Abstract
Metaphase spindles assemble to a steady state in length by mechanisms that involve microtubule dynamics and motor proteins, but they are incompletely understood. We found that Xenopus extract spindles recapitulate the length of egg meiosis II spindles, by using mechanisms intrinsic to the spindle. To probe these mechanisms, we perturbed microtubule polymerization dynamics and opposed motor proteins and measured effects on spindle morphology and dynamics. Microtubules were stabilized by hexylene glycol and inhibition of the catastrophe factor mitotic centromere-associated kinesin (MCAK) (a kinesin 13, previously called XKCM) and destabilized by depolymerizing drugs. The opposed motors Eg5 and dynein were inhibited separately and together. Our results are consistent with important roles for polymerization dynamics in regulating spindle length, and for opposed motors in regulating the relative stability of bipolar versus monopolar organization. The response to microtubule destabilization suggests that an unidentified tensile element acts in parallel with these conventional factors, generating spindle shortening force.
INTRODUCTION
A mitotic, or meiotic, spindle at metaphase can maintain a steady state in size and shape for prolonged periods, despite rapid turnover of subunits, movement of internal components and dissipation of free energy. In this article, we address the mechanisms that govern metaphase spindle length. Length is important for spindle function, because it influences the distance over which chromosomes are segregated. Furthermore, probing the factors that govern length provide information on assembly and force-producing mechanisms. Metaphase spindle length tends to be relatively constant within a given cell type, but it varies considerably between species and between cell types in an organism. Spindle length typically increases with cell size and genome size, but this relationship can break down in specialized cells. In large eggs, female meiotic spindles are typically small compared with the egg cell. This is appropriate for meiosis biology; the egg meiotic spindle segregates one set of chromosomes a short distance into a polar body, retaining the other set near the cortex. In Xenopus laevis, the egg is ∼1000 μm in diameter, and unfertilized eggs arrest at metaphase of meiosis II, containing a spindle ∼25 μm in length attached at one pole to the cortex at the top of the egg (Cha et al., 1998). Spindles assembled in cytoplasmic extracts made from unfertilized Xenopus eggs recapitulate meiosis II morphology (Sawin and Mitchison, 1991a), but their length has not been systematically studied.
Spindle length per se has received relatively little attention, but many models have been proposed for how forces on chromosomes and poles are generated. These forces are thought to also govern spindle length, with steady-state length arising from a balance of pushing and pulling forces. Force balance models can be divided into those that highlight the role of microtubule polymerization dynamics (Inoue and Sato, 1967; Margolis and Wilson, 1981; Mitchison et al., 1986; Inoue and Salmon, 1995); those that highlight action of ATPase motor proteins (McIntosh et al., 1969; Hoyt et al., 1993; Gaglio et al., 1996; Sharp et al., 2000; Nedelec, 2002; Cytrynbaum et al., 2003); and those that highlight the role of the “spindle matrix”, a hypothetical, nonmicrotubule, tensile element (Pickett-Heaps et al., 1997). Recently, a different type of model was proposed, in which spindle length is set not by a balance of forces, but by a concentration gradient of morphogens diffusing from a source at chromatin to a global sink in the cytoplasm (Karsenti and Vernos, 2001). Most of these models have in common that they seek to explain spindle length with the microtubule system, including dynamics regulators, motors, and cross-linkers, as the sole mechanochemical element. Exceptions are the original polymerization dynamics model that preceded the discovery of tubulin (Inoue and Sato, 1967), and spindle matrix models, that explicitly propose a nonmicrotubule, tensile element (Pickett-Heaps et al., 1997). The most widely discussed models have been those in which spindle length is governed by some combination of polymerization dynamics and opposed motor proteins, and the purpose of this study was to critically evaluate these models in Xenopus extract spindles.
A useful distinction in considering models for spindle length regulation is between mechanisms that act extrinsic to the spindle, versus intrinsic mechanisms. Potential extrinsic mechanisms include limiting amounts of some subunit, and forces generated at the cell cortex. Intrinsic mechanisms include balanced forces within the spindle and a possible morphogen gradient emanating from chromatin. In mammalian tissue culture mitosis, the spindle incorporates ∼50% of the cell's tubulin (Zhai and Borisy, 1994), suggesting component limitation is a significant factor. Pulling forces from the cortex acting on astral microtubules are known to play a significant role in length regulation in several mitotic systems (Sharp et al., 2000). Thus, extrinsic and intrinsic factors probably act in concert to govern the length of typical mitotic spindles. In contrast, extrinsic mechanisms are probably much less important in egg meiosis. The meiotic spindle is small compared with the egg (∼10-5 of the egg volume in Xenopus) and presumably does not deplete a significant fraction of the egg's tubulin. Pulling from the cortex operates mainly on one spindle pole in egg meiotic spindles (Lutz et al., 1988) and is probably a minor factor in governing spindle length. In this study, we formally demonstrate that the length of spindles assembled in Xenopus egg extracts is governed by intrinsic mechanisms, and we investigate these mechanisms by perturbation experiments.
MATERIALS AND METHODS
Xenopus egg extract sand spindles with replicated DNA were made by standard methods (Desai et al. 1999a) and used within 90 min of assembly. Spindle assembly and imaging were performed at 19–20°C. For polarization imaging, we used a Nikon (Melville, NY) TE-300 inverted microscope equipped for differential interference contrast using de Senarmont compensation, a 20× long working distance objective, a heat reflection filter, and a cooled charge-coupled device camera. The Wollaston prisms were removed, and the polarizer set slightly away from extinction. A chamber was made by drilling a 22-mm-round hole in a 1-mm-thick sheet of stainless steel the shape of a microscope slide. A 25-mm-round coverslip was cemented under the hole with valap (33% paraffin wax, 33% beeswax, and 33% lanolin). Three to 10 μl of extract containing spindles was deposited on the coverslip and smeared out into a disk 5–10 mm in width with the pipette tip. The spread extract was immediately covered with 250 μl of mineral oil. Length measurements were made by capturing images of many random fields and measuring all spindles, ignoring multipolar structures. Perturbing reagents were added and mixed immediately before spreading the extract in the chamber. Microscopes for wide-field and confocal fluorescence imaging, and fluorescent probes for tubulin and kinetochores have been described previously (Desai et al. 1999a; Maddox et al., 2003). Optimal concentrations of X-rhodamine tubulin for confocal speckle imaging were determined empirically for each extract, with ∼50 nM typical. Affinity-purified, inhibitory antibody to Xenopus mitotic centromere-associated kinesin (MCAK) was made and characterized as described previously (Walczak et al., 1996). Affinity-purified anti-nuclear mitotic apparatus (NuMA) IgG was made by immunization with the C-terminal peptide (C)TAKSPRASNKLFERKQQRNK coupled to keyhole limpet hemocyanin, and affinity purified using the same peptide coupled to agarose, by using the methods described in Field et al. (1998). Specificity was tested by immunoprecipitation as described in the legend to Supplemental Figure 1. Anti-NuMA pulled down primarily a band of the expected molecular weight, whose identity was confirmed by Western blotting of the immunoprecipitate (Supplemental Figure 1). Western blotting of whole extract was negative for NuMA, presumably because the high protein concentration interfered with transfer to nitrocellulose, a problem we have noticed with other antibodies. For imaging, the IgG was labeled with Alexa488-NHS ester (Molecular Probes, Eugene, OR) according the manufacturer's recommendations. p50 dynamitin was made as described previously (Heald et al., 1997). Immunodepletion with magnetic beads was performed as described previously (Funabiki and Murray, 2000).
The microtubule-depolymerizing drug N-(2-napthyl)-3-trifluoromethylbenzene sulfonamide (105D) was tested for effects on pure tubulin polymerization as described in the legend to Supplemental Figure 2. Its caged derivative had no effect in this assay. Synthesis of 105D and its caged derivative are described in the legend for Supplemental Figure 3. Both were >95% pure by thin layer chromatography (TLC) and liquid chromatography/mass spectrometry (LC/MS) and gave the expected molecular ions. On photolysis in methanol by using a hand-lamp, caged 105D released 105D with ∼100% yield by TLC and LC/MS. For experiments in extracts, caged 105D was dissolved in dimethyl sulfoxide as a 200 mM stock by warming to 60°C and added to extracts to a final concentration of 400 μM. This concentration is approximately the solubility limit, and it was chosen so that only a fraction of the compound had to be photocleaved to cause microtubule depolymerization, thus limiting the UV dose delivered to spindles. The caged drug had no discernible effect on spindle assembly in the absence of UV light. To measure the rate of uncaging by using a microscope in extracts, we collected sequential images with a DAPI filter set. 105D is weakly fluorescent in the UV, whereas its caged derivative is not, so the field gets brighter as uncaging proceeds (Supplemental Figure 4). We estimated a half-time for photorelease in extract of ∼5 s with 360 nm illumination from a 100W Hg bulb through a 4,6-diamidino-2-phenylindole (DAPI) filter cube to a 60×/1.4 numerical aperture PlanApo objective in a Nikon 800e upright microscope. For spindle depolymerization experiments, caged 105D was added to preformed spindles in extracts that also contained X-rhodamine tubulin (∼200 nM) and Alexa488-anti-CenpA (∼1 μg/ml; Maddox et al., 2003). Spindles were located using dim rhodamine illumination and time-lapse imaging initiated with a double label filter cube (wide-field) or no filter cube (confocal). After a few preuncaging frames had been collected, a DAPI filter cube was brought into the epi path, and the field illuminated with 360-nm light for 1–2 s. Then, the cube was changed back and the time-lapse sequence continued. One to 2 s of UV illumination in extracts containing 400 μM caged 105D generated sufficient free105D to depolymerize spindles in the field and had no effect on spindles when the caged drug was not present. The effect of uncaging was remarkably local. A spindle in the microscope field (∼200-μm circle) subject to 360-nm illumination with caged-105D present rapidly disassembled, whereas spindles outside the field were unaffected. Thus, we were able to trigger and follow depolymerization of several spindles in each slide-coverslip preparation. To determine why the effects of photoreleasing 105D are local and persistent, we imaged the drug diffusing away from a UV-illuminated area, by using its intrinsic fluorescence, and by limiting the observation light to minimize further photorelease. We observed that 105D partitions into membranes and moves only very slowly through the extract after photorelease (Supplemental Figure 4), explaining its local effects on minute time scales.
RESULTS
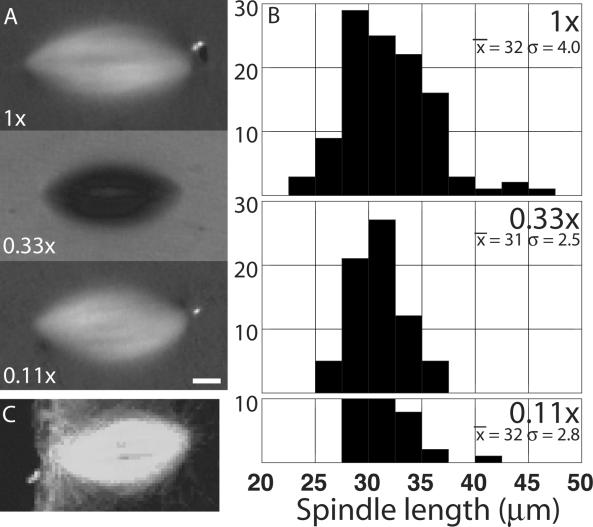
To measure spindle length in a convenient and nonperturbing way, mitotic extract (3–10 μl) containing spindles with replicated chromosomes (“cycled spindles”, Desai et al. 1999a) was spread in a thick (∼100 μm) layer under mineral oil and imaged by polarization microscopy (Figure 1A). Spindles were remarkably stable in this preparation, and retained a constant length for at least 30 min by time-lapse imaging (our unpublished data). The spindles usually rotated and translated slowly, showing they were free of interactions with the substrate that might influence their length. To test whether the amount of any extract component is limiting for spindle length, we assembled spindles in parallel at three different concentrations of added sperm and then measured the distribution of spindle lengths (Figure 1, A and B). The morphology, birefringence, mean length, and length distribution were similar in each case. A confocal fluorescence image of a meiosis II spindle at similar magnification in an unfertilized egg is shown for comparison (Figure 1C). The average length of extract spindles varied slightly from preparation to preparation, in part due to variability in the extent to which spindles fused. Extract spindles are, on average, a little longer than egg meiosis II spindles (∼25 μm; Cha et al., 1998), but overall the extract system does a good job of recapitulating meiosis II morphology. Because spindle length was independent of spindle concentration, and spindles seemed not to interact with the substrate, we conclude that length is regulated by mechanisms intrinsic to the spindle.
Figure 1.
Length of in vitro spindles is independent of spindle density. Cycled spindles were assembled at a final density of 50, 16.7, and 5.5 sperm nuclei/μl and imaged in oil overlay chambers by polarization microscopy 90–120 min after addition of cytostatic factor (CSF) extract. (A) Representative images at each dilution. Bar, 5 μm. (B) Histograms of spindle lengths at each dilution measured from polarization images. The mean and SD (in micrometers) is written on each histogram. Note they are similar at each dilution. (C) Immunofluorescence image of the meiosis II spindle in an unfertilized egg, at a comparable magnification, for comparison. The animal cortex of the egg is to the left. Anti-tubulin staining and laser confocal imaging. Image kindly provided by David Gard (University of Utah) (see Cha et al., 1998).
To test whether protein polymerization dynamics plays an important role in governing spindle length in Xenopus extracts, as they do in other spindles (Inoue and Sato, 1967), we first measured the effect of an agent that nonspecifically promotes protein assembly. We used 2-methypentane-2,4-diol, called “hexylene glycol,” in the mitosis literature (suggested by Robert Palazzo). This solvent promotes aggregation of many proteins and is used for this purpose in crystallography. It also promotes microtubule polymerization and stabilizes spindles and asters (Rebhun et al., 1975; Harris and Clason, 1992). At 3% (vol/vol) and above, hexylene glycol promoted rapid nucleation of microtubule asters throughout the extract. At 2%, it only slowly promoted nucleation, but it had a remarkable effect on preformed spindles, causing them to increase in birefringent retardation (a measure of spindle microtubule density) and to grow progressively in length and volume (Figure 2A and Movie M1). Spindle length increased at a constant rate of ∼1.7 μm/min, at least to the point where length doubled (Figure 2B). To probe the mechanics of elongation in hexylene glycol, we imaged fluorescent tubulin at speckle levels by spinning disk confocal microscopy (Figure 2C and Movie M2). Visual inspection and kymograph analysis (Figure 2D) showed that speckles throughout the spindle moved polewards at a rate similar to the rate of pole separation. We conclude that spindles elongate in hexylene glycol by anti-parallel sliding between the two half-spindles, with little or no microtubule depolymerization. This situation is reminiscent of poleward flux with depolymerization blocked. The antiparallel sliding component of poleward flux can be blocked by adenyl-5′-yl imidodiphosphate (AMPPNP) (Sawin and Mitchison, 1991b) and Eg5 inhibitors, including monastrol (Miyamoto et al., 2004), so we tested the effect of these agents on spindle elongation. Monastrol caused spindle collapse when added alone, but this collapse could be suppressed by also adding p50 dynamitin (discussed below). Hexylene glycol-induced elongation was blocked by both agents (Figure 2B). We conclude that spindle elongation in hexylene glycol occurs by antiparallel sliding between the half spindles, most likely driven by Eg5. Consistent with this view, extract spindles also elongate by antiparallel sliding when pole organization is disrupted (Gaetz and Kapoor, 2004), dependent on Eg5 activity (Shirasu-Hiza et al., 2004). The rate of spindle expansion in hexylene glycol (∼1.7 μm/min) is slower than that that rate of antiparallel sliding during flux in control spindles (∼4 μm/min). Eg5 motors seem to work against an unknown mechanical load to drive flux (Miyamoto et al., 2004), and we suspect that hexylene glycol increases this load.
Figure 2.
Hexylene glycol promotes spindle expansion. Hexylene glycol [2% (vol/vol)] was added to cycled spindles from a 20% stock in water immediately before imaging. (A) Polarization images from a time-lapse sequence. Elapsed time shown in minutes and seconds. Hexylene glycol was added ∼1 min before the first time point. Note the growth in spindle length and width. Time elapsed shown as minutes:seconds. Length of time box, 10 μm. See Movie M1 (B) Spindle length as a function of time after addition of hexylene glycol alone (2%; black circles), hexylene glycol plus AMPPNP (1.5 mM; open squares), or hexylene glycol plus monastrol (400 μM) and p50 dynamitin (0.9 mg/ml; gray diamonds). (C) Spinning disk confocal fluorescence images of a spindle containing speckle level tubulin to which 2% hexylene glycol has been added. The line in the second panel was used to make the kymograph in D. See Movie M2. Bar, 5 μm. (D) Kymograph through the upper pole of the spindle shown in C. Time is vertical, distance along the line horizontal. Note the movement of the pole away from the spindle equator, which is to the right in the panel. Note also that many thin lines, which are speckle trajectories, parallel the movement of the pole (black lines highlight examples). Near the equator, speckles also are moving in the other direction (white line), in parallel with the opposite pole.
We next tested a more specific microtubule stabilizing agent, inhibitory antibody to Xenopus MCAK (a kinesin 13, formerly called XKCM1). This kinesin promotes microtubule catastrophes in an ATP-dependent reaction (Desai et al. 1999b). It is the most potent known catastrophe factor in Xenopus egg extract (Tournebize et al., 2000), and removing or inhibiting it induces massive microtubule polymerization in M-phase extracts (Walczak et al., 1996). Plus ends in extract spindles are thought to undergo bounded dynamic instability, meaning that they do not grow indefinitely, but rather catastrophe frequently enough to have a defined average length (Verde et al., 1992). Titrating a catastrophe inhibitor into extract is predicted to first increase this average length and then cause a transition to the unbounded regime, where plus ends grow indefinitely. We titrated anti-MCAK into extract, adding the inhibitor well before spindle assembly, and fixing at two time points, to ensure we were measuring spindle length at steady state. Increasing concentrations of anti-MCAK up to 10 μg/ml caused a small, dose-dependent increase in spindle length, and a larger, dose-dependent increase in total microtubules per spindle (Figure 3). At 15 μg/ml (Figure 3) and above (Figure 4), microtubules elongated dramatically, forming large asters. When MCAK was inhibited to this extent before spindle assembly, disorganized structures formed whose length could not be defined (Figure 3). We infer that decreasing the catastrophe rate while staying in the bounded regime modestly increased spindle length. Decreasing catastrophe to the point of entering the unbounded regime resulted in disorganization, and not a dramatic increase in length.
Figure 3.
Titration of anti-MCAK antibody effects. Affinity-purified, inhibitory antibody to MCAK (Walczak et al., 1996) was added to spindle assembly reactions containing X-rhodamine tubulin 20 min after bring the extract back into M phase after replication, well before spindle assembly. Samples were squash-fixed at 80 and 120 min after bringing the extract into M phase. (A) Representative spindles fixed at 80 min as a function of final antibody concentration. Note the massive microtubule growth out of the spindle at 15 μg/ml (and higher; our unpublished data), indicative of transition to unbounded dynamic instability. Bar, 10 μm. (B) Quantification of spindle length in this experiment. Open bars are 80- and black 120-min samples. The error bars indicate one SD above and below the mean, and the number in each bar the sample size. (C) Quantification of total microtubules per spindle measured as integrated fluorescence intensity minus local background signal. Labeling as in B.
Figure 4.
Inhibition of MCAK promotes microtubule outgrow and pole curling, but not spindle elongation. Affinity-purified, inhibitory antibody to MCAK was added at 150 μg/ml final to cycled spindles immediately before imaging. (A) Polarization images from a time-lapse sequence. Note the dramatic outgrowth of microtubules from the spindle (arrows in second panel). There is some alteration of spindle shape and pole structure over the sequence, but pole-to-pole distance remains approximately constant. Elapsed times shown in minutes:seconds. Antibody was added ∼1 min before the first time point. Time bar, 18 μm. See Movie M3. (B) Spinning disk confocal fluorescence images of a spindle containing speckle level X-rhodamine tubulin (green) and Alexa488-labeled antibodies to NUMA at 5 μg/ml as a pole marker (red). Note that the upper pole, visualized by anti-NUMA localization (white arrow), is curled over and apparently attached to the body of the spindle. By the last time point, the pole has moved toward the spindle equator, and become partially disorganized. In the tubulin channel, the movements of the microtubules that are associated with this pole movement can be visualized (blue arrows). In the center of the spindle, microtubules slide apart in both directions. In the upper part, most of the flow is upwards. At the top of the spindle, the flow curls over, correlating with movement of the pole back toward the equator. Bar, 5 μm. See Movie M4.
To better understand the effects of unbounding microtubule length, we imaged preformed spindles live after adding saturating amounts of anti-MCAK. Microtubules rapidly extended away from the spindle, converting them into large asters (Figure 4A and Movie M3). During this outgrowth, the spindle itself did not seem to elongate, and in some cases the poles even moved slightly closer together. Although the principle effect of inhibiting MCAK is inhibiting plus end catastrophes, this protein also has the biochemical capability to depolymerize minus ends at poles (Desai et al. 1999b), and other kinesin 13 family members have been implicated in this activity (Gaetz and Kapoor, 2004; Rogers et al., 2004). To probe effects on poles, we imaged a fluorescent spindle pole marker as well as tubulin speckles after inhibiting MCAK (Figure 4B and Movie M4). The pole marker was affinity-purified antibody raised to a carboxy-terminal peptide from Xenopus NuMA, labeled with Alexa488. NuMA accumulates at the poles of Xenopus extract spindles by a dynein–dynactin-dependent mechanism and has often been used as a pole marker (Merdes et al., 1996). Our antibody was specific by immunoprecipitation (Supplemental Figure 1), gave the reported localization for NuMA in live and fixed spindles and had no discernible effect on spindle assembly or dynamics when added to extracts or on the response to inhibiting MCAK as judged by comparing effects in the tubulin channel with and without the probe. Confocal imaging confirmed massive outgrowth of microtubules from the spindle starting a few minutes after adding anti-MCAK. Anti-parallel microtubules sliding in the center of the spindle that is characteristic of poleward flux continued (Movie M4). By tubulin imaging alone, and more informatively by tubulin + NuMA imaging, we observed progressive disorganization of poles. In about half the spindles, this disorganization takes the form of the pole elongating and curling backward toward the equator, seeming to track back along the outer surface of the spindle (Figure 4B and Movie M4). Tubulin speckles follow this pole movement, moving out from the main body of the spindle, and curling around with the moving pole (Figure 4B, blue arrows). Due to the disorganization of pole structure, and lack of a direct assay for depolymerization, we were unable to quantify microtubule depolymerization at poles in this experiment.
To further probe the role of microtubule dynamics in governing spindle length, we rapidly depolymerized microtubules. We added 20 μM nocodazole to an aliquot of extract containing spindles on a microscope slide, mixed them, put on a coverslip, and initiated imaging by using a dry 40× lens to facilitate rapid location of spindles. Spindles shortened and depolymerized completely in 2–3 min with this treatment, and it was necessary to locate them within ∼10 s of drug addition to obtain useful information on early events. Spindle shortening in nocodazole was previously argued to occur by pulling at kinetochores (Cassimeris et al., 1990), so we used a nonperturbing kinetochore probe (Alexa488-anti-CenpA IgG; Maddox et al., 2003) to observe possible action of such forces. As expected from previous work (Inoue and Sato, 1967; Salmon et al., 1984; Cassimeris et al., 1990), the microtubule density dropped rapidly, and the pole-to-pole distance decreased (Figure 5A and Movie M5). Note that the images in Figure 5 and Movie M5 are normalized to peak intensity, optimizing visualization of remaining structures, but giving a misleading impression of microtubule density, which is quantified as total tubulin fluorescence in Figure 5B. We expected to see stretched kinetochores pulling the poles inwards in this experiment, and we were surprised to observe that the distance between sister kinetochores invariably decreased shortly after drug addition, indicating loss of tension (Figure 5A, red dots; Figure 5B, triangles; and Movie M5). All kinetochores visualized experienced this relaxation (>50 kinetochore pairs in 13 spindles in 10 sequences). In some cases, kinetochores seemed to experiencing compression during spindle collapse. This was evident from lateral movement away from the spindle axis, twisting of the kinetochore pair (Figure 5A, 119 s), and apparent curving or buckling of kinetochore microtubules (Figure 5A, 119 s, note microtubule bundle connected to the upper sister of the pair marked with blue lines; also see Movie M5).
Figure 5.
Rapid microtubule depolymerization by using nocodazole. Cycled spindles containing labeled tubulin (green) and a kinetochore marker (anti-CenpA; red) were mixed with nocodazole (20 μM final) and wide-field time-lapse imaging was initiated ∼10 s after mixing by using a 40× dry objective. (A) Imaging of spindle collapse. Elapsed time is shown in seconds relative to the first image; tubulin fluorescence is normalized to peak brightness in each image, to highlight the organization of remaining microtubules. Insets show 3× magnification of marked kinetochores. Note the sister kinetochores are initially well separated. The spindle rapidly looses fluorescence, shortens, and the sister kinetochore move together. Note some evidence of buckling of kinetochore microtubules at 59, 119 s. Bar, 5 μm (main panels), 1.7 μm (insets). See Movie M5. (B) Quantification of the sequence in A, showing distance between the marked kinetochores in A (k-k), pole to pole distance (p-p) and integrated tubulin fluorescence after subtracting local background signal. Note that k-k distance drops faster (in percent terms) than p-p distance.
Further investigation of the forces in collapsing spindles required high-resolution imaging, which was difficult using nocodazole addition because it took too long to find and focus on a spindle. Therefore, we synthesized a photochemically “caged'' microtubule-depolymerizing drug that allowed us to find and image a spindle before and after triggering depolymerization (Figure 6A). We started with 105D, a depolymerizing drug that was found by phenotypic screening of a combinatorial library (Mitchison, 2003). It arrests cells in mitosis in tissue culture cells with a phenotype similar to nocodazole and with an IC50 of ∼3 μM (∼20-fold less potent than nocodazole). It inhibited polymerization of pure tubulin (Supplemental Figure 2) and depolymerized extract spindles with an IC50 of ∼20 μM, again ∼20-fold less potent than nocodazole. 105D has several advantages for making a caged derivative. It is simple to synthesize and modify (Supplemental Figure 3). The caged drug had no detectable effect on microtubules or extract spindles at its solubility limit (∼400 μM). 105D is weakly fluorescent in the DAPI channel, whereas its caged derivative is not. Thus, photorelease and subsequent movement of the drug can be imaged and quantified by fluorescence microscopy (Supplemental Figure 4).
Figure 6.
Microtubule depolymerization using a caged drug. Cycled spindles containing labeled tubulin, kinetochore, and pole markers in some cases, and caged 105D were imaged by time-lapse wide-field (B and C) or confocal (D) fluorescence. At t = 0, 105D was photoreleased by 1- to 2-s illumination with a UV filter set. All times are in seconds relative to photorelease. Bar, 7.5 μm (B), 6 μm (C), 5 μm (D), and 1.25 μm (D, insets). (A) Structure of caged 105D and the photochemical reaction that releases active drug. (B) Example of a spindle before and after photorelease of 105D (wide-field). Tubulin fluorescence is presented with brightness and contrast held constant throughout the sequence to highlight the rapid decrease in microtubule density. This sequence also contained a fluorescent kinetochore probe; see Movie M6. (C) Second widefield example. Tubulin fluorescence is normalized to peak brightness in each image, to highlight the organization of remaining microtubules. Note the decrease in spindle length, and buckling of remaining stable microtubules. See Movie M7. (D) Example of photorelease of 105D (confocal). Probes are for tubulin (green), kinetochore (anti-CenpA; red), and poles (anti-NUMA; red). Tubulin fluorescence is normalized to peak brightness in each image. Insets show marked kinetochore pairs at 4× magnification. The focal plane was changed several times during this sequence, and different kinetochore pairs are shown in each panel. At late time points, most of the remaining stable microtubules connect to kinetochores. Note that the poles (large red dots) move progressively together. Sister kinetochores first move together and then twist and move away from the spindle axis, whereas their attached microtubules either buckle or move outward, suggesting kinetochore fibers experience compression from the collapsing poles. The spindle was optically sectioned twice during this sequence, and the images at 941 and 978 s are two focal planes from one through-focal series. Note the absence of straight microtubule bundles directly connecting the poles at any time point or focal plane.
Time-lapse imaging of spindles before and after photorelease of 105D showed effects broadly similar to nocodazole addition. The intensity of the tubulin signal rapidly decreased after photorelease (Figure 6, B and D); the spindle shortened (Figure 6, B–D, and Movies M6 and M7); and sister kinetochores moved together and twisted (Figure 6D and Movie M7). 105D did not depolymerize spindle microtubules as efficiently as nocodazole, and a subset of microtubule bundles was stable for several minutes after photorelease. Most of these stable microtubules terminated at kinetochores (Figure 6D and Movie M7), implying they are kinetochore microtubules, which are known to be selectively resistant to depolymerizing drugs in other systems (Cassimeris et al., 1990). In nocodazole, kinetochore fibers eventually disappeared (Figure 5A), but after uncaging 105D they did not, perhaps because 105D partially stabilizes kinetochore microtubules. Other tubulin drugs are known to have stabilizing as well as destabilizing effects (Wilson et al., 1999). Both wide-field (Figure 6C) and confocal (Figure 6D) sequences suggested that the spindle poles are pulled (or pushed) together after photorelease, resulting in compression of attached microtubules. These forces caused buckling of selectively stable kinetochores fibers, movement of sister kinetochores toward each other, and twisting or sideways movement of the sister pair. If the poles were pulled together by microtubules, we would expect to see straight bundles of microtubules connecting them. By through-focus imaging of >10 collapsing and fully collapsed spindles with wide-field and confocal microscopy, we were unable to find any straight bundles of microtubules connecting poles or any sister kinetochore pairs still under tension. Figure 6D, 941 and 978 s, shows two images from a through-focus confocal image series where all remaining microtubule bundles were either buckled or pushed out sideways from the spindle axis, and all kinetochore pairs are close together. We tentatively conclude that when overlap microtubules are rapidly removed with a drug, the poles are pulled (or pushed) together by something other than microtubules. Spindle collapse is unlikely to depend on F-actin, because our extracts routinely included cytochalasin D (∼3 μM), and addition of latrunculin B to 30 μM (in addition to cytochalasin D) had no effect on the speed, extent, or morphology of collapse induced by photorelease of 105D (our unpublished data). The photorelease experiment made it possible to collect before and after data on spindle length and microtubule density. For seven representative spindles, the peak rate of shortening (measured pole to pole) averaged 7 μm/min (range 2–13 μm/min). Length plateaued after 350 s on average (n = 6, range 300–450 s), when the spindle was 47% on average of its initial length (range 31–57%). Total tubulin fluorescence plateaued ∼300 s after initiating depolymerization, at an average of 8% of the initial fluorescence (range 5–11%). The half-time between initiating depolymerization and reaching the plateau in total fluorescence was 55 s (n = 7, range = 53–60 s).
We next probed the role of opposed motor proteins in governing spindle length, focusing on Eg5 and dynein. Eg5 is essential for bipolarity in extract spindles (Sawin et al., 1992; Kapoor et al., 2000), where it drives antiparallel sliding associated with poleward flux and spindle elongation (Figure 2; Miyamoto et al., 2004; Shirasu-Hiza et al., 2004). Dynein works together with dynactin and NuMA to organize and focus the poles (Merdes et al., 1996) and seems to be the dominant minus end-directed motor in Xenopus extract spindles on the basis of inhibition experiments (Heald et al., 1997). Using polarization microscopy, we confirmed the effects of inhibiting Eg5 with monastrol, and dynactin with excess p50 dynamitin, (Figure 7, B and C). Unexpectedly, when both these inhibitors were added together, they counteracted each other (Figure 7D). Almost all spindles were now bipolar, and their poles were more organized than with p50 alone. We quantified length and morphology for spindles in the presence of inhibitors, adding them before and after spindle assembly (Table 1). p50 almost completely rescued the effect of monastrol on bipolarity, whether it was added before or after assembly. Monastrol partially rescued the effect of p50 on poles morphology. When both inhibitors were added before spindle assembly, length was almost completely rescued (32 vs. 39 μm in controls; Table 1). This was less true when both inhibitors were added after assembly (23 vs. 37 μm in controls; Table 1), probably reflecting reduced effectiveness of p50 when added after assembly. Although the double-inhibited spindles had relatively normal morphology and length by polarization imaging, they were much more fragile than control spindles. Unlike control spindles, they were easily damaged by squashing between a slide and coverslip or by touching with microneedles, and their average length in replicate experiments was more variable than with controls. We note that the concentrations of p50 we used was probably insufficient to completely block pole organization. Complete inhibition of pole organization tends to increase spindle length (Gaetz and Kapoor, 2004; Shirasu-Hiza et al., 2004), perhaps due to displacement of a kinesin 13 depolymerization factor from the poles (Gaetz and Kapoor, 2004).
Figure 7.
Eg5 and dynein/dynactin play antagonistic roles in spindle assembly. Cycled spindles were assembled in the presence of no drug, the dynactin inhibitor p50 dynamitin (0.9 mg/ml), the Eg5 inhibitor monastrol (200 μM), or both. Drugs were added at the time of CSF addition, and spindles were imaged live in oil overlay chambers by polarization microscopy 90–120 min later. A panel of representative images is shown for each condition. Note the expected appearance of p50 spindles with unfocussed poles and monopolar monastrol spindles. When both drugs were added, bipolarity was completely rescued, and pole focusing was partially rescued. Bar, 10 μm. See Table 1 for quantitation.
Table 1.
Affect of motor protein perturbation on spindle morphology and length scored by polarization microscopy
| % Bipolar spindles | Avg pole focus index (bipolar spindles) | Avg pole to pole length (μm) | SD of pole to pole length (μm) | No. scored | |
|---|---|---|---|---|---|
| Agents added before spindle assembly | |||||
| None | 100 | 1.8 | 39 | 4 | 115 |
| p50 | 98 | 0.1 | 37 | 9 | 59 |
| Monastrol | 2 | * | 1 | 4 | 100 |
| p50 + monastrol | 100 | 0.8 | 32 | 5 | 104 |
| Agents added after spindle assembly | |||||
| None | 100 | 1.9 | 37 | 7 | 107 |
| p50 | 100 | 1.0 | 43 | 4 | 65 |
| Monastrol | 6 | * | 1 | 3 | 81 |
| p50 + monastrol | 79 | 1.3 | 23 | 2 | 87 |
Spindle assembly, drug concentrations, and polarization imaging are as per Figure 7. Motor-perturbing agents were added either before spindle assembly, at the time of CSF add-back, with images taken 90–120 min later, or 60 min after CSF add-back, when steady-state spindles were already assembled, with images taken 30–60 min later. Spindles were scored as monopolar if they appeared as a single aster at 20×; otherwise, they were bipolar. Multipolar and aggregated structures represented <20% of the total in all cases and were not counted. Any bipolar spindles with poles separated by less than ∼3 μm would be scored as monopolar. To quantify pole focusing, each spindle was assigned a score of 2 if both poles were well focused, 1 if both poles were partially focused or one pole was focused and the other unfocussed, and 0 if neither pole was focused. Pole focusing was scored only for bipoles.
Too few bipoles present to reliably calculate the index. Pole to pole length was averaged for all structures and counted as zero for monpoles
Eg5 is still present in spindles after monastrol treatment (Kapoor et al., 2001) and might influence the response to dynein inhibition even when inhibited. We therefore tested whether p50 also could rescue the effect of Eg5 depletion. Depletion of Eg5 to <5% of normal resulted in assembly of almost entirely monopolar spindles as reported previously, and addition of p50 before assembly rescued this effect (Figure 8). Eg5 depleted p50 spindles were mostly bipolar, their poles were more focused than in p50 alone, and they were approximately the right length. We conclude that although inhibition or removal of Eg5 caused spindles to collapse to monopoles, coinhibition of dynein reversed that effect and allowed assembly of spindles that are physically fragile but nevertheless able to achieve an approximately normal steady-state length. Rescue of bipolarity in Eg5 inhibited spindles by p50, discovered here, was a key technical advance for probing the role of Eg5 in flux (Miyamoto et al., 2004).
Figure 8.
Antagonizing dynein/dynactin rescues the effects of Eg5 depeletion. Extracts were depleted of Eg5 by using affinity-purified antibody and magnetic beads. Three rounds of depletion were used to remove all Eg5 that could be detected on Western blots (>95%). (A) Western blot analysis of Eg5 and mock-depleted extracts. Eg5 is removed to below the detection limit in the depleted extract. (B) Quantitation of spindle morphology. Eg5 and mock-depleted extracts were used to assemble cycled spindles in the presence or absence of p50 dynamitin (0.9 mg/ml). In Eg5 depletion alone, the majority of spindles were monopolar asters. Addition of p50 promoted assembly of mostly bipolar spindles. (C) Typical spindle assembled in mock depleted extract. (D) Typical spindle assembled in mock-depleted extract + p50. Note bipolar organization with splayed poles. (E) Typical spindle-assembled in Eg5-depleted extract. Note monopolar organization. (F) Typical spindle assembled in Eg5 depleted extract + p50. Note rescue of both bipolarity and pole morphology. Bar, 5 μm.
DISCUSSION
In this article, we begin a systematic experimental attack on the mechanisms that govern spindle length in the Xenopus extract system. Extract spindles, and by implication egg meiosis II spindles, achieve a steady state in length and mass by purely intrinsic mechanisms (Figure 1). We performed perturbation experiments to test standard models for length regulation based on polymerization dynamics and opposed motor proteins, finding they can account for some, but not all, of the results. To account for the response to rapid microtubule depolymerization, we propose adding a non-microtubule tensile element. Because egg meiotic spindles are small relative to the cell than contains them, and their assembly is largely chromatin driven rather than microtubule organizing center driven (Karsenti and Vernos, 2001), they may use length-governing mechanisms that are different from somatic mitotic spindles.
Polymerization dynamics models predict that increasing microtubule length by increasing polymerization or decreasing depolymerization should cause spindles to elongate. They can account for the response of extract spindles to hexylene glycol (Figure 2) and also for the slight increase in spindle length observed when the catastrophe factor MCAK is partially inhibited, but dynamic instability is still bounded (Figure 3). They fail to account for the response of spindles to stronger MCAK inhibition, when microtubules go into unbounded growth and plus ends leave the spindle, but the spindle poles do not separate further, and rather curl back toward the equator (Figure 4). This curling phenomenon might be due to inhibition of minus end depolymerization at poles by anti-MCAK or simply to disorganization of poles by misdirected motor activity (discussed below). We currently lack an assay for measuring depolymerization at poles that is required to distinguish these possibilities. Kymographs of tubulin speckles do not provide a reliable assay for depolymerization at poles, because converting sliding rates into depolymerization rates requires knowing whether minus ends are static or moving, which has not been measured in extract spindles. Previous interpretation of kymographs assumed static minus ends depolymerizing at poles during metaphase (Sawin and Mitchison, 1991; Rogers et al., 2004), and Kinesin 13 family members (previously called KinI kinesins) were implicated in depolymerization (Gaetz and Kapoor, 2004; Rogers et al., 2004). However, an alternative model can be proposed, in which stable minus ends are distributed throughout the spindle. These ends move poleward at the flux rate without depolymerizing and loose stability when they reach the poles, leading to loss of the microtubule from the plus end. To distinguish these models, we need to localize minus ends in extract spindles and to measure their dynamic behavior. Perhaps the largest discrepancy from standard polymerization dynamics models was the response to microtubule-depolymerizing drugs. In previous work, drug- or pressure-induced spindle shortening was interpreted as a consequence of pulling forces at kinetochores generated by microtubule depolymerization (Cassimeris et al., 1990; Inoue and Salmon, 1995). Instead, we found that depolymerizing drugs induced a switch from tension at kinetochores in unperturbed spindles (Maddox et al., 2003) to compression, arguing that kinetochores were not pulling the poles together. In fact, our imaging suggested that the poles are pulled together by something other than microtubules, as discussed below.
Opposed motor models predict that when the zone of microtubule overlap in the spindle is increased by microtubule polymerization, more plus end-directed motors are recruited, increasing the sliding forces between antiparallel microtubules, and elongating the spindle (Sharp et al., 2000; Cytrynbaum et al., 2003). Such models predict spindle elongation in hexylene glycol, its sensitivity to AMPPNP and Eg5 inhibitors, and elongation with partial inhibition of MCAK. A motor model might also account for the effect of complete MCAK inhibition on pole morphology (Figure 4B). Minus end-directed motor complexes at the pole faced with large numbers of invading microtubules of the wrong polarity may track toward the minus ends of the invaders, causing curling of the poles. We tried to test this by inhibiting dynein and MCAK at the same time, but the results were ambiguous due to disorganization of poles. Opposed motor models predict opposite effects of inhibiting Eg5 and dynein/dynactin, and compensating effects when both are inhibited (Hoyt et al., 1993; Gaglio et al., 1996; Sharp et al., 2000). In motor inhibition experiments, we tended to see either relatively normal length bipoles, or monopoles, rather than intermediate length bipoles (Table 1, inhibitors added before assembly). This suggests that opposed motor activity controls the relative stability of monopolar versus bipolar organization as a switch-like transition, rather than controlling spindle length as a continuous variable. More work is required to address this point experimentally. In a systematic investigation of different classes of opposed motor models, Nedelec (2002) found several stable bipolar solutions. In all cases, the relative stability of bipolar versus monopolar organization depended on motor activities, but in most cases motor activities did not control pole-to-pole distance, which was instead governed by microtubule polymerization dynamics. One of the functions of Eg5 in bipolar spindles is to drive the sliding components of poleward flux. At the monastrol concentration used in Figure 7 and Table 1 (200 μM), rate of antiparallel sliding at metaphase is reduced to ∼10% of control values (near the detection limit; Miyamoto et al., 2004), yet average spindle length in spindles treated with p50 plus monastrol is reduced by <2-fold. Insensitivity of steady-state length to the rate of antiparallel sliding is surprising, because sliding should tend to increase length, and spindles indeed elongate when microtubule destabilization at poles is inhibited (Gaetz and Kapoor, 2004; Rogers et al., 2004; Shirasu-Hiza et al., 2004). To account for these observations, either minus end depolymerization rate must decrease in concert with sliding rate after Eg5 inhibition/removal, or spindle length must be governed by a mechanism that is insensitive to sliding rate. New methods for probing minus end dynamics are required to distinguish these hypotheses.
The most surprising observation in our study was the response of spindles to microtubule-depolymerizing drugs, which indicated that poles can be pulled (or pushed) together by something other than microtubules. It is possible that our through-focus imaging missed a small number of straight microtubules running pole to pole that pulled poles together in response to 105D, but we consider this unlikely because kinetochore bundles remained buckled for several minutes after photorelease, implying the pulling factor does not depolymerize. We also consider it unlikely that 105D causes microtubules to curve on their own, because it did not have this action on pure microtubules, and the response to nocodazole also showed evidence for kinetochore compression and microtubule buckling. We hypothesize that an unidentified tensile element pulls the poles together and that this element also opposes elongation in unperturbed spindles. We consider two possible candidates for this element: external membranes and an internal matrix. Extract spindles are surrounded by a sheath of membranous organelles, including mitochondria and endoplasmic reticulum, that seem physically connected in thin section electron microscopy images (Coughlin and Mitchison, unpublished data). Membranes may be important for spindle assembly, because it failed in high-speed supernatants of Xenopus extracts, unless they were supplemented with purified membranes (Shirasu-Hiza and Mitchison, unpublished data). Membranes also surround meiotic spindles in Drosophila oocytes, where their importance for spindle assembly was revealed by genetics (Kramer and Hawley, 2003). Perhaps membranes serve as a tensile element encapsulating the spindle, tending to oppose spindle expansion, and driving collapse when microtubules are depolymerized. Alternatively, spindles may contain some internal polymer or gel that exerts tension between the poles, as proposed in the spindle matrix hypothesis (reviewed in Pickett-Heaps et al., 1997). Detergent-treated, isolated sea urchin embryo spindles contracted when microtubules were removed using calcium (Salmon and Segall, 1980), an observation more consistent an internal matrix than tension from membranes. A molecular candidate for an internal matrix is poly(ADP-ribose), a nonprotein macro-molecule that is required for bipolar organization of Xenopus extract spindles, and that seems to turn over much less rapidly than microtubules (Chang et al., 2004).
In Figure 9, we combine polymerization dynamics, motors, and a hypothetical tensile element to try and account for all our data. The model is more explicit in molecular terms than the data justify, but we hope it provokes discussion and attempts at experimental falsification. We envisage the matrix as a cross-linked gel that attaches to poles and plus end-directed motors (Kapoor and Mitchison, 2001), and thus becomes stretched, storing elastic energy and pulling the poles inward at steady state. Hexylene glycol, a nonspecific protein-aggregating reagent, promotes recruitment of more matrix in addition to stabilizing microtubules, resulting in balanced growth and increased spindle length while retaining approximately normal morphology. MCAK inhibition leads to increased tubulin polymerization without a parallel increase in matrix assembly and to unbounded growth of plus ends through the poles and out of the spindle. Although plus end-directed motors try to push the elongated half spindles apart, this is opposed by matrix stretched between the poles and by curling of poles back toward the equator driven by dynein-containing complexes moving on microtubules of the wrong polarity that invaded the poles (orange arrows). Drug-induced depolymerization causes microtubules to disassemble faster than the matrix. Tensile forces focus onto remaining kinetochore microtubules, causing spindle collapse with buckling of kinetochore fibers. Dynactin inhibition by p50 leads to splaying out of poles and detachment of matrix. Partial destruction of poles by p50 (this study) did not increase spindle length, perhaps because some matrix remains attached. Complete destruction of poles cause spindles elongation (Gaetz and Kapoor, 2004; Shirasu-Hiza et al., 2004). Eg5 inhibition promotes movement of the poles together by a combination of matrix contraction and dynein pulling. Inhibition of both dynactin and Eg5 results in bipolar spindle that are physically fragile and lack poleward flux (Miyamoto et al., 2004) but are relatively normal in length (Table 1). We propose these lack matrix as well as the opposed motor systems and that they regulate length by dynamic instability alone. Figure 9 does not address other potentially important processes in spindle length regulation, including signals diffusing from chromatin and poleward flux, and new experiments are required to integrate these processes into a complete model. The spindle matrix hypothesis has long been controversial, but the experiments we report should help in the design of future experiments to test molecular candidates.
Figure 9.
Interpretation of results. See text for details.
Acknowledgments
We thank other members of the Marine Biological Laboratory Cell Division Group and our winter laboratories for comments, and David Gard for information on the egg meiosis II spindle. This work was funded by National Institutes of Health Grants GM-39565 (to T.J.M.), GM-24364 and GM-606780 (to E.D.S.), and GM-65933 (to T.M.K.), and by Marine Biological Laboratory fellowships from Universal Imaging and Nikon.
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E05-02-0174) on March 23, 2005.
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
References
- Cassimeris, L., Rieder, C. L., Rupp, G., and Salmon, E. D. (1990). Stability of microtubule attachment to metaphase kinetochores in PtK1 cells. J. Cell Sci. 96, 9-15. [DOI] [PubMed] [Google Scholar]
- Cha, B. J., Error, B., and Gard, D. L. (1998). XMAP230 is required for the assembly and organization of acetylated microtubules and spindles in Xenopus oocytes and eggs. J. Cell Sci. 111, 2315-2327. [DOI] [PubMed] [Google Scholar]
- Chang, P., Jacobsen, M. K., and Mitchison, T. J. (2004). Poly(ADP-ribose) is required for spindle assembly and structure. Nature 432, 645-649. [DOI] [PubMed] [Google Scholar]
- Cytrynbaum, E. N., Scholey, J. M., and Mogilner, A. (2003). A force balance model of early spindle pole separation in Drosophila embryos. Biophys. J. 84, 757-769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai, A., Murray, A., Mitchison, T. J., and Walczak, C. E. (1999a). The use of Xenopus egg extracts to study mitotic spindle assembly and function in vitro. Methods Cell Biol. 61, 385-412. [DOI] [PubMed] [Google Scholar]
- Desai, A., Verma, S., Mitchison, T. J., and Walczak, C. E. (1999b). Kin I kinesins are microtubule-destabilizing enzymes. Cell 96, 69-78. [DOI] [PubMed] [Google Scholar]
- Field, C. M., Oegema, K., Zheng, Y., Mitchison, T. J., and Walczak, C. E. (1998). Purification of cytoskeletal proteins using peptide antibodies. Methods Enzymol. 298, 525-541. [DOI] [PubMed] [Google Scholar]
- Funabiki, H., and Murray, A. W. (2000). The Xenopus chromokinesin Xkid is essential for metaphase chromosome alignment and must be degraded to allow anaphase chromosome movement. Cell 102, 411-424. [DOI] [PubMed] [Google Scholar]
- Gaetz, J., and Kapoor, T. M. (2004). Dynein/dynactin regulate metaphase spindle length by targeting depolymerizing activities to spindle poles. J. Cell Biol. 166, 465-471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaglio, T., Saredi, A., Bingham, J. B., Hasbani, M. J., Gill, S. R., Schroer, T. A., and Compton, D. A. (1996). Opposing motor activities are required for the organization of the mammalian mitotic spindle pole. J. Cell Biol. 135, 399-414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris, P. J., and Clason, E. L. (1992). Conditions for assembly of tubulin-based structures in unfertilized sea urchin eggs. Spirals, monasters and cytasters. J. Cell Sci. 102, 557-567. [DOI] [PubMed] [Google Scholar]
- Heald, R., Tournebize, R., Habermann, A., Karsenti, E., and Hyman, A. (1997). Spindle assembly in Xenopus egg extracts: respective roles of centrosomes and microtubule self-organization. J. Cell Biol. 138, 615-628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyt, M. A., He, L., Totis, L., and Saunders, W. S. (1993). Loss of function of Saccharomyces cerevisiae kinesin-related CIN8 and KIP1 is suppressed by KAR3 motor domain mutations. Genetics 135, 35-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue, S., and Sato, H. (1967). Cell motility by labile association of molecules. The nature of mitotic spindle fibers and their role in chromosome movement. J. Gen. Physiol. Suppl. 50, 259-292. [PMC free article] [PubMed] [Google Scholar]
- Inoue, S., and Salmon, E. D. (1995). Force generation by microtubule assembly/disassembly in mitosis and related movements. Mol. Biol. Cell 6, 1619-1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor, T. M., Mayer, T. U., Coughlin, M. L., and Mitchison, T. J. (2000). Probing spindle assembly mechanisms with monastrol, a small molecule inhibitor of the mitotic kinesin, Eg5. J. Cell Biol. 150, 975-988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor, T. M., and Mitchison, T. J. (2001). Eg5 is static in bipolar spindles relative to tubulin: evidence for a static spindle matrix. J. Cell Biol. 154, 1125-1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karsenti, E., and Vernos, I. (2001). The mitotic spindle: a self-made machine. Science 294, 543-547. [DOI] [PubMed] [Google Scholar]
- Kramer, J., and Hawley, R. S. (2003). The spindle-associated transmembrane protein Axs identifies a membranous structure ensheathing the meiotic spindle. Nat. Cell Biol. 5, 261-263. [DOI] [PubMed] [Google Scholar]
- Lutz, D. A., Hamaguchi, Y., and Inoue, S. (1988). Micromanipulation studies of the asymmetric positioning of the maturation spindle in Chaetopterus sp. oocytes: I. Anchorage of the spindle to the cortex and migration of a displaced spindle. Cell Motil. Cytoskeleton 11, 83-96. [DOI] [PubMed] [Google Scholar]
- Maddox, P., Straight, A., Coughlin, P., Mitchison, T. J., and Salmon, E. D. (2003). Direct observation of microtubule dynamics at kinetochores in Xenopus extract spindles: implications for spindle mechanics. J. Cell Biol. 162, 377-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto, D. T., Perlman, Z. E., Burbank, K. S., Groen, A. C., and Mitchison, T. J. (2004). The kinesin Eg5 drives poleward microtubule flux in Xenopus extract spindles. J. Cell Biol. 167, 813-818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolis, R. L., and Wilson, L. (1981). Microtubule treadmills–possible molecular machinery. Nature 293, 705-711. [DOI] [PubMed] [Google Scholar]
- McIntosh, J. R., Hepler, R. K., and vanWie, D. G. (1969). Model for mitosis. Nature 224, 659-663. [Google Scholar]
- Merdes, A., Ramyar, K., Vechio, J. D., and Cleveland, D. W. (1996). A complex of NuMA and cytoplasmic dynein is essential for mitotic spindle assembly. Cell 87, 447-458. [DOI] [PubMed] [Google Scholar]
- Mitchison, T. J. (2004). Probing cell division with “chemical genetics.” In: The Harvey Lectures, Vol. 98, New York: Wiley–Liss, 19-41. [PubMed] [Google Scholar]
- Mitchison, T., Evans, L., Schulze, E., and Kirschner, M. (1986). Sites of microtubule assembly and disassembly in the mitotic spindle. Cell 45, 515-527. [DOI] [PubMed] [Google Scholar]
- Nedelec, F. (2002). Computer simulations reveal motor properties generating stable antiparallel microtubule interactions. J. Cell Biol. 158, 1005-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickett-Heaps, J. D., Forer, A., and Spurck, T. (1997). Traction fibre: toward a “tensegral” model of the spindle. Cell Motil. Cytoskeleton 37, 1-6. [DOI] [PubMed] [Google Scholar]
- Rebhun, L. I., Jemiolo, D., Ivy, N., Mellon, M., and Nath, J. (1975). Regulation of the In Vivo mitotic apparatus by glycols and metabolic inhibitors. Ann. N.Y. Acad. Sci. 253, 362-377. [DOI] [PubMed] [Google Scholar]
- Rogers, G. C., Rogers, S. L., Schwimmer, T. A., Ems-McClung, S. C., Walczak, C. E., Vale, R. D., Scholey, J. M., and Sharp, D. J. (2004). Two mitotic kinesins cooperate to drive sister chromatid separation during anaphase. Nature 427, 364-370. [DOI] [PubMed] [Google Scholar]
- Salmon, E. D., and Segall, R. R. (1980). Calcium-labile mitotic spindles isolated from sea urchin eggs (Lytechinus variegatus). J. Cell Biol. 86, 355-365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmon, E. D., McKeel, M., and Hays, T. (1984). Rapid rate of tubulin dissociation from microtubules in the mitotic spindle in vivo measured by blocking polymerization with colchicine. J. Cell Biol. 99, 1066-1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawin, K. E., LeGuellec, K., Philippe, M., and Mitchison, T. J. (1992). Mitotic spindle organization by a plus-end-directed microtubule motor. Nature 359, 540-543. [DOI] [PubMed] [Google Scholar]
- Sawin, K. E., and Mitchison, T. J. (1991a). Mitotic spindle assembly by two different pathways in vitro. J. Cell Biol. 112, 925-940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawin, K. E., and Mitchison, T. J. (1991b). Poleward microtubule flux mitotic spindles assembled in vitro. J. Cell Biol. 112, 941-954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp, D. J., Rogers, G. C., and Scholey, J. M. (2000). Microtubule motors in mitosis. Nature 407, 41-47. [DOI] [PubMed] [Google Scholar]
- Shirasu-Hiza, M., Perlman, Z. E., Wittmann, T., Karsenti, E., and Mitchison, T. J. (2004). Eg5 causes elongation of meiotic spindles when flux-associated microtubule depolymerization is blocked. Curr. Biol. 14, 1941-1945. [DOI] [PubMed] [Google Scholar]
- Tournebize, R., Popov, A., Kinoshita, K., Ashford, A. J., Rybina, S., Pozniakovsky, A., Mayer, T. U., Walczak, C. E., Karsenti, E., and Hyman, A. A. (2000). Control of microtubule dynamics by the antagonistic activities of XMAP215 and XKCM1 in Xenopus egg extracts. Nat. Cell Biol. 2, 13-19. [DOI] [PubMed] [Google Scholar]
- Verde, F., Dogterom, M., Stelzer, E., Karsenti, E., and Leibler, S. (1992). Control of microtubule dynamics and length by cyclin A- and cyclin B-dependent kinases in Xenopus egg extracts. J. Cell Biol. 118, 1097-1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walczak, C. E., Mitchison, T. J., and Desai, A. (1996). XKCM 1, a Xenopus kinesin-related protein that regulates microtubule dynamics during mitotic spindle assembly. Cell 84, 37-47. [DOI] [PubMed] [Google Scholar]
- Wilson, L., Panda, D., and Jordan, M. A. (1999). Modulation of microtubule dynamics by drugs: a paradigm for the actions of cellular regulators. Cell Struct. Funct. 24, 329-335. [DOI] [PubMed] [Google Scholar]
- Zhai, Y., and Borisy, G. G. (1994). Quantitative determination of the proportion of microtubule polymer present during the mitosis-interphase transition. J. Cell Sci. 107, 881-890. [DOI] [PubMed] [Google Scholar]