Abstract
Microtubules regulate diverse cellular processes, including chromosome segregation, nuclear positioning, and cytokinesis. In many organisms, microtubule nucleation requires γ-tubulin and associated proteins present at specific microtubule organizing centers (MTOCs). In fission yeast, interphase cytoplasmic microtubules originate from poorly characterized interphase MTOCs and spindle pole body (SPB), and during late anaphase from the equatorial MTOC (EMTOC). It has been previously shown that Mto1p (Mbo1p/Mod20p) function is important for the organization/nucleation of all cytoplasmic microtubules. Here, we show that Mto2p, a novel protein, interacts with Mto1p and is important for establishing a normal interphase cytoplasmic microtubule array. In addition, mto2Δ cells fail to establish a stable EMTOC and localize γ-tubulin complex members to this medial structure. As predicted from these functions, Mto2p localizes to microtubules, the SPB, and the EMTOC in an Mto1p-dependent manner. mto2Δ cells fail to anchor the cytokinetic actin ring in the medial region of the cell and under conditions that mildly perturb actin structures, these rings unravel in mto2Δ cells. Our results suggest that the Mto2p and the EMTOC are critical for anchoring the cytokinetic actin ring to the medial region of the cell and for proper coordination of mitosis with cytokinesis.
INTRODUCTION
Microtubules are assembled both in the cytoplasm and the nucleus in yeast. The mitotic spindle formed in the nucleus ensures efficient and high-fidelity chromosome segregation. On the other hand, cytoplasmic microtubules are critical for proper nuclear positioning, nuclear migration, cell polarity, and cell motility. In most eukaryotic cells, microtubules are nucleated from specific structures called the microtubule organizing centers (MTOCs; Urbani and Stearns, 1999). The minus-ends of the microtubules are anchored to the MTOCs, and this anchorage determines the polarity of these microtubules (Bornens, 2002). The centrosome constitutes a major MTOC in animal cells.
In Schizosaccharomyces pombe, there are three different MTOCs (Hagan, 1998). First, the interphase cytoplasmic array of microtubules is organized from interphase MTOCs (iMTOCs). These structures lie adjacent to the nucleus where the antiparallel microtubules overlap (Drummond and Cross, 2000; Tran et al., 2001; Sagolla et al., 2003). Second, the spindle pole body (SPB) organizes the intranuclear mitotic spindle. Third, during late anaphase, an equatorial MTOC (EMTOC) is formed that organizes a postanaphase array (PAA) of microtubules that disassembles after septation and entry into the next cell cycle. Formation of the EMTOC depends on the activity of a GTPase signaling cascade known as the septation initiation network (SIN; Heitz et al., 2001). The SIN triggers cytokinesis in S. pombe and coordinates septation with nuclear division (Le Goff et al., 1999a; McCollum and Gould, 2001). In addition, the cell cycle-specific E3 ubiquitin ligase, the anaphase promoting complex (APC), and the cytokinetic actin ring (CAR) are essential for EMTOC formation (Heitz et al., 2001). Recent evidence has linked the EMTOC to the formation of iMTOCs (Zimmerman et al., 2004) because EMTOC components seem to move from the EMTOC to the iMTOC as satellites after cytokinesis. Failure to disassemble the EMTOC in the rsp1-1 mutant leads to a defect in the organization of interphase cytoplasmic microtubules (Zimmerman et al., 2004).
The MTOC in higher eukaryotes contains a multiprotein complex called the γ-tubulin ring complex (γ-TuRC; Gunawardane et al., 2000). This complex, which contains γ-tubulin, is responsible for microtubule nucleation from the MTOC (Horio et al., 1991; Zheng et al., 1991; Oakley, 2000). Although there are two different forms of the γ-tubulin complex present in higher eukaryotes, centrosomal γ-TuRC and cytosolic γ-tubulin small complex (γ-TuSC), it seems that only the γ-TuRC possesses robust microtubule nucleating capacity on its own in vitro (Oegema et al., 1999; Zhang et al., 2000). The budding yeast (Saccharomyces cerevisiae) Tub4p complex is similar in composition to the γ-TuSC and includes two highly conserved proteins Spc97p and Spc98p, in addition to γ-tubulin (Geissler et al., 1996; Knop et al., 1997; Vinh et al., 2002). Like the γ-TuSC, Tub4p complex reconstituted from insect cells exhibits low microtubule nucleation activity, suggesting that additional components are necessary for robust nucleation of microtubules by this complex (Oegema et al., 1999; Vinh et al., 2002). The S. pombe γ-tubulin complex includes the highly conserved proteins Tug1p (γ-tubulin), Alp4p (homologous to Spc97p), and Alp6p (homologous to Spc98p (Horio et al., 1991; Vardy and Toda, 2000) but unlike the S. cerevisiae Tub4p complex, additional members of the γ-TuC, Alp16p and Gfh1p, have been identified in S. pombe. These proteins share regions of similarity with hGCP6 and hGCP4, respectively (Fujita et al., 2002; Venkatram et al., 2004). γ-TuC mutants in S. pombe display defects in both spindle and cytoplasmic microtubule organization, and all γ-TuC components localize to the SPB both in interphase and mitotic cells and also to the EMTOC (Horio et al., 1991; Paluh et al., 2000; Vardy and Toda, 2000; Fujita et al., 2002; Venkatram et al., 2004). Conditional inactivation of the essential components Tug1p, Alp4p, and Alp6p affects the function of all microtubules. In contrast, the loss of Alp16p or Gfh1p function specifically affects different aspects of cytoplasmic microtubule function and has little or no effect on the function of the mitotic spindle (Vardy et al., 2002; Venkatram et al., 2004). This suggests that different members of the γ-TuC play specific roles in microtubule organization and might do so by binding to distinct proteins either on the cytoplasmic or the nuclear face of the SPB.
Mbo1p/Mod20p is a protein that copurifies with the S. pombe γ-TuC (Sawin et al., 2004; Venkatram et al., 2004). Based on its role in microtubule organization, it has been renamed microtubule organizer (Mto)1p. Mto1p is related to human centrosomin, S. cerevisiae Spc110p and S. pombe Pcp1p and localizes to the SPB, the EMTOC, and also along cytoplasmic microtubules (Sawin et al., 2004; Venkatram et al., 2004). Interestingly, mto1Δ cells exhibit several defects that are indicative of its inability to organize cytoplasmic microtubules: reduced number of microtubule bundles, reduced efficiency of regenerating microtubules after a transient depolymerization, lack of astral microtubules during mitosis, and lack of an EMTOC structure during late mitosis (Sawin et al., 2004; Venkatram et al., 2004). As a result of their inability to efficiently organize cytoplasmic microtubules, mto1Δ cells exhibit defects in cell polarity, nuclear positioning, and proper specification of the cleavage plane at the medial region of the cell. mto1Δ cells have very little defect, if any, associated with the mitotic spindle, indicating that it might associate with the γ-TuC at the cytoplasmic face of the SPB and not at the nuclear face.
To understand the role of Mto1p, we reasoned that it is important to identify other proteins that might specifically affect its function. To that end, we focused on identifying additional proteins that might bind to it. We present here the identification and characterization of one such protein, Mto2p, which like Mto1p, plays a key role in the organization of interphase cytoplasmic microtubules and is critical for proper EMTOC formation. The lack of a functional EMTOC structure in cells lacking mto2+ has allowed us to confirm a role for this structure in anchoring the CAR during cytokinesis.
MATERIALS AND METHODS
Sequence Comparison
Identification of coiled-coil domains in Mto2p was performed using the program COILS (http://www.ch.embnet.org/software/COILS_form.html).
Yeast Strains and Genetic Methods
S. pombe strains used in this study (Table 1) were grown in yeast extract (YE) or minimal medium with appropriate supplements (Moreno et al., 1991). Crosses were performed in glutamate medium, and double-mutant strains were constructed by tetrad analysis. Yeast transformations were performed by electroporation method (Prentice, 1992). Regulated expression of genes from different strengths of nmt1 promoters (Basi et al., 1993; Maundrell, 1993) was achieved by shifting cells grown in media containing thiamine (promoter repressed) to media that lacks thiamine (promoter nonrepressed) after three washes with thiamine-free media.
Table 1.
Strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| KGY 246 | h-ade6-M210 leu1-32 ura4-D18 | Our stock |
| KGY 247 | h+ade6-M210 leu-32 ura4-D18 | Our stock |
| KGY 249 | h+ade6-M216 leu-32 ura4-D18 | Our stock |
| KGY795 | h+cdc11-CFP::kanR ade6-M210 leu-32 ura4-D18 | This study |
| KGY2153 | h-cps-191 ade6-M210 leu-32 ura4-D18 lys-131 | Our stock |
| KGY2430 | h-mto1-myc::kanR ade6-M210 leu-32 ura4-D18 | Our stock |
| KGY3240 | h-mto2-GFP::kanR ade6-M210 leu-32 ura4-D18 | This study |
| KGY3245 | h-mto2-YFP::kanR ade6-M210 leu1-32 ura4-D18 | This study |
| KGY3274 | h-mto2::URA4 ade6-M210 leu1-32 ura4-D18 | This study |
| KGY3310 | h+mto1::URA4 mto2-GFP::kanR ade6-M210 leu1-32 ura4-D18 | This study |
| KGY3311 | h-mto2::URA4 mto1-GFP::kanR ade6-M210 leu1-32 ura4-D18 | This study |
| KGY3344 | h+cdc15-GFP::kanR ade6-M210 leu1-32 ura4-D18 | Our stock |
| KGY4333 | h+cdc25-22 mto2-GFP::kanR ade6-M210 leu1-32 ura4-D18 | This study |
| KGY4540 | h-mto2::URA4 alp6-GFP::kanR ade6-M210 leu1-32 ura4-D18 | This study |
| KGY4563 | h+mto2::URA4 alp4-GFP::kanR ade6-M210 leu1-32 ura4-D18 | This study |
| KGY4773 | h+alp4-1891 mto2-GFP::kanR ade6-M210 leu1-32 ura4-D18 | This study |
| KGY4873 | h-mto2::URA4 cdc15-GFP::kanR ade6-M210 leu1-32 ura4-D18 | This study |
| KGY4895 | h+mto2-GFP::kanR ade6-M210 leu1-32 ura4-D18 | This study |
| KGY4908 | h+alp16::URA4 mto2-GFP::kanR ade6-M210 leu1-32 ura4-D18 | This study |
| KGY4909 | h-gfh1::URA4 mto2-GFP::kanR ade6-M210 leu1-32 ura4-D18 | This study |
| KGY5009 | h-cdc11-CFP::kanR mto2-YFP::kanR ade6-M210 leu1-32 ura4-D18 | This study |
| KGY5126 | h-cps1-191 mto2::URA4 ade6-M210 leu1-32 ura4-D18 lys1-131 | This study |
| KGY5210 | h-mto1-myc::kanR mto2-GFP::kanR ade6-M210 leu1-32 ura4-D18 | This study |
Epitope Tagging of cdc11+ and mto2+
mto2+ was tagged at its chromosomal locus at its 3′ end with sequences coding for 13 copies of the Myc epitope, or green fluorescent protein (GFP), or the yellow fluorescent protein (YFP) or the tandem affinity purification (TAP) tag (Tasto et al., 2001) by a PCR-mediated strategy as described previously (Bahler et al., 1998). cdc11+ was tagged at its chromosomal locus at its 3′ end with sequences coding for cyan fluorescent protein (CFP) as described above. Proper integration of these epitope cassettes was confirmed by PCR.
Cloning and Expression of mto2+
The SPBC902.06 open reading frame (ORF) was amplified from S. pombe genomic DNA by PCR. To facilitate cloning and expression of this ORF by using the nmt1 promoters, an NdeI site and an SmaI site were added to the 5′ and 3′ ends of this PCR fragment. After restriction enzyme digestion, the fragment was cloned into pREP1 (Basi et al., 1993) plasmid to facilitate overexpression studies. The clone was sequenced in its entirety to confirm that the SPBC902.06 ORF was cloned.
Deletion of mto2+
Deletion of mto2+ ORF was achieved by PCR-based one-step homologous recombination according to Bahler et al. (1998). ura4+ was amplified by PCR from plasmid pKG358 by using a forward oligonucleotide corresponding to 80 base pairs (bp) upstream of the ATG codon and a reverse oligonucleotide corresponding to 80 bp downstream of the STOP codon. The amplified fragment was transformed into a diploid strain, and stable integrants were selected. Deletion of one copy of mto2+ in such diploid strains was confirmed by PCR. Sporulation of the heterozygous diploids and tetrad dissection analyses revealed that mto2+ was dispensable for vegetative growth.
Protein Methods
Total cell extracts of S. pombe were prepared in NP-40 buffer (Gould et al., 1991), and immunoprecipitations were carried out using either anti-GFP or 9E10 (anti-Myc) antibodies as described previously (McDonald et al., 1999). Protein transfer, blotting (Venkatram et al., 2004), and detection was performed using the Odyssey system according to manufacturer's instructions (Li-Cor, Lincoln, NE). Protein complexes were obtained using TAP strategy as described previously (Tasto et al., 2001) with one variation: the lysates were clarified at 3000 rpm on a tabletop GS-6R centrifuge in lieu of ultracentrifugation. TAP pellets were subjected to mass spectrometric analyses as described previously (Venkatram et al., 2004).
Lysate Binding Assay
DNA sequence encoding Mto2p was amplified by PCR from wild-type S. pombe genomic DNA. The PCR product contained XmaI and SalI restriction sites at the 5′ and 3′ ends, respectively. The product was cut with these restriction enzymes and cloned into similarly cut pGEX4T-1 to create a glutathione S-transferase (GST)-Mto2p fusion. This fusion protein along with GST was expressed and purified as described previously (Venkatram et al., 2004). The GST and GST-Mto2p proteins bound to glutathione-Sepharose beads were incubated with either control S. pombe cell lysate or lysate containing Mto1p-MYC, and the bound complexes were analyzed as described previously (Morrell et al., 2004). For in vitro binding, DNA sequence encoding Mto2p was amplified as described above into pBluescript vector. Mto2p was then expressed using the TnT-coupled transcription-translation system (Promega, Madison, WI). The MBP and MBP-Mto1p proteins bound to amylose beads were incubated with equal amounts in vitro transcribed-translated Mto2p. Binding of Mto2p, washing the beads, and detection were carried out as described previously (Venkatram et al., 2004).
Physiological Methods
Microtubule regrowth experiments were performed similarly to those described in Sawin et al. (2004). Briefly, cultures grown in YE medium were chilled in flasks in an ice water slurry for 30 min and then returned to shaking water baths at 32°C. Samples at various times as indicated were quickly removed from the culture and fixed in ice-cold ethanol for 10 min, and later processed for immunofluorescence. Microtubules were stained with TAT1 anti-tubulin antibodies (a gift from Prof. Keith Gull, Oxford University, Oxford, United Kingdom) as described previously (Balasubramanian et al., 1997). For cdc25-22 block and release experiments, logarithmically growing cdc25-22 cells in YE medium were arrested in G2 by shifting cells to 36°C for 3 h 45 min followed by shifting them back to 25°C. Samples were taken and processed for live imaging as described below.
Latrunculin A (LatA) Treatment and Cdc15p-GFP Visualization
Treatment of S. pombe cells with low doses of LatA was performed essentially as described previously (Mishra et al., 2004). Briefly, cdc15-GFP and mto2Δcdc15-GFP strains in logarithmic phase of growth at 25°C were synchronized using lactose gradients as described previously (Lieberman, 1995). The cells were then released into fresh medium containing either 0.2 μM LatA or an equal volume of dimethyl sulfoxide (DMSO) for control. Samples were taken every 30 min for imaging and quantification of Cdc15p-GFP ring structures. Visualization of Cdc15p-GFP was performed as described generally under microscopic analyses. The percentage of cells with a Cdc15p-GFP unraveling from the medial region was calculated (n = 200 for each time point).
Microscopy Analyses
For indirect immunofluorescence analyses, S. pombe cells were fixed with 70% ethanol. For anti-actin staining, mouse monoclonal actin antibodies (clone N350; Amersham Biosciences, Piscataway, NJ) were used at a dilution of 1:100 with phosphate-buffered saline/bovine serum albumin. Strains producing chromosomal CFP/GFP/YFP-fusion proteins were grown in YE medium and subjected to live imaging as described previously (Venkatram et al., 2004). Staining of DNA with 4,6-diamidino-2-phenylindole (DAPI) was performed and analyzed as described previously (Tasto et al., 2003). For time-lapse experiments, cells were placed on a hanging drop glass slide containing either YE agar (for Mto2p-GFP) or thiamine-free minimal medium agar (for Atb2p-GFP) and covered with a coverslip. Time-lapse and static images were taken at a spacing of 0.5 μM. The time-lapse images were obtained at intervals of 45 or at 20 s and processed as described previously (Venkatram et al., 2004).
RESULTS
Purification of Mto1p-binding Proteins and Identification of Mto2p
To identify the proteins that might bind to Mto1p and regulate its function, we modified the mto1+ locus to enable the expression of a C-terminally TAP-tagged versions of Mto1p (Venkatram et al., 2004). The mto1-TAP strain grew normally, suggesting that the epitope did not compromise the function of this protein. Tandem affinity purification steps were then carried out from this strain, and the protein composition of a portion of each TAP complex was analyzed by silver staining (our unpublished data) with the remainder analyzed by tandem mass spectrometry (Venkatram et al., 2004). Proteins identified from such a purification at >10% sequence coverage that were absent from the complex purified from untagged cells or from unrelated TAP purifications (our unpublished data) were considered for further analysis. As expected, we identified known γ-TuC components and another novel polypeptide encoded by the ORF SPBC902.06 of molecular mass 44 kDa (at a coverage of 19.8% by mass). We have named this protein microtubule organizer 2 (Mto2p). Sequence analyses of this protein revealed no apparent homologues in other genomes. In addition, the only recognizable motif present in this protein is a coiled-coil domain, spanning from amino acid 110 to 150.
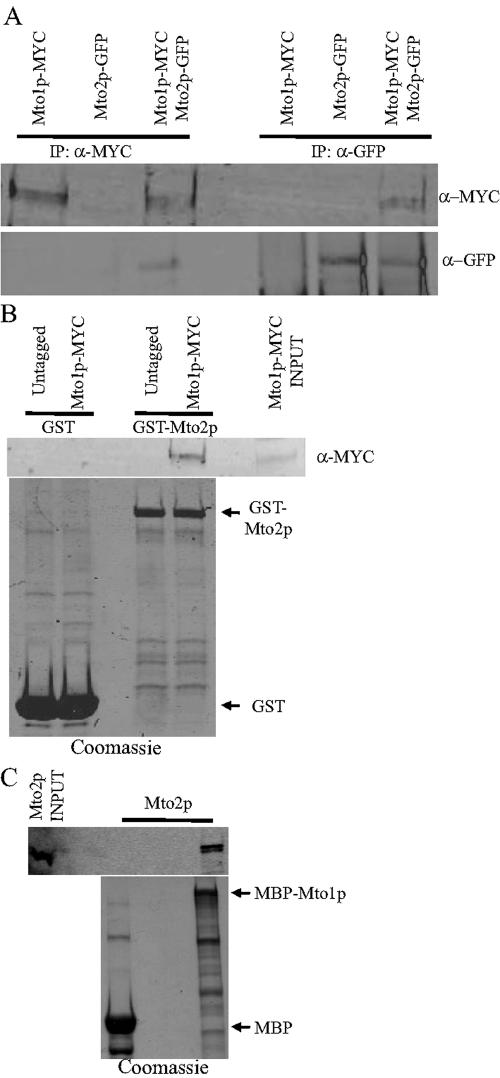
To validate our TAP results, we performed a reciprocal purification from a strain expressing Mto2p-TAP and found Mto1p in the protein complex reproducibly, albeit at a low sequence coverage (4.8, 7.2, and 7% by mass). To further support our TAP results, we sought to determine whether these proteins are present in the same immune complex. To accomplish this, we modified the locus encoding Mto2p to create an mto2-gfp strain. The tagged mto2+ allele was combined with mto1-myc allele to create double-tagged strains. In an anti-MYC immunoprecipitate from mto1-myc mto2-gfp strains but not from single-tagged strains, both Mto1p-MYC and Mto2p-GFP were detected (Figure 1A, IP:α-MYC). Similar specific complex formation was detected when the same strains were immunoprecipitated with anti-GFP antibodies (Figure 1A, IP:α-GFP). Further evidence of association was that bacterially produced GST-Mto2p specifically bound to Mto1p-MYC from S. pombe lysates (Figure 1B). Quantitation of the band intensity corresponding to Mto1p-MYC present in the INPUT lane and that bound to GST-Mto2p indicated that ∼1/10 of Mto1p-MYC present in the lysate bound to GST-Mto2p. Collectively, these data establish that Mto1p and Mto2p exist in a physical complex in vivo. To determine whether these two proteins interact directly, we incubated MBP or MBP-Mto1p fusion proteins with in vitro transcribed-translated Mto2p. Mto2p bound to MBP-Mto2p but not MBP control (Figure 1C), demonstrating a specific and direct interaction between Mto1p and Mto2p.
Figure 1.
Mto1p binds to Mto2p. (A) Protein lysates were prepared from cells expressing tagged alleles of mto1+ and mto2+. These were subjected to immunoprecipitation using either 9E10 (IP:α-MYC) or anti-GFP (IP:α-GFP) antibodies. The immunoprecipitates were resolved by SDS-PAGE and immunoblotted with either α-MYC or α-GFP antibodies as indicated. (B) Protein lysates prepared from either control cells (untagged) or cells expressing a MYC-tagged allele of mto1+ were prepared and added to either GST or GST-Mto2p beads. After incubation, the beads were washed and bound proteins were resolved by SDS-PAGE and immunoblotted with α-MYC antibodies as indicated. A parallel gel was run and stained with Coomassie Blue to visualize the GST proteins. The arrows in the bottom panel indicate GST and GST-Mto2p. (C) MBP and MBP-Mto1p on beads were incubated with equal amounts of Mto2p made in vitro by using coupled transcription/translation. After being washed, the proteins were detected by autoradiography (top) or Coomassie staining (bottom). The arrows indicate MBP and MBP-Mto1p. A sample (15%) of in vitro translated Mto2p is shown in the INPUT lane.
Mto2p Localizes to the MTOC and along Interphase Microtubules
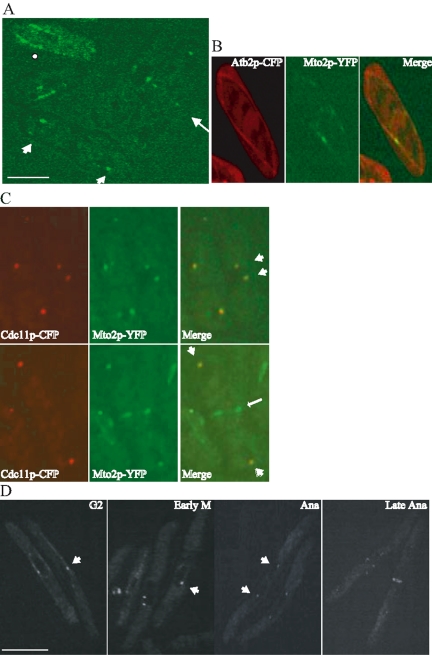
Given that Mto2p exists in a complex with Mto1p and that Mto1p localizes to MTOCs and along cytoplasmic microtubules, we wanted to determine whether Mto2p exhibited a similar localization pattern. To this end, we created a strain in which a GFP or a YFP tag was fused to the 3′ end of the mto2+ locus to produce Mto2p-GFP or Mto2p-YFP from its endogenous promoter. The mto2-GFP and mto2-YFP strains exhibited normal morphology, indicating that the addition of the tags did not affect Mto2p function (see below). In an asynchronous population of cells expressing Mto2p-GFP, we observed several localization patterns (Figure 2A). Mto2p-GFP was found along linear arrays reminiscent of microtubules and upon higher resolution imaging, discrete “dot”-like localization was observed (Figure 2A). In addition, in both interphase and mitotic cells, Mto2p-GFP was found in dots juxtaposed to the nucleus, which presumably represent SPBs (Figure 2A). Also, in late mitotic cells, a ring of Mto2p-GFP was seen in the medial region of the cell resembling that of an EMTOC (Figure 2A).
Figure 2.
Mto2p localizes to the SPB and the EMTOC. (A) Cells expressing Mto2p-GFP were subjected to live imaging by using a GFP filter set. The fusion protein localizes along cytoplasmic filaments likely representing microtubules (indicated by a dot); to the SPB (indicated by arrowheads) and to the medial EMTOC structure (indicated by an arrow). (B) Cells expressing CFP-Atb2p and Mto2p-YFP were subjected to live imaging by using a CFP/YFP filter set. (C) Cells expressing Cdc11p-CFP and Mto2p-YFP were subjected to live imaging by using a CFP/YFP filter set. The localization of the CFP and the YFP fusion proteins to the SPBs are indicated by block arrows and the EMTOC localization of Mto2p-GFP by a thin arrow. (D) cdc25-22 cells expressing Mto2p-GFP were subjected to live imaging by using a GFP filter set. The localization of Mto2p-GFP to SPBs and the EMTOC are indicated by arrows. The cell cycle stage is indicated in the top right corner of each panel. Bars, 5 μm.
To ascertain that Mto2p localizes along cytoplasmic microtubules, we performed colocalization analyses in strains that expressed CFP-Atb2p and Mto2p-YFP by live imaging. As shown in Figure 2B, in interphase cells, Mto2p-YFP localized along cytoplasmic microtubules. To confirm that Mto2p indeed localizes to the SPB, we colocalized Mto2p with a known constitutive component of the SPB. Cdc11p is a scaffold for the SIN and a constitutive component of the SPB (Krapp et al., 2001; Tomlin et al., 2002). Hence, for colocalization analyses we generated strains that expressed Cdc11p-CFP and Mto2p-YFP and performed live imaging on logarithmically growing cells. As shown in Figure 2C (top), in early and midmitotic cells, Mto2p-YFP colocalized with Cdc11p-CFP at the SPB. However, in late mitotic cells (Figure 2C, bottom) that have separated their nuclei to the opposite ends of the cell, an extra signal of Mto2p-YFP was found in the medial region of the cell that did not colocalize with the Cdc11p-CFP signal. This clearly establishes that Mto2p is similar to Mto1p in its localization pattern and that it localizes to the SPB, EMTOC, and likely along cytoplasmic microtubules. Genome-wide localization analyses of proteins fused to GFP indicated that a portion of Mto2p localizes to the SPB and the EMTOC, an observation consistent with our results (Ding et al., 2000).
To better document the changes in Mto2p-GFP localization during mitosis, we analyzed its localization in cells that were synchronously passing through mitosis. To this end, we performed a block and release experiment using a cdc25-22 strain expressing Mto2p-GFP. In cells arrested in G2 owing to Cdc25p inactivation, Mto2p-GFP was found along cytoplasmic microtubules and at the SPB (Figure 2D, G2). As these cells entered mitosis, Mto2p-GFP was found only on duplicated/separated SPBs, and cytoplasmic staining was lost (Figure 2D, early M and Ana). As the cells traversed into late anaphase, Mto2p-GFP was found at the EMTOC structure in addition to the SPBs (Figure 2D, late Ana). Thus, Mto2p localizes to the EMTOC as this structure is formed. Together with the biochemical data, this establishes that Mto2p likely exists in a complex with Mto1p throughout the cell cycle.
mto2Δ Cells Lack an EMTOC
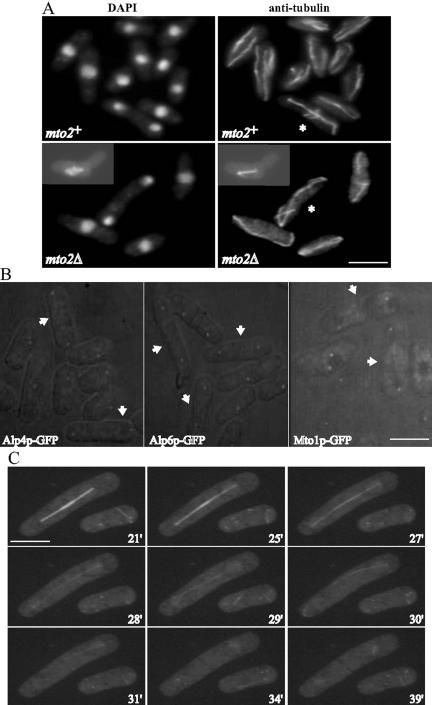
To gain insight into the function of mto2+, we deleted one copy of this gene in a diploid strain and replaced it with a copy of ura4+ by homologous recombination. After sporulation, tetrads were dissected and such analyses revealed that mto2+ was not essential for vegetative growth (our unpublished data; Figure 3A). However, an exponentially growing population of mto2Δ cells exhibited several abnormal phenotypes, some indicative of microtubule defects. First, about one-third (38%) of the cells were bent (Figure 3A). In addition, most interphase cells exhibited fewer microtubule arrays (83% of mto2Δ cells contained 2 or fewer microtubule arrays, compared with 12% of mto2+ cells) than wild-type cells, and most cells (63%) had microtubules bent around the edge of the cell (Figures 3A and 4). Notably, although mitotic cells contained a normal spindle, an EMTOC was not detected in late anaphase cells (Figure 3, A and C). As a result, septating cells did not contain a PAA of microtubules. These microtubule defects are reminiscent of those observed in mto1Δ cells (Sawin et al., 2004; Venkatram et al., 2004). One notable difference, however, was that mto2Δ cells formed astral microtubules (Figure 3A and Figure 3video2).
Figure 3.
mto2Δ cells are viable but exhibit polarity and microtubule defects. (A) Exponentially growing mto2+ and mto1Δ cells were fixed and stained with either DAPI or anti-tubulin antibodies to visualize DNA and microtubules, respectively. The asterisks indicate anaphase cells. The inset illustrates a cell with a short spindle with astral microtubules. (B) Live imaging of exponentially growing mto2Δ cells expressing Alp4p-GFP, Alp6p-GFP, or Mto1p-GFP as indicated. The arrows in each panel indicate late anaphase cells. (C) Time-lapse confocal microscopy of live mto2Δ cells expressing GFP-tubulin. The numbers on each panel correspond to the time (in minutes) elapsed since the capturing of the first frame. Bars, 5 μm.
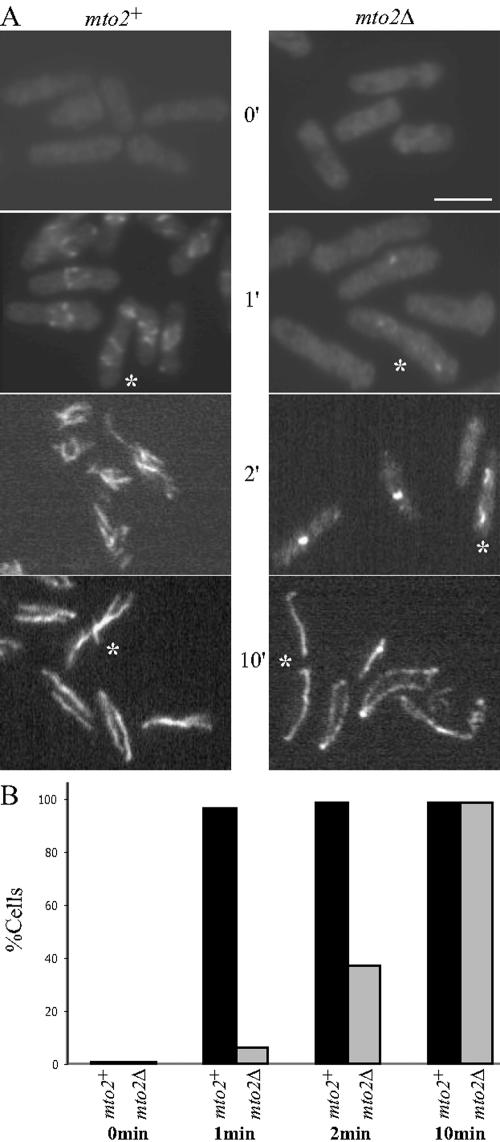
Figure 4.
Mto2p is required for efficient de novo microtubule nucleation. (A) Exponentially growing mto2+ and mto2Δ cells were incubated on ice for 30 min and then released to 32°C. Samples were collected at various time points before (0 min) and after the release, fixed, and stained with anti-tubulin antibodies as indicated. The asterisks indicate late anaphase cells. Bar, 5 μm. (B) Quantification of cells with microtubule structures. Cells (n ≥ 300) from the above-described experiment were counted for the presence of microtubule filaments even if they seemed aberrant.
Given that the EMTOC was not observed in mto2Δ cells, we analyzed the localization of individual γ-TuC components in mto2Δ cells. Whereas the SPB localization of γ-TuC members was not affected, in late mitotic cells, medial localization of these proteins was not observed (Figure 3B; our unpublished data). Hence, we conclude that mto2Δ cells are defective in the localization of the γ-TuC to the EMTOC. Previously, we and others have shown that mto1Δ cells exhibit a similar defect (Sawin et al., 2004; Venkatram et al., 2004). Hence, we sought to determine whether Mto2p was required for the EMTOC localization of Mto1p. As shown Figure 3B, Mto1p-GFP did not localize to the medial region of mto2Δ cells in late anaphase. This indicates that Mto2p, like Mto1p, is required to recruit and/or stabilize the γ-TuC in the medial region of the cell to establish a functional EMTOC structure. To verify that this is the case, we performed live confocal imaging of mto2Δ cells expressing GFP-α-tubulin. In wild-type cells, the EMTOC forms during anaphase and can be detected before spindle disassembly (Heitz et al., 2001; Venkatram et al., 2004; Fig 3video1). An EMTOC was not assembled in any of 17 mitotic mto2Δ cells examined, even though spindle disassembly was readily observed (Figure 3C and Fig 3video2 and Fig 3video3).
Mto2p Is Required for the Efficient De Novo Microtubule Nucleation
The observed defects in the interphase microtubule cytoskeleton in mto2Δ cells prompted us to test whether Mto2p function was required for microtubule nucleation and/or organization as was determined previously for Mto1p (Sawin et al., 2004). Transient incubation of wild-type and mto2Δ cells on ice for 30 min depolymerized all microtubules in vivo (Figure 4A, 0′). When these cells were returned to 32°C, discrete microtubules around the nucleus were observed in almost all wild-type cells and at this time, in late anaphase cells, a medial microtubule structure was observed (Figure 4A, 1′). In most cells microtubule growth occurred rapidly and by 10 min, all wild-type cells exhibited normal arrays of microtubules, including PAAs (Figure 4A, 2′ and 10′). In contrast, very few mto2Δ cells, if any, contained microtubules before the 10-min time point (Figure 4A). Moreover, in late anaphase cells, an EMTOC structure failed to form (compare cells marked with *). Although by 10 min most mto2Δ cells had regrown their microtubules, these structures were aberrant and fewer in number (Figure 4, A and B). They were bent and wrapped around the cell ends (Figures 3A and 4A). These results indicate that Mto2p function is required for efficient de novo nucleation of microtubules, EMTOC formation, and proper organization of cytoplasmic microtubules.
Mto1p Function Is Required for Proper Mto2p Localization
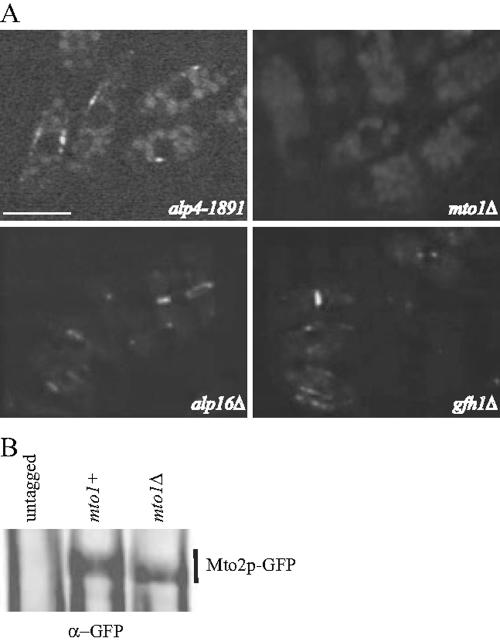
The results presented above indicate a close physical and functional relationship exists between Mto1p and Mto2p. To understand whether Mto1p also regulates Mto2p function, we started by analyzing the localization of Mto2p-GFP in γ-TuC mutants. Mto2p-GFP was present at all its normal locations in alp16Δ and gfh1Δ mutant cells indicating that these proteins do not influence Mto2p localization (Figure 5A). In alp4-1891 cells, Mto2p-GFP was detected at SPBs, indicating that γ-TuC function is not required for the proper localization of Mto2p-GFP to SPBs (Figure 5A). However, we were not able to detect any specific Mto2p-GFP signal in mto1Δ cells, indicating that Mto1p is critical for Mto2p localization (Figure 5A). This is not due to reduced levels of Mto2p in the absence of Mto1p because immunoblot analysis showed that an equal amount of Mto2p-GFP was present in both mto1+ and mto1Δ cells (Figure 5B). The difference in the mobility of Mto2p-GFP in mto1+ and mto1Δ cells probably reflects a difference in a posttranslational modification of Mto2p. Collectively, these results indicate that Mto1p and Mto2p are mutually dependent for their proper localization: Mto2p for the EMTOC localization of Mto1p and Mto1p for the proper localization of Mto2p to all MTOC structures.
Figure 5.
Proper localization of Mto2p-GFP is dependent on Mto1p function. (A) Cells expressing Mto2p-GFP in different genetic backgrounds as indicated, were subjected to live imaging using a GFP filter set. The localization of the fusion protein was observed at 36°C in alp4-1891 cells and at 27°C in other mutant cells. Bar, 5 μm. (B) Protein lysates prepared from either control cells (untagged) or mto1+ and mto1Δ cells expressing Mto2p-GFP were prepared and subjected to immunoprecipitation by using anti-GFP antibodies. The immunoprecipitates were washed, resolved by SDS-PAGE, and immunoblotted with α-GFP antibodies as indicated. The band corresponding to Mto2p-GFP is indicated.
Mto2p and the EMTOC Play a Role in Anchoring the CAR
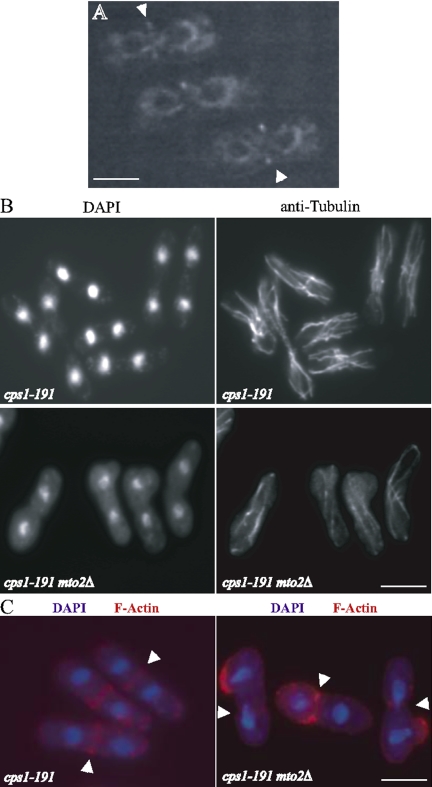
Because mto2Δ cells lack an EMTOC structure, we used this strain to address potential roles of the EMTOC in fission yeast cytokinesis. A temperature-sensitive mutant in the β-glucan synthase encoding cps1+ (cps1-191) arrests in G2 after a failed cytokinesis with two nuclei and a fully assembled CAR (Le Goff et al., 1999b; Liu et al., 2000; Pardo and Nurse, 2003). cps1-191-arrested cells also contain a PAA of microtubules, having failed to reorganize their microtubule cytoskeleton as the cells exit mitosis (Liu et al., 1999; Pardo and Nurse, 2003; Figure 6B, top row). The G2 arrest of cps1-191 cells is due to the activation of a cytokinesis checkpoint that includes the Wee1p kinase, the Clp1p/Flp1p phosphatase, and the SIN (Le Goff et al., 1999b; Liu et al., 2000; Cueille et al., 2001; Trautmann et al., 2001). Given that cps1-191 mutants arrest with a PAA of microtubules, we expected that they arrested with an EMTOC and established this by analyzing Mto2p-GFP (Figure 6A). Because cps1-191 mutants arrest with a PAA of microtubules and elicit a checkpoint-dependent arrest, we asked whether the formation of PAA microtubules (and hence the EMTOC) is required for this arrest by examining the arrest phenotype of a cps1-191 mto2Δ double mutant. Although cps1-191mto2Δ double mutant cells did not arrest with a PAA of microtubules (Figure 6B, bottom row), 94% of them still arrested as binucleate cells comparable with the 96% binucleate arrest of the single cps1-191 mutant. The maintenance of the cell cycle arrest indicates that the EMTOC and/or PAA is not required for the cytokinesis checkpoint. Interestingly, whereas cps1-191 cells contained only PAA microtubules, the double mutant formed an interphase array of microtubules that were fewer in number, bent, and often wrapped around the cell tips.
Figure 6.
Mto2p function is required for medial anchoring of the CAR in a cps1-191 mutant. (A) cps1-191 cells expressing Mto2p-GFP were shifted to 36°C for 4 h and were subjected to live imaging by using a GFP filter set. Mto2p-GFP localization to the EMTOC is indicated by arrowheads. (B) Exponentially growing cps1-191 or cps1-191mto2Δ cells were shifted to 36°C for 4 h. These cells were then fixed and processed for staining with DAPI and anti-tubulin antibodies to visualize DNA and microtubules, respectively. (C) Exponentially growing cps1-191 or cps1-191mto2Δ cells was shifted to 36°C for 4 h. These cells were then fixed and processed for staining with DAPI and anti-actin antibodies to visualize DNA and the CAR, respectively. The DNA is false-colored with blue, whereas actin is false-colored with red. The merge images are shown, and the position of the CAR is indicated by arrowheads. Bars, 5 μm.
Transient depolymerization of microtubules in cps1-191 mutant cells leads to the movement of the CAR from the medial region of these cells (Pardo and Nurse, 2003). Because cps1-191 mutant cells arrest with a PAA of microtubules, this result suggested a role for these microtubules or the EMTOC in anchoring the CAR to the medial region. We therefore examined whether loss of Mto2p function had a similar effect on the CAR in cps1-191 and cps1-191 mto2Δ cells. Staining of actin structures indicated that cps1-191 single mutant cells arrested with a CAR at the medial region (Figure 6C). However, in cps1-191 mto2Δ double mutant cells, the CAR was displaced from the medial region and in some cases was positioned over one of the nuclei (Figure 6C). Together, our results are consistent with a role for the EMTOC and/or PAA in medial anchoring of the CAR during a prolonged arrest.
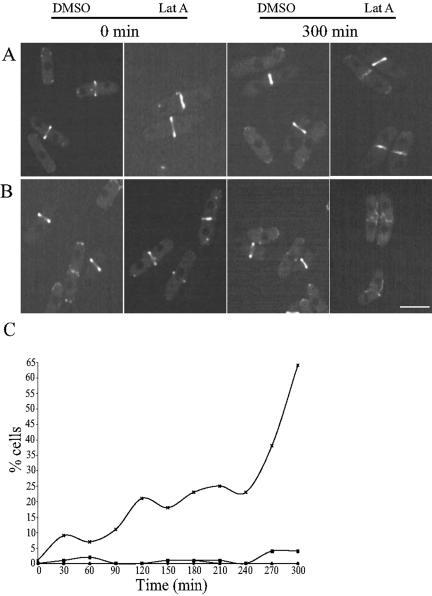
Low doses of LatA greatly delay cytokinesis and in wild-type cells, the CAR is maintained and does not constrict even for as long as 300 min (Mishra et al., 2004). We therefore asked whether the CAR could be maintained in mto2Δ cells that lack the EMTOC and PAA by using Cdc15p-GFP to visualize the CAR. After cells were synchronized in G2 and allowed to enter mitosis, Cdc15p rings formed normally in both wild-type and mto2Δ cells (Figure 7C). Even after 300 min of treatment with 0.2 μM LatA (or DMSO solvent control), Cdc15p-GFP rings remained intact and were properly positioned in wild-type cells (Figure 7, A and C). Although treating mto2Δ cdc15-GFP cells with DMSO had no effect on the CAR, LatA treatment had a dramatic effect. Cdc15-GFP was often visualized to unravel from the medial region of the cell (Figure 7, B and C). This phenotype was found to gradually increase with time and at 300 min, about two-thirds (64%) of the cells exhibited rings that were unraveled (Figure 7C). This shows that Mto2p function, presumably through its role in establishing the EMTOC and PAA, is required for preserving the integrity of and anchoring the CAR when cytokinesis is delayed.
Figure 7.
Mto2p function is required for medial anchoring and maintenance of the Cdc15p-GFP ring upon LatA treatment. (A and B) mto2+ (A) or mto2Δ (B) cells expressing Cdc15p-GFP were treated with either 0.2 μM of LatA or an equal volume of DMSO as indicated and were subjected live imaging by using a GFP filter set. Representative images at 0 and 300 min after treatment are shown. Bar, 5 μm. (C) Quantification of cells with unraveled Cdc15p-GFP rings. Cells (n = 200) from the above-described experiment were counted to determine the percentage of cells with Cdc15p-GFP unraveled from the medial region.
DISCUSSION
The S. pombe γ-TuC is essential for the formation of a mitotic spindle, and for the formation and proper organization of longitudinal arrays of cytoplasmic microtubules and astral microtubules during mitosis. In addition, the γ-TuC localizes to the medial region in late anaphase as part of the EMTOC structure and organizes the PAA of microtubules. In this article, we report our identification of Mto2p, a protein that is specifically required for the formation of normal cytoplasmic microtubule arrays and the EMTOC. Our analyses of mto2Δ cells provide another illustration of the role played by the EMTOC and/or PAA in anchoring the CAR, at least during a prolonged cell cycle delay.
Mto2p and Cytoplasmic Microtubule Organization
Mto2p seems to influence the ability of the γ-TuC to nucleate specific sets of cytoplasmic microtubules, and in two respects the phenotype of mto2Δ cells is similar to cells lacking Mto1p. This is not surprising given our evidence that these proteins physically associate with each other, mirror each other in localization pattern, and depend upon each other for proper localization. Previously, we were able to demonstrate that Mto1p was physically associated with the γ-TuC. We have not been able to do the same with Mto2p. Mto2p was not identified in Alp4p–TAP or Alp6p–TAP complexes nor were components of the γ-TuC identified in the Mto2p–TAP complex. Moreover, Mto2p sedimented away from γ-TuC components in gel filtration analyses (our unpublished data). Although our inability to detect an Mto2p-γ-TuC association might reflect an unknown technical problem, it is also possible that Mto1p exists in two complexes, one containing the γ-TuC and another containing Mto2p.
Akin to the lack of Mto1p function, mto2Δ cells exhibit a reduced number of improperly organized interphase cytoplasmic microtubules. This suggests that Mto2p is required for γ-tubulin complex function at particular sites in the cytoplasm. Although it is possible that this regulation is direct, it is also possible that Mto2p acts through Mto1p to perform this function. Along these lines, it is interesting to compare the differences between mto1Δ and mto2Δ cells in their ability to nucleate microtubules de novo. Whereas mto2Δ cells were able to nucleate aberrant microtubules after 10 min, mto1Δ cells did not do so until 40 min (Figure 4; Sawin et al., 2004). Therefore, we conclude that mto1Δ cells are more defective in this respect than mto2Δ cells. This difference could reflect the fact that Mto1p-GFP still localizes to SPBs in mto2Δ cells, although not readily detectable along microtubules or at the EMTOC. Hence, SPB-localized Mto1p probably contributes to microtubule nucleation in the absence of Mto2p. It will be of interest to determine the SPB binding partner(s) of Mto1p and to determine its roles in Mto1p function and cytoplasmic microtubule organization. It is clear that Mto1p and Mto2p localize along cytoplasmic microtubules, although the functional significance of this localization is not clear. It is conceivable that microtubule-localized Mto1p and Mto2p recruit other microtubule-associated proteins such as motor proteins to regulate microtubule dynamics and organization and/or the γ-TuC to nucleate microtubules along preexisting bundles.
A major phenotype associated with ablation of Mto1p function is the absence of astral microtubules in mitotic cells (Sawin et al., 2004; Venkatram et al., 2004). However, astral microtubules are formed in mitotic mto2Δ cells (Figure 3). Again, this difference is likely explained by the presence of Mto1p at the SPB in these cells. Hence, we propose that Mto1p is a multifunctional protein that promotes astral microtubule nucleation and other types of microtubule nucleation through different mechanisms and that only one of these is dependent upon Mto2p. Structure–function analyses of Mto1p should help define the regions of the protein that direct it to the various parts of the cell and that regulate different microtubule nucleation events.
Mto2p and the EMTOC Function
The EMTOC is formed during late anaphase in S. pombe cells and establishes a postanaphase array of microtubules (Hagan and Petersen, 2000). Because Mto1p is required for the EMTOC localization of other γ-TuC members, we suggested previously that Mto1p might act as a progenitor of this structure (Venkatram et al., 2004). However, the lack of proper Mto1p localization to the EMTOC in mto2Δ cells suggests that Mto2p might play this key role or that these proteins work together to accomplish this task. As candidate progenitors, it is possible that their functions are regulated during anaphase. Given that the EMTOC is formed only during late anaphase after chromosome segregation in an APC-dependent manner (Heitz et al., 2001), EMTOC formation is probably linked to cyclin-dependent kinase (CDK) inactivation. Cells lacking Clp1p/Flp1p phosphatase function, which have sustained CDK activity (Esteban et al., 2004; Wolfe and Gould, 2004), fail to form an EMTOC (our unpublished data), providing another correlation between CDK down-regulation and EMTOC formation. Hence, it would be important to determine whether Mto2p and/or Mto1p function is regulated by CDK inactivation. It also will be important to determine the role of the Polo kinase in Mto2p and Mto1p function because overexpression of Polo kinase in S. pombe cells induces EMTOC formation in a small percentage of cells (Heitz et al., 2001).
The function of the EMTOC is not clear, although it has recently been proposed to be analogous to the metazoan midbody structure and to play a role in tethering the CAR between the divided DNA masses during cytokinesis (Pardo and Nurse, 2003). The formation of an EMTOC is dependent on the formation of a CAR, the SIN, and the APC (Heitz et al., 2001). Given that all these functions are essential, it has not been possible to determine the role of the EMTOC in isolation. Previous analyses of mto1Δ cells indicated that the EMTOC is not an essential structure. However, although mto1Δ cells lack an EMTOC, they have additional defects not shared with mto2Δ cells, indicating that mto2Δ cells provide a better model to study the consequences of EMTOC loss.
Previous work has shown that the depolymerization of microtubules in cps1-191 mutants, which arrest with a stable CAR and a PAA, leads to loss of medial anchoring of the CAR (Pardo and Nurse, 2003). This led to the proposal that the EMTOC and/or PAA play a role in anchoring the CAR (Pardo and Nurse, 2003). We tested this proposal directly by visualizing the CAR in cps1-191mto2Δ cells and conclude that the EMTOC is required for the medial anchoring of the CAR. A second approach has recently been used to stabilize the CAR. Treatment of fission yeast cells with LatA stabilizes the CAR and delays cytokinesis (Mishra et al., 2004). Treatment of mto2Δ cells with LatA and subsequent visualization of the CAR independently verified the proposal that the EMTOC is required for CAR anchoring. Hence, our analyses clearly establish that the EMTOC functions to anchor/maintain the CAR, at least under conditions where cytokinesis is delayed. We have entertained the possibility that Mto2p might perform a similar function during an unperturbed cell cycle and have observed a delay, albeit minor, in the completion of cytokinesis in mto2Δ cells (our unpublished data). Collectively, these data raise the possibility that the EMTOC and the CAR are intimately linked, maybe, even physically. For example, it is possible that the EMTOC interacts with a component of the CAR and this interaction both stabilizes and anchors the CAR until it constricts during an unperturbed cell cycle. Additionally (not mutually exclusive), it is possible that the EMTOC regulates the organization of sterol-rich membrane domains in the medial region of the cell, because these domains have been implicated in the positioning and maintenance of the CAR (Wachtler et al., 2003). Together, the role of the EMTOC in CAR positioning/maintenance is consistent with the established role of the spindle midzone and the midbody structures in positioning the cleavage furrow (Glotzer, 2004). Hence, our studies provide more credence to the hypothesis that the EMTOC might perform some function carried out by the metazoan midbody structure.
Both Cps1p inactivation and LatA treatment prevent cytokinesis by eliciting the cytokinesis checkpoint. In this study, we show that cps1-191mto2Δ cells do not contain PAA microtubules but still arrest as binucleate cells, indicating that the EMTOC and PAA are not part of the cytokinesis checkpoint. Given that mto2Δ cells are checkpoint-proficient and the CAR is not anchored at the medial region in these cells, we conclude that neither Mto2p function nor the anchoring of the CAR plays a role in the cytokinesis checkpoint. Indeed, recent work on Lsk1p protein kinase has provided evidence that the maintenance of the cytokinesis checkpoint and the integrity of the CAR are independent (Karagiannis et al., 2005). Further analyses of Mto2p are likely to provide additional insight into the role of the EMTOC, and perhaps the γ-TuC, in regulating CAR function and cytokinesis.
Note added in proof. While this work was in revision, similar results were reported by Samejima et al. (2005) and Janson et al (2005) regarding the role of Mto2p in S. pombe microtubule nucleation.
Supplementary Material
Acknowledgments
We thank Joseph Tasto and Anna Feoktistova for technical help and Ken Sawin and Phong Tran for sharing unpublished results. We also thank all Gould laboratory members, especially Rachel Roberts, for discussions during the course of this work and Mohan Balasubramanian for suggesting the LatA experiment. This work was supported by the Howard Hughes Medical Institute of which K.L.G. is an investigator.
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E04-12-1043) on March 30, 2005.
Abbreviations used: CAR, cytokinetic actin ring; EMTOC, equatorial microtubule organizing center; LatA, latrunculin A; MTOC, microtubule organizing center; PAA, postanaphase array; SPB, spindle pole body.
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
References
- Bahler, J., Wu, J. Q., Longtine, M. S., Shah, N. G., McKenzie, A., 3rd, Steever, A. B., Wach, A., Philippsen, P., and Pringle, J. R. (1998). Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14, 943-951. [DOI] [PubMed] [Google Scholar]
- Balasubramanian, M. K., McCollum, D., and Gould, K. L. (1997). Cytokinesis in fission yeast Schizosaccharomyces pombe. Methods Enzymol. 283, 494-506. [DOI] [PubMed] [Google Scholar]
- Basi, G., Schmid, E., and Maundrell, K. (1993). TATA box mutations in the Schizosaccharomyces pombe nmt1 promoter affect transcription efficiency but not the transcription start point or thiamine repressibility. Gene 123, 131-136. [DOI] [PubMed] [Google Scholar]
- Bornens, M. (2002). Centrosome composition and microtubule anchoring mechanisms. Curr. Opin. Cell Biol. 14, 25-34. [DOI] [PubMed] [Google Scholar]
- Cueille, N., Salimova, E., Esteban, V., Blanco, M., Moreno, S., Bueno, A., and Simanis, V. (2001). Flp1, a fission yeast orthologue of the S. cerevisiae CDC14 gene, is not required for cyclin degradation or rum1p stabilisation at the end of mitosis. J. Cell Sci. 114, 2649-2664. [DOI] [PubMed] [Google Scholar]
- Ding, D. Q., Tomita, Y., Yamamoto, A., Chikashige, Y., Haraguchi, T., and Hiraoka, Y. (2000). Large-scale screening of intracellular protein localization in living fission yeast cells by the use of a GFP-fusion genomic DNA library. Genes Cells 5, 169-190. [DOI] [PubMed] [Google Scholar]
- Drummond, D. R., and Cross, R. A. (2000). Dynamics of interphase microtubules in Schizosaccharomyces pombe. Curr. Biol. 10, 766-775. [DOI] [PubMed] [Google Scholar]
- Esteban, V., Blanco, M., Cueille, N., Simanis, V., Moreno, S., and Bueno, A. (2004). A role for the Cdc14-family phosphatase Flp1p at the end of the cell cycle in controlling the rapid degradation of the mitotic inducer Cdc25p in fission yeast. J. Cell Sci. 117, 2461-2468. [DOI] [PubMed] [Google Scholar]
- Fujita, A., Vardy, L., Garcia, M. A., and Toda, T. (2002). A fourth component of the fission yeast gamma-tubulin complex, Alp16, is required for cytoplasmic microtubule integrity and becomes indispensable when gamma-tubulin function is compromised. Mol. Biol. Cell 13, 2360-2373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geissler, S., Pereira, G., Spang, A., Knop, M., Soues, S., Kilmartin, J., and Schiebel, E. (1996). The spindle pole body component Spc98p interacts with the gamma-tubulin-like Tub4p of Saccharomyces cerevisiae at the sites of microtubule attachment. EMBO J. 15, 3899-3911. [PMC free article] [PubMed] [Google Scholar]
- Glotzer, M. (2004). Cleavage furrow positioning. J. Cell Biol. 164, 347-351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould, K. L., Moreno, S., Owen, D. J., Sazer, S., and Nurse, P. (1991). Phosphorylation at Thr167 is required for Schizosaccharomyces pombe p34cdc2 function. EMBO J. 10, 3297-3309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunawardane, R. N., Lizarraga, S. B., Wiese, C., Wilde, A., and Zheng, Y. (2000). gamma-Tubulin complexes and their role in microtubule nucleation. Curr. Top Dev. Biol. 49, 55-73. [DOI] [PubMed] [Google Scholar]
- Hagan, I. M. (1998). The fission yeast microtubule cytoskeleton. J. Cell Sci. 111, 1603-1612. [DOI] [PubMed] [Google Scholar]
- Hagan, I. M., and Petersen, J. (2000). The microtubule organizing centers of Schizosaccharomyces pombe. Curr. Top Dev. Biol. 49, 133-159. [DOI] [PubMed] [Google Scholar]
- Heitz, M. J., Petersen, J., Valovin, S., and Hagan, I. M. (2001). MTOC formation during mitotic exit in fission yeast. J. Cell Sci. 114, 4521-4532. [DOI] [PubMed] [Google Scholar]
- Horio, T., Uzawa, S., Jung, M. K., Oakley, B. R., Tanaka, K., and Yanagida, M. (1991). The fission yeast gamma-tubulin is essential for mitosis and is localized at microtubule organizing centers. J. Cell Sci. 99, 693-700. [DOI] [PubMed] [Google Scholar]
- Janson, M. E., Setty, T. G., Paoletti, A., and Tran, P. T. (2005). Efficient formation of bipolar microtuble bundles requires microtubule-bound γ-tubulin complexes. J. Cell. Biol. (in press). [DOI] [PMC free article] [PubMed]
- Karagiannis, J., Bimbo, A., Rajagopalan, S., Liu, J., and Balasubramanian, M. K. (2005). The nuclear kinase Lsk1p positively regulates the septation initiation network and promotes the successful completion of cytokinesis in response to perturbation of the actomyosin ring in Schizosaccharomyces pombe. Mol. Biol. Cell 16, 358-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knop, M., Pereira, G., Geissler, S., Grein, K., and Schiebel, E. (1997). The spindle pole body component Spc97p interacts with the gamma-tubulin of Saccharomyces cerevisiae and functions in microtubule organization and spindle pole body duplication. EMBO J. 16, 1550-1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krapp, A., Schmidt, S., Cano, E., and Simanis, V. (2001). S. pombe cdc11p, together with sid4p, provides an anchor for septation initiation network proteins on the spindle pole body. Curr. Biol. 11, 1559-1568. [DOI] [PubMed] [Google Scholar]
- Le Goff, X., Utzig, S., and Simanis, V. (1999a). Controlling septation in fission yeast: finding the middle, and timing it right. Curr. Genet. 35, 571-584. [DOI] [PubMed] [Google Scholar]
- Le Goff, X., Woollard, A., and Simanis, V. (1999b). Analysis of the cps1 gene provides evidence for a septation checkpoint in Schizosaccharomyces pombe. Mol. Gen. Genet. 262, 163-172. [DOI] [PubMed] [Google Scholar]
- Lieberman, H. B. (1995). Extragenic suppressors of Schizosaccharomyces pombe rad9 mutations uncouple radioresistance and hydroxyurea sensitivity from cell cycle checkpoint control. Genetics 141, 107-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, J., Wang, H., and Balasubramanian, M. K. (2000). A checkpoint that monitors cytokinesis in Schizosaccharomyces pombe. J. Cell Sci. 113, 1223-1230. [DOI] [PubMed] [Google Scholar]
- Liu, J., Wang, H., McCollum, D., and Balasubramanian, M. K. (1999). Drc1p/Cps1p, a 1,3-beta-glucan synthase subunit, is essential for division septum assembly in Schizosaccharomyces pombe. Genetics 153, 1193-1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maundrell, K. (1993). Thiamine-repressible expression vectors pREP and pRIP for fission yeast. Gene 123, 127-130. [DOI] [PubMed] [Google Scholar]
- McCollum, D., and Gould, K. L. (2001). Timing is everything: regulation of mitotic exit and cytokinesis by the MEN and SIN. Trends Cell. Biol. 11, 89-95. [DOI] [PubMed] [Google Scholar]
- McDonald, W. H., Ohi, R., Smelkova, N., Frendewey, D., and Gould, K. L. (1999). Myb-related fission yeast cdc5p is a component of a 40S snRNP-containing complex and is essential for pre-mRNA splicing. Mol. Cell. Biol. 19, 5352-5362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra, M., Karagiannis, J., Trautmann, S., Wang, H., McCollum, D., and Balasubramanian, M. K. (2004). The Clp1p/Flp1p phosphatase ensures completion of cytokinesis in response to minor perturbation of the cell division machinery in Schizosaccharomyces pombe. J. Cell Sci. 117, 3897-3910. [DOI] [PubMed] [Google Scholar]
- Moreno, S., Klar, A., and Nurse, P. (1991). Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 194, 795-823. [DOI] [PubMed] [Google Scholar]
- Morrell, J. L., et al. (2004). Sid4p-Cdc11p assembles the septation initiation network and its regulators at the S. pombe SPB. Curr. Biol. 14, 579-584. [DOI] [PubMed] [Google Scholar]
- Oakley, B. R. (2000). gamma-Tubulin. Curr. Top Dev. Biol. 49, 27-54. [DOI] [PubMed] [Google Scholar]
- Oegema, K., Wiese, C., Martin, O. C., Milligan, R. A., Iwamatsu, A., Mitchison, T. J., and Zheng, Y. (1999). Characterization of two related Drosophila γ-tubulin complexes that differ in their ability to nucleate microtubules. J. Cell Biol. 144, 721-733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paluh, J. L., Nogales, E., Oakley, B. R., McDonald, K., Pidoux, A. L., and Cande, W. Z. (2000). A mutation in gamma-tubulin alters microtubule dynamics and organization and is synthetically lethal with the kinesin-like protein pkl1p. Mol. Biol. Cell 11, 1225-1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardo, M., and Nurse, P. (2003). Equatorial retention of the contractile actin ring by microtubules during cytokinesis. Science 300, 1569-1574. [DOI] [PubMed] [Google Scholar]
- Prentice, H. L. (1992). High efficiency transformation of Schizosaccharomyces pombe by electroporation. Nucleic Acids Res. 20, 621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagolla, M. J., Uzawa, S., and Cande, W. Z. (2003). Individual microtubule dynamics contribute to the function of mitotic and cytoplasmic arrays in fission yeast. J. Cell Sci. 116, 4891-4903. [DOI] [PubMed] [Google Scholar]
- Samejima, I., Lourenco, P. C., Snaith, H. A., and Sawln, K. E. (2005). Fission yeast mto2p regulates microtubule nucleation by the centrosomin-related protein mto1p. Mol. Biol. Cell (in press). [DOI] [PMC free article] [PubMed]
- Sawin, K. E., Lourenco, P. C., and Snaith, H. A. (2004). Microtubule nucleation at non-spindle pole body microtubule-organizing centers requires fission yeast centrosomin-related protein mod20p. Curr. Biol. 14, 763-775. [DOI] [PubMed] [Google Scholar]
- Tasto, J. J., Carnahan, R. H., McDonald, W. H., and Gould, K. L. (2001). Vectors and gene targeting modules for tandem affinity purification in Schizosaccharomyces pombe. Yeast 18, 657-662. [DOI] [PubMed] [Google Scholar]
- Tasto, J. J., Morrell, J. L., and Gould, K. L. (2003). An anillin homologue, Mid2p, acts during fission yeast cytokinesis to organize the septin ring and promote cell separation. J. Cell Biol. 160, 1093-1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomlin, G. C., Morrell, J. L., and Gould, K. L. (2002). The spindle pole body protein cdc11p links sid4p to the fission yeast septation initiation network. Mol. Biol. Cell 13, 1203-1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran, P. T., Marsh, L., Doye, V., Inoue, S., and Chang, F. (2001). A mechanism for nuclear positioning in fission yeast based on microtubule pushing. J. Cell Biol. 153, 397-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trautmann, S., Wolfe, B. A., Jorgensen, P., Tyers, M., Gould, K. L., and McCollum, D. (2001). Fission yeast Clp1p phosphatase regulates G2/M transition and coordination of cytokinesis with cell cycle progression. Curr. Biol. 11, 931-940. [DOI] [PubMed] [Google Scholar]
- Urbani, L., and Stearns, T. (1999). The centrosome. Curr. Biol. 9, R315-R317. [DOI] [PubMed] [Google Scholar]
- Vardy, L., Fujita, A., and Toda, T. (2002). The gamma-tubulin complex protein Alp4 provides a link between the metaphase checkpoint and cytokinesis in fission yeast. Genes Cells 7, 365-373. [DOI] [PubMed] [Google Scholar]
- Vardy, L., and Toda, T. (2000). The fission yeast gamma-tubulin complex is required in G(1) phase and is a component of the spindle assembly checkpoint. EMBO J. 19, 6098-6111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatram, S., Tasto, J. J., Feoktistova, A., Jennings, J. L., Link, A. J., and Gould, K. L. (2004). Identification and characterization of two novel proteins affecting fission yeast gamma-tubulin complex function. Mol. Biol. Cell 15, 2287-2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinh, D. B., Kern, J. W., Hancock, W. O., Howard, J., and Davis, T. N. (2002). Reconstitution and characterization of budding yeast gamma-tubulin complex. Mol. Biol. Cell 13, 1144-1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wachtler, V., Rajagopalan, S., and Balasubramanian, M. K. (2003). Sterol-rich plasma membrane domains in the fission yeast Schizosaccharomyces pombe. J. Cell Sci. 116, 867-874. [DOI] [PubMed] [Google Scholar]
- Wolfe, B. A., and Gould, K. L. (2004). Fission yeast Clp1p phosphatase affects G(2)/M transition and mitotic exit through Cdc25p inactivation. EMBO J. 23, 919-929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, L., Keating, T. J., Wilde, A., Borisy, G. G., and Zheng, Y. (2000). The role of Xgrip210 in gamma-tubulin ring complex assembly and centrosome recruitment. J. Cell Biol. 151, 1525-1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, Y., Jung, M. K., and Oakley, B. R. (1991). Gamma-Tubulin is present in Drosophila melanogaster and Homo sapiens and is associated with the centrosome. Cell 65, 817-823. [DOI] [PubMed] [Google Scholar]
- Zimmerman, S., Tran, P. T., Daga, R. R., Niwa, O., and Chang, F. (2004). Rsp1p, a J domain protein required for disassembly and assembly of microtubule organizing centers during the fission yeast cell cycle. Dev. Cell 6, 497-509. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.