Abstract
From an insertional mutagenesis screen, we isolated a novel gene, mto2+, involved in microtubule organization in fission yeast. mto2Δ strains are viable but exhibit defects in interphase microtubule nucleation and in formation of the postanaphase microtubule array at the end of mitosis. The mto2Δ defects represent a subset of the defects displayed by cells deleted for mto1+ (also known as mod20+ and mbo1+), a centrosomin-related protein required to recruit the γ-tubulin complex to cytoplasmic microtubule-organizing centers (MTOCs). We show that mto2p colocalizes with mto1p at MTOCs throughout the cell cycle and that mto1p and mto2p coimmunoprecipitate from cytoplasmic extracts. In vitro studies suggest that mto2p binds directly to mto1p. In mto2Δ mutants, although some aspects of mto1p localization are perturbed, mto1p can still localize to spindle pole bodies and the cell division site and to “satellite” particles on interphase microtubules. In mto1Δ mutants, localization of mto2p to all of these MTOCs is strongly reduced or absent. We also find that in mto2Δ mutants, cytoplasmic forms of the γ-tubulin complex are mislocalized, and the γ-tubulin complex no longer coimmunoprecipitates with mto1p from cell extracts. These experiments establish mto2p as a major regulator of mto1p-mediated microtubule nucleation by the γ-tubulin complex.
INTRODUCTION
The γ-tubulin complex is a large, conserved multisubunit protein complex involved in microtubule nucleation in eukaryotic cells (Stearns and Kirschner, 1994; Zheng et al., 1995; for reviews, see Gunawardane et al., 2000; Oakley, 2000; Schiebel, 2000; Job et al., 2003). In a variety of cell types, the γ-tubulin complex is found both localized to microtubule-organizing centers (MTOCs) and also in significant levels in cytoplasmic pools. Although much of the characterization of γ-tubulin function has involved work on soluble complexes, most MTOCs in vivo are associated with largely insoluble compartments or subcellular organelles, and as a result we currently understand very little of the mechanisms controlling the intracellular localization and activity of the γ-tubulin complex and how these mechanisms might be regulated. Recently, a small number of proteins have been identified that may interact with the γ-tubulin complex and recruit it to structures such as the centrosome, the Golgi apparatus, or other MTOCs (Takahashi et al., 2002; Terada et al., 2003; Kawaguchi and Zheng, 2004; Rios et al., 2004; Sawin et al., 2004; Thompson et al., 2004; Venkatram et al., 2004; Zimmerman et al., 2004a,b). However, in this area much still remains to be learned.
The fission yeast Schizosaccharomyces pombe has recently become a useful model system for understanding microtubule nucleation and dynamics. Three distinct patterns of microtubule nucleation take place during the mitotic cell cycle in fission yeast (Hagan, 1998). During interphase, dynamic microtubules tend to run along the long axis of the cylindrically shaped cells and terminate at cell tips. These microtubules are nucleated from cytoplasmic interphase MTOCs (iMTOCs) in the middle of the cell that may be associated with the nuclear envelope or with microtubule bundles themselves (Drummond and Cross, 2000; Tran et al., 2001; Sawin et al., 2004; Zimmerman et al., 2004a). Nucleation sites on the surface of the nucleus are easily recognized in microtubule regrowth experiments after microtubule depolymerization, but whether they exist in the same form during steady-state growth is unclear. By contrast, although de novo nucleation of microtubules from microtubule bundles has not yet been directly observed in growing cells, components of the γ-tubulin complex are seen not only at the spindle pole body (SPB, the yeast centrosome equivalent) but also in the form of small satellite particles moving along microtubules (Sawin et al., 2004; Zimmerman et al., 2004a). Against this background of incomplete understanding, in this work we apply the term “iMTOC” somewhat freely, both to nuclear surface nucleation sites and to the aforesaid microtubule-associated particles. During mitosis, duplicated SPBs nucleate an intranuclear mitotic spindle, as well as cytoplasmic astral microtubules, which may help to position the spindle and/or coordinate mitotic progression with cytokinesis (Gachet et al., 2001; Oliferenko and Balasubramanian, 2002; Gachet et al., 2004). At the end of mitosis, a specialized microtubule structure, termed the postanaphase array (PAA), is nucleated from an equatorial MTOC (eMTOC) at the cell division site (Heitz et al., 2001; Pardo and Nurse, 2003).
There is good evidence that γ-tubulin plays a role in microtubule nucleation at all fission yeast MTOCs (Horio et al., 1991; Paluh et al., 2000; Heitz et al., 2001; Sawin et al., 2004; Venkatram et al., 2004; Zimmerman et al., 2004a). Thus far, nearly all of the subunits of the higher eukaryotic γ-tubulin complex have been identified in fission yeast (Horio et al., 1991; Stearns et al., 1991; Vardy and Toda, 2000; Fujita et al., 2002; Venkatram et al., 2004), and recent efforts have begun to identify fission yeast proteins regulating γ-tubulin function. The nonessential protein mod20p/mbo1p seems to play a particularly important role in γ-tubulin complex-mediated microtubule nucleation (Sawin et al., 2004; Venkatram et al., 2004). To establish a common nomenclature, from hereon in this article we refer to mod20p/mbo1p as microtubule organizer 1 (mto1p). In mto1Δ mutants, new microtubule nucleation by iMTOCs is profoundly crippled, such that only the SPB is (poorly) active for microtubule nucleation. In addition, during mitosis, mto1Δ mutants fail to nucleate cytoplasmic astral microtubules, although the assembly of the intranuclear mitotic spindle seems to be normal. Moreover, at the end of mitosis, when wild-type cells form a PAA, mto1Δ mutants fail to nucleate any microtubules from the cell division site. The role of mto1p in these various manifestations of spatially restricted microtubule nucleation seems to be to recruit the γ-tubulin complex to MTOCs, as components of the γ-tubulin complex are coimmunoprecipitated with mto1p, and in mto1Δ cells, the γ-tubulin complex is specifically absent from the sites at which microtubule nucleation should but does not occur.
Potential homologues of mto1p are found in fungi and in higher eukaryotes. These are all large proteins, with extensive regions of predicted α-helical coiled-coil, and they share in their amino-terminus a roughly 60-amino acid region of limited sequence conservation. Some of these proteins, such as Aspergillus nidulans apsB and Drosophila centrosomin (cnn), have been characterized functionally (Suelmann et al., 1998; Megraw et al., 1999; Terada et al., 2003), and this analysis is consistent with the proposed function of mto1p. Other proteins, such as human myomegalin/PDE4DIP or CDK5RAP2, are to date only little characterized (Ching et al., 2000; Verde et al., 2001; Andersen et al., 2003), but they are likely, even on the basis of limited analysis, to be equally interesting. Amino acid sequence analysis also suggests that in some organisms, mto1p-like proteins may exist as pairs of paralogs. For example, in fission yeast, the putative mto1p paralog pcp1p, which may be orthologous to budding yeast Spc110p (Knop and Schiebel, 1998; Flory et al., 2002), has been hypothesized to be responsible for intranuclear mitotic spindle microtubule nucleation, which is independent of mto1p (Sawin et al., 2004; Venkatram et al., 2004).
From this perspective, a deeper understanding of how mto1p localization and activity are regulated should provide further insights into the mechanisms controlling microtubule nucleation by the γ-tubulin complex in fission yeast. Here, we describe a novel nonessential gene, mto2+, which is involved in mto1p-mediated microtubule nucleation. Mto2p is physically associated with mto1p and colocalizes with mto1p throughout the cell cycle. Interestingly, mto2Δ mutants display a subset of the microtubule-nucleation–defective phenotypes seen in mto1Δ mutants. Collectively, our results suggest that mto2p promotes the ability of mto1p to recruit the γ-tubulin complex to a subset of MTOCs away from the SPB.
MATERIALS AND METHODS
Yeast Strain Construction and Growth Conditions
Standard yeast genetic techniques were used throughout (Moreno et al., 1991; Alfa et al., 1993). Insertional mutagenesis was performed as described previously (Snaith and Sawin, 2003). Deletion and tagging of genes at both N and C termini was achieved using polymerase chain reaction (PCR)-based targeting methods (Bahler et al., 1998), and deletions and tagging were confirmed by PCR and Western blotting as appropriate. See Table 1 for a list of strains used. The cyan fluorescent protein (CFP)-atb2 plasmid pRL72 (Glynn et al., 2001) was used to transform strains for the experiments shown in Figure 7. Growth conditions were as described previously (Moreno et al., 1991), except that sodium glutamate was used as nitrogen source for minimal medium. Although this is unrelated to mto2+ function per se, we note for interested investigators that the mto2+ locus is closely linked (distance ∼10 kb) to alp6+, an essential component of the γ-tubulin complex (Vardy and Toda, 2000).
Table 1.
Fission yeast strains used in this study
| Strain no. | Genotype | Source |
|---|---|---|
| KS515 | h+ ade6-216 leu1-32 ura4-D18 | Laboratory stock |
| KS516 | h- ade6-210 leu1-32 ura4-D18 | Laboratory stock |
| KS819 | h+ mto1-GFP:kanMX6 ade6-216 leu1-32 ura4-D18 | Laboratory stock (Sawin et al., 2004) |
| KS968 | h+ mto2Δ::kanMX6 kanMX6 nmt41:GFP-atb2 ade6-216 | This study |
| KS976 | h- mto2Δ::kanMX6 ade6-216 leu1-32 ura4-D18 | This study |
| KS1014 | h- mto1Δ::kanMX6 mto2Δ::kanMX6 ade6-216 leu1-32 ura4-D18 | This study |
| KS1138 | h- alp4-3HA:kanMX6 ade6 his leu1-32 ura4-D18 | T. Toda (Vardy et al., 2000) |
| KS1158 | h- mto1Δ::kanMX6 ade6-210 leu1-32 ura4-D18 | Laboratory stock (Sawin et al., 2004) |
| KS1236 | h- kanMX6:nmt81:GFP-atb2 ade6-210 leu1-32 ura4-D18 | Laboratory stock (Sawin et al., 2004) |
| KS1238 | h- nda3-KM311 ade6-210 leu1-32 ura4-D18 | Laboratory stock (Hiraoka et al. 1984) |
| KS1368 | h+ alp4-GFP:kanMX6 ade6-216 leu1-32 ura4-D18 | T. Toda (Vardy et al., 2000) |
| KS1370 | h- alp4-3HA:kanMX6 ade6-216 leu1-32 ura4-D18 | This study |
| KS1389 | h- kanMX6:nmt81:GFP-mto1 ade6-210 leu1-32 ura4-D18 | This study |
| KS1407 | h- mto1-GFP:kanMX6 mto2Δ::kanMX6 ade6-216 leu1-32 ura4-D18 | This study |
| KS1408 | h+ mto1-GFP:kanMX6 mto2Δ::kanMX6 ade6-216 leu1-32 ura4-D18 | This study |
| KS1409 | h- mto2Δ::kanMX6 kanMX6:nmt81:GFP-atb2 ade6-216 leu1-32 ura4-D18 | This study |
| KS1431 | h- kanMX6:nmt81:GFP-mto1 mto2Δ::kanMX6 ade6-210 leu1-32 ura4-D18 | This study |
| KS1459 | h+ mto1Δ::kanMX6 mto2-GFP:kanMX6 ade6-216 leu1-32 ura4-D18 | This study |
| KS1478 | h- kanMX6:nmt41:GFP-mto2 ade6-210 leu1-32 ura4-D18 | This study |
| KS1507 | h+ mto1-13myc:kanMX6 ade6-216 leu1-32 ura4-D18 | Laboratory stock (Sawin et al., 2004) |
| KS1517 | h- alp4-3HA:kanMX6 mto1-13myc:kanMX6 ade6-216 leu1-32 ura4-D18 | Laboratory stock (Sawin et al., 2004) |
| KS1607 | h- mto2Δ::kanMX6 nda3-KM311 ade6-210 leu1-32 ura4-D18 | This study |
| KS1615 | h- mto1(1-285)-13myc:kanMX6 | This study |
| KS1616 | h- mto1(1-500)-13myc:kanMX6 ade6-210 leu1-32 ura4-D18 | This study |
| KS1617 | h- mto1(1-800)-13myc:kanMX6 ade6-210 leu1-32 ura4-D18 | This study |
| KS1619 | h- mto1(1-1051)-13myc:kanMX6 ade6-210 leu1-32 ura4-D18 | This study |
| KS1640 | h- mto1-13myc:kanMX6 mto2Δ::kanMX6 ade6-216 leu1-32 ura4-D18 | This study |
| KS1889 | h+ mto2-GFP:kanMX6 ade6-216 leu1-32 ura4-D18 | This study |
| KS2120 | h+ mto1-YFP:kanMX6 ade6-216 leu1-32 ura4-D18 | This study |
| KS2131 | h- kanMX6:nmt41:GFP-mto2 kanMX6:nmt81:mto1 ade6-210 leu1-32 ura4-D18 | This study |
| KS2156 | h- mto2-CFP:kanMX6 ade6-210 leu1-32 ura4-D18 | This study |
| KS2169 | h- mto1-13myc:kanMX6 mto2Δ::kanMX6 alp4-3HA:kanMX6 ade6-216 leu1-32 ura4-D18 | This study |
| KS2180 | h- mto1-YFP:kanMX6 mto2-CFP:kanMX6 ade6-216 leu1-32 ura4-D18 | This study |
| KS2220 | h+ mto1-YFP:kanMX6 mto2Δ::kanMX6 ade6-216 leu1-32 ura4-D18 | This study |
| KS2248 | h+ alp4-GFP:natMX6 ade6-216 leu1-32 ura4-D18 | This study |
| KS2258 | h+ alp4-GFP:natMX6 mto2Δ::kanMX6 ade6-216 leu1-32 ura4-D18 | This study |
Figure 7.
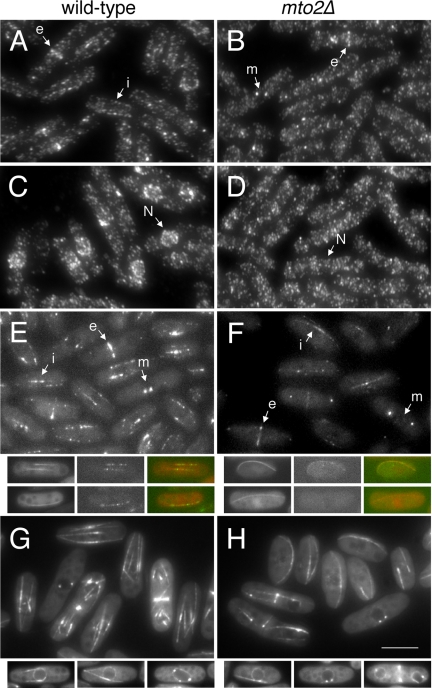
Mto1p localization in mto2Δ mutants. (A–D) Mto1p does not redistribute to the nuclear surface upon cold-shock in mto2Δ mutants. Immunofluorescent staining with affinity-purified anti-mto1p antibody. Wild-type and mto2Δ cells were fixed and stained (A and B) before and (C and D) after 30 min cold-shock at 0°C. Note interphase (i), mitotic SPB (m) and eMTOC (e) localization of mto1p. Nuclear position also is marked (N). (E and F) Localization of mto1-YFP in living wild-type and mto2Δ cells. Projections of Z-sections through the entire cell volume. Smaller panels show single Z-section examples of mto1-YFP in cells coexpressing CFP-atb2 tubulin. Note that not all microtubules in mto2Δ cells have associated mto1-YFP. Same labeling convention as above. (G and H) Localization of mildly overexpressed nmt81:GFP-mto1p in wild-type and mto2Δ cells. Projections of Z-sections through the entire cell volume. Small panels show examples of single Z-sections in interphase and mitotic cells, allowing visualization of the GFP-mto1p localized to the nuclear envelope that is obscured in projections. In all panels, images comparing wild-type and mto2Δ cells were always acquired and processed under identical conditions, allowing direct comparison of intensities. Bar, 5 μm (large panels); 7 μm (small panels).
Antibodies
Antibodies to mto2p were raised in sheep against recombinant glutathione S-transferase (GST)-mto2p purified from Escherichia coli. Anti-mto2p antibody was affinity purified from serum by using standard methods (Sawin et al., 1992). First, anti-GST antibodies were removed by passing serum through a column of GST coupled to Affigel (Bio-Rad, Hercules, CA) until all anti-GST reactivity was removed. Anti-mto2p antibodies were then affinity purified on a column of GST-mto2p coupled to Affigel and eluted at pH 3.0. The anti-mto1p polyclonal antibody and TAT1 anti-tubulin (Woods et al., 1989), 9E10 anti-myc (Evan et al., 1985), and GTU-88 anti-γ-tubulin (Sigma-Aldrich, St. Louis, MO) monoclonal antibodies were used as described previously (Sawin et al., 2004). Secondary antibodies were Alexa labeled (Molecular Probes, Eugene, OR) for immunofluorescence and horseradish peroxidase labeled (Amersham Biosciences, Piscataway, NJ; and Sigma-Aldrich) for Western blotting.
Immunofluorescence and Physiology Experiments
For anti-tubulin and anti-mto1p immunofluorescence, cells were fixed in methanol at –70°C and processed exactly as described previously (Sawin and Nurse, 1998; Sawin et al., 2004).
Cell morphology assays after growth to stationary phase on YE5S agar plates were performed as described previously (Snaith and Sawin, 2003). Cells were washed off plates and fixed in 3% formaldehyde before imaging. Assays of microtubule regrowth after cold-shock were as described previously (Sawin et al., 2004). Exponentially growing cells were chilled in ice water bath for 30 min and transferred to a prewarmed flask and incubated at 32°C for the specified time before collection by filtration and fixation. Mitotic block-and- release experiments by using reversible cold-sensitive nda3 mutants also were exactly as described previously (Sawin et al., 2004).
Biochemical Methods
For coimmunoprecipitation experiments of mto2p with mto1p, yeast cells were disrupted by bead-beating by using 0.5-mm zirconium beads in a buffer containing 50 mM Tris, pH 8.0, 150 mM NaCl, 1 mM EDTA, and 1 mM phenylmethylsulfonyl fluoride (PMSF). Cleared cell extract was incubated with anti-myc antibody (9E10) or affinity-purified anti-mto2p antibody for 30 min. Protein A- or protein G-Sepharose was added to the cell extract/antibody mixture and incubated for a further 30 min at 4°C. The Sepharose beads were washed four times with the buffer before being boiled in 2× SDS PAGE buffer and used for SDS-PAGE and Western blotting.
For coimmunoprecipitation experiments of mto1p with components of the γ-tubulin complex, frozen yeast cells were disrupted by grinding with a mortar and pestle. These and further manipulations were carried out exactly as described previously (Sawin et al., 2004).
A TnT T7 coupled in vitro transcription/translation kit (Promega, Madison, WI) was used to synthesize mto1p polypeptides for use in GST pull-down assays. GST-mto2p was expressed in E. coli BL21(DE3)pLysS and purified on glutathione (GSH)-Sepharose (Amersham Biosciences) following manufacturer's instructions. Truncated mto1 genes were amplified by PCR from a plasmid template, and the resulting PCR products were used as the template for in vitro transcription/translation. All 5′ primers start with a common 40-base sequence (5′-CCGCGGGGCCCTAATACGACTCACTATAGGGAGAACCATG-3′, which contains T7 primer, Kozak sequence, and an initiation methionine codon) followed by the appropriate specific mto1 sequence. Fifteen microliters of a 50-μl reaction was incubated with GST-mto2p bound to GSH-Sepharose (Sigma-Aldrich) in 500 μl of GST binding buffer (50 mM Tris, pH 7.5, 100 mM NaCl, 1% Triton X-100, 10% glycerol, and 1 mM PMSF) at 4°C for 1 h. Beads were washed four times with the GST binding buffer before boiling in 2× SDS-PAGE buffer.
In Vivo Fluorescence Imaging
Single time-point and time-lapse imaging of green fluorescent protein (GFP)-, CFP-, and yellow fluorescent protein (YFP)-fusion proteins in fission yeast was essentially as described previously (Snaith and Sawin, 2003; Sawin et al., 2004). Cells were mounted on medium-agarose pads before imaging and sealed with VALAP. Images were collected on a Nikon TE300 inverted microscope with automated filter and z-axis control, running MetaMorph software (Universal Imaging, Downingtown, PA). Appropriate neutral density filters were used to reduce photobleaching and photodamage. Some sequences were further deconvolved using Softworx (Applied Precision, Issaquah, WA).
RESULTS
Aberrant Cell Shape and Microtubule Organization in mto2 Mutants
From an insertional mutagenesis screen designed to identify genes regulating cell polarity in fission yeast (Snaith and Sawin, 2003), we isolated a mutant that produces a curved cell shape upon return to growth from stationary phase (Figure 1A). Sequencing of the disrupted gene showed it to be SPBC902.06, a previously uncharacterized open reading frame, which we have termed mto2+. Database searches did not reveal any obvious eukaryotic homologues of mto2p at the primary sequence level. Targeted deletion of the mto2+ open reading frame yielded viable cells (mto2Δ) with a phenotype identical to the original disruption mutant, and the mto2Δ strain was used for all subsequent experiments.
Figure 1.
Phenotype of mto2Δ cells. (A) Morphology of wild-type and mutant cells 3 h after return to fresh medium from stationary phase. Only wild-type cells are straight. Bar, 10 μm. (B) Microtubule organization in wild-type and mutant cells, as shown by anti-tubulin immunofluorescent staining of asynchronous cultures. Note that mutants tend to have fewer microtubule bundles, and bundles often curve around cell tips. Bar, 2 μm.
The cell shape phenotype of mto2Δ mutants closely resembled that of deletion mutants of the recently characterized gene mto1+ (Sawin and Snaith, 2004; Venkatram et al., 2004). Mto1p is required for microtubule nucleation from non-SPB sites within the cell as well as for the nucleation of cytoplasmic (but not intranuclear) microtubules from the SPB, and mto1p acts to recruit the γ-tubulin complex to these sites (Sawin and Snaith, 2004; Venkatram et al., 2004). We therefore examined both cell shape and microtubule organization in mto2Δ mutants relative to mto1Δ single mutants and to mto1Δ mto2Δ double mutants (Figure 1, A and B). Cell shape defects were not more severe in the double mutant relative to either single mutant, suggesting that mto1p and mto2p gene products may act in the same genetic pathway. Similar results also were obtained with regard to microtubule organization, in which both single and double mutants showed a range of microtubule defects normally not seen in wild-type cells, including fewer (and often thicker) microtubule bundles per cell and microtubules curving around cell tips.
We next examined the behavior of mto2Δ mutants in microtubule regrowth assays after a cold shock at 0°C. In these experiments, wild-type interphase cells rapidly renucleate microtubules from numerous independent sites in the cell middle, probably in contact with the nuclear surface, whereas mto1Δ mutants renucleate microtubules extremely slowly and then only from the SPB (Figure 2; Sawin et al., 2004). Comparing wild-type, mto1Δ, and mto2Δ cells in this assay, we found that mto2Δ mutants showed a phenotype distinct from both wild-type cells and mto1Δmutants. Although, as in mto1Δ mutants, microtubule nucleation in mto2Δ mutants was restricted to a single bundle growing from the SPB, the rate of regrowth was considerably faster in mto2Δ mutants, which reached a steady-state distribution ∼10 min after warming, in contrast to 30–60 min for mto1Δ cells and ∼4 min for wild-type cells (Figure 2). In these experiments, mto1Δ mto2Δ double mutants showed the strong defects characteristic of mto1Δ single mutants, further supporting the notion that mto1p and mto2p act in a common genetic pathway.
Figure 2.
Microtubule regrowth after cold shock in wild-type and mutant cells. Antitubulin staining of fixed cells. Microtubules were depolymerized by a 30-min cold shock and then fixed at the indicated times after being returned to 32°C. Note that all mutants initially nucleate a single microtubule bundle, but mto2Δ mutants nucleate much faster and reach steady state more quickly than mto1Δ and mto2Δ mto1Δ mutants. The position of the nucleus in cells can be identified by faint negative staining. Bar, 2 μm.
The defects in interphase microtubule nucleation observed in mto2Δ mutants in microtubule regrowth experiments after cold shock also were observed during live-cell imaging of mto2Δ cells expressing the GFP-tubulin fusion protein GFP-atb2 from the low-strength nmt81 promoter. In contrast to wild-type cells, which nucleate microtubules from multiple independent sites (see Movie 01; Brunner and Nurse, 2000; Drummond and Cross, 2000; Tran et al., 2001; Sawin et al., 2004), mto2Δ mutants nucleated microtubules from only a single site on the nuclear surface, which we interpret to be the SPB (see Movies 02–05). mto2Δ mutants also contained fewer microtubule bundles, in particular when expressing GFP-atb2 from the medium-strength nmt41 promoter (see Movie 05). In addition, microtubules in mto2Δ mutants displayed unusual dynamics not seen in wild-type cells, including bending around the cell tip and then sometimes breaking, generating a new microtubule bundle as a result (see Movies 02–05). Similar bend-breakage behavior has been observed previously in mto1Δ mutants (Sawin et al., 2004; see Discussion).
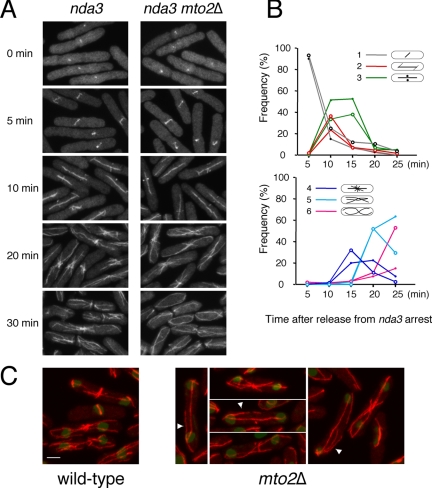
To investigate a role for mto2+ in mitotic microtubule nucleation, we examined microtubule regrowth after release from a mitotic metaphase arrest, by using the cold-sensitive β-tubulin mutant nda3-km311 (Hiraoka et al., 1984) in wild-type (i.e., mto2+) and mto2Δ backgrounds (Figure 3). Previous work showed that mto1Δ nda3 mutants reassemble and elongate an intranuclear mitotic spindle at the same rate as nda3 single mutants but are completely defective in the growth of cytoplasmic astral microtubules from the SPB and in the formation of a postanaphase array from the eMTOC at the end of mitosis (Sawin et al., 2004; Venkatram et al., 2004). By contrast, we found that under the same regime, mto2Δ nda3 mutants were able to nucleate not only spindle microtubules but also cytoplasmic astral microtubules and PAA microtubules (Figure 3A). Interestingly, in these experiments the PAAs of mto2Δ nda3 mutants seemed to be slightly less well focused than those of nda3 single mutants and to reorganize slightly more rapidly (Figure 3B). We therefore investigated the organization of PAAs in asynchronous cultures. By immunofluorescence, we found that although PAAs were visible in actively cycling mto2Δ (i.e., nda3+) mutants, they varied widely in morphology and were often marked by fewer, longer microtubules, and mutant cells often contained long astral microtubules and/or additional microtubules not apparently focused at the eMTOC (Figure 3C).
Figure 3.
PAA formation of microtubules in mto2Δ mutants. Anti-tubulin staining of fixed cells. (A) Mitotic spindle, PAA, and interphase microtubule organization in wild-type and mto2Δ cells after release from nda3 mitotic arrest. Note that spindles and PAAs occur at similar times in both strains, but mto2Δ mutants reach an interphase organization more quickly. (B) Quantitation of early (top) and late (bottom) microtubule structures at different times after release from the nda3 arrest in the experiment in A. Filled circles, wild-type cells; open circles, mto2Δ. Early structures (classes 1–3) appear and disappear with comparable kinetics in the two strains, whereas well defined PAAs (class 4) are not sustained in mto2Δ mutants and are more quickly replaced by more disorganized microtubules (classes 5 and 6). (C) PAAs of microtubules in asynchronous wild-type and mto2Δ cells. Note aberrant PAA and cytoplasmic astral microtubules (arrowheads) in mto2Δ mutants. Bar, 2 μm.
To further understand the basis for this complexity, we followed the appearance of PAAs in live mto2Δ cells expressing nmt81:GFP-atb2 (see Movies 01–05). Although wild-type cells showed robust PAAs with continuous microtubule nucleation from eMTOCs (see Movie 01), mto2Δ mutants typically showed obvious but less frequent nucleation from the eMTOC, such that PAAs were either weak or highly transient. In addition, astral microtubules were clearly observed in mto2Δ mutants, and after nucleation, both PAA and astral microtubules were typically more stable dynamically, with astral microtubules often making long excursions into their sister cell before septation (see Movies 02–04). As with interphase microtubule dynamics, these effects of mto2+ deletion seemed somewhat stronger in cells expressing nmt41: GFP-atb2 (see Movie 05). Collectively, these results suggest that, although mto2p is not absolutely required for PAA formation, it may nevertheless contribute to the stability of the eMTOC and PAA formation. The apparent stability of the PAA in mto2Δ nda3 mutants may be a consequence of an extended mitotic arrest (see Discussion).
Together, the above-mentioned analyses of interphase and mitotic microtubule nucleation suggest that mto2p is involved in the regulation of microtubule nucleation in the same genetic network as mto1p and that mto2p is required for a specific subset of mto1p-dependent microtubule nucleation functions.
Localization of mto2p to Microtubule-organizing Centers
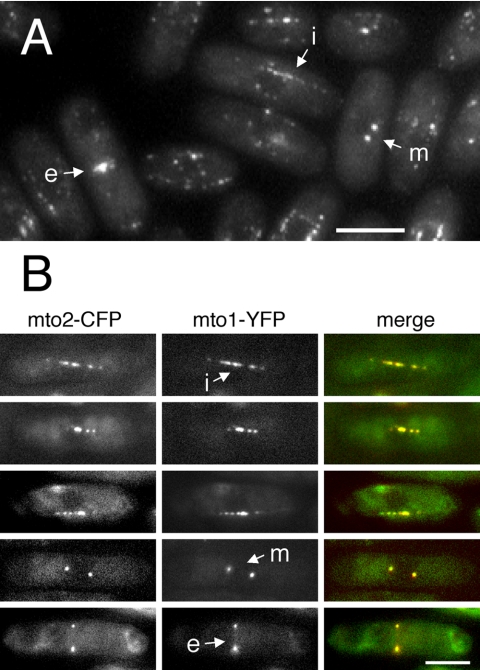
To determine the intracellular localization of mto2p, we tagged the carboxy-terminus of the endogenous mto2+ open reading frame with GFP, such that the resulting fusion protein is expressed from its endogenous promoter and is the sole source of mto2p in the cell (see, for example, Figure 6). In living cells, mto2-GFP was localized to the SPBs during both interphase and mitosis, to the eMTOC, and to satellite particles of varying intensity on cytoplasmic interphase microtubules (Figure 4A; see Movie 06). A similar localization has been previously observed for mto1-GFP, with mto1-GFP satellite particles showing rapid back-and-forth movements on microtubules (Sawin et al., 2004; Venkatram et al., 2004). Mto2-GFP satellites showed similar movements by time-lapse videomicroscopy, on both interphase and PAA microtubules (see Movie 07). Attempts to localize untagged mto2p in fixed cells with affinity-purified anti-mto2p antibodies (Figure 5) were unsuccessful, due to high cytoplasmic background (our unpublished data).
Figure 6.

Mto1p and mto2p protein levels in wild-type and mutant strains. Extracts from wild-type cells (lane 1), as well as mto1-GFP (lane 2), mto2Δ (lane 3), mto1Δ (lane 4), mto1-GFP mto2Δ (lane 5), mto2-GFP mto1Δ (lane 6), and mto2-GFP (lane 7) were blotted and probed with antibodies to mto1p (top) and mto2p (middle). α-Tubulin also was probed, as a loading control (bottom). Note that deletion of mto2p does not affect mto1p levels, or vice-versa. Also note that GFP-tagging of mto1p slightly destabilizes the resulting fusion protein, whereas GFP-tagging of mto2p may slightly stabilize the resulting fusion.
Figure 4.
Mto2p localization in wild-type cells. (A) Mto2-GFP in live cells. Projection of Z-sections through the entire cell volume. Note localization to mitotic SPBs (arrow m) and eMTOC (arrow e) in mitotic and postmitotic cells, and satellite particles in interphase cells (arrow i). (B) Coexpression of mto2-CFP and mto1-YFP in live cells. Single Z-sections. The two signals colocalize during interphase (top three rows) as well as during mitosis (fourth row) and cell division (bottom row). Labeling convention as in A. Bars, 5 μm.
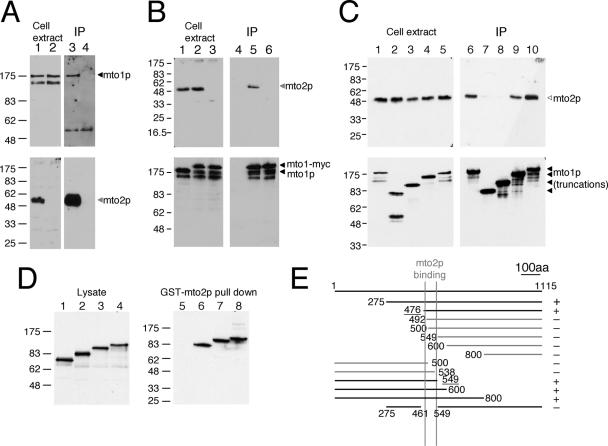
Figure 5.
Physical interactions between mto2p and mto1p. (A) Immunoprecipitation of mto1p with mto2p from cell extracts. Affinity-purified anti-mto2p antibody was used for immunoprecipitation. Cell extracts and immunoprecipitates of wild-type (lanes 1 and 3) and mto2Δ (lanes 2 and 4) were blotted and probed with antibodies to mto1p (top) and mto2p (bottom). (B) Immunoprecipitation of mto2p with mto1-myc. Anti-myc antibody was used for immunoprecipitation. Cell extracts and immunoprecipitates of mto1+ (i.e., no tag on mto1p; lanes 1 and 4), mto1-myc (lanes 2 and 5), and mto1-myc mto2Δ (lanes 3 and 6) were blotted and probed with antibodies to mto2p (top) and mto1p (bottom). (C) Some truncated mto1 proteins cannot immunoprecipitate mto2p. Anti-myc antibody was used to immunoprecipitate carboxyl-terminal truncations of mto1-myc. Cell extracts and immunoprecipitates of full-length mto1-myc (1-1115; lanes 1 and 6), mto1(1-285)-myc (lanes 2 and 7), mto1(1-500)-myc (lanes 3 and 8), mto1(1-800)-myc (lanes 4 and 9), and mto1(1-1051)-myc (lanes 5 and 10) were blotted and probed with antibodies to mto2p (top) and mto1p (bottom). Immunoprecipitate lanes represent ∼20× extract-equivalent loading relative to total cell extract lanes. (D) In vitro binding of mto1p truncations to GST-mto2p by pull-down assays. Mto1 proteins expressed in reticulocyte lysate (left) were tested for binding to GST-mto2 (right). Reticulocyte lysates and pull-downs of mto1(1-500) (lanes 1 and 5), mto1(1-600) (lanes 2 and 6), mto1(1-700) (lanes 3 and 7), and mto1(1-800) (lanes 4 and 8) were blotted and probed with antibodies to mto1p. (E) Summary of results of additional GST-mto2p pull-down assays, showing central region of mto1p required for binding to mto2p.
The similarity of mto2-GFP localization and movement to that of mto1-GFP suggested that the two proteins might colocalize. In live cells coexpressing mto2-CFP and mto1-YFP, we observed extensive colocalization of CFP and YFP signals at the SPB, at the cell division site and at interphase satellites (Figure 4B). These results suggest that mto2p and mto1p colocalize to MTOCs constitutively throughout the cell cycle.
Mto2p Interacts Directly with mto1p
Given the colocalization of mto2p with mto1p at the light microscope level, we investigated whether mto2p and mto1p interact physically (Figure 5). We found that affinity-purified anti-mto2p antibodies were able to coimmunoprecipitate mto1p with mto2p in native fission yeast extracts (Figure 5A) and that anti-myc antibodies were able to coimmunoprecipitate mto2p with myc-tagged mto1p (Figure 5B). These results indicate that mto1p and mto2p are likely to exist in a common complex in vivo. To further define the regions of mto1 required for interaction with mto2p, we repeated coimmunoprecipitation experiments by using strains containing C-terminal truncations of myc-tagged mto1p. Both mto1(1-800)-myc and mto1(1-1051)-myc were able to coimmunoprecipitate mto2p, whereas mto1(1-285)-myc and mto1(1-500)-myc were not (Figure 5C). This suggests that a central portion of mto1p is likely to be important for its association with mto2p (full-length mto1p is 1115 amino acids; Figure 5E).
To determine whether the observed interaction between mto2p and mto1p is direct and to further map the regions of mto1p required for interaction with mto2p, we performed GST pull-down experiments by using bacterially expressed GST-mto2p and in vitro-translated mto1p. Initial experiments showed that GST-mto2p was able to pull down mto1(1-600), mto1(1-700), and mto1(1-800) but not mto1(1-500), suggesting that mto1p and mto2p bind directly and also that the GST pull-down experiments reproduced the nature of the interaction observed in our coimmunoprecipitation experiments (Figure 5D). A series of amino- and carboxyl-terminal deletions of in vitro-translated mto1p further showed that an ∼90-amino acid sequence in the central portion of mto1p, between amino acids 461 and 549, is required for interaction with mto2p (Figure 5E; additional data not shown); this is distinct from the region of mto1p conserved among centrosomin-related proteins (mto1p amino acids 249–308).
Role of mto2p in mto1p Localization
Together with the physical association between mto2p and mto1p, the observation that mto2Δ mutants display only a subset of the microtubule nucleation defects seen in mto1Δ suggested that the association of mto2p with mto1p may be required to regulate specific aspects of mto1p function. We therefore investigated possible mechanisms by which mto2p might exert this effect.
We first explored the possibility that mto1p protein levels might be altered in mto2Δ mutants, because interphase and PAA microtubule nucleation might be more sensitive than intranuclear spindle microtubule nucleation to mto1p levels. However, mto1p levels were not altered in mto2Δ mutants nor were mto2p levels altered in mto1Δ mutants (Figure 6).
Our previous work showed that during regrowth of interphase microtubules after cold shock, the nucleation from multiple sites around the cell nucleus that occurs in wild-type cells (Figure 2) is driven by a redistribution of mto1p to the nuclear surface, as a specific result of the cold treatment (Sawin et al., 2004). Because microtubule regrowth in mto2Δ mutants is restricted to the SPB in this assay, we examined mto1p localization upon cold-shock in mto2Δ mutants. Before cold-shock, anti-mto1p staining revealed mto1p at SPBs and eMTOCs in both wild-type and mto2Δ cells (Figure 7, A and B). Mto1p was also visible along microtubules in wild-type cells (Figure 7A; Sawin et al., 2004), although this was less apparent in mto2Δ cells (Figure 7B). On cold shock, in contrast to wild-type cells, no accumulation of mto1p was observed at the nuclear surface in mto2Δ mutants (Figure 7D), consistent with the mto2Δ microtubule nucleation phenotype. This suggested that deletion of mto2+ might affect the behavior of mto1p in vivo. We therefore further analyzed whether normal mto1p localization depended on mto2p.
Because mto1-GFP is localized to SPBs, eMTOCs, and dynamic microtubule-associated interphase satellites in living cells (Sawin et al., 2004), we sought to image mto1-GFP in mto2Δ mutants. Initial experiments suggested that mto1-GFP was at SPBs and eMTOCs in mto2Δ mutants, but cellular autofluorescence interfered with our ability to detect potentially faint mto1-GFP signals from interphase satellites (our unpublished data). We therefore focused our analysis on mto1-YFP localization, which is better separated spectrally from the autofluorescence (our unpublished data). In mto2Δ mutants, mto1-YFP was observed at SPBs and eMTOCs (Figure 7, E and F; see Movies 08 and 09), consistent with observed nucleation of cytoplasmic astral microtubules from the SPB and PAA microtubules from the eMTOC in mto2Δ mutants. However, mto1-YFP signal at eMTOCs was reduced in mto2Δ mutants relative to wild-type cells; this may contribute to the reduced PAA nucleation characteristic of mto2Δ mutants.
We also observed mto1-YFP interphase satellite particles in mto2Δ mutants, although relative to wild-type cells, the mto1-YFP signal was faint and prone to photobleaching, and its localization to microtubules was more easily recognized in single sections than in projections encompassing the entire cell volume (compare Figure 7 with Movie 09). We confirmed this microtubule localization by coexpressing CFP-atb2 in double-label experiments (Figure 7, E and F); in some mto2Δ cells, microtubules without associated mto1-YFP were seen (compare also Movies 08 and 09). Under optimal imaging conditions, we observed dynamic movement of mto1-YFP interphase satellites on microtubules in mto2Δ mutants, although this was difficult to detect routinely and was not easily seen in all cells (Movies 10 and 11).
We also examined the localization of moderately overexpressed GFP-mto1p in wild-type and mto2Δ strains (Figure 7, G and H). Expressed at three to four times wild-type levels from the nmt81 promoter (our unpublished data), GFP-mto1p showed localization to the SPB and eMTOC, as well as noticeable localization to the nuclear envelope (Figure 7G) and apparently uniform decoration of cytoplasmic microtubules, in contrast to the satellite-particle localization observed when mto1-GFP is expressed from its endogenous promoter. Interestingly, none of these localizations required the presence of mto2p (Figure 7H).
Together, the above-mentioned results indicate that mto2p is not absolutely required for localization of mto1p to the SPB or to eMTOCs or for localization to interphase cytoplasmic microtubules; however, the degree to which mto1p localizes to eMTOCs and interphase microtubules does seem to depend partially on mto2p. Moreover, in the cold shock assay, relocalization of mto1p to the nuclear surface depends on mto2p. These results suggest that although mto2p may affect some aspects of mto1p localization under specific experimental conditions, this is unlikely to be the sole cause of all of the observed microtubule nucleation defects in mto2Δ mutants (see Discussion for further details).
Mto2p Is Required for Coimmunoprecipitation of the γ-Tubulin Complex with mto1p and for Proper In Vivo Localization of the γ-Tubulin Complex
Because the major molecular role for mto1p in microtubule nucleation seems to be to bind and recruit the γ-tubulin complex to different MTOCs (Sawin et al., 2004; Venkatram et al., 2004), we tested whether mto1p could still bind to the γ-tubulin complex in the absence of mto2p. Strikingly, whereas cytoplasmic extracts prepared from cells expressing mto1-myc could coimmunoprecipitate mto2p, γ-tubulin, and hemagglutinin (HA)-tagged alp4p (an essential component of the γ-tubulin complex; Vardy and Toda, 2000) with mto1-myc, neither γ-tubulin nor alp4-HA was coimmunoprecipitated with mto1-myc to any extent in mto2Δ cells, even after very long exposures of Western blots (Figure 8; additional data not shown). This indicates that soluble forms of mto1p are not associated with the γ-tubulin complex in mto2Δ mutants; we address this further below in relation to mto1Δ and mto2Δ phenotypes (see Discussion).
Figure 8.

The γ-tubulin complex is no longer coimmunoprecipitated with mto1p in mto2Δ cells. Anti-myc antibody was used for immunoprecipitation. Cell extracts and immunoprecipitates of mto1-myc alp4-HA (lanes 1 and 4), negative control mto1+ alp4-HA (i.e., no tag on mto1p; lanes 2 and 5), and mto1-myc alp4-HA mto2Δ (lanes 3 and 6) were blotted and probed with antibodies to myc (top), mto2p (second), γ-tubulin (third), and HA (bottom). Asterisk marks the position of anti-myc IgG heavy chain in immunoprecipitates; γ-tubulin is the band underneath. Immunoprecipitate lanes represent ∼40× extract equivalent loading relative to total cell extract lanes.
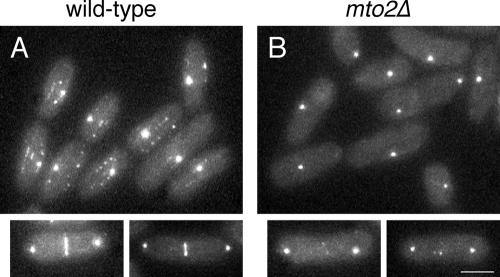
As a corollary to the coimmunoprecipitation experiments, we examined the localization of GFP-tagged alp4p in wild-type and mto2Δ cells in vivo. In wild-type cells, alp4-GFP was found at SPBs throughout the cell cycle, at eMTOCs at the end of mitosis, and on microtubule-associated satellite particles during interphase (Vardy and Toda, 2000; Zimmerman et al., 2004a; Figure 9A). By contrast, in mto2Δ cells, alp4-GFP interphase satellite particles were not observed, and only barely detectable amounts of alp4-GFP were seen at eMTOCs, although alp4-GFP remained associated with SPBs throughout the cell cycle (Figure 9B). These results indicate that mto2p is required for the correct cytoplasmic localization of the γ-tubulin complex at non-SPB sites; in conjunction with our immunoprecipitation data, this provides a straightforward mechanistic explanation for the range of microtubule nucleation defects observed in mto2Δ mutants.
Figure 9.
The γ-tubulin complex is not properly localized to eMTOCs or interphase satellite particles in mto2Δ mutants. Alp4-GFP localization in live wild-type (A) and mto2Δ mutant (B) cells. Large panels show interphase cells (with one just-divided cell pair in B); small panels show dividing cells. Bar, 5 μm.
Mto1p Is Required for mto2p Localization
We also examined mto2-GFP localization in mto1Δ mutants. Strikingly, mto2-GFP interphase satellites were no longer observed in mto1Δ cells, and mto2-GFP localization to SPBs and eMTOCs was almost abolished (Figure 10B). Approximately one-third of cells showed a faint GFP signal suggestive of the SPB, and dividing cells showed only a very faint signal at the septum, or no signal at all. The faint signals were always small and punctate rather than band-like (compare Figure 10, A and B), suggesting that this small amount of mto2-GFP might be recruited to the division site only at the very end of septation; alternatively, it is possible that low amounts of mto2-GFP are present throughout septation in mto1Δ mutants but are visible only when concentrated into a small volume. Because mto1p and mto2p interact, we tried to test more directly whether mto2p localization to microtubules occurs specifically via its interaction with mto1p. We found that in mto1+ cells, overexpressed GFP-mto2p (nmt41: GFP-mto2p) was easily visualized at SPBs and at eMTOCs but was not seen on microtubules above levels of cytoplasmic GFP-mto2p background fluorescence; some protein aggregation also was observed (Figure 10C). However, when nmt81:mto1p was simultaneously overexpressed in the GFP-mto2p–overexpressing cells, GFP-mto2p now localized to interphase microtubules, as well as to the nuclear envelope, mirroring the localization of nmt81:GFP-mto1p (Figure 10D; compare with Figure 7G). These results indicate not only that mto1p is required for normal mto2p localization to most subcellular structures but also that mto1p protein levels are limiting for mto2p localization, suggesting that mto2p localization to microtubules occurs in the form of a mto1p/mto2p protein complex in which mto1p provides the microtubule targeting.
Figure 10.
Mto1p is required for mto2p localization. (A and B) Localization of mto2-GFP in live wild-type (A) and mto1Δ (B) cells. Projections of Z-sections through the entire cell volume. Images in A and B were acquired and processed under identical conditions, allowing direct comparison of intensities. (C and D) Localization of overexpressed nmt41:GFP-mto2p in wild-type cells (C) and cells mildly overexpressing nmt81:mto1+ (i.e., untagged mto1p) (D). Projections of Z-sections through the entire cell volume. Smaller panels show single Z-section examples of nmt41:GFP-mto2p localizing to the nuclear envelope only when mto1p is overexpressed (arrow). Images in C and D were acquired and processed under identical conditions, allowing direct comparison of intensities. Bar, 5 μm.
DISCUSSION
Model for mto2p Function
In this work, we have identified and characterized a novel protein, mto2p, which plays an important role in the regulation of fission yeast microtubule nucleation. Most microtubule nucleation in eukaryotic cells is thought to be mediated by the γ-tubulin complex (Gunawardane et al., 2000; Oakley, 2000; Schiebel, 2000; Job et al., 2003). In fission yeast, normal cytoplasmic microtubule nucleation from the SPB, eMTOC, and iMTOCs requires the centrosomin-related protein mto1p to recruit the γ-tubulin complex to these MTOCs (Sawin et al., 2004; Venkatram et al., 2004). Given the spatio-temporal diversity of fission yeast MTOCs (Hagan, 1998; Drummond and Cross, 2000; Tran et al., 2001; Sawin et al., 2004; Venkatram et al., 2004; Zimmerman et al., 2004a), it might be expected that regulators of mto1p exist within cells. Here, we have shown that mto2p is required for cytoplasmic microtubule nucleation from iMTOCs but not from the SPB. Mto2p also contributes significantly to eMTOC nucleation. In conjunction with our biochemical and localization studies, these data suggest a provisional model in which mto2p regulates microtubule nucleation mediated by mto1p and the γ-tubulin complex. We propose the following four main points, which are discussed in more detail below: 1) Mto2p is a stable component of an mto1p-containing protein complex that recruits the γ-tubulin complex to MTOCs. 2) Mto1p interphase satellite particles, which may represent the soluble population of mto1p complexes that can be probed by immunoprecipitation, do not bind the γ-tubulin complex in the absence of mto2p. 3) Mto1p complexes at SPBs and eMTOCs, which may not be identifiable in immunoprecipitation experiments because they are largely insoluble, can associate with the γ-tubulin complex even in the absence of mto2p. 4) Mto2p is not required for the general ability of the mto1p complex to be targeted to SPB, eMTOC, and interphase satellite iMTOCs but may contribute to the efficient recruitment of mto1p to the eMTOC and the behavior and/or dynamics of mto1p satellites.
With regard to the first point, we have shown that mto1p and mto2p are very closely colocalized at MTOCs throughout the cell cycle and that mto2p is likely to bind directly to a central portion of mto1p. In addition, mto2p is coimmunoprecipitated with mto1-myc much more efficiently than the γ-tubulin complex (Figure 8). This suggests to us that it may be most appropriate to think of γ-tubulin complex recruitment to MTOCs as involving an interaction between the γ-tubulin complex and an mto1p/mto2p complex.
The second two points of the model raise several related issues concerning protein–protein interactions between mto1p and the γ-tubulin complex. The γ-tubulin complex is no longer found in mto1-myc immunoprecipitates from cytoplasmic extracts in mto2Δ strains. This could be interpreted to mean that in this instance, absolutely no mto1p in the cell is capable of interacting with the γ-tubulin complex. However, our current understanding of the role of γ-tubulin in fission yeast microtubule nucleation (Horio et al., 1991; Stearns et al., 1991; Paluh et al., 2000; Vardy and Toda, 2000; Heitz et al., 2001; Fujita et al., 2002; Zimmerman et al., 2004a) and previous and current analyses of mto1Δ and mto2Δ mutant phenotypes (Sawin et al., 2004; Venkatram et al., 2004) lead us to suggest that small amounts of solid-phase mto1p interacting with the γ-tubulin complex at SPBs and eMTOCs may be sufficient for the nucleation activities observed in cells. This view is supported by our observation of extremely low levels of alp4-GFP at eMTOCs in mto2Δ mutants, which correlates with their reduced microtubule nucleation activity. In general, essentially all biochemical work on γ-tubulin in eukaryotes has been done on soluble complexes, and very little is known about solid-phase γ-tubulin complexes in eukaryotes (see, for example, Salas, 1999; Terada et al., 2003; Zimmerman et al., 2004b).
A second issue concerns the role of mto2p in promoting association of mto1p with the γ-tubulin complex. One possibility is that mto2p forms a “bridge” between an mto1p/mto2p complex and the γ-tubulin complex. If this were the case, we might expect a functional analog of mto2p to be similarly associated with mto1p at SPBs and eMTOCs, in the place of mto2p. Although this has not yet been investigated, we note that mto2p seems to remain associated with mto1p at all MTOCs, throughout the cell cycle (even when, in our view, it may not be very important for mto1p function). Thus, we speculate that the role of mto2p in promoting γ-tubulin complex binding to mto1p may be more indirect, possibly allosteric, and that supplementary mechanisms may promote mto2p-independent γ-tubulin complex binding to mto1p at SPBs and eMTOCs. Future experiments to dissect the molecular interactions governing localization of mto1p and γ-tubulin complex association will help to resolve this issue.
The final point of the model concerns targeting of an mto1p/mto2p complex to MTOCs and its consequences for nucleation. Overall, our data from GFP and YFP fusion proteins suggest that mto1p targeting is independent of mto2p and that mto2p targeting depends on mto1p. However, in the two cases where mto2Δ microtubule nucleation is not completely normal (i.e., iMTOC and eMTOC nucleation), levels of mto1-YFP are somewhat reduced and/or more difficult to detect at the corresponding MTOCs. This could be due to unrecognized defects in the mto1p complex itself or alternatively to the defects in microtubule organization known to exist in mto2Δ mutants. Aberrantly organized microtubules might fail to provide an optimal distribution of tracks or loading sites for mto1p complexes (for example, in the form of microtubule minus ends). In either case, it is worth noting that achieving a full complement of mto1p at eMTOCs could depend on microtubule-based movement of mto1p on PAA microtubules, which are themselves abnormal in mto2Δ mutants. Further work toward understanding the nature of satellite particle movements, particularly using in vitro systems, will help to clarify these issues.
Complicating our understanding of the role of mto2p in the subcellular targeting of mto1p is the fact that mto1p does not redistribute properly to the nuclear surface in cold shock experiments in mto2Δ mutants. This result stands in apparent opposition to our observations with GFP- and YFP-fusions to mto1p in live mto2Δ cells, especially with regard to nmt81:GFP-mto1p localization, for reasons which remain unclear. The fact that a significant fraction of mto1p may not remain in situ after fixation (Sawin et al., 2004) may be a contributory factor. In any case, the microtubule nucleation defects seen in mto2Δ mutants both in the cold shock experiment and during live cell analysis are most easily understood by the failure of cytoplasmic mto1p complexes to associate with the γ-tubulin complex, as shown in our immunoprecipitation and alp4-GFP localization studies.
Microtubule Nucleation and Dynamics in mto2Δ Mutants
Under steady-state growth conditions, mto1Δ mutants do not nucleate new interphase microtubules, and during recovery from cold shock they nucleate microtubules very poorly, and only from the SPB (Sawin et al., 2004; Venkatram et al., 2004). By contrast, mto2Δ cells efficiently nucleate interphase microtubules both during steady-state growth and after cold shock, but in both cases apparently only from the SPB. How do we account for these different microtubule nucleation phenotypes relative to each other and to wild-type cells? In fission yeast, the discoid SPB is associated with the nuclear envelope (Ding et al., 1997). It has been proposed previously that mto1p might be localized to the cytoplasmic face of the SPB in wild-type cells and that the poor SPB-mediated microtubule nucleation seen in mto1Δ mutants after cold shock may be driven by an mto1p paralog, pcp1p, from the nucleoplasmic face of the SPB (Sawin et al., 2004; Venkatram et al., 2004). We suggest that the efficient nucleation of cytoplasmic microtubules from the SPB in mto2Δ mutants is driven not by pcp1p but by active mto1p localized to the cytoplasmic face of the SPB. This mto2p-independent localization of mto1p also would account for the nucleation of cytoplasmic astral microtubules seen during mitosis in mto2Δ mutants and not mto1Δ mutants. In wild-type (i.e., mto2+) interphase cells, the additional ability to recruit the γ-tubulin complex to mto1p satellites would drive non-SPB–mediated microtubule nucleation.
At the end of mitosis in asynchronous cultures, mto1Δ mutants completely fail to form a PAA (Sawin et al., 2004; Venkatram et al., 2004), whereas mto2Δ mutants nucleate PAA microtubules but at a reduced frequency relative to wild-type cells. This can be explained by the presence of active mto1p at eMTOCs in mto2Δ mutants, but at a reduced level (see comments above). In contrast, after release from an nda3 mitotic arrest, mto2Δ cells show a more robust PAA, similar to wild-type cells. To explain this, we suggest that during an extended mitotic arrest in nda3 mto2Δ mutants, additional mto1p may be able to slowly accumulate at the cell division site. The mechanism of mto1p targeting to the division site is not known but may involve an interaction with the cytokinetic actin ring (Heitz et al., 2001; Pardo and Nurse, 2003; Sawin et al., 2004), which is formed and maintained during the nda3 arrest (Chang et al., 1996).
Finally, we note that the dynamics of interphase, PAA, and astral microtubules in mto2Δ mutants are different from wild-type cells. In particular, mto2Δ interphase microtubule dynamics look like those in mto1Δ cells, with abnormally bundled microtubules curling around cell tips and bend-breakage events generating new microtubule fragments (Sawin et al., 2004). We have previously suggested for mto1Δ mutants that these phenomena may represent an indirect consequence of altering the number of microtubule nucleation sites in a system with a fixed volume and tubulin concentration (see Sawin et al., 2004 for a further discussion). We believe an identical case can be made for mto2Δ mutants. Although we will not repeat the detailed arguments here, we note that theoretical calculations based on a simplified model of dynamic instability in a closed system arrive at conclusions consistent with this view (Mitchison and Kirschner, 1987).
Supplementary Material
Acknowledgments
We thank A. Douglas and P. Sasajala for contributions to this work during summer projects; F. Chang, K. Gull, I. Stancheva, and T. Toda for antibodies, strains, and plasmids; H. Ohkura for comments and discussion; and K. Gould, S. Venkatram, and P. Tran for alerting us to similar work on mto2p in the respective laboratories. K.E.S. is a Wellcome Trust Senior Research Fellow in Basic Biomedical Sciences. H.A.S. is a Caledonian Research Fellow. This work was supported by the Wellcome Trust.
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E04-11-1003) on January 19, 2005.
Abbreviations: eMTOC, equatorial microtubule-organizing center; iMTOC, interphase microtubule-organizing center; PAA, postanaphase array; SPB, spindle pole body.
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
References
- Alfa, C., Fantes, P., Hymas, J., McLeod, M., and Warbrick, E. (1993). Experiments with Fission Yeast: A Laboratory Course Manual, Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
- Andersen, J. S., Wilkinson, C. J., Mayor, T., Mortensen, P., Nigg, E. A., and Mann, M. (2003). Proteomic characterization of the human centrosome by protein correlation profiling. Nature 426, 570-574. [DOI] [PubMed] [Google Scholar]
- Bahler, J., Wu, J. Q., Longtine, M. S., Shah, N. G., McKenzie, A., 3rd, Steever, A. B., Wach, A., Philippsen, P., and Pringle, J. R. (1998). Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14, 943-951. [DOI] [PubMed] [Google Scholar]
- Brunner, D., and Nurse, P. (2000). CLIP170-like tip1p spatially organizes microtubular dynamics in fission yeast. Cell 102, 695-704. [DOI] [PubMed] [Google Scholar]
- Chang, F., Woollard, A., and Nurse, P. (1996). Isolation and characterization of fission yeast mutants defective in the assembly and placement of the contractile actin ring. J. Cell Sci. 109, 131-142. [DOI] [PubMed] [Google Scholar]
- Ching, Y. P., Qi, Z., and Wang, J. H. (2000). Cloning of three novel neuronal Cdk5 activator binding proteins. Gene 242, 285-294. [DOI] [PubMed] [Google Scholar]
- Ding, R., West, R. R., Morphew, D. M., Oakley, B. R., and McIntosh, J. R. (1997). The spindle pole body of Schizosaccharomyces pombe enters and leaves the nuclear envelope as the cell cycle proceeds. Mol. Biol. Cell 8, 1461-1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond, D. R., and Cross, R. A. (2000). Dynamics of interphase microtubules in Schizosaccharomyces pombe. Curr. Biol. 10, 766-775. [DOI] [PubMed] [Google Scholar]
- Evan, G. I., Lewis, G. K., Ramsay, G., and Bishop, J. M. (1985). Isolation of monoclonal antibodies specific for human c-myc proto-oncogene product. Mol. Cell. Biol. 5, 3610-3616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flory, M. R., Morphew, M., Joseph, J. D., Means, A. R., and Davis, T. N. (2002). Pcp1p, an Spc110p-related calmodulin target at the centrosome of the fission yeast Schizosaccharomyces pombe. Cell Growth Differ. 13, 47-58. [PubMed] [Google Scholar]
- Fujita, A., Vardy, L., Garcia, M. A., and Toda, T. (2002). A fourth component of the fission yeast gamma-tubulin complex, Alp16, is required for cytoplasmic microtubule integrity and becomes indispensable when gamma-tubulin function is compromised. Mol. Biol. Cell 13, 2360-2373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gachet, Y., Tournier, S., Millar, J. B., and Hyams, J. S. (2001). A MAP kinase-dependent actin checkpoint ensures proper spindle orientation in fission yeast. Nature 412, 352-355. [DOI] [PubMed] [Google Scholar]
- Gachet, Y., Tournier, S., Millar, J. B., and Hyams, J. S. (2004). Mechanism controlling perpendicular alignment of the spindle to the axis of cell division in fission yeast. EMBO J. 23, 1289-1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glynn, J. M., Lustig, R. J., Berlin, A., and Chang, F. (2001). Role of bud6p and tea1p in the interaction between actin and microtubules for the establishment of cell polarity in fission yeast. Curr. Biol. 11, 836-845. [DOI] [PubMed] [Google Scholar]
- Gunawardane, R. N., Lizarraga, S. B., Wiese, C., Wilde, A., and Zheng, Y. (2000). gamma-Tubulin complexes and their role in microtubule nucleation. Curr. Top. Dev. Biol. 49, 55-73. [DOI] [PubMed] [Google Scholar]
- Hagan, I. M. (1998). The fission yeast microtubule cytoskeleton. J. Cell Sci. 111, 1603-1612. [DOI] [PubMed] [Google Scholar]
- Heitz, M. J., Petersen, J., Valovin, S., and Hagan, I. M. (2001). MTOC formation during mitotic exit in fission yeast. J. Cell Sci. 114, 4521-4532. [DOI] [PubMed] [Google Scholar]
- Hiraoka, Y., Toda, T., and Yanagida, M. (1984). The NDA3 gene of fission yeast encodes beta-tubulin: a cold-sensitive nda3 mutation reversibly blocks spindle formation and chromosome movement in mitosis. Cell 39, 349-358. [DOI] [PubMed] [Google Scholar]
- Horio, T., Uzawa, S., Jung, M. K., Oakley, B. R., Tanaka, K., and Yanagida, M. (1991). The fission yeast gamma-tubulin is essential for mitosis and is localized at microtubule organizing centers. J. Cell Sci. 99, 693-700. [DOI] [PubMed] [Google Scholar]
- Job, D., Valiron, O., and Oakley, B. (2003). Microtubule nucleation. Curr. Opin. Cell Biol. 15, 111-117. [DOI] [PubMed] [Google Scholar]
- Kawaguchi, S., and Zheng, Y. (2004). Characterization of a Drosophila centrosome protein CP309 that shares homology with Kendrin and CG-NAP. Mol. Biol. Cell 15, 37-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knop, M., and Schiebel, E. (1998). Receptors determine the cellular localization of a gamma-tubulin complex and thereby the site of microtubule formation. EMBO J. 17, 3952-3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megraw, T. L., Li, K., Kao, L. R., and Kaufman, T. C. (1999). The centrosomin protein is required for centrosome assembly and function during cleavage in Drosophila. Development 126, 2829-2839. [DOI] [PubMed] [Google Scholar]
- Mitchison, T. J., and Kirschner, M. W. (1987). Some thoughts on the partitioning of tubulin between monomer and polymer under conditions of dynamic instability. Cell Biophys. 11, 35-55. [DOI] [PubMed] [Google Scholar]
- Moreno, S., Klar, A., and Nurse, P. (1991). Molecular analysis of the fission yeast Schizosaccharomyces pombe. Methods Enzymol. 194, 795-823. [DOI] [PubMed] [Google Scholar]
- Oakley, B. R. (2000). gamma-Tubulin. Curr. Top. Dev. Biol. 49, 27-54. [DOI] [PubMed] [Google Scholar]
- Oliferenko, S., and Balasubramanian, M. K. (2002). Astral microtubules monitor metaphase spindle alignment in fission yeast. Nat. Cell Biol. 4, 816-820. [DOI] [PubMed] [Google Scholar]
- Paluh, J. L., Nogales, E., Oakley, B. R., McDonald, K., Pidoux, A. L., and Cande, W. Z. (2000). A mutation in gamma-tubulin alters microtubule dynamics and organization and is synthetically lethal with the kinesin-like protein pkl1p. Mol. Biol. Cell 11, 1225-1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardo, M., and Nurse, P. (2003). Equatorial retention of the contractile actin ring by microtubules during cytokinesis. Science 300, 1569-1574. [DOI] [PubMed] [Google Scholar]
- Rios, R. M., Sanchis, A., Tassin, A. M., Fedriani, C., and Bornens, M. (2004). GMAP-210 recruits gamma-tubulin complexes to cis-Golgi membranes and is required for Golgi ribbon formation. Cell 118, 323-335. [DOI] [PubMed] [Google Scholar]
- Salas, P. J. (1999). Insoluble gamma-tubulin-containing structures are anchored to the apical network of intermediate filaments in polarized CACO-2 epithelial cells. J. Cell Biol. 146, 645-658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawin, K. E., Lourenco, P. C., and Snaith, H. A. (2004). Microtubule nucleation at non-spindle pole body microtubule-organizing centers requires fission yeast centrosomin-related protein mod20p. Curr. Biol. 14, 763-775. [DOI] [PubMed] [Google Scholar]
- Sawin, K. E., Mitchison, T. J., and Wordeman, L. G. (1992). Evidence for kinesin-related proteins in the mitotic apparatus using peptide antibodies. J. Cell Sci. 101, 303-313. [DOI] [PubMed] [Google Scholar]
- Sawin, K. E., and Nurse, P. (1998). Regulation of cell polarity by microtubules in fission yeast. J. Cell Biol. 142, 457-471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawin, K. E., and Snaith, H. A. (2004). Role of microtubules and tea1p in establishment and maintenance of fission yeast cell polarity. J. Cell Sci. 117, 689-700. [DOI] [PubMed] [Google Scholar]
- Schiebel, E. (2000). gamma-Tubulin complexes: binding to the centrosome, regulation and microtubule nucleation. Curr. Opin. Cell Biol. 12, 113-118. [DOI] [PubMed] [Google Scholar]
- Snaith, H. A., and Sawin, K. E. (2003). Fission yeast mod5p regulates polarized growth through anchoring of tea1p at cell tips. Nature 423, 647-651. [DOI] [PubMed] [Google Scholar]
- Stearns, T., Evans, L., and Kirschner, M. (1991). Gamma-tubulin is a highly conserved component of the centrosome. Cell 65, 825-836. [DOI] [PubMed] [Google Scholar]
- Stearns, T., and Kirschner, M. (1994). In vitro reconstitution of centrosome assembly and function: the central role of gamma-tubulin. Cell 76, 623-637. [DOI] [PubMed] [Google Scholar]
- Suelmann, R., Sievers, N., Galetzka, D., Robertson, L., Timberlake, W. E., and Fischer, R. (1998). Increased nuclear traffic chaos in hyphae of Aspergillus nidulans: molecular characterization of apsB and in vivo observation of nuclear behaviour. Mol. Microbiol. 30, 831-842. [DOI] [PubMed] [Google Scholar]
- Takahashi, M., Yamagiwa, A., Nishimura, T., Mukai, H., and Ono, Y. (2002). Centrosomal proteins CG-NAP and Kendrin provide microtubule nucleation sites by anchoring gamma-tubulin ring complex. Mol. Biol. Cell 13, 3235-3245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terada, Y., Uetake, Y., and Kuriyama, R. (2003). Interaction of Aurora-A and centrosomin at the microtubule-nucleating site in Drosophila and mammalian cells. J. Cell Biol. 162, 757-763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, H. M., Cao, H., Chen, J., Euteneuer, U., and McNiven, M. A. (2004). Dynamin 2 binds gamma-tubulin and participates in centrosome cohesion. Nat. Cell Biol. 6, 335-342. [DOI] [PubMed] [Google Scholar]
- Tran, P. T., Marsh, L., Doye, V., Inoue, S., and Chang, F. (2001). A mechanism for nuclear positioning in fission yeast based on microtubule pushing. J. Cell Biol. 153, 397-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vardy, L., and Toda, T. (2000). The fission yeast gamma-tubulin complex is required in G(1) phase and is a component of the spindle assembly checkpoint. EMBO J. 19, 6098-6111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatram, S., Tasto, J. J., Feoktistova, A., Jennings, J. L., Link, A. J., and Gould, K. L. (2004). Identification and characterization of two novel proteins affecting fission yeast gamma-tubulin complex function. Mol. Biol. Cell 15, 2287-2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verde, I., Pahlke, G., Salanova, M., Zhang, G., Wang, S., Coletti, D., Onuffer, J., Jin, S. L., and Conti, M. (2001). Myomegalin is a novel protein of the Golgi/centrosome that interacts with a cyclic nucleotide phosphodiesterase. J. Biol. Chem. 276, 11189-11198. [DOI] [PubMed] [Google Scholar]
- Woods, A., Sherwin, T., Sasse, R., MacRae, T. H., Baines, A. J., and Gull, K. (1989). Definition of individual components within the cytoskeleton of Trypanosoma brucei by a library of monoclonal antibodies. J. Cell Sci. 93, 491-500. [DOI] [PubMed] [Google Scholar]
- Zheng, Y., Wong, M. L., Alberts, B., and Mitchison, T. (1995). Nucleation of microtubule assembly by a gamma-tubulin-containing ring complex. Nature 378, 578-583. [DOI] [PubMed] [Google Scholar]
- Zimmerman, S., Tran, P. T., Daga, R. R., Niwa, O., and Chang, F. (2004a). Rsp1p, a J domain protein required for disassembly and assembly of microtubule organizing centers during the fission yeast cell cycle. Dev. Cell 6, 497-509. [DOI] [PubMed] [Google Scholar]
- Zimmerman, W. C., Sillibourne, J., Rosa, J., and Doxsey, S. J. (2004b). Mitosis-specific anchoring of gamma tubulin complexes by pericentrin controls spindle organization and mitotic entry. Mol. Biol. Cell 15, 3642-3657. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.