Abstract
Incoming adenovirus type 2 (Ad2) and Ad5 shuttle bidirectionally along microtubules, biased to the microtubule-organizing center by the dynein/dynactin motor complex. It is unknown how the particles reach the nuclear pore complex, where capsids disassemble and viral DNA enters the nucleus. Here, we identified a novel link between nuclear export and microtubule-mediated transport. Two distinct inhibitors of the nuclear export factor CRM1, leptomycin B (LMB) and ratjadone A (RJA) or CRM1-siRNAs blocked adenovirus infection, arrested cytoplasmic transport of viral particles at the microtubule-organizing center or in the cytoplasm and prevented capsid disassembly and nuclear import of the viral genome. In mitotic cells where CRM1 is in the cytoplasm, adenovirus particles were not associated with microtubules but upon LMB treatment, they enriched at the spindle poles implying that CRM1 inhibited microtubule association of adenovirus. We propose that CRM1, a nuclear factor exported by CRM1 or a protein complex containing CRM1 is part of a sensor mechanism triggering the unloading of the incoming adenovirus particles from microtubules proximal to the nucleus of interphase cells.
INTRODUCTION
Molecular crowding restricts the diffusion of macromolecules in the cytoplasm. Many cargo complexes use motors for effective cytoplasmic trafficking (reviewed in Luby-Phelps, 2000; Ploubidou and Way, 2001; Smith and Enquist, 2002). Long-range cargo transport is typically mediated by microtubules (MTs) and MT-associated motors of the kinesin and dynein families. These motors transport mRNAs for cell fate and polarity determinations (reviewed in Saxton, 2001), transcriptional regulators, such as the NF kappa B inhibitor IκB (Crepieux et al., 1997), the tumor suppressor protein p53 (Giannakakou et al., 2000; Giannakakou et al., 2002), the developmental factors β-catenin or adenomatous poliposis coli protein (APC, reviewed in Bienz, 2002), the apoptosis effector bim (Puthalakath et al., 1999), and stress-related signaling kinases of the ERK and JNK families (reviewed in Verhey and Rapoport, 2001). Many of these factors act in the nucleus but it is not known how they are released from the MTs to gain access to the nucleoplasm.
Microorganisms and many viruses prominently abuse the MT shuttling system (Sodeik, 2000; Ploubidou and Way, 2001; Smith and Enquist, 2002; Grieshaber et al., 2003). During entry, viruses take advantage of the dynein/dynactin motor complex for directional transport to the MT minus ends organized at the centrosome, the MT-organizing center (Bornens, 2002). This has been demonstrated for adenoviruses (Suomalainen et al., 1999; Leopold et al., 2000; Suomalainen et al., 2001; Mabit et al., 2002; Kelkar et al., 2004), herpes viruses (Dohner et al., 2002; Douglas et al., 2004), the lentivirus human immunodeficiency virus 1 (McDonald et al., 2002), the retroviruses human foamy virus 13 and Mason-Pfizer monkey virus (Petit et al., 2003; Sfakianos et al., 2003), canine parvovirus (Suikkanen et al., 2003), endosomal influenza virus (Lakadamyali et al., 2003), African swine fever virus (Alonso et al., 2001; Jouvenet et al., 2004), and the P-protein polymerase complex of rabies virus (Jacob et al., 2000; Raux et al., 2000; Finke et al., 2004). Like for cellular cargoes, it is unknown how the motor interactions with MTs are regulated and how the cargo is released from the motors at the final destination.
Adenovirus enters the nucleus by binding to the cytoplasmic fibril protein CAN/Nup214 of the nuclear pore complex, dismantles the capsid and releases the DNA into the nucleoplasm (Greber et al., 1996; Trotman et al., 2001; Martin-Fernandez et al., 2004). In this study, we asked if nuclear protein export provides a link between the nucleus and the perinuclear cytoplasm to inform the virus about the nuclear position in the cytoplasm. The nuclear export factor CRM1 (chromosome region maintenance 1) predominantly localizes to the nucleus and exports leucine-rich nuclear export sequence (NES) containing proteins (Fornerod et al., 1997a). The loading of CRM1 to proteins containing physiological NESs in the nucleus is assisted by Ran:GTP, whereas cargo discharge occurs in the cytoplasm or the cytoplasmic face of the NPC upon stimulation of Ran:GTP hydrolysis by Ran-GAP (Mahajan et al., 1997; Gorlich and Kutay, 1999). Our results indicate that the targeting of incoming adenovirus from MTs to the NPC requires CRM1 activity. Treating a variety of different cells with CRM1-specific siRNAs or inhibitors of CRM1, such as leptomycin B (LMB) or ratjadone A (RJA) strongly reduced adenovirus infection and blocked the nuclear targeting of viral particles, the disassembly of capsids and the import of viral DNA. In most cell types tested, LMB or RJA blocked the cytoplasmic transport of adenovirus at the MTOC and in one cell line the block occurred in the cytoplasm, precluding viral attachment to the NPC. We suggest that in normal cells CRM1 or a nuclear factor exported by CRM1 dissociates adenovirus particles from MTs in the perinuclear region and thus enables viral binding to NPCs and infection.
MATERIALS AND METHODS
Cells, Transfection, and Viruses
TC7 (African green monkey kidney) cells were obtained from J. Bulinski (Columbia University, New York) and used as described (Suomalainen et al., 1999). HeLa cells (obtained from American Type Culture Collection, Rockville, MD), human lung carcinoma A549 cells, Ad5-E1 transfected human embryonic retinoblast 911 cells (Fallaux et al., 1998), human epithelial KB cells (obtained from American Type Culture Collection), human epithelial carcinoma cells A431 (obtained from J. Pavlovic, Institute of Medical Virology, University of Zürich, Switzerland), African green monkey kidney cells CV1 (American Type Culture Collection) and COSN cells, a subline of COS7 cells containing origin-defective SV40 DNA (Gluzman, 1981) obtained from S. Hemmi (Institute of Molecular Biology, University of Zürich, Switzerland; Hemmi et al., 1994) were grown in 10% fetal bovine serum or 7% clone III serum (Hyclone, PerBio Science, Lausanne, Switzerland) on alcian blue-coated glass coverslips (Suomalainen et al., 1999). Freshly isolated, primary human umbilical vein endothelial cells (HUVEC) were a gift of L. Jornot (Respiratory Division, University Hospital Geneva, Switzerland) and were grown in RPMI 1640 plus 15% fetal calf serum and 15 μg/ml endothelial growth factor and 90 μg/ml heparin (Invitrogen, Basel, Switzerland). Primary human fibroblasts isolated from human foreskin were provided by S. Hemmi (Institute of Molecular Biology, University of Zürich, Switzerland). Plasmid DNAs were transfected into 50% confluent HeLa or TC7 cells grown on 12-mm coverslips using FuGENE 6 (Roche, Indianapolis, IN) according to the manufacture's protocol. Wild-type Ad2 and the mutant temperature sensitive (ts) Ad2 ts1 were grown, isolated, and labeled with TR as published (Greber et al., 1998; Nakano and Greber, 2000). Ad5-luc was used as described (Mabit et al., 2002).
Antibodies and Chemicals
Rabbit anti-hexon R70 antibodies were provided by M. Horwitz (Albert Einstein College of Medicine, New York), and rabbit anti-protein VII antibodies were from U. Pettersson (Uppsala University, Sweden) and used as described (Greber et al., 1993, 1997; Trotman et al., 2001). γ-tubulin was visualized using the mouse monoclonal antibody TU-30 from P. Draber (Institute of Molecular Genetics, Prague, Czech Republic) used as described (Suomalainen et al., 1999). Cells were fixed in 3% pFA for 10 min and extracted with methanol at -20°C for 5 min or were directly fixed and extracted in chilled methanol at -20°C for 4 min (Suomalainen et al., 1999). The anti-tyrosinated α-tubulin antibody 1A2 (Kreis, 1987) was used on PHEMO fixed cells as described (Mabit et al., 2002). Rabbit anti-CRM1 was obtained from M. Fornerod (Fornerod et al., 1997b) and rabbit antinuclear lamins A, B, and C peptide antibody 8188 was supplied by L. Gerace (Scripps Research Institute, La Jolla, CA; Greber et al., 1997). Rabbit anti-calnexin antibody was provided by A. Helenius (ETH Zürich, Switzerland). Goat IgG against mouse IgG coupled to Alexa 350, 488, or 594 were from Molecular Probes (Leiden, The Netherlands) and goat anti-mouse coupled to Cy5 from Jackson ImmunoResearch (West Grove, PA). DAPI (Molecular Probes) was used as indicated by the manufacturer. HRP conjugated-goat anti-rabbit antibody was from Sigma (Sigma, Fluka, Buchs, Switzerland). LMB, a generous gift of B. Wolff (Novartis Forschungsinstitut, Vienna, Austria), was dissolved in dimethyl sulfoxide (DMSO) and kept at -20°C until use. Nocodazole, thymidine and RJA were purchased from Sigma. Control treatments included DMSO alone at the carrier concentration of <0.1% (vol/vol).
siRNA Experiments
CRM1 targeting siRNA (sense UGUGGUGAAUUGCUUAUAC·d(TT); antisense GUAUAAGCAAUUCACCACA·d(TT); Lund et al., 2004) and nonsilencing control siRNA were obtained from Qiagen (Hilden, Germany). HeLa cells were transfected with 16.6 or 80 pmol of siRNA at a confluency of 40–50% in 24-well dishes on 12-mm coverslips or six-well dishes, respectively, using Oligofectamine and Opti-MEM reduced serum medium (Invitrogen), and infected with Ad5-eGFP 46 h posttransfection.
Western Blot Analysis
HeLa cells grown in 35-mm dishes were washed with phosphate-buffered saline (PBS) and lysed in 300 μl of 2% hot SDS. The lysate was passed through a 20-gauge needle several times and heated to 95°C for 30 s. After centrifugation at 16,000 × g for 10 min, 150 μl of the supernatant was mixed with 50 μl of sample buffer (200 mM Tris/HCl, pH 6.8, 8% SDS, 0.4% bromphenol blue, 40% glycerol, 167 mM dithiothreitol) and heated to 95°C for 10 min. Extracts were separated on 10% SDS-PAGE, transferred to Hybond-ECL nitrocellulose membrane (Amersham Biosciences, Zurich, Switzerland), and blocked with 5% (wt/vol) dried milk in 50 mM Tris/100 mM sodium chloride/0.1% Tween, pH 7.4 (TNT). After immunological probing HRP-conjugated antibodies were detected with ECL Plus reagents (Amersham Biosciences). Filters were stripped with 100 mM β-mercaptoethanol, 2% SDS, 62.5 mM Tris/HCl, pH 6.7, at 50°C for 30 min, washed extensively with TNT, blocked with 5% dried milk, and reprobed with an anti-calnexin antibody.
Transferrin Uptake
HeLa cells were serum starved for 4 h in DMEM-bovine serum albumin medium and incubated with 20 μg/ml human transferrin–labeled with Alexa647 (Molecular Probes) for 30 min, washed briefly and chased in transferrin-free medium for 10 min, fixed in 3% paraformaldehyde, and processed for immunofluorescence using CRM1-specific antibodies.
Metabolic Labeling
TC7 cells grown in 35-mm-diameter dishes were treated with or without 20 nM LMB and infected with Ad5-luc. One hour before lysis cells were starved in methionine-free, serum-free medium (Life Technologies, Invitrogen) at 37°C for 20 min and pulse-labeled with 5 μCi [35S]methionine for 40 min as described (Imelli et al., 2004). Cells were washed with PBS, lysed in Ripa buffer (20 mM Tris/HCl, pH 7.4, 130 mM NaCl, 2 mM EDTA, 0.1% SDS, 0.5% deoxycholate, 1% Triton X-100) and protease inhibitors (1 mM phenylmethylsulfonyl fluoride and 1 μg each of chymostatin, leupeptin, aprotinin, and pepstatin/ml), precipitated with trichloroacetic acid, and incorporated radioactivity was analyzed in a liquid scintillation counter (Beckman Coulter, Krefeld, Germany) as described (Greber et al., 1993).
Cell Cycle Synchronization
Exponentially growing cells were treated with 2 mM thymidine (Sigma) for 16 h, washed twice with serum-free medium, and released in normal medium for 12 h 10 min (adopted from Fang et al., 1998). Cells were treated with 20 nM LMB for 20 min and infected with Ad2-TR for 1 h, fixed in PHEMO, and processed for immunofluorescence as described above.
Ad5-luc and Ad5-eGFP Measurements
Triplicate sets of cells were grown on 24-well dishes in DMEM in the presence of 7% clone III, treated with drugs for 20 min, incubated with Ad5-luc on ice for 60 min, and warmed for 4 h. Cells were washed with PBS and lysed in 400 μl of lysis buffer (Luciferase Assay System, Promega, Madison, WI). Lysate, 20 μl, was mixed with 50 μl luciferase substrate and chemiluminescence was measured as described (Mabit et al., 2002). In parallel dishes the cell numbers were determined by crystal violet staining and spectrophotometry at 590 nm. For eGFP measurements, HeLa cells grown in six-well dishes were infected with 2 μg of Ad5-eGFP 46 h posttransfection of siRNA. Cells were washed with PBS and detached from the dish by trypsinization 5 h postinfection. Cells were permeabilized with 0.25% Triton X-100, blocked with 10% goat serum cells, and immunolabeled with a primary CRM1-specific antibody (Fornerod et al., 1997b), followed by a Cy5-conjugated secondary antibody (Jackson ImmunoResearch). Samples were analyzed by flow cytometry in a Beckman Coulter FC500 Cytometer equipped with a 488-nm Argon laser and a 638-nm solid state laser.
Microscopy of Infected Cells
Cells grown on 12-mm coverslips were treated with drugs for 20 min and incubated with 0.2–0.5 μg Ad2-TR or 50 μg Ad2 per 0.25 ml cold RPMI-bovine serum albumin binding medium for 45–60 min in the presence or absence of drugs. Unbound virus was washed off and cells were incubated in DMEM-bovine serum albumin with or without drug at 37°C for indicated times, fixed in pFA (or as otherwise indicated), quenched with 25 mM NH4Cl in PBS at room temperature for 5 min, and permeabilized with 0.5% Triton X-100 in PBS for 5 min. Samples were mounted in 3 μl mounting medium (DAKO, Carpinteria, CA) and analyzed by confocal laser scanning microscopy on a Leica SP2 microscope (Heerbrugg, Switzerland; 63× oil-immersion objectives) using UV excitation 351 and 364 nm, FITC 468 nm, TR 568 nm, Cy5 647 nm and long-pass emission filters in sequential recording modes at 0.5-μm section thickness. Fluorescence in situ hybridizations were carried out as described (Greber et al., 1997). Quantification of the subcellular localization of Ad2-TR was performed as described (Nakano and Greber, 2000). Electron microscopy and Ad2 particle quantifications were carried out as described (Nakano et al., 2000).
RESULTS
CRM1-mediated Nuclear Export Required for Adenovirus Transduction
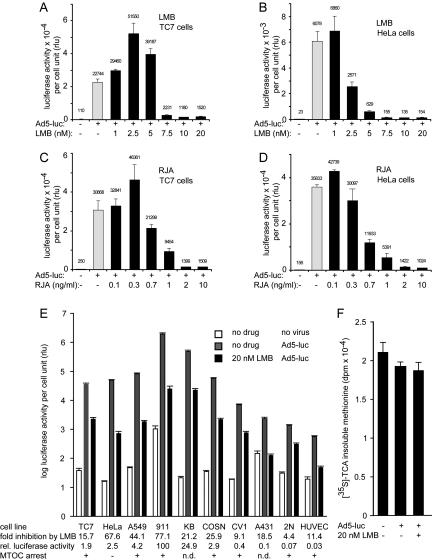
Nuclear export is a key process in viral infections and cell communication, e.g., directly involved in HIV genomic RNA assembly in the cytoplasm (for reviews, see Komeili and O'Shea, 2000; Cullen, 2003). We examined if nuclear export was required for adenovirus infection of cultured epithelial cells. HeLa or TC7 cells were treated with different concentrations of LMB, which covalently binds to CRM1 and suppresses CRM1 binding to hydrophobic NESs (Fukuda et al., 1997; Ossareh-Nazari et al., 1997; Wolff et al., 1997; Koster et al., 2003). We also used RJA, another specific inhibitor of CRM1 (Koster et al., 2003). Cells were treated with inhibitors for 30 min, inoculated with luciferase expressing transgenic adenovirus (Ad-luc) in the presence or absence of the drugs, and scored for luciferase activity 4 h postinfection. In both cell types, we measured a dose-dependent inhibition of luciferase activity with maximal effects of 20–50-fold at concentrations larger than 7.5 nM LMB and 2 ng·ml-1 RJA, respectively (Figure 1, A–D). Nuclear export of the indicator protein 2xNES-eGFP was strongly suppressed in TC7 and HeLa cells (unpublished data) using, for example, 20 nM LMB or 10 ng·ml-1 RJA, indicating that the drugs were effective as expected (see Figure 3). Interestingly, at lower doses of LMB (∼1 nM) and RJA (0.1–0.3 ng·ml-1), we observed an increased Ad-luc expression in both cell types, in the range of 0.1–2-fold compared with no-drug–treated cells (Figure 1, A–D). LMB or RJA concentrations blocking nuclear export, however, inhibited Ad-luc expression (Figure 1, A and C), ranging from 4.4- to 77-fold, depending on the cells tested (Figure 1E). The strongest effects were obtained in cells that expressed the highest levels of luciferase, i.e., human embryonic retinoblast 911 cells, followed by KB, HeLa, A549, COSN, and TC7 cells. The weakest LMB inhibitions were in the range of 4–10-fold in cells that were poorly infected with Ad5-luc, i.e., CV1 cells, the primary human foreskin fibroblasts (2N), and HUVECs. To control for cell integrity, we assessed the metabolic state of the LMB-treated cells. A dose of 20 nM LMB for 5 h did not induce any cytopathic effects indicated by viability/cytotoxicity live cell assays (unpublished data) and metabolic incorporation of [35S]methionine (Figure 1F), confirming the selectivity of the drugs.
Figure 1.
LMB and RJA inhibit adenovirus-mediated gene expression. TC7 (A and C) or HeLa (B and D) cells were treated with LMB (A and B) or RJA (C and D), incubated with Ad5-luc in the cold for 60 min (2.9 × 104 particles per cell), washed, and analyzed for luciferase activity 4 h postinfection. The mean values of triplicate samples from one typical experiment are shown as relative light units (rlu), normalized to the number of cells, including the SEs of the mean from triplicate samples. (E) The average fold inhibition of normalized luciferase activity in TC7, HeLa, A549, 911, KB, COSN, CV1, A431, normal human foreskin fibroblasts (2N), and HUVEC cells treated with 20 nM LMB, compared with non–drug-treated cells. The mean values of triplicate samples are shown. Independently, the subcellular localization of Ad2-TR in the 20 nM LMB-treated cells was scored: (+) indicating MTOC-arrest, (-) random cytoplasmic distribution and not determined phenotypes (n.d.). The viability of the cells treated with 20 nM LMB and infected with Ad5-luc was measured by metabolic labeling with [35S]methionine (F).
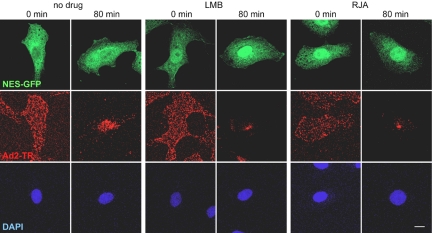
Figure 3.
LMB and RJA inhibit the nuclear export of 2xNES-eGFP and nuclear targeting of adenovirus. TC7 cells transfected with plasmid DNA encoding 2xNES-eGFP for 24 h were treated or not treated with 20 nM LMB or 10 ng·ml-1 RJA, incubated with Ad2-TR in the cold, and warmed for 0 or 80 min. Entire stacks of CLSM images are shown for 2xNES-eGFP (NES-GFP, green) and Ad2-TR (red), and sections across the cell center for DAPI stainings of DNA. Bar, 10 μm.
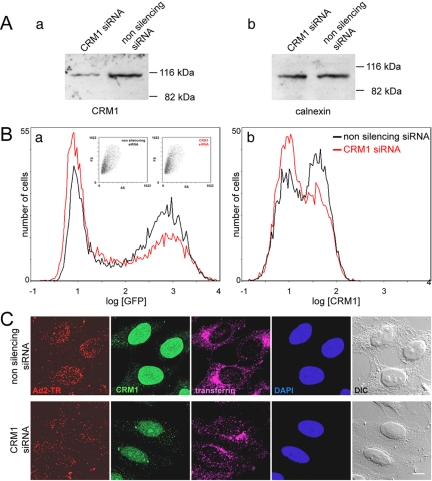
We next tested if the inhibition of adenovirus transduction by LMB and RJA was specifically due to CRM1 inactivation. HeLa cells were treated with CRM1 specific siRNAs for 46 h, infected with GFP-expressing adenovirus (Ad5-eGFP) for 5 h and analyzed for eGFP and CRM1 expression by flow cytometry (Figure 2). The CRM1 levels were reduced by ∼40% in the CRM1 siRNA-treated cells as indicated by Western blotting and flow cytometry (Figure 2, A and B), and eGFP expression was significantly lower than that in the control siRNA-treated cells, whereas the number of noninfected cells increased (Figure 2B, left peak). We observed no signs of toxicity, and this was confirmed by immunofluorescence analyses of cells labeled with transferrin-Alexa647- and Texas Red-labeled Ad2 (Ad2-TR, Figure 2C). The CRM1 siRNA-treated cells had significantly less CRM1 that controls treated with nonsilencing siRNA. Both populations readily internalized transferrin and Ad2-TR, as indicated by perinuclear endosomes and cytoplasmic viral puncta. Strikingly, the nuclear accumulation of adenovirus was strongly reduced in cells treated with CRM1-specific siRNA, similar to the LMB-treated cells (Figure 1). We also noticed that reduced nuclear targeting of adenovirus was only observed in cells with significant CRM1 silencing effects and that prolonged treatments with the CRM1 siRNAs reduced cell viability, consistent with previous observations (Lund et al., 2004). Together, this data indicated a specific requirement of CRM1 for nuclear targeting and infection of adenovirus.
Figure 2.
Knockdown of cellular CRM1 by siRNA reduces nuclear targeting of Ad2. (A) HeLa cells were transfected with siRNA specific for CRM1 or nonsilencing siRNA. Cell lysates were analyzed by Western blotting using anti-CRM1 (a) and anti-calnexin antibodies (b), respectively. (B) CRM1 specific siRNA (red) or nonsilencing siRNA (black) transfected HeLa cells were infected with eGFP-expressing Ad at 46 h posttransfection, immunolabeled for CRM1 detection, and analyzed by flow cytometry of eGFP fluorescence (a) and CRM1 (b). (C) CRM1-siRNA and nonsilencing siRNA-transfected cells were incubated with Ad2-TR in the cold for 1 h, warmed for 40 min, pulsed with transferrin-647, fixed 80 min postinfection, and immunolabeled for CRM1. The nuclei are stained with DAPI and DIC images are included for reference. Projections of entire stack of CLSM images are shown. Bar, 10 μm.
LMB Arrests Microtubule-dependent Cytoplasmic Transport of Adenovirus at the MTOC
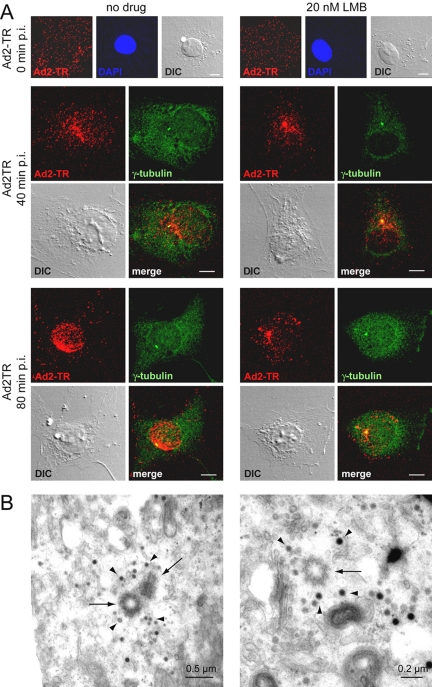
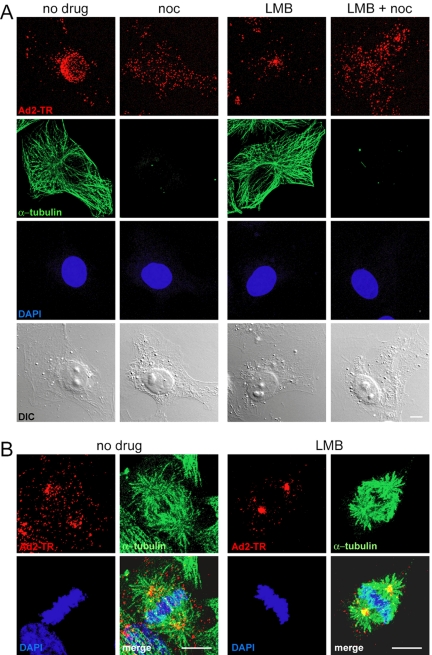
Because LMB or RJA blocked CRM1 more efficiently than the CRM1 siRNAs, we used short treatments of TC7 cells with LMB to knock down CRM1 and investigate the nature of the CRM1 block on adenovirus entry. In both control and CRM1-inhibited cells, Ad2-TR was found to be evenly distributed across the cells 0 min after warming, whereas virus was enriched only at the nucleus of the control cells 80 min postinfection (Figure 3). The incoming Ad2-TR was clustered in the LMB- or RJA-treated cells predominantly at a single spot near the nucleus. The efficacy of LMB and RJA in these cells was controlled by transfection of the nuclear export sequence bearing GFP (2xNES-eGFP) indicator protein showing a clear reduction of NES-GFP signal in the cytoplasm of LMB- or RJA-treated cells (Figure 3). These reductions were not complete but were identical to a previous report reflecting the nature of the NES-eGFP construct (Saydam et al., 2001). The enrichment of incoming adenovirus at the perinuclear punctum was time-dependent, already visible at 40 min and increased at 80 min postinfection (Figure 4). This effect was also confirmed by subcellular quantification of Ad2-TR fluorescence (unpublished data). Subsequent immunolabeling and EM experiments identified the cytoplasmic puncta of the arrested viruses. An antibody against γ-tubulin, an integral component of the centrosome (Joshi et al., 1992) strongly colocalized with LMB-arrested Ad2-TR (Figure 4A). High-resolution thin section EM of LMB-treated cells infected with Ad2 for 120 min confirmed that virus particles were enriched in a pericentriolar region, near two centrioles (Figure 4B). A similar centrosomal localization of incoming adenovirus was found in LMB-treated A549, 911, COSN, CV1, normal fibroblasts, and HUVECs (see Figure 1E) and rat kangaroo PtK2 and Vero cells using light microscopy (unpublished data). The adenovirus enrichment at the centrosome was completely blocked in nocodazole-treated cells lacking MTs (Figure 5A), indicating that LMB intercepted the incoming virus on MTs. This effect was specific for cytosolic virus, because LMB did not affect the formation of the MT network after cold-induced depolymerization, or the trafficking of transferrin (see Figure 2) or endosomal Ad2 mutant ts1 (unpublished data; Greber et al., 1996). Accordingly, Ad2-TR moved bidirectionally to and from the MTOC in both LMB and control TC7 cells (see Supplementary Movies S1 and S1). LMB-treated cells did, however, not contain any viruses on their nuclei at 30 min postinfection, unlike control cells. This effect was due to inhibition of a cellular target rather than the virus proper, because a centrosomal arrest of Ad2 was not observed if the particles but not the cells were treated with LMB (unpublished data).
Figure 4.
Adenovirus arrests at the MTOC of LMB-treated TC7 cells. (A) Cells were pretreated with or without 20 nM LMB in growth medium for 30 min, exposed to Ad2-TR in the cold for 60 min, warmed for indicated times with or without LMB, and fixed with pFA (at 0 min postinfection pi) or methanol (40 and 80 min postinfection). The MTOC was immunolabeled by with an γ-tubulin antibody. The projections of the complete stacks of confocal sections are shown. DIC images are included as a reference. Bar, 10 μm. (B) Thin-section electron micrographs across the centrosomes of Ad2-infected LMB-treated TC7 cells 120 min postinfection. Note the localization of viral particles (arrowheads) in the pericentriolar matrix near the centrioles (arrows).
Figure 5.
The MTOC arrest of adenovirus in LMB-treated TC7 cells requires MTs and also occurs in mitotic cells. (A) TC7 cells were treated with or without LMB (20 nM) and nocodazole (noc, 20 μM), infected with Ad2-TR for 80 min, fixed with PHEMO fix, and immunostained for α-tubulin (green) and DAPI (blue). DIC images are included as a reference. Bar, 10 μm. (B) Synchronized mitotic cells treated with or without LMB were infected with Ad2-TR (red) for 60 min, fixed with PHEMO fix, and stained for α-tubulin (green) and DNA (DAPI, blue). Whole-cell CLSM analyses are shown. Note the enrichment of Ad2-TR at spindle pole bodies of LMB-treated mitotic cells. Bar, 10 μm.
We next analyzed mitotic TC7 cells either treated or not treated with LMB. TC7 cells were synchronized by a thymidine block into S-phase, released and treated with LMB for 20 min, and infected with Ad2-TR for 1 h. The LMB-treated cells enriched cytosolic adenovirus particles at both spindle poles, unlike the control cells (Figure 5B). Importantly, the LMB treated cells showed no obvious cytotoxicity or mitotic defects and had normal spindle poles as judged by light microscopy. In the untreated mitotic cells, the viruses were dispersed throughout the cytoplasm not colocalizing with MTs or the spindle poles (Figure 5B and unpublished data). This stands in contrast to interphase TC7 cells where more than 95% of Ad2-TR colocalized with MTs 40 min postinfection (Stidwill and Greber, 2000). The reason for the defect of MT colocalization of adenovirus in mitotic cells is not due to an irreversible impairment of the viruses by mitotic factors, because Ad2-TR readily engaged with MTs after the cells had completed mitosis (unpublished data). The data show that MTs of normal mitotic cells fail to transport adenoviruses to the spindle poles, consistent with the notion that CRM1 or a CRM1-associated factor blocks viral attachment to MTs in the mitotic cytoplasm, including the spindle MTs and the poles. Interestingly, CRM1 immunoreactivity was found throughout the cytosol and on the spindle of mitotic TC7 cells in both control and LMB-treated cells (unpublished data), suggesting that the mitotic localization of CRM1 is independent of its NES-binding function. Nonetheless, the inhibition of mitotic CRM1 by LMB blocked viral transport at the spindle poles.
CRM1 Is Required for Ad2 Disassembly at the NPC and Nuclear Import
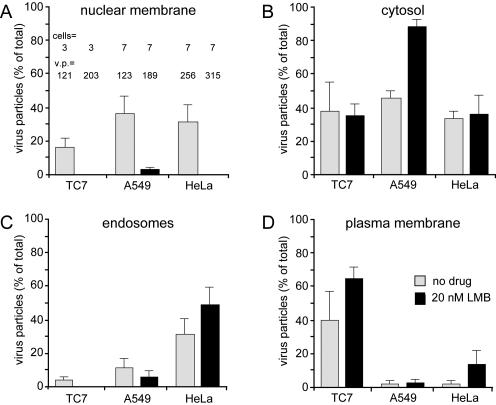
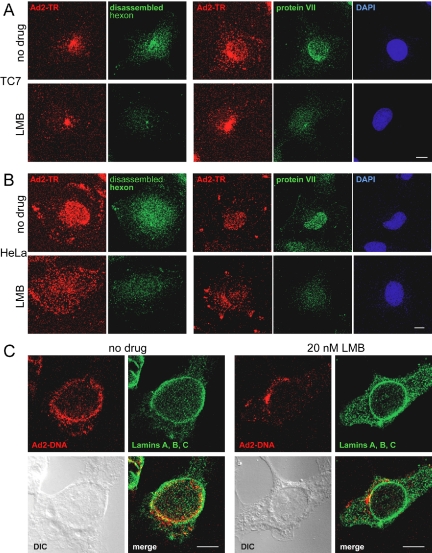
Infection requires capsid disassembly and disassembly in turn depends on the association of capsids with the NPC (Trotman et al., 2001). We therefore characterized the subcellular localizations of Ad2 in interphase cells lacking functional CRM1 in three different cell lines, TC7, A549, and HeLa cells, 60 min postinfection. Quantitative EM analyses indicated that LMB strongly reduced the number of virus particles at the nuclear membrane of all cell types, but did not significantly affect the number of cytosolic-, endosomal-, and plasma membrane–associated particles, indicating that LMB did not affect virus entry into the cytosol (Figure 6). To assess the status of the cytosolic capsids that failed to associate with the NPC, we analyzed capsid disassembly in both TC7 and HeLa cells using the disassembly-specific anti-hexon antibody R70 (Baum et al., 1972; Trotman et al., 2001). The results show that the MTOC-arrested Ad2-TR capsids were largely devoid of R70 epitopes in the LMB-treated TC7 cells but were strongly R70-positive in the control cells (Figure 7A). Likewise, LMB strongly inhibited the R70 staining of the randomly dispersed cytosolic Ad2 in HeLa cells (Figure 7B). These results were confirmed by immunostainings of the viral DNA-associated protein VII, which is protected in the intact capsid and not accessible to antibodies by immunofluorescence analyses, unless detached from the capsid (Greber et al., 1997). LMB strongly blocked the appearance of protein VII in the nucleus of infected TC7 and HeLa cells (Figure 7). This result was further confirmed by fluorescence in situ hybridizations monitoring the incoming viral genome. In LMB-treated TC7 cells the viral DNA was absent from the nucleoplasm but enriched in a perinuclear punctum and smaller cytoplasmic dots reinforcing the notion that CRM1 is necessary for targeting functional adenoviruses from the MTs to the NPC and presenting them to the disassembly machinery.
Figure 6.
Inhibition of nuclear envelope targeting of adenovirus in TC7, A549, and HeLa cells. Quantitative subcellular analyses of Ad2 by thin-section EM in LMB-treated and control TC7, A549, and HeLa cells. Viral particles (v.p.) were counted at the nuclear membrane (A), in the cytosol (B), endosomes (C), and at the plasma membrane (D). Results indicate the means and SE from the number of analyzed cells and v.p.
Figure 7.
LMB inhibits adenovirus capsid disassembly and nuclear import of protein VII and viral DNA. LMB-treated and control TC7 (A) and HeLa (B) cells were infected with Ad2 for 150 min, fixed, and immunostained for the disassembled capsid protein hexon and the DNA-associated core protein VII. The merged stacks of the entire set of CLSM sections are shown. Bar, 10 μm. (C) LMB blocks Ad2 DNA import into the nucleus. TC7 cells were infected with Ad2 in the presence or absence of LMB for 180 min, processed for in situ hybridization, and immunostained for lamins A/B/C. Single CLSM sections across a central plane of the cells are shown including DIC. Bar, 10 μm.
DISCUSSION
Viral invasion of the nucleus depends on the coordination of many host functions. This facilitates the propagation of, for example, adenoviruses in almost all vertebrates (Davison et al., 2003). Of the more than 50 human adenovirus serotypes, the respiratory viruses Ad2 and Ad5 cause severe infections in immunocompromised patients (Horwitz, 2001). In modified forms these vectors are widely used for gene transfer (Russell, 2000). They enter epithelial cells by clathrin-mediated endocytosis, activate macropinocytosis, escape to the cytosol in an integrin-dependent manner, and efficiently deliver their DNA genome into the nucleus (Meier et al., 2002; Imelli et al., 2004). Cytosolic particles are transported on preferentially stable MTs by the dynein/dynactin motor complex toward the MTOC, often located near the nucleus (Suomalainen et al., 1999; Leopold et al., 2000; Giannakakou et al., 2002). The first viruses reaching the nucleus are those trafficking near the MTOC, suggesting that this region is critical to hand over the viruses from the MTs to the nuclear membrane. It is unknown how viruses are detached from MTs.
Here we provide evidence linking two previously unconnected pathways, nuclear export and cytoplasmic transport on microtubules. Our in vivo results provide a molecular explanation for the observation that enucleated human epithelial cells arrested Ad5 particles at the MTOC (Bailey et al., 2003). Using CRM1-specific drugs and siRNAs, we show that the export factor CRM1 acts as a positional indicator of the nucleus for the incoming adenovirus. If CRM1 function is blocked, the detachment of adenoviruses from the MTs fails and viral binding to the nuclear membrane is prevented. In normal cells, CRM1 predominantly localizes to the nucleus and functions to export NES-containing proteins from the nucleus to the cytoplasm (reviewed in Cullen, 2003; Greber and Fornerod, 2005). It is potently inhibited by the bacterial polyketides LMB and RJA. These inhibitors are highly specific because 1) an Schizosaccharomyces pombe strain expressing an LMB-resistant CRM1 was found to be insensitive to LMB and 2) LMB bound specifically to CRM1 in the low nanomolar range (Neville et al., 1997; Kudo et al., 1999; Koster et al., 2003), exactly matching our experimental conditions for blocking CRM1. Both LMB and RJA blocked the transport of incoming adenoviruses at the MTOC in many different cell types or apparently randomly in the HeLa cytoplasm. A similar nuclear transport block was established in HeLa cells by siRNA-mediated knock down of CRM1. In the absence of functional CRM1, the adenoviruses failed to detach from the MTs and the MTOC, did not bind to NPCs, and did not disassemble and import the DNA into the nucleus, which is normally mediated by the NPC and associated proteins (Saphire et al., 2000; Trotman et al., 2001). Thus, LMB and RJA are novel viral entry inhibitors, establishing a previously unknown postentry block of infections. They are among a small number of compounds restricting viral entry (McKinlay et al., 1992; Greber et al., 1994).
Host restriction factors are part of innate immunity reactions against incoming retroviruses and are responsible for the narrow host range of these viruses (Goff, 2004). Restrictions are targeted against the capsid protein before reverse transcription. HIV, for example, overcomes these restrictions by incorporating cyclophilin A, a modulator of the viral sensitivity to innate immunity factors (Towers et al., 2003). In addition, viruses that are targeted to the MTOC by the dynein/dynactin motor complex, such as the adenovirus and HIV could be subject to another postentry host restriction point, degradation by the aggresomal pathway that collects misfolded or otherwise abnormal proteins (Kopito, 2000). By recruiting CRM1 adenovirus could escape from this restriction and target to the NPC for uncoating. Whether CRM1 acts as a gatekeeper or is mobilized to the virus is not known. We can envision several modes how CRM1 overcomes an entry block. Normally, CRM1 mediates nuclear export of a large variety of leucine-rich export sequence containing substrates by forming a complex with the GTP-bound form of the small GTPase Ran in the nucleus (Petosa et al., 2004). It is possible that either CRM1 alone, a short-lived CRM1-NES-Ran:GTP complex or an unknown NES-containing nuclear protein interact with adenovirus. If CRM1 alone interacted with adenovirus this interaction could be of supraphysiological affinity independent of Ran (Engelsma et al., 2004). In this case, CRM1 would stay bound to the capsid until disassembly at the NPC had occurred. A cytosolic CRM1-NES-Ran:GTP complex on the other hand would be a regulated MT dislodging trigger, because it has a limited life time owing to the hydrolysis of GTP activated by the cytoplasmic RanGAP protein (Bednenko et al., 2003; Quimby and Dasso, 2003). The chromatin-bound GTP/GDP exchange factor RCC1 in turn activates the complex by loading Ran:GTP and establishes a gradient across the nuclear membrane in intact cells and also around mitotic chromosomes in cell-free systems (Kalab et al., 2002; Macara, 2002; Weis, 2002). This regulated scenario is attractive because it would account for our observation that adenoviruses were prevented from association with both spindle and astral MTs of mitotic cells. This block was relieved by LMB leading to virus accumulation at the spindle poles. In fact, experiments with Xenopus egg extracts have suggested that CRM1 is in a complex with Ran:GTP proximal to mitotic chromatin (Kalab et al., 2002), although it is not known if a Ran:GTP gradient exists around chromosomes of epithelial cells (Gorlich et al., 2003). Regardless, even if Ran:GTP were randomly distributed in mitotic TC7 cells, it could account for dislodging adenovirus particles from MTs. On exit of the cells from mitosis, CRM1 would be retrieved to the newly formed nucleus together with Ran:GTP and the cytosolic adenoviruses could be transported to the nucleus using microtubules, as observed in our studies. The scenario that a NES-containing cargo alone unloads the incoming adenovirus from the MTs is perhaps less likely because in mitotic cells with mixed nuclear and cytoplasmic contents, LMB was very effective at clustering viruses at the spindle poles, very similar to MTOC clustering in interphase cells. This suggests that the export function of CRM1 is not needed for dislodging adenovirus from MTs.
There is yet another possibility to carry CRM1 to perinuclear MTs of interphase cells, namely via the nucleoporins RanBP2 or Nup214/CAN, as suggested by the observation of RanBP2 antigens at the MTOC (Salina et al., 2003). Nucleoporin extensions might be possible via extended conformations of the FG domains, which could amount to some hundred nanometers. Regardless, the requirement of CRM1 for nuclear targeting of adenovirus is strong, because adenoviruses are retained at the MTOC even if the MTOC is positioned in immediate vicinity to the nuclear membrane. Interestingly, CRM1 has been found at low concentrations at the MTOC and it was postulated to be required to control centriole synthesis (Forgues et al., 2003). It is thus feasible that CRM1 is part of a centrosomal network, including kinases, phosphatases, scaffolding proteins, and nuclear envelope proteins. Indeed, a small fraction of Ran is tightly associated with the centrosome throughout the cell cycle (Zimmerman and Doxsey, 2000; Keryer et al., 2003). Other centrosomal proteins such as centrin and pericentrin shuttle between the nucleus and the centrosome in an LMB-dependent manner. This suggests that centrosomal activities can be regulated by CRM1-dependent nuclear export involving specific cargo proteins and Ran. It is thus possible that viral infectivity is both positively and negatively regulated by centrosomal or cytosolic CRM1, because very low concentrations of LMB or RJA increased Ad5 gene expressions. Future experiments will investigate how CRM1 interacts with cytosolic adenovirus or affects an active transport system carrying perinuclear viruses. This will be crucial to address the issues of innate immunity and host restrictions as well as turnover and immunogenicity of vectors.
Acknowledgments
We thank B. Wolff (Novartis, Vienna) for generous gift of LMB, S. Güttinger and U. Kutay (ETH Zürich, Switzerland) for CRM1 siRNA and advice, and M. Fornerod, F. Melchior, C. Burckhardt, and D. Püntener for discussions. This work was supported by the Swiss National Science Foundation and the Kanton Zürich (U.F.G.).
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E05-02-0121) on April 6, 2005.
Abbreviations used: Ad, adenovirus; DAPI, 4′,6-diamidino-2-phenylindole dihydrochloride; DIC, differential interference contrast; LMB, leptomycin B; MT, microtubule; MTOC, microtubule organizing center; NPC, nuclear pore complex; RJA, ratjadone A; TR, Texas Red.
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
References
- Alonso, C., Miskin, J., Hernaez, B., Fernandez-Zapatero, P., Soto, L., Canto, C., Rodriguez-Crespo, I., Dixon, L., and Escribano, J. M. (2001). African swine fever virus protein p54 interacts with the microtubular motor complex through direct binding to light-chain dynein. J. Virol. 75, 9819-9827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey, C. J., Crystal, R.G., and Leopold, P. L. (2003). Association of adenovirus with the microtubule organizing center. J. Virol. 77, 13275-13287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum, S. G., Horwitz, M. S., and J. V. Maizel, J. (1972). Studies of the mechanism of enhancement of human adenovirus infection in monkey cells by simian virus 40. J. Virol. 10, 211-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bednenko, J., Cingolani, G., and Gerace, L. (2003). Nucleocytoplasmic transport: navigating the channel. Traffic 4, 127-135. [DOI] [PubMed] [Google Scholar]
- Bienz, M. (2002). The subcellular destinations of APC proteins. Nat. Rev. Mol. Cell. Biol. 3, 328-338. [DOI] [PubMed] [Google Scholar]
- Bornens, M. (2002). Centrosome composition and microtubule anchoring mechanisms. Curr. Opin. Cell Biol. 14, 25-34. [DOI] [PubMed] [Google Scholar]
- Crepieux, P., Kwon, H., Leclerc, N., Spencer, W., Richard, S., Lin, R., and Hiscott, J. (1997). I kappaB alpha physically interacts with a cytoskeleton-associated protein through its signal response domain. Mol. Cell. Biol. 17, 7375-7385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen, B. R. (2003). Nuclear mRNA export: insights from virology. Trends Biochem. Sci. 28, 419-424. [DOI] [PubMed] [Google Scholar]
- Davison, A. J., Benko, M., and Harrach, B. (2003). Genetic content and evolution of adenoviruses. J. Gen. Virol. 84, 2895-2908. [DOI] [PubMed] [Google Scholar]
- Dohner, K., Wolfstein, A., Prank, U., Echeverri, C., Dujardin, D., Vallee, R., and Sodeik, B. (2002). Function of dynein and dynactin in herpes simplex virus capsid transport. Mol. Biol. Cell 13, 2795-2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas, M. W., Diefenbach, R. J., Homa, F. L., Miranda-Saksena, M., Rixon, F. J., Vittone, V., Byth, K., and Cunningham, A. L. (2004). Herpes simplex virus type 1 capsid protein VP26 interacts with dynein light chains RP3 and Tctex1 and plays a role in retrograde cellular transport. J. Biol. Chem. 279, 28522-28530. [DOI] [PubMed] [Google Scholar]
- Engelsma, D., Bernad, R., Calafat, J., and Fornerod, M. (2004). Supraphysiological nuclear export signals bind CRM1 independently of RanGTP and arrest at Nup358. EMBO J. 23, 3643-3652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fallaux, F. J. et al. (1998). New helper cells and matched early region 1-deleted adenovirus vectors prevent generation of replication-competent adenoviruses. Hum. Gene Ther. 9, 1909-1917. [DOI] [PubMed] [Google Scholar]
- Fang, G., Yu, H., and Kirschner, M. W. (1998). Direct binding of CDC20 protein family members activates the anaphase-promoting complex in mitosis and G1. Mol. Cell 2, 163-171. [DOI] [PubMed] [Google Scholar]
- Finke, S., Brzozka, K., and Conzelmann, K. K. (2004). Tracking fluorescence-labeled rabies virus: enhanced green fluorescent protein-tagged phosphoprotein P supports virus gene expression and formation of infectious particles. J. Virol. 78, 12333-12343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forgues, M., Difilippantonio, M. J., Linke, S. P., Ried, T., Nagashima, K., Feden, J., Valerie, K., Fukasawa, K., and Wang, X. W. (2003). Involvement of Crm1 in hepatitis B virus X protein-induced aberrant centriole replication and abnormal mitotic spindles. Mol. Cell. Biol. 23, 5282-5292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornerod, M., Ohno, M., Yoshida, M., and Mattaj, I. W. (1997a). CRM1 is an export receptor for leucine-rich nuclear export signals. Cell 90, 1051-1060. [DOI] [PubMed] [Google Scholar]
- Fornerod, M., van Deursen, J., van Baal, S., Reynolds, A., Davis, D., Murti, K. G., Fransen, J., and Grosveld, G. (1997b). The human homologue of yeast CRM1 is in a dynamic subcomplex with CAN/Nup214 and a novel nuclear pore component Nup88. EMBO J. 16, 807-816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda, M., Asano, S., Nakamura, T., Adachi, M., Yoshida, M., Yanagida, M., and Nishida, E. (1997). CRM1 is responsible for intracellular transport mediated by the nuclear export signal. Nature 390, 308-311. [DOI] [PubMed] [Google Scholar]
- Giannakakou, P., Nakano, M. Y., Nicolaou, K. C., O'Brate, A., Yu, J., Blagosklonny, M. V., Greber, U. F., and Fojo, T. (2002). Enhanced microtubule-dependent trafficking and p53 nuclear accumulation by suppression of microtubule dynamics. Proc. Natl. Acad. Sci. USA 99, 10855-10860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannakakou, P., Sackett, D. L., Ward, Y., Webster, K. R., Blagosklonny, M. V., and Fojo, T. (2000). p53 is associated with cellular microtubules and is transported to the nucleus by dynein. Nat. Cell Biol. 2, 709-717. [DOI] [PubMed] [Google Scholar]
- Gluzman, Y. (1981). SV40-transformed simian cells support the replication of early SV40 mutants. Cell 23, 175-182. [DOI] [PubMed] [Google Scholar]
- Goff, S. P. (2004). Retrovirus restriction factors. Mol. Cell 16, 849-859. [DOI] [PubMed] [Google Scholar]
- Gorlich, D., and Kutay, U. (1999). Transport between the cell nucleus and the cytoplasm. Annu. Rev. Cell Dev. Biol. 15, 607-660. [DOI] [PubMed] [Google Scholar]
- Gorlich, D., Seewald, M. J., and Ribbeck, K. (2003). Characterization of Randriven cargo transport and the RanGTPase system by kinetic measurements and computer simulation. EMBO J. 22, 1088-1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greber, U. F., and Fornerod, M. (2005). Nuclear import in viral infections. Curr. Top. Microbiol. Immunol. 285, 109-138. [DOI] [PubMed] [Google Scholar]
- Greber, U. F., Nakano, M. Y., and Suomalainen, M. (1998). Adenovirus entry into cells: a quantitative fluorescence microscopy approach. In: Adenovirus Methods and Protocols, Methods in Molecular Medicine, Vol. 21, ed. W.S.M. Wold, Totowa, NJ: Humana Press, 217-230. [Google Scholar]
- Greber, U. F., Singh, I., and Helenius, A. (1994). Mechanisms of virus uncoating. Trends Microbiol. 2, 52-56. [DOI] [PubMed] [Google Scholar]
- Greber, U. F., Suomalainen, M., Stidwill, R. P., Boucke, K., Ebersold, M., and Helenius, A. (1997). The role of the nuclear pore complex in adenovirus DNA entry. EMBO J. 16, 5998-6007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greber, U. F., Webster, P., Weber, J., and Helenius, A. (1996). The role of the adenovirus protease in virus entry into cells. EMBO J. 15, 1766-1777. [PMC free article] [PubMed] [Google Scholar]
- Greber, U. F., Willetts, M., Webster, P., and Helenius, A. (1993). Stepwise dismantling of adenovirus 2 during entry into cells. Cell 75, 477-486. [DOI] [PubMed] [Google Scholar]
- Grieshaber, S. S., Grieshaber, N. A., and Hackstadt, T. (2003). Chlamydia trachomatis uses host cell dynein to traffic to the microtubule-organizing center in a p50 dynamitin-independent process. J. Cell Sci. 116, 3793-3802. [DOI] [PubMed] [Google Scholar]
- Hemmi, S., Bohni, R., Stark, G., Di Marco, F., and Aguet, M. (1994). A novel member of the interferon receptor family complements functionality of the murine interferon gamma receptor in human cells. Cell 76, 803-810. [DOI] [PubMed] [Google Scholar]
- Horwitz, M. S. (2001). Adenoviruses. In: Fields Virology, eds. D. M. Knipe and P. M. Howley, Philadelphia, PA: Raven Press, 2301-2326.
- Imelli, N., Meier, O., Boucke, K., Hemmi, S., and Greber, U. F. (2004). Cholesterol is required for endocytosis and endosomal escape of adenovirus type 2. J. Virology 78, 3089-3098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob, Y., Badrane, H., Ceccaldi, P. E., and Tordo, N. (2000). Cytoplasmic dynein LC8 interacts with lyssavirus phosphoprotein. J. Virol. 74, 10217-10222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi, H. C., Palacios, M. J., McNamara, L., and Cleveland, D. W. (1992). Gamma-tubulin is a centrosomal protein required for cell cycle-dependent microtubule nucleation. Nature 356, 80-83. [DOI] [PubMed] [Google Scholar]
- Jouvenet, N., Monaghan, P., Way, M., and Wileman, T. (2004). Transport of African swine fever virus from assembly sites to the plasma membrane is dependent on microtubules and conventional kinesin. J. Virol. 78, 7990-8001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalab, P., Weis, K., and Heald, R. (2002). Visualization of a Ran-GTP gradient in interphase and mitotic Xenopus egg extracts. Science 295, 2452-2456. [DOI] [PubMed] [Google Scholar]
- Kelkar, S. A., Pfister, K. K., Crystal, R. G., and Leopold, P. L. (2004). Cytoplasmic dynein mediates adenovirus binding to microtubules. J. Virol. 78, 10122-10132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keryer, G., Di Fiore, B., Celati, C., Lechtreck, K. F., Mogensen, M., Delouvee, A., Lavia, P., Bornens, M., and Tassin, A. M. (2003). Part of Ran is associated with AKAP450 at the centrosome: involvement in microtubule-organizing activity. Mol. Biol. Cell 14, 4260-4271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komeili, A., and O'Shea, E. K. (2000). Nuclear transport and transcription. Curr. Opin. Cell Biol. 12, 355-360. [DOI] [PubMed] [Google Scholar]
- Kopito, R. R. (2000). Aggresomes, inclusion bodies and protein aggregation. Trends Cell Biol. 10, 524-530. [DOI] [PubMed] [Google Scholar]
- Koster, M., Lykke-Andersen, S., Elnakady, Y.A., Gerth, K., Washausen, P., Hofle, G., Sasse, F., Kjems, J., and Hauser, H. (2003). Ratjadones inhibit nuclear export by blocking CRM1/exportin 1. Exp. Cell Res. 286, 321-331. [DOI] [PubMed] [Google Scholar]
- Kreis, T. E. (1987). Microtubules containing detyrosinated tubulin are less dynamic. EMBO J. 6, 2597-2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudo, N., Taoka, H., Toda, T., Yoshida, M., and Horinouchi, S. (1999). A novel nuclear export signal sensitive to oxidative stress in the fission yeast transcription factor Pap1. J. Biol. Chem. 274, 15151-15158. [DOI] [PubMed] [Google Scholar]
- Lakadamyali, M., Rust, M. J., Babcock, H. P., and Zhuang, X. (2003). Visualizing infection of individual influenza viruses. Proc. Natl. Acad. Sci. USA 100, 9280-9285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leopold, P. L., Kreitzer, G., Miyazawa, N., Rempel, S., Pfister, K. K., Rodriguez-Boulan, E., and Crystal, R. G. (2000). Dynein- and microtubule-mediated translocation of adenovirus serotype 5 occurs after endosomal lysis. Hum. Gene. Ther. 11, 151-165. [DOI] [PubMed] [Google Scholar]
- Luby-Phelps, K. (2000). Cytoarchitecture and physical properties of cytoplasm: volume, viscosity, diffusion, intracellular surface area. Int. Rev. Cytol. 192, 189-221. [DOI] [PubMed] [Google Scholar]
- Lund, E., Guttinger, S., Calado, A., Dahlberg, J. E., and Kutay, U. (2004). Nuclear export of microRNA precursors. Science 303, 95-98. [DOI] [PubMed] [Google Scholar]
- Mabit, H., Nakano, M. Y., Prank, U., Saam, B., Döhner, K., Sodeik, B., and Greber, U. F. (2002). Intact microtubules support Adenovirus and Herpes simplex virus infections. J. Virol. 76, 9962-9971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macara, I. G. (2002). Why FRET about Ran? Dev. Cell 2, 379-380. [DOI] [PubMed] [Google Scholar]
- Mahajan, R., Delphin, C., Guan, T., Gerace, L., and Melchior, F. (1997). A small ubiquitin-related polypeptide involved in targeting RanGAP1 to nuclear pore complex protein RanBP2. Cell 88, 97-107. [DOI] [PubMed] [Google Scholar]
- Martin-Fernandez, M., Longshaw, S. V., Kirby, I., Santis, G., Tobin, M. J., Clarke, D. T., and Jones, G. R. (2004). Adenovirus Type-5 entry and disassembly followed in living cells by FRET, fluorescence anisotropy, and FLIM. Biophys. J. 87, 1316-1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald, D., Vodicka, M. A., Lucero, G., Svitkina, T. M., Borisy, G. G., Emerman, M., and Hope, T. J. (2002). Visualization of the intracellular behavior of HIV in living cells. J. Cell Biol. 159, 441-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinlay, M. A., Pevear, D. C., and Rossmann, M. G. (1992). Treatment of the picornavirus common cold by inhibitors of viral uncoating and attachment. Annu. Rev. Microbiol. 46, 635-654. [DOI] [PubMed] [Google Scholar]
- Meier, O., Boucke, K., Vig, S., Keller, S., Stidwill, R. P., Hemmi, S., and Greber, U. F. (2002). Adenovirus triggers macropinocytosis and endosomal leakage together with its clathrin mediated uptake. J. Cell Biol. 158, 1119-1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano, M. Y., Boucke, K., Suomalainen, M., Stidwill, R. P., and Greber, U. F. (2000). The first step of adenovirus type 2 disassembly occurs at the cell surface, independently of endocytosis and escape to the cytosol. J. Virol. 74, 7085-7095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano, M. Y., and Greber, U. F. (2000). Quantitative microscopy of fluorescent adenovirus entry. J. Struct. Biol. 129, 57-68. [DOI] [PubMed] [Google Scholar]
- Neville, M., Stutz, F., Lee, L., Davis, L. I., and Rosbash, M. (1997). The importin-beta family member Crm1p bridges the interaction between Rev and the nuclear pore complex during nuclear export. Curr. Biol. 7, 767-775. [DOI] [PubMed] [Google Scholar]
- Ossareh-Nazari, B., Bachelerie, F., and Dargemont, C. (1997). Evidence for a role of CMR1 in signal-mediated nuclear protein export. Science 278, 141-144. [DOI] [PubMed] [Google Scholar]
- Petit, C., Giron, M. L., Tobaly-Tapiero, J., Bittoun, P., Real, E., Jacob, Y., Tordo, N., De The, H., and Saib, A. (2003). Targeting of incoming retroviral Gag to the centrosome involves a direct interaction with the dynein light chain 8. J. Cell Sci. 116, 3433-3442. [DOI] [PubMed] [Google Scholar]
- Petosa, C., Schoehn, G., Askjaer, P., Bauer, U., Moulin, M., Steuerwald, U., Soler-Lopez, M., Baudin, F., Mattaj, I. W., and Muller, C. W. (2004). Architecture of CRM1/Exportin1 suggests how cooperativity is achieved during formation of a nuclear export complex. Mol. Cell 16, 761-775. [DOI] [PubMed] [Google Scholar]
- Ploubidou, A., and Way, M. (2001). Viral transport and the cytoskeleton. Curr. Opin. Cell Biol. 13, 97-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puthalakath, H., Huang, D. C., O'Reilly, L. A., King, S. M., and Strasser, A. (1999). The proapoptotic activity of the Bcl-2 family member Bim is regulated by interaction with the dynein motor complex. Mol. Cell 3, 287-296. [DOI] [PubMed] [Google Scholar]
- Quimby, B. B., and Dasso, M. (2003). The small GTPase Ran: interpreting the signs. Curr. Opin. Cell Biol. 15, 338-344. [DOI] [PubMed] [Google Scholar]
- Raux, H., Flamand, A., and Blondel, D. (2000). Interaction of the rabies virus P protein with the LC8 dynein light chain. J. Virol. 74, 10212-10216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell, W. C. (2000). Update on adenovirus and its vectors. J. Gen. Virol. 81, 2573-2604. [DOI] [PubMed] [Google Scholar]
- Salina, D., Enarson, P., Rattner, J. B., and Burke, B. (2003). Nup358 integrates nuclear envelope breakdown with kinetochore assembly. J. Cell Biol. 162, 991-1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saphire, A.C.S., Guan, T. L., Schirmer, E. C., Nemerow, G. R., and Gerace, L. (2000). Nuclear import of adenovirus DNA in vitro involves the nuclear protein import pathway and hsc70. J. Biol. Chem. 275, 4298-4304. [DOI] [PubMed] [Google Scholar]
- Saxton, W. M. (2001). Microtubules, motors, and mRNA localization mechanisms: watching fluorescent messages move. Cell 107, 707-710. [DOI] [PubMed] [Google Scholar]
- Saydam, N., Georgiev, O., Nakano, M. Y., Greber, U. F., and Schaffner, W. (2001). Nucleo-cytoplasmic trafficking of metal-regulatory transcription factor 1 is regulated by diverse stress signals. J. Biol. Chem. 276, 25487-25495. [DOI] [PubMed] [Google Scholar]
- Sfakianos, J. N., LaCasse, R. A., and Hunter, E. (2003). The M-PMV cytoplasmic targeting-retention signal directs nascent Gag polypeptides to a pericentriolar region of the cell. Traffic 4, 660-670. [DOI] [PubMed] [Google Scholar]
- Smith, G. A., and Enquist, L. W. (2002). Break ins and break outs: viral interactions with the cytoskeleton of mammalian cells. Annu. Rev. Cell Dev. Biol. 18, 135-161. [DOI] [PubMed] [Google Scholar]
- Sodeik, B. (2000). Mechanisms of viral transport in the cytoplasm. Trends Microbiol. 8, 465-472. [DOI] [PubMed] [Google Scholar]
- Stidwill, R. S., and Greber, U. F. (2000). Intracellular virus trafficking reveals physiological characteristics of the cytoskeleton. News Physiol. Sci. 15, 67-71. [DOI] [PubMed] [Google Scholar]
- Suikkanen, S., Aaltonen, T., Nevalainen, M., Valilehto, O., Lindholm, L., Vuento, M., and Vihinen-Ranta, M. (2003). Exploitation of microtubule cytoskeleton and dynein during parvoviral traffic toward the nucleus. J. Virol. 77, 10270-10279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suomalainen, M., Nakano, M. Y., Boucke, K., Keller, S., and Greber, U. F. (2001). Adenovirus-activated PKA and p38/MAPK pathways boost microtubule-mediated nuclear targeting of virus. EMBO J. 20, 1310-1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suomalainen, M., Nakano, M. Y., Boucke, K., Keller, S., Stidwill, R. P., and Greber, U. F. (1999). Microtubule-dependent minus and plus end-directed motilities are competing processes for nuclear targeting of adenovirus. J. Cell Biol. 144, 657-672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towers, G. J., Hatziioannou, T., Cowan, S., Goff, S. P., Luban, J., and Bieniasz, P. D. (2003). Cyclophilin A modulates the sensitivity of HIV-1 to host restriction factors. Nat. Med. 9, 1138-1143. [DOI] [PubMed] [Google Scholar]
- Trotman, L. C., Mosberger, N., Fornerod, M., Stidwill, R. P., and Greber, U. F. (2001). Import of adenovirus DNA involves the nuclear pore complex receptor CAN/Nup214 and histone H1. Nat. Cell Biol. 3, 1092-1100. [DOI] [PubMed] [Google Scholar]
- Verhey, K. J., and Rapoport, T. A. (2001). Kinesin carries the signal. Trends. Biochem. Sci. 26, 545-550. [DOI] [PubMed] [Google Scholar]
- Weis, K. (2002). Nucleocytoplasmic transport: cargo trafficking across the border. Curr. Opin. Cell Biol. 14, 328-335. [DOI] [PubMed] [Google Scholar]
- Wolff, B., Sanglier, J. J., and Wang, Y. (1997). Leptomycin B is an inhibitor of nuclear export: inhibition of nucleo-cytoplasmic translocation of the human immunodeficiency virus type 1 (HIV-1) Rev protein and Rev-dependent mRNA. Chem. Biol. 4, 139-147. [DOI] [PubMed] [Google Scholar]
- Zimmerman, W., and Doxsey, S. J. (2000). Construction of centrosomes and spindle poles by molecular motor-driven assembly of protein particles. Traffic 1, 927-934. [PubMed] [Google Scholar]