguish the two catalytic subunits. DIABLO/Smac is one of only a few specific proteins identified as substrates for the IMP complex in the mitochondrial intermembrane space.
INTRODUCTION
Programmed cell death is a means whereby metazoans can remove unwanted cells, with failure of programmed cell death enabling cancer and autoimmune disease, and inappropriate cell death contributing to neurodegenerative disease. Several of the proteins that regulate cell death, including Bcl-2 family members, signal-transduction receptors, effector proteases and DIABLO/Smac, are specifically enclosed within subcellular structures allowing precise control over the commitment of a cell to die. Understanding where and how these key regulators are localized is crucial in understanding their normal biological function and their role in the pathogenesis of malignant diseases.
A family of intracellular proteases called caspases implement programmed cell death (Ekert et al., 1999). The activity of caspases is regulated by a family of inhibitor of apoptosis proteins (IAPs) that bind and neutralize active caspases (Deveraux and Reed, 1999). For example, the inhibitor MIHA/XIAP/hILP/BIRC4 can bind and inhibit processed caspases 3, 7, and 9 (Duckett et al., 1996; Liston et al., 1996; Uren et al., 1996; Deveraux et al., 1997, 1998). This damping of caspase activity provides a layer of regulation over the cell death-promoting activities of this family of effector proteases.
In mammalian cells, signals for cell death can lead to rupture of the mitochondrial outer membrane, and under these conditions the inhibition of caspases can be antagonized by the mitochondrial proteins DIABLO/Smac (Du et al., 2000; Verhagen et al., 2000) and HtrA2/Omi (Suzuki et al., 2001; Martins et al., 2002; Verhagen et al., 2002). Structural studies (Chai et al., 2000; Liu et al., 2000; Wu et al., 2000) have shown that purified DIABLO is a homodimer (Chai et al., 2000), and each DIABLO dimer can bind to the BIR domains of inhibitor proteins via contacts made to the N-terminal residues of DIABLO (Liu et al., 2000; Wu et al., 2000). The avid interaction DIABLO makes with the inhibitor MIHA competes the inhibitor away from active caspase 9, freeing caspase 9 to proteolytically activate downstream caspases (Ekert et al., 2001).
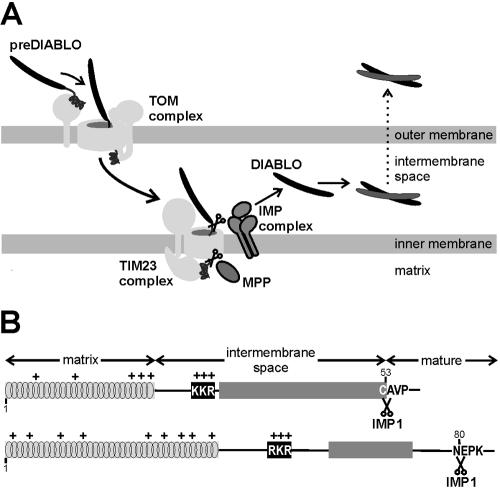
Mouse DIABLO is translated as a 237-residue precursor protein (preDIABLO) with an N-terminal presequence that must be cleaved to generate the mature form with the amino-terminal sequence A54VPI (Du et al., 2000; Verhagen et al., 2000). Here, we provide evidence that the N-terminal pre-sequence of DIABLO precursor is a bipartite, stop-transfer type targeting signal. We suggest that the precursor form of DIABLO is recognized and translocated through the mitochondrial outer membrane via the translocase in the outer mitochondrial membrane (TOM) complex and in the intermembrane space is transferred to the translocase in the inner mitochondrial membrane (TIM23) complex. The stop-transfer sequence is cleaved by the IMP complex, an oligomeric inner membrane peptidase. We have identified metazoan homologues of the catalytically active subunits of the IMP complex, hitherto only identified in yeast, and show the mammalian orthologues function to process, and thereby activate, DIABLO/Smac in mitochondria.
MATERIALS AND METHODS
Plasmids, Yeast Strains, and Media
DNA fragments corresponding to MmImp1 (AK015978) and MmImp2 (AF359564) were amplified by PCR by using primers that generated in-frame restriction sites. PCR products were subcloned in front of green fluorescent protein (GFP)-S65T under the control of the MET25 promoter (George et al., 1998) for analysis by confocal microscopy or into pYADE4 under the control of the ADH2 promoter (Brunelli and Pall, 1993) for the complementation assays. FLAG-tagged MmImp1 and MmImp2 cDNAs were generated by introducing an in frame XbaI restriction site into the 3′ end of the coding regions by PCR, effectively removing the stop codon of each cDNA.
To generate yeast mutants lacking the IMP1 gene or the SOM1 gene, PCR-mediated gene disruption (Wach et al., 1997) was used with the plasmid pFA6a-HIS3MX6 as template. For the knockin strain, MmImp1 was first cloned into a vector in front of a HIS3 gene marker. This vector was then used for PCR-mediated gene disruption. Yeast plasmids Caspase-3-LacZ, pADH-(TRP1)-MIHA, and pGALL-(HIS3)-Diablo54-237 and empty vector controls have been described previously (Hawkins et al., 2001). pGALL-(HIS3)-Diablo1-237 was generated by subcloning full-length mouse DIABLO with XhoI and NotI into pGALL-(HIS3).
Strains of Saccharomyces cerevisiae were grown at 30°C on YPAD [2% (wt/vol) glucose, 1%(wt/vol) yeast extract, and 2% (wt/vol) peptone supplemented with adenine sulfate)], grown until late log phase, and harvested by centrifugation; or grown on solid media containing 2% agar in YPAD or YPEG [2% (vol/vol) ethanol, 2% (wt/vol) glycerol, 1%(wt/vol) yeast extract, and 2% (wt/vol) peptone) or YPGal (2% (wt/vol) galactose, 1%(wt/vol) yeast extract, and 2% (wt/vol) peptone supplemented with adenine sulfate].
To determine expression levels of the DIABLO constructs, cytosolic extracts of transformed yeast were prepared according to Cartwright et al. (1997) and analyzed by SDS-PAGE and immunoblotting.
To rupture the mitochondrial outer membrane in vivo, yeast cells were cultured transiently on media containing 120 mM acetic acid. Acetic acid is not metabolized by glucose-repressed yeast cells, enters cells in the protonated form, but, if the extracellular pH is lower than the intracellular pH, deprotonation leads to transient acidification of the cytosol and some rupture of the relatively fragile mitochondrial outer membrane (Ludovico et al., 2002).
Functional IAP Antagonism Assay
Transformations were performed as described previously (Hawkins et al., 2000) and grown in selective minimal media with glucose overnight, recovered, and washed three times in 10 mM Tris-HCl, pH 8.0, EDTA 1 mM (TE), and the cell suspensions were standardized (as determined from OD600). After incubation for 8 h at room temperature in selective minimal media containing galactose, pH 3.0, lacking or containing 20 mM acetic acid, yeast was recovered, washed once with TE, and resuspended in TE. The yeast suspensions were equalized, and 5-μl drops of serial fivefold dilutions were spotted onto selective minimal media containing either glucose (to repress expression of caspase 3 and DIABLO) or galactose (to induce their expression).
Preparation of Mitochondria and Protease Sensitivity
Mitochondria were isolated according to Daum et al. (1982). Osmotic shock treatment, to produce rupture the outer membrane in purified mitochondria, was as described by Glick et al. (1992b), and trypsin treatments were performed as described previously (Beilharz et al., 1998). Samples of mitochondrial protein (100 μg) were separated by Tris-glycine SDS-PAGE, and Western blots were carried out according to published methods (Lithgow et al., 1994; Beilharz et al., 1998; Sambrook and Russell, 2001).
Prediction of the Mitochondrial Targeting Sequence in DIABLO
The mitochondrial targeting sequences of hundreds of proteins are now known and are usually rich in positively charged residues and with tendencies to form two to three turns of a helix with amphipathic character (von Heijne, 1986). Predotar (http://www.inra.fr/predotar/) is a neural network predictor trained to find such extensions and predicts a high score (0.984) for the likelihood that the amino terminus of DIABLO is a mitochondrial (matrix) targeting sequence. MitoProtII (ftp://ftp.ens.fr/pub/molbio) calculates a probability for a protein being mitochondrial based on physicochemical properties, including a mesohydrophobicity score (Claros et al., 1995; Claros and Vincens, 1996). Mitoprot scores DIABLO as 98.99% likely to be a mitochondrial protein. We recently showed that a combined prediction from both Predotar and MitoprotII is an excellent indicator for mitochondrial location (Lucattini et al., 2004).
Fluorescence Microscopy
For fluorescence microscopy, cells were visualized directly or after staining with MitoTracker (MitoTracker Red CM-H2 × Ros) according to the standard protocol from Molecular Probes (Eugene, OR). All fluorescence images were captured using an MRC1024 confocal scanning laser microscope (Bio-Rad, Hercules, CA) mounted on an Axioskop (Carl Zeiss, Jena, Germany).
RESULTS
The precursor Form of DIABLO Is Targeted to Mitochondria by Its Amino Terminus
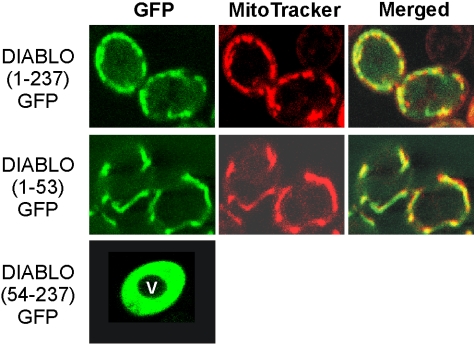
Sequence analysis suggested that the previously reported mitochondrial localization of DIABLO in mammalian cells was due to the presence of a mitochondrial targeting sequence at the amino terminus of the protein. This would imply that DIABLO also should be targeted to mitochondria in nonmammalian cells. To determine whether this was true, three GFP reporter proteins were constructed for expression in yeast (Figure 1). GFP was fused to the full preDIABLO sequence [DIABLO(1-237)GFP], to the 53 N-terminal residues of DIABLO [DIABLO(1-53)GFP], or to DIABLO from which the first 53 amino acids had been removed [DIABLO(54-237)GFP]. Analysis of yeast cells costained with MitoTracker Red revealed that preDIABLO and DIABLO(1-53)GFP were exclusively localized to mitochondria, whereas DIABLO(54-237)GFP was distributed throughout the cytosol (Figure 1). This indicates that the 53-amino acid presequence of DIABLO is necessary and sufficient for targeting of proteins to mitochondria in yeast.
Figure 1.
DIABLO is targeted to mitochondria in yeast. Yeast cells expressing preDIABLO [DIABLO(1-237)GFP], DIABLO(1-53)GFP, or DIABLO(54-237)GFP were costained with the fluorescent dye MitoTracker Red and viewed by confocal microscopy. Filters selective for the green fluorescence of GFP (left) or the red fluorescence of MitoTracker Red (middle) were used. Green and red fluorescence pictures merged are shown (right) (V, cell vacuole).
Isolation of mitochondria from yeast cells expressing preDIABLO showed that much of the precursor form of DIABLO had been processed (Figure 2). As in mammalian cells expressing DIABLO (Du et al., 2000; Verhagen et al., 2000), some unprocessed preDIABLO was still partially exposed on the mitochondrial surface, because it was degraded when mitochondria were incubated with trypsin. The processed form, however, was protected from trypsin cleavage within the organelle, as is an apparent processing intermediate. Consistent with this conclusion, when the outer membrane was ruptured by osmotic shock, processed DIABLO was degraded by trypsin, as was the intermembrane space protein cytochrome (Cyt) b2. Matrix-located proteins, such as the mitochondrial 70-kDa heat-shock protein (mtHSP70) remained protected from trypsin by the inner mitochondrial membrane, even after the outer membrane was ruptured (Figure 2). The small fraction of DIABLO that is not degraded in this sample remains resistant to protease even in the presence of Triton X-100 (our unpublished data) that solubilizes the inner membrane, suggesting this fraction of the protein is aggregated into a protease inaccessible form. These experiments show that DIABLO is targeted by its amino terminal targeting sequence to the mitochondria, where the targeting peptide is removed liberating DIABLO in the intermembrane space.
Figure 2.
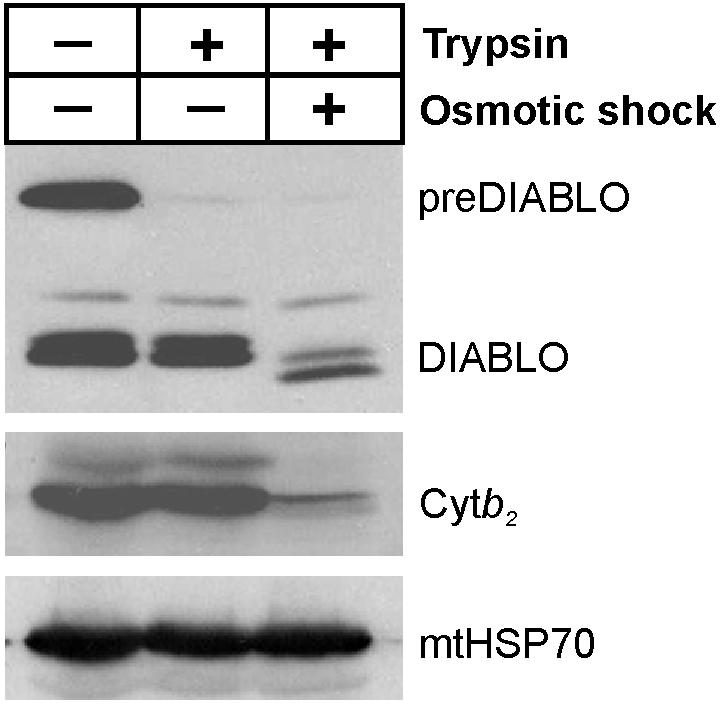
DIABLO is localized to the intermembrane space of mitochondria. Mitochondria (100 μg of protein) were isolated from yeast cells expressing preDIABLO and incubated in the presence (+) or absence (-) of 1.5 μg of trypsin with (+) and without (-) first rupturing the outer membrane by osmotic shock. After separation by SDS-PAGE, samples were analyzed by immunoblotting with antibodies recognizing DIABLO, the intermembrane space protein Cytb2 and the matrix-located mtHSP70.
Processing of preDIABLO Is Required for Antagonism of IAP Function In Vivo
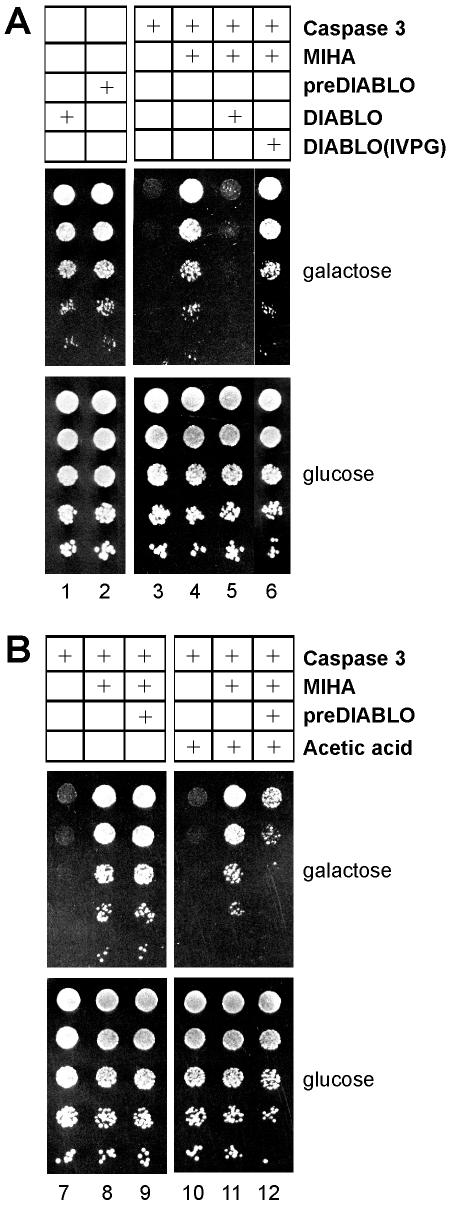
Mature DIABLO can antagonize the caspase inhibitory properties of MIHA in yeast (Hawkins et al., 2001), and we exploited this cellular system to explore the impact of the amino terminal region of DIABLO on its ability to antagonize IAP activity. Yeast tolerated expression of either the precursor or mature forms of DIABLO (Figure 3A, lanes 1 and 2). Expression of autoactivating caspase 3 was toxic (lane 3), unless inhibited by coexpression of MIHA (lane 4). Coexpression of the mature form of DIABLO [corresponding to DIABLO(54-237)] killed the cells due to liberation of active caspase 3 from MIHA (Figure 3A, lane 5).
Figure 3.
DIABLO expressed in yeast inhibits the antiapoptotic effect of MIHA/XIAP. Yeast was transformed with the plasmids coding for the indicated proteins or with the appropriate control vectors. Where indicated, cultures of transformed cells were incubated with or without acetic acid for8hat room temperature. Serial dilutions were made from samples with equivalent cell numbers, and 5 μl of each dilution was spotted onto expression-inducing (galactose) or -repressing (glucose) solid media. The top-most row in each panel corresponds to the most concentrated yeast suspension and the serial dilutions were spotted vertically down the plate. Colony size indicates growth rate and colony number cell viability.
To be certain that the death of yeast cells is directly a result of the DIABLO-IAP interaction and requires the correctly processed N terminus of DIABLO, we designed and tested a mutant form (IVPG) of DIABLO. In the crystal structure of the DIABLO-IAP complex (PDB accession 1G73; the N-terminal residues are A54, V55, P56, and I57), residues A54 and I57 are buried at the interface with the IAP BIR3 domain, in small and large pockets, respectively. The V55 and P56 side chains largely project up and out of the DIABLO-IAP interface. The mutations A54L and L57G were made on the basis that the leucine residue would have difficulty packing in the small pocket in BIR3 that accommodates the small alanine residue, and the presence of a glycine will eliminate all the favorable packing that was contributed by the large isoleucine residue in the large pocket of the IAP. In addition, the mutant sequence IVPG at the N terminus of DIABLO should favor the formation of a type-II beta-turn at VPGA58, which should further destabilize the interaction with IAP. These otherwise conservative mutations, A54L and L57G, do not effect the expression of the mutant protein as judged by Western blotting of cytosolic extracts (our unpublished data), but the mutant DIABLO (IVPG) is ineffective at promoting the caspase-mediated death of yeast cells (Figure 3B, lane 6).
Yeast cells expressing the precursor form of DIABLO, which is targeted to mitochondria (Figure 1), together with autoactivating caspase 3 and MIHA survived (Figure 3B, lane 9) because the matured, active form is compartmentalized (within the mitochondrial intermembrane space) from MIHA and caspase 3. Nevertheless, rupturing the outer membrane of mitochondria by treatment of intact cells with 120 mM acetic acid (Ludovico et al., 2002) was able to antagonize the antiapoptotic effect of MIHA (Figure 3B, lanes 11 and 12). Because inhibition of MIHA requires the correctly processed AVPI amino terminus of DIABLO (Figure 3A, lane 6), preDIABLO must have been cleaved, in vivo, at the same processing site in yeast as it is in mammalian cells.
preDIABLO Is Processed by the IMP Complex
In yeast, few proteins have been identified that are proteolytically processed for release into the intermembrane space. The three best characterized, Cytb2, Cytc1, and Mcr1 (Gakh et al., 2002), each first dock with the TIM23 complex and are then processed by the IMP complex. The IMP complex is a hetero-oligomer integrated in the mitochondrial inner membrane, and deletion of any one subunit leads to destabilization and loss of the other subunits (Nunnari et al., 1993; Gakh et al., 2002). Two catalytic subunits, Imp1 and Imp2, are similar in sequence but seem to have nonoverlapping substrate specificities (Nunnari et al., 1993; Gakh et al., 2002). A third noncatalytic subunit, Som1 has been shown to assist substrate recognition by the catalytic Imp1 subunit (Esser et al., 1996; Jan et al., 2000). The oligomeric IMP complex processes mitochondrial presequences releasing matured proteins to the intermembrane space. To determine whether DIABLO is a substrate of the IMP complex, purified mitochondria of yeast strains expressing preDIABLO, but lacking one of the three subunits of the Imp complex, were assayed in Western blots for the presence or absence of the mature form of DIABLO (Figure 4).
Figure 4.
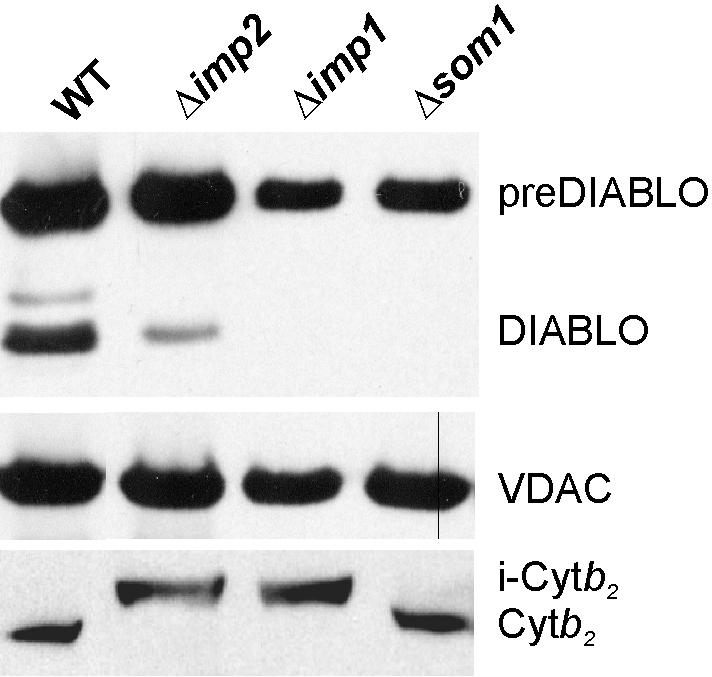
Yeast mutants lacking Imp1 cannot process preDIABLO. Mitochondria (100 μg of protein) isolated from wild-type cells expressing preDIABLO, or the indicated yeast mutants expressing preDIABLO, were compared after SDS-PAGE separation of proteins. Western blot replicas of the samples were probed with anti-sera recognizing DIABLO, the voltage-dependent anion channel (VDAC), and Cytb2. Position of i-Cytb2 and mature (Cytb2) forms of cytochrome b2 (Glick et al., 1992b; Lithgow et al., 1994) are shown.
The model substrate Cytb2 is processed in two steps: initially by the matrix peptidase MPP to generate an intermediate (i-Cytb2; Figure 4), and this intermediate is then substrate for the IMP complex. In wild-type or in Δsom1 mutants, processing to the mature form of Cytb2 occurred. The Imp1 subunit is primarily responsible for Cytb2 processing (Nunnari et al., 1993), and Figure 4 shows processing of Cytb2 is blocked completely in Δimp1 cells. Because yeast Δimp2 mutants express decreased levels of the Imp1 subunit (Nunnari et al., 1993), Cytb2 processing also is also blocked in Δimp2 cells. Analogously, processing of preDIABLO was partially compromised in Δimp2 mutants and failed to occur at all in either the Δimp1 and Δsom1 mutants. We conclude the IMP complex processes preDIABLO to its mature form, and it is likely the Imp1 catalytic site at which this occurs.
Identification of Mouse Homologues for Yeast Imp1 and Imp2 Proteases
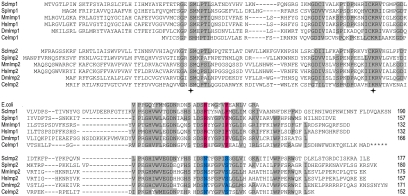
An IMP complex has not yet been functionally characterized in organisms other than S. cerevisiae. However, iterative BLAST analyses revealed genes predicted to encode proteins similar to Imp1 and Imp2 in all animals and fungi for which substantial genome sequence data exist (Figure 5). The catalytic serine and lysine residues and the structurally important arginine and aspartate residues from the yeast Imp1 and Imp2 (Chen et al., 1999) are conserved across all species (Figure 5, stars). In addition, we found a sequence motif that distinguishes Imp1(RX5P) from Imp2(NX5S). The sequence motif defining the Imp1 and Imp2 subunits sits close by structurally important aspartate residues thought to stabilize the shape of the substrate-binding cleft in the bacterial leader peptidase (Paetzel et al., 1998).
Figure 5.
Multiple sequence alignment of the Imp family of proteases. Iterative BLAST analysis was undertaken with the Imp2 sequence from S. cerevisiae. The Imp1 and Imp2 sequences were aligned with ClustalW, and a representative selection is shown (Sc, S. cerevisiae; Sp, Schizosaccharomyces pombe, Ce, Caenorhabditis elegans; Dm, Drosophila melanogaster; Hs, Homo sapiens; Mm, M. musculus). Asterisks designate a possible C-terminal extension to CeImp1. Amino acid residues conserved across at least six species are highlighted, and the total number of residues shown. Imp1-specific residues (RX5P) are colored pink and Imp2-specific residues (NX5S) are colored blue. Catalytic residues in Imp1 and Imp2 (analogous to Ser90 and K145 of E. coli leader peptidase) are designated with stars.
MmImp1 and MmImp2 Function as Catalytic Subunits of an IMP Complex
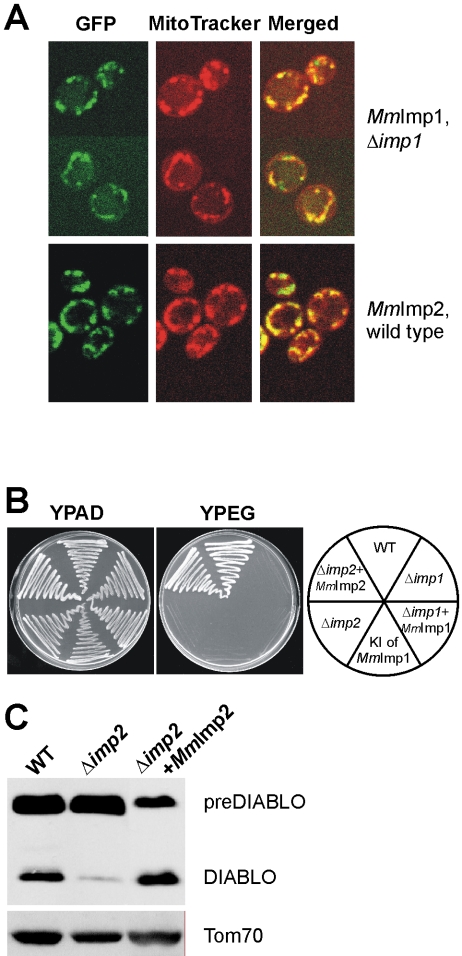
The cDNAs encoding the putative mouse proteases were cloned and expressed in yeast cells to determine their intracellular localization. Both MmImp1 (MmImp1 refers to the Mus musculus form of Imp1 according to the nomenclature of Pfanner et al. (1996) and MmImp2 localize to mitochondria (Figure 6A). Like yeast Imp1, MmImp1 could be stably expressed in Δimp1 yeast (Figure 6A) but not in wild-type (i.e., Imp1+) yeast cells (our unpublished data). The cytochrome substrates of the IMP complex are key components of the mitochondrial electron transport chain, so that neither Δimp1 mutants nor Δimp2 yeast mutants can grow on nonfermentable carbon sources. We exploited this phenotype to test for complementation, which would imply functional homology by the proposed mouse counterparts of the yeast IMP genes.
Figure 6.
MmImp1 and MmImp2 are mouse subunits of the IMP complex. (A) GFP was fused to the C terminus of MmImp1 and MmImp2 and the fusion proteins expressed in Δimp1 or wild-type yeast cells. Cells were costained with the fluorescent dye MitoTracker Red and visualized by confocal microscopy. Z-sections through representative cells are shown. Green fluorescence of GFP in groups of cells (left), red fluorescence (middle), and the merged pictures of green and red fluorescence (right). (B) Wild-type yeast cells and the Δimp1 or Δimp2 mutants were transformed with the respective mouse homologue to be tested for complementation of the growth defect on the nonfermentable carbon sources glycerol and ethanol (YPEG). In addition, the open reading frame in the yeast gene encoding Imp1 (YMR150c) was directly replaced with the MmImp1 open-reading frame through homologous recombination (KIMmImp1). (C) Mitochondria expressing preDIABLO isolated from wild type, yeast mutant lacking Imp2, and yeast mutant lacking Imp2 but expressing MmImp2 were compared after SDS-PAGE separation of proteins. Amounts of DIABLO and the control protein Tom70 were compared by Western blot analysis.
The MmImp2 subunit restored activity in the Δimp2 strain, allowing it to process cytochromes and grow on the nonfermentable carbon sources ethanol and glycerol (Figure 6B). Based on sequence similarities, correct mitochondrial location and functional complementation, we suggest an IMP complex exists in mammalian cells for the processing and activation of proteins such as preDIABLO. That MmImp1 does not complement the growth defect of Δimp1 yeast cells (Figure 6B) suggests additional factors such as a mammalian homolog of Som1 might be needed for full activity of the MmImp1 subunit.
The loss of Imp2 destabilizes the IMP complex so that Δimp2 yeast cells process preDIABLO only poorly. However, this impairment was reversed by expression of MmImp2 in the Δimp2 yeast (Figure 6C). We conclude that preDIABLO is proteolytically activated to DIABLO by the IMP complex and that the mammalian Imp1 and Imp2 sequences represent functional homologues of the yeast IMP complex subunits.
DISCUSSION
In healthy cells, mature DIABLO is encapsulated within mitochondria, where it cannot interact with IAPs. Here, we have shown that after preDIABLO is translated in the cytosol and transported across the mitochondrial outer membrane, it is processed by the IMP complex in the intermembrane space to produce the potent, mature form. Activated DIABLO then remains in the intermembrane space until an apoptotic signal is received (Figure 7A).
Figure 7.
A stop-transfer pathway for the import and activation of DIABLO by mitochondria. (A) The precursor form of DIABLO (black) is synthesized in the cytosol and the N-terminal presequence recognized by the TOM complex and thereby transferred across the outer membrane. Engaged in the TIM23 complex, DIABLO might be processed by the matrix-located peptidase, such as mitochondrial processing peptidase, and is processed by the IMP complex composed of Imp1, Imp2, and Som1. Release of processed DIABLO into the intermembrane space allows for its assembly into a dimer and its availability for release into the cytosol only if the outer membrane is ruptured. (B) The presequences of DIABLO (top schematic) and Cytb2 (bottom schematic) show similar regions corresponding to a basic amphipathic helix with the matrix targeting information (gray) and the intermembrane sorting sequence, containing a core of hydrophobic residues (dark gray) preceded by a cluster of three positively charged residues (black) important for recognition of the sorting sequence. For DIABLO the IMP1 processing site is designated between C53 and A54 and for Cytb between N80 and E81.
Two well-studied mitochondrial proteins, Cytb2 and Cytc1, have bipartite targeting sequences consisting of a short region of basic, amphipathic helix followed by a processing site for the matrix processing peptidase and an additional sorting signal. The sorting signal dictates arrest of newly imported Cytb2 and Cytc1 in the inner membrane TIM23 complex, providing access to the IMP complex for proteolytic release of Cytb2 and Cytc1 from their presequences (Glick et al., 1992a,b; van Loon and Schatz, 1987). The presequences of Cytb2 and Cytc1 resemble those in the targeting sequence of preDIABLO. A short region at the N terminus of preDIABLO is predicted to form a basic amphipathic helix that might direct the protein to the TIM23 complex (Figure 7B; see Materials and Methods). We have demonstrated that the final processing of DIABLO is carried out by the IMP complex, making its import pathway into mitochondria (Figure 7A) analogous to the stop-transfer pathway traveled by Cytb2 and Cytc1 (Glick et al., 1992a,b).
Imp1 and Imp2 are members of the signal peptidase family of proteases. In Escherichia coli, a signal peptidase removes the presequence of proteins translocated across the bacterial membrane (Paetzel et al., 2002). Two proteins related to the IMP subunits are present in all animals and fungi where complete genome information is available, and a diptych of amino acid residues corresponding to the motif RX5P in Imp1 and NX5S in Imp2 seems to be diagnostic for Imp1 or Imp2. The RX5P motif of Imp1 and its surrounding residues are conserved in the E. coli leader peptidase (Figure 5), and as part of a thorough examination of the structure of crystallized leader peptidase, Paetzel et al. (1998, 2002) suggested that arginine282 (in the RX5P motif) is important in stabilizing the active site region and positioning of surrounding amino acids). We note from their data that the proline288 residue seems to be involved in another structurally important position, in a turn between two beta-sheets (Strahm and Lithgow, unpublished observations). The distinguishing motif in Imp2(NX5S) and the context provided by surrounding residues might influence the structure around the active site enough to broaden the range of substrates that can be processed by the IMP complex. That the RX5P and NX5S sequence motifs are each so widely conserved through evolution shows they are fundamentally important for IMP complex activity and makes them the diagnostic motif in distinguishing Imp1 from Imp2.
Other Substrates of the Mammalian IMP Complex
A number of proteins that might potentiate apoptosis are released from mitochondria (van Gurp et al., 2003; Saelens et al., 2004). None of these have the hallmarks that would suggest activation by the IMP complex. A candidate substrate, AIF is a peripheral component of the mitochondrial inner membrane (Arnoult et al., 2002) made as a precursor protein, with a presequence of 101 residues processed after import into mitochondria (Susin et al., 1999). It is not clear yet which protease is responsible for AIF processing, but the presequence of AIF shows no obvious similarity to a stop-transfer sequence, and the processing site has none of the residues suggested to be important for recognition by the IMP complex (Gakh et al., 2002).
Cytochrome c is imported into the intermembrane space via the TOM complex without the participation of the TIM complex and without any proteolytic processing (Diekert et al., 2001; Wiedemann et al., 2003). HtrA2/Omi is targeted to the intermembrane space of mitochondria, but it is processed autocatalytically in mammalian cells (Seong et al., 2004) and in yeast (Verhagen and Silke, unpublished results), and its role in apoptosis might be secondary to its role as a molecular chaperone for other mitochondrial proteins (Vaux and Silke, 2003).
Endonuclease G (EndoG) is a mitochondrial protein, released during cell death, that contributes to nuclear DNA fragmentation in the terminal stages of apoptosis (Li et al., 2001). EndoG is encoded in the nucleus, translated as an ∼33-kDa precursor in the cytosol, translocated across the mitochondrial membranes with the presequence and then cleaved to yield the ∼28-kDa mature nuclease (Schafer et al., 2004). EndoG had been tentatively suggested to be located in the intermembrane space (Ohsato et al., 2002), but EndoG must be located in the mitochondrial matrix for it to generate the primers needed for mitochondrial DNA replication (Cote and Ruiz-Carrillo, 1993) and would therefore be processed by matrix-located proteases (Gakh et al., 2002).
Discrete defects in mitochondrial protein import and sorting can lead to human disease. Mohr-Tranenberg syndrome, a deafness-dystonia disorder, results through mutations that effect assembly of the Tim8/Tim13 complex, thereby inhibiting protein sorting to the TIM22 complex (Roesch et al., 2002; Binder et al., 2003). Chromosomal mapping of patients suffering from Gilles de la Tourette syndrome, another neurological condition, has revealed that one of the genes located at a breakpoint region (7q31) that is associated with symptom development encodes the human ortholog of the protein we designate here as MmImp2 (Petek et al., 2001; Gakh et al., 2002). Speculation, based on sequence similarity the authors noted to the yeast Imp2, suggested that defects in respiratory chain complexes might impact on the etiology of Tourette syndrome and other neuropsychiatric disorders (Petek et al., 2001; Gakh et al., 2002). Our findings on the role of the IMP complex in processing non-cytochrome substrates such as DIABLO, suggest that if defects in the IMP complex contribute to Tourette syndrome and other conditions, it could be through downstream influences on cell development and function.
Acknowledgments
We thank Miha Pakusch and Katherine Vascotto for technical assistance, and Andrew Perry, Terry Mulhern, Paul Gooley, and James Whisstock for critical discussions. This work was supported by a fellowship from the Swiss National Science Foundation (to L. B.), an early career research grant from the University of Melbourne (to L. B.), a Center Grant from the Leukemia and Lymphoma Society (to D.L.V.), and a grant from the Australian Research Council (to T. L.).
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E04-12-1086) on April 6, 2005.
Abbreviations used: IAP, inhibitor of apoptosis protein; IMP, inner membrane peptidase; TIM23, translocase of the inner mitochondrial membrane; TOM, translocase of the outer mitochondrial membrane.
References
- Arnoult, D., Parone, P., Martinou, J. C., Antonsson, B., Estaquier, J., and Ameisen, J. C. (2002). Mitochondrial release of apoptosis-inducing factor occurs downstream of cytochrome c release in response to several proapoptotic stimuli. J. Cell Biol. 159, 923-929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beilharz, T., Suzuki, C. K., and Lithgow, T. (1998). A toxic fusion protein accumulating between the mitochondrial membranes inhibits protein assembly in vivo. J. Biol. Chem. 273, 35268-35272. [DOI] [PubMed] [Google Scholar]
- Binder, J., Hofmann, S., Kreisel, S., Wohrle, J. C., Bazner, H., Krauss, J. K., Hennerici, M. G., and Bauer, M. F. (2003). Clinical and molecular findings in a patient with a novel mutation in the deafness-dystonia peptide (DDP1) gene. Brain 126, 1814-1820. [DOI] [PubMed] [Google Scholar]
- Brunelli, J. P., and Pall, M. L. (1993). A series of yeast shuttle vectors for expression of cDNAs and other DNA sequences. Yeast 9, 1299-1308. [DOI] [PubMed] [Google Scholar]
- Cartwright, P., Beilharz, T., Hansen, P., Garrett, J., and T. Lithgow. (1997). Mft52, an acid-bristle protein in the cytosol that delivers precursor proteins to yeast mitochondria. J. Biol. Chem. 272, 5320-5325. [DOI] [PubMed] [Google Scholar]
- Chai, J., Du, C., Wu, J. W., Kyin, S., Wang, X., and Shi, Y. (2000). Structural and biochemical basis of apoptotic activation by Smac/DIABLO. Nature 406, 855-862. [DOI] [PubMed] [Google Scholar]
- Chen, X., Van Valkenburgh, C., Fang, H., and Green, N. (1999). Signal peptides having standard and nonstandard cleavage sites can be processed by Imp1p of the mitochondrial inner membrane protease. J. Biol. Chem. 274, 37750-37754. [DOI] [PubMed] [Google Scholar]
- Claros, M. G., Perea, J., Shu, Y., Samatey, F. A., Popot, J. L., and Jacq, C. (1995). Limitations to in vivo import of hydrophobic proteins into yeast mitochondria. The case of a cytoplasmically synthesized apocytochrome b. Eur. J. Biochem. 228, 762-771. [PubMed] [Google Scholar]
- Claros, M. G., and Vincens, P. (1996). Computational method to predict mitochondrially imported proteins and their targeting sequences. Eur. J. Biochem. 241, 779-786. [DOI] [PubMed] [Google Scholar]
- Cote, J., and Ruiz-Carrillo, A. (1993). Primers for mitochondrial DNA replication generated by endonuclease G. Science 261, 765-769. [DOI] [PubMed] [Google Scholar]
- Daum, G., Gasser, S. M., and Schatz, G. (1982). Import of proteins into mitochondria. Energy-dependent, two-step processing of the intermembrane space enzyme cytochrome b2 by isolated yeast mitochondria. J. Biol. Chem. 257, 13075-13080. [PubMed] [Google Scholar]
- Deveraux, Q. L., and Reed, J. C. (1999). IAP family proteins-suppressors of apoptosis. Genes Dev. 13, 239-252. [DOI] [PubMed] [Google Scholar]
- Deveraux, Q. L., Roy, N., Stennicke, H. R., Van Arsdale, T., Zhou, Q., Srinivasula, S. M., Alnemri, E. S., Salvesen, G. S., and Reed, J. C. (1998). IAPs block apoptotic events induced by caspase-8 and cytochrome c by direct inhibition of distinct caspases. EMBO J. 17, 2215-2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deveraux, Q. L., Takahashi, R., Salvesen, G. S., and Reed, J. C. (1997). X-linked IAP is a direct inhibitor of cell-death proteases. Nature 388, 300-304. [DOI] [PubMed] [Google Scholar]
- Diekert, K., de Kroon, A. I., Ahting, U., Niggemeyer, B., Neupert, W., de Kruijff, B., and Lill, R. (2001). Apocytochrome c requires the TOM complex for translocation across the mitochondrial outer membrane. EMBO J. 20, 5626-5635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du, C., Fang, M., Li, Y., Li, L., and Wang, X. (2000). Smac, a mitochondrial protein that promotes cytochrome c-dependent caspase activation by eliminating IAP inhibition. Cell 102, 33-42. [DOI] [PubMed] [Google Scholar]
- Duckett, C. S., Nava, V. E., Gedrich, R. W., Clem, R. J., Van Dongen, J. L., Gilfillan, M. C., Shiels, H., Hardwick, J. M., and Thompson, C. B. (1996). A conserved family of cellular genes related to the baculovirus iap gene and encoding apoptosis inhibitors. EMBO J. 15, 2685-2694. [PMC free article] [PubMed] [Google Scholar]
- Ekert, P. G., Silke, J., Hawkins, C. J., Verhagen, A. M., and Vaux, D. L. (2001). DIABLO promotes apoptosis by removing MIHA/XIAP from processed caspase 9. J. Cell Biol. 152, 483-490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekert, P. G., Silke, J., and Vaux, D. L. (1999). Caspase inhibitors. Cell Death Differ. 6, 1081-1086. [DOI] [PubMed] [Google Scholar]
- Esser, K., Pratje, E., and Michaelis, G. (1996). SOM 1, a small new gene required for mitochondrial inner membrane peptidase function in Saccharomyces cerevisiae. Mol. Gen. Genet. 252, 437-445. [DOI] [PubMed] [Google Scholar]
- Gakh, O., Cavadini, P., and Isaya, G. (2002). Mitochondrial processing peptidases. Biochim. Biophys. Acta 1592, 63-77. [DOI] [PubMed] [Google Scholar]
- George, R., Beddoe, T., Landl, K., and Lithgow, T. (1998). The yeast nascent polypeptide-associated complex initiates protein targeting to mitochondria in vivo. Proc. Natl. Acad. Sci. USA 95, 2296-2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glick, B. S., Beasley, E. M., and Schatz, G. (1992a). Protein sorting in mitochondria. Trends Biochem. Sci. 17, 453-459. [DOI] [PubMed] [Google Scholar]
- Glick, B. S., Brandt, A., Cunningham, K., Muller, S., Hallberg, R. L., and Schatz, G. (1992b). Cytochromes c1 and b2 are sorted to the intermembrane space of yeast mitochondria by a stop-transfer mechanism. Cell 69, 809-822. [DOI] [PubMed] [Google Scholar]
- Hawkins, C. J., Silke, J., Verhagen, A. M., Foster, R., Ekert, P. G., and Ashley, D. M. (2001). Analysis of candidate antagonists of IAP-mediated caspase inhibition using yeast reconstituted with the mammalian Apaf-1-activated apoptosis mechanism. Apoptosis 6, 331-338. [DOI] [PubMed] [Google Scholar]
- Hawkins, C. J., Wang, S. L., and Hay, B. A. (2000). Monitoring activity of caspases and their regulators in yeast Saccharomyces cerevisiae. Methods Enzymol. 322, 162-174. [DOI] [PubMed] [Google Scholar]
- Jan, P. S., Esser, K., Pratje, E., and Michaelis, G. (2000). Som1, a third component of the yeast mitochondrial inner membrane peptidase complex that contains Imp1 and Imp2. Mol. Gen. Genet. 263, 483-491. [DOI] [PubMed] [Google Scholar]
- Li, L. Y., Luo, X., and Wang, X. (2001). Endonuclease G is an apoptotic DNase when released from mitochondria. Nature 412, 95-99. [DOI] [PubMed] [Google Scholar]
- Liston, P., et al. (1996). Suppression of apoptosis in mammalian cells by NAIP and a related family of IAP genes. Nature. 379, 349-353. [DOI] [PubMed] [Google Scholar]
- Lithgow, T., Junne, T., Wachter, C., and Schatz, G. (1994). Yeast mitochondria lacking the two import receptors Mas20p and Mas70p can efficiently and specifically import precursor proteins. J. Biol. Chem. 269, 15325-15330. [PubMed] [Google Scholar]
- Liu, Z., Sun, C., Olejniczak, E. T., Meadows, R. P., Betz, S. F., Oost, T., Herrmann, J., Wu, J. C., and Fesik, S. W. (2000). Structural basis for binding of Smac/DIABLO to the XIAP BIR3 domain. Nature 408, 1004-1008. [DOI] [PubMed] [Google Scholar]
- Lucattini, R., Likic, V. A., and Lithgow, T. (2004). Bacterial proteins predisposed for targeting to mitochondria. Mol. Biol. Evol. 21, 652-658. [DOI] [PubMed] [Google Scholar]
- Ludovico, P., Rodrigues, F., Almeida, A., Silva, M. T., Barrientos, A., and Corte-Real, M. (2002). Cytochrome c release and mitochondria involvement in programmed cell death induced by acetic acid in Saccharomyces cerevisiae. Mol. Biol. Cell 13, 2598-2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins, L. M., et al. (2002). The serine protease Omi/HtrA2 regulates apoptosis by binding XIAP through a reaper-like motif. J. Biol. Chem. 277, 439-444. [DOI] [PubMed] [Google Scholar]
- Nunnari, J., Fox, T. D., and Walter, P. (1993). A mitochondrial protease with two catalytic subunits of nonoverlapping specificities. Science 262, 1997-2004. [DOI] [PubMed] [Google Scholar]
- Ohsato, T., Ishihara, N., Muta, T., Umeda, S., Ikeda, S., Mihara, K., Hamasaki, N., and Kang, D. (2002). Mammalian mitochondrial endonuclease G. Digestion of R-loops and localization in intermembrane space. Eur. J. Biochem. 269, 5765-5770. [DOI] [PubMed] [Google Scholar]
- Paetzel, M., Dalbey, R. E., and Strynadka, N. C. (1998). Crystal structure of a bacterial signal peptidase in complex with a beta-lactam inhibitor. Nature 396, 186-190. [DOI] [PubMed] [Google Scholar]
- Paetzel, M., Karla, A., Strynadka, N. C., and Dalbey, R. E. (2002). Signal peptidases. Chem. Rev. 102, 4549-4580. [DOI] [PubMed] [Google Scholar]
- Petek, E., Windpassinger, C., Vincent, J. B., Cheung, J., Boright, A. P., Scherer, S. W., Kroisel, P. M., and Wagner, K. (2001). Disruption of a novel gene (IMMP2L) by a breakpoint in 7q31 associated with Tourette syndrome. Am. J. Hum. Genet. 68, 848-858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfanner, N., Douglas, M. G., Endo, T., Hoogenraad, N. J., Jensen, R. E., Meijer, M., Neupert, W., Schatz, G., Schmitz, U. K., and Shore, G. C. (1996). Uniform nomenclature for the protein transport machinery of the mitochondrial membranes. Trends Biochem. Sci. 21, 51-52. [PubMed] [Google Scholar]
- Roesch, K., Curran, S. P., Tranebjaerg, L., and Koehler, C. M. (2002). Human deafness dystonia syndrome is caused by a defect in assembly of the DDP1/TIMM8a-TIMM13 complex. Hum. Mol. Genet. 11, 477-486. [DOI] [PubMed] [Google Scholar]
- Saelens, X., Festjens, N., Walle, L. V., van Gurp, M., van Loo, G., and Vandenabeele, P. (2004). Toxic proteins released from mitochondria in cell death. Oncogene 23, 2861-2874. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., and Russell, D. W. (2001). Molecular Cloning. A Laboratory Manual, 3rd ed., Cold Spring Harbor, NY: Cold Harbor Laboratory Press.
- Schafer, P., Scholz, S. R., Gimadutdinow, O., Cymerman, I. A., Bujnicki, J. M., Ruiz-Carrillo, A., Pingoud, A., and Meiss, G. (2004). Structural and functional characterization of mitochondrial EndoG, a sugar non-specific nuclease which plays an important role during apoptosis. J. Mol. Biol. 338, 217-228. [DOI] [PubMed] [Google Scholar]
- Seong, Y. M., Choi, J. Y., Park, H. J., Kim, K. J., Ahn, S. G., Seong, G. H., Kim, I. K., Kang, S., and Rhim, H. (2004). Autocatalytic processing of HtrA2/Omi is essential for induction of caspase-dependent cell death through antagonizing XIAP. J. Biol. Chem. 279, 37588-37596. [DOI] [PubMed] [Google Scholar]
- Susin, S. A., et al. (1999). Molecular characterization of mitochondrial apoptosis-inducing factor. Nature 397, 441-446. [DOI] [PubMed] [Google Scholar]
- Suzuki, Y., Imai, Y., Nakayama, H., Takahashi, K., Takio, K., and Takahashi, R. (2001). A serine protease, HtrA2, is released from the mitochondria and interacts with XIAP, inducing cell death. Mol. Cell. 8, 613-621. [DOI] [PubMed] [Google Scholar]
- Uren, A. G., Pakusch, M., Hawkins, C. J., Puls, K. L., and Vaux, D. L. (1996). Cloning and expression of apoptosis inhibitory protein homologs that function to inhibit apoptosis and/or bind tumor necrosis factor receptor-associated factors. Proc. Natl. Acad. Sci. USA 93, 4974-4978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Gurp, M., Festjens, N., van Loo, G., Saelens, X., and Vandenabeele, P. (2003). Mitochondrial intermembrane proteins in cell death. Biochem. Biophys. Res. Commun. 304, 487-497. [DOI] [PubMed] [Google Scholar]
- van Loon, A. P., and Schatz, G. (1987). Transport of proteins to the mitochondrial intermembrane space: the `sorting' domain of the cytochrome c1 presequence is a stop-transfer sequence specific for the mitochondrial inner membrane. EMBO J. 6, 2441-2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaux, D. L., and Silke, J. (2003). HtrA2/Omi, a sheep in wolf's clothing. Cell 115, 251-253. [DOI] [PubMed] [Google Scholar]
- Verhagen, A. M., Ekert, P. G., Pakusch, M., Silke, J., Connolly, L. M., Reid, G. E., Moritz, R. L., Simpson, R. J., and Vaux, D. L. (2000). Identification of DIABLO, a mammalian protein that promotes apoptosis by binding to and antagonizing IAP proteins. Cell 102, 43-53. [DOI] [PubMed] [Google Scholar]
- Verhagen, A. M., et al. (2002). HtrA2 promotes cell death through its serine protease activity and its ability to antagonize inhibitor of apoptosis proteins. J. Biol. Chem. 277, 445-454. [DOI] [PubMed] [Google Scholar]
- von Heijne, G. (1986). Mitochondrial targeting sequences may form amphiphilic helices. EMBO J. 5, 1335-1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wach, A., Brachat, A., Alberti-Segui, C., Rebischung, C., and Philippsen, P. (1997). Heterologous HIS3 marker and GFP reporter modules for PCR-targeting in Saccharomyces cerevisiae. Yeast 13, 1065-1075. [DOI] [PubMed] [Google Scholar]
- Wiedemann, N., Kozjak, V., Prinz, T., Ryan, M. T., Meisinger, C., Pfanner, N., and Truscott, K. N. (2003). Biogenesis of yeast mitochondrial cytochrome c: a unique relationship to the TOM machinery. J. Mol. Biol. 327, 465-474. [DOI] [PubMed] [Google Scholar]
- Wu, G., Chai, J., Suber, T. L., Wu, J. W., Du, C., Wang, X., and Shi, Y. (2000). Structural basis of IAP recognition by Smac/DIABLO. Nature 408, 1008-1012. [DOI] [PubMed] [Google Scholar]