Abstract
Bread wheat (Triticum aestivum) is a globally dominant crop and major source of calories and proteins for the human diet. Compared with its wild ancestors, modern bread wheat shows lower genetic diversity, caused by polyploidisation, domestication and breeding bottlenecks1,2. Wild wheat relatives represent genetic reservoirs, and harbour diversity and beneficial alleles that have not been incorporated into bread wheat. Here we establish and analyse extensive genome resources for Tausch’s goatgrass (Aegilops tauschii), the donor of the bread wheat D genome. Our analysis of 46 Ae. tauschii genomes enabled us to clone a disease resistance gene and perform haplotype analysis across a complex disease resistance locus, allowing us to discern alleles from paralogous gene copies. We also reveal the complex genetic composition and history of the bread wheat D genome, which involves contributions from genetically and geographically discrete Ae. tauschii subpopulations. Together, our results reveal the complex history of the bread wheat D genome and demonstrate the potential of wild relatives in crop improvement.
Subject terms: Plant genetics, Agricultural genetics, Genome evolution, Population genetics, Evolutionary genetics
Analysis of 46 newly sequenced or re-sequenced Tausch’s goatgrass (Aegilops tauschii) accessions establishes the origin of the bread wheat (Triticum aestivum) D genome from genetically and geographically discrete Ae. tauschii subpopulations.
Main
Bread wheat (T. aestivum) is one of the most widely cultivated and most successful crop species worldwide, and has a pivotal role in the global food system. Modern bread wheat shows a remarkably wide geographical distribution and adaptability to various climatic conditions1. Current yield gains, however, might be insufficient to meet future bread wheat demands3, which calls for concerted efforts to diversify and intensify wheat breeding to further raise yields. Bread wheat is an allohexaploid species (2n = 6x = 42, AABBDD genome) whose evolution involved the hybridization of three wild grass species. An initial hybridization between the A genome donor Triticum urartu (2n = 2x = 14) and an unknown B genome donor related to the goatgrass Aegilops speltoides gave rise to tetraploid wild emmer wheat (Triticum turgidum subsp. dicoccoides, 2n = 4x = 28, AABB genome) 0.5–0.8 million years ago4. The second hybridization event happened between a domesticated tetraploid wheat and the D genome progenitor Tausch’s goatgrass (Ae. tauschii; 2n = 2x = 14, DD genome). This hybridization that gave rise to bread wheat most probably occurred along the southern shores of the Caspian Sea 8,000–11,000 years ago5,6. Polyploidization and domestication events such as the origin of bread wheat represent extreme genetic bottlenecks1,2,7,8. In the case of bread wheat, recurrent hybridizations with wild wheat relatives and other domesticated wheat species have significantly increased genetic diversity following domestication2,9–13. The underlying gene flow contributed to the adaptability of bread wheat to diverse climatic conditions outside the Fertile Crescent, the geographical region where wheat was domesticated. Compared with the A and B genomes, however, D genome diversity in bread wheat remains low because the above gene flow has predominantly involved tetraploid species with an AB genome2,6,10,14.
Here we establish a comprehensive set of genomic resources for the bread wheat D genome progenitor Ae. tauschii, including whole-genome sequencing (WGS) data of a large Ae. tauschii diversity panel and chromosome-scale assemblies representing the three Ae. tauschii lineages. The genomic resources proved useful for haplotype and gene discovery and enabled us to unravel the composition and evolution of the bread wheat D genome.
Genomic resources for Ae. tauschii
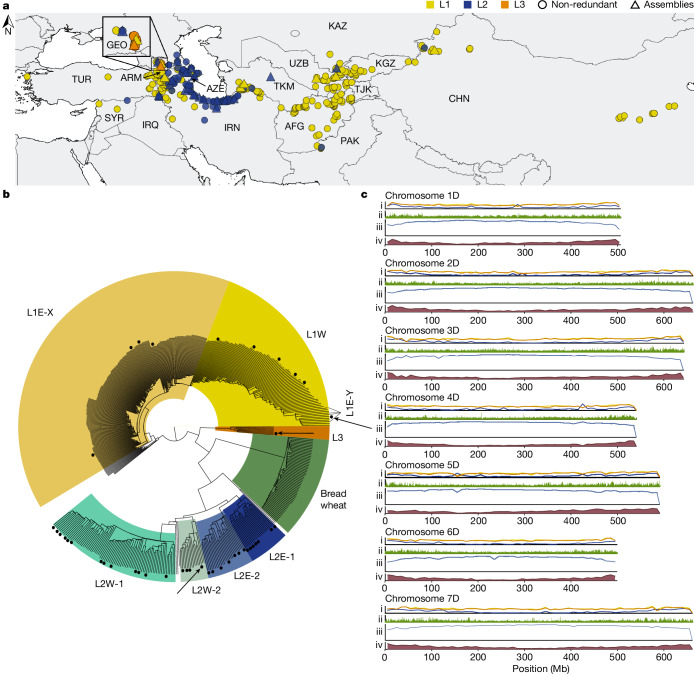
To comprehensively assess genetic diversity in Ae. tauschii, we first compiled a presence–absence k-mer matrix from a diversity panel comprising 920 sequenced Ae. tauschii accessions (Supplementary Table 1 and Supplementary Note 1). We optimized the k-mer matrix workflow for large diversity panels (Supplementary Note 2) using WGS data from this and previous studies14,15 (Supplementary Table 1). The k-mer analysis revealed 493 non-redundant Ae. tauschii accessions, whereas the remaining accessions shared at least 96% of their k-mers with a given non-redundant accession (Supplementary Table 2). The non-redundant diversity panel spanned the geographical range of Ae. tauschii from northwestern Turkey to eastern China (Fig. 1a) and defined a phylogeny demarcated by the 3 basal lineages, with 335 accessions for lineage 1 (L1), 150 accessions for lineage 2 (L2) and 8 accessions for lineage 3 (L3) (Fig. 1b).
Fig. 1. The Ae. tauschii diversity panel and genomes.
a, Geographical distribution of the 493 non-redundant Ae. tauschii accessions in the diversity panel. Accessions selected to generate high-quality assemblies are indicated by triangles coloured according to their respective lineage. AFG, Afghanistan; ARM, Armenia; AZE, Azerbaijan; CHN, China; GEO, Georgia; IRQ, Iraq; IRN, Islamic Republic of Iran; KAZ, Kazakhstan; KGZ, Kyrgyzstan; PAK, Pakistan; SYR, Syrian Arab Republic; TJK, Tajikistan; TKM, Turkmenistan; TUR, Türkiye; UZB, Uzbekistan. b, SNP-based phylogeny of the non-redundant Ae. tauschii accessions showing the subpopulations within the three lineages as labelled on the tree. Accessions sequenced with PacBio HiFi are indicated by black dots next to the tree branches. The three reference accessions TA10171 (L1), TA1675 (L2) and TA2576 (L3) are indicated by black arrows. The D subgenome from 59 wheat landraces is shown in relation to Ae. tauschii. c, Linear chromosome representation showing structural variants, nucleotide diversity and annotation features across the Ae. tauschii panel and genomes relative to the TA1675 L2 reference assembly. The tracks show (i) mean structural variant density in 10 Mb windows for L1 (yellow), L2 (blue) and L3 (orange) accessions of the 46 high-quality assemblies (range 0–800 structural variants), (ii) nucleotide diversity in 10-kb windows across the diversity panel of 493 non-redundant Ae. tauschii accessions (π = 0–0.0045), (iii) repeat density in TA1675 in 10-Mb windows (range 1–10 million repeat-masked nucleotides), and (iv) gene density in TA1675 in 10-Mb windows (range 0–350 high-confidence genes).
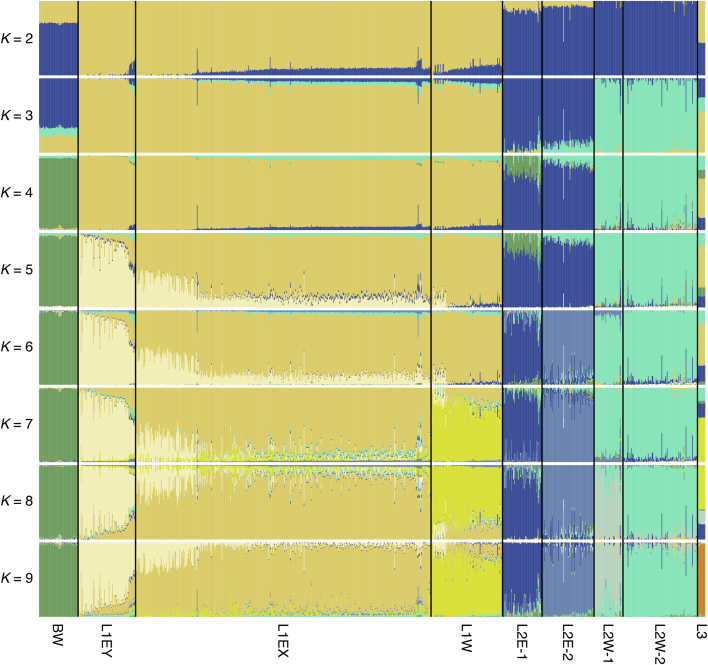
We performed single nucleotide polymorphism (SNP)-based phylogenetic (Fig. 1b and Extended Data Fig. 1a) and ancestry analyses (Extended Data Fig. 2) on the diversity panel, which defined four geographically distinct subpopulations for Ae. tauschii L2, referred to as L2E-1 (southwestern Caspian Sea), L2E-2 (southeastern Caspian Sea), L2W-1 (Caucasus) and L2W-2 (Turkmenistan and northern Iran), in accordance with the literature5,15. Group L2E from the southern Caspian Sea (representing subpopulations L2E-1 and L2E-2 here) was previously identified as the main contributor of the bread wheat D genome5. Of the 150 non-redundant Ae. tauschii L2 accessions, we could assign 133 to one of the four L2 subpopulations on the basis of an ancestry threshold of greater than or equal to 70% (Supplementary Table 3). The remaining 17 L2 accessions were considered admixed.
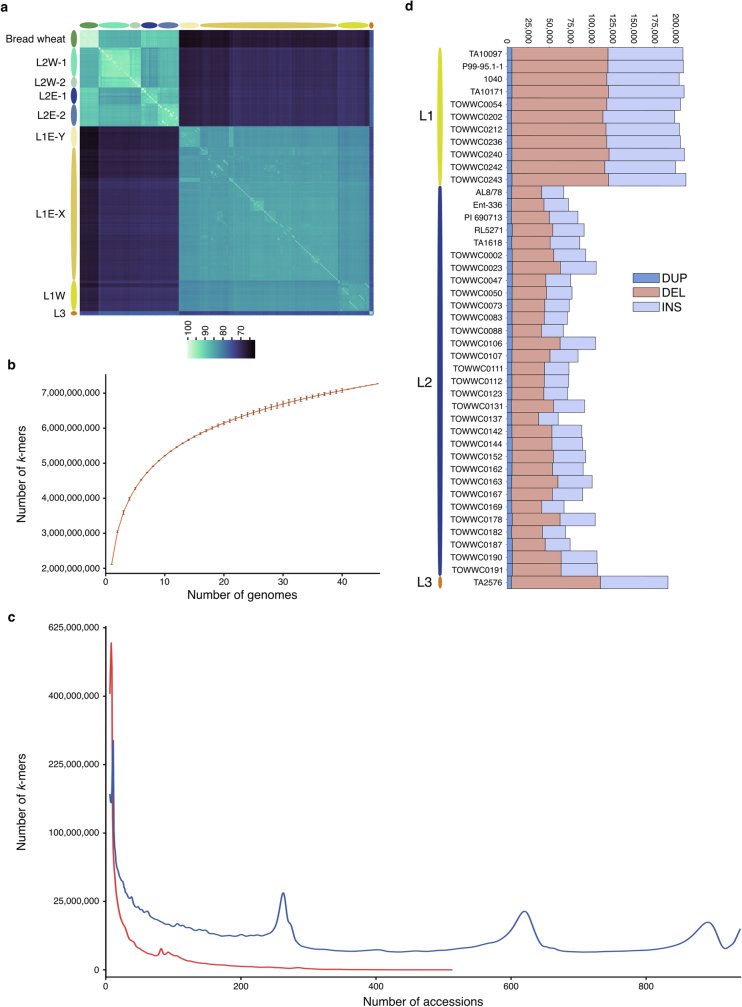
Extended Data Fig. 1. Aegilops tauschii genomic resources.
a, Clustered heatmap showing SNP-based pairwise identity across 957 Ae. tauschii accessions and 59 bread wheat landraces. The different Ae. tauschii subpopulations are indicated on the left. b, Logarithmic curve fit to k-mer accumulation across the 46 Ae. tauschii accessions selected for high-quality genome assemblies. The vertical bars show the standard deviation. c, k-mer frequency distributions across 920 Ae. tauschii accessions. The red curve shows k-mers that are absent in the 46 accessions selected for high-quality genome assemblies. The blue curve shows k-mers present in the 46 accessions. The peaks at ~250 and ~600 correspond to L2 and L1-specific k-mers, respectively. A square root function was applied to the y-axis for better visualization. d, Number of structural variants across Ae. tauschii accessions from lineages 1, 2 and 3 relative to the chromosome-scale assembly of L2 accession TA1675. Shown are duplications (DUP), deletions (DEL), and insertions (INS) ranging from 50 bp to 100 kb.
Extended Data Fig. 2. Ae. tauschii population structure from K = 2 to K = 9.
Each vertical bar represents an accession and the bars are filled by colours representing the proportion of each ancestry. The subpopulation designations are described in the main text. BW = bread wheat.
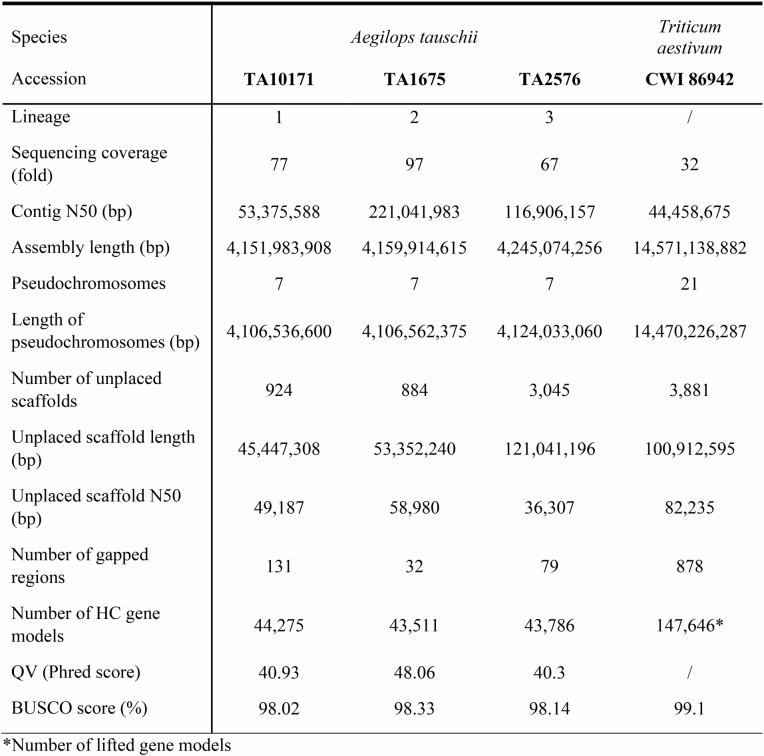
Using genetic, geographical and phenotypic diversity, we selected 46 accessions to construct high-quality genome assemblies, comprising 11 L1 accessions, 34 L2 accessions and 1 L3 accession (Fig. 1a,b and Supplementary Table 4). The 46 accessions captured 72.5% of the genetic diversity present in the Ae. tauschii diversity panel based on k-mer analysis. The majority of the k-mers that were not captured in the 46 high-quality assemblies are rare and were found in fewer than 5% of the accessions that make up the Ae. tauschii diversity panel (Extended Data Fig. 1b,c and Supplementary Table 5). We sequenced the selected 46 accessions by PacBio circular consensus sequencing16 to a median genome coverage of 23-fold (18- to 47-fold) and generated primary contig-level assemblies with contig N50 values ranging from 15.02 Mb to 263.79 Mb (median 45.26 Mb) (Supplementary Table 6). Phred quality scores ranged from 34.9 to 48.3 (median 45.5) and k-mer completeness scores ranged from 95.1 to 99.8% (median 99.4%) based on 21-mer content comparison with short-read WGS data (Supplementary Table 6). We calculated the benchmarking universal single-copy orthologues (BUSCO) scores for each accession17, returning values between 98.0% and 98.6% (Supplementary Table 6), indicating high contiguity, accuracy and completeness of the assemblies. We selected one representative accession per lineage to generate de novo annotated pseudochromosome assemblies, namely TA10171 (L1), TA1675 (L2) and TA2576 (L3). For these three accessions, we increased the sequencing coverage to 67- to 97-fold, generated assemblies with contig N50 values of 53.38 Mb (L1), 221.04 Mb (L2) and 116.91 Mb (L3) (Supplementary Table 6), and used Hi-C chromatin conformation capture18 to scaffold the assemblies into pseudomolecules (Extended Data Table 1 and Extended Data Fig. 3a–c). De novo annotation of the three chromosome-scale assemblies revealed 43,511 to 44,275 protein-coding genes (Extended Data Table 1). We included five previously generated high-quality Ae. tauschii assemblies15,19 and the bread wheat D genome20 for a gene cluster analysis. In total, we identified 52,722 clusters, of which 18,835 and 33,953 were core and dispensable, respectively (Supplementary Table 7). We then scaffolded the remaining 43 L1 and L2 contig-level assemblies using their respective L1 and L2 chromosome-scale references as guides (Supplementary Table 6).
Extended Data Table 1.
Assembly statistics for the three chromosome-scale Aegilops tauschii references and wheat landrace CWI 86942
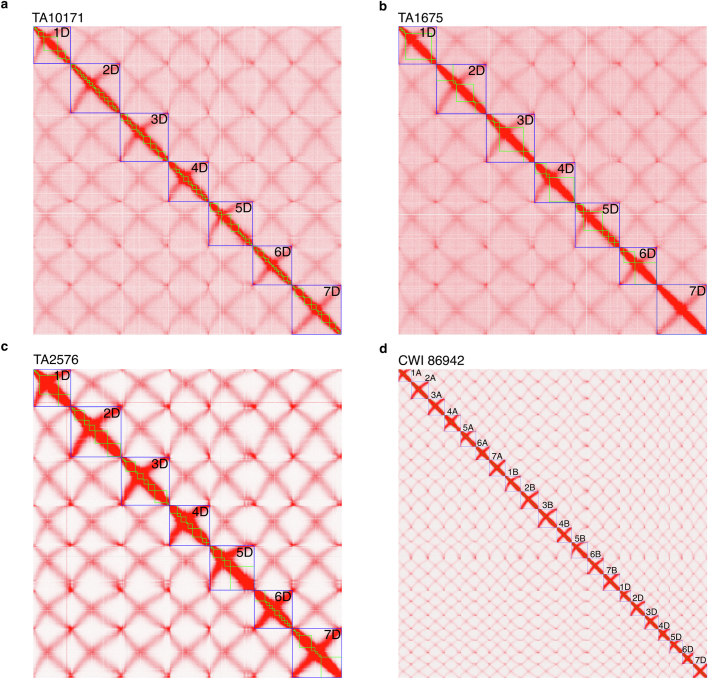
Extended Data Fig. 3. Chromosome contact maps of Ae. tauschii accessions TA10171 (a), TA1675 (b), TA2576 (c), and bread wheat accession CWI 86942 (d).
Green boxes represent individual PacBio contigs. Blue boxes indicate chromosomes. Chromosome 7D of TA1675 was assembled as a single PacBio contig.
In accordance with previous observations5,14,21, we detected increased nucleotide diversity in L2 (π = 0.00038) compared with L1 and L3 (L1, π = 0.00021; L3, π = 0.00024) (Fig. 1c and Supplementary Table 8). Structural variants were called across the high-quality assemblies relative to the TA1675 (L2) reference assembly (Fig. 1c). L1 accessions showed a similar distribution of structural variants to that in the L3 accession TA2576 (Extended Data Fig. 1d), with a median of 205,856 and 191,179 structural variants per accession for L1 and L3, respectively (Supplementary Table 9). The L2 accessions had a median of 85,401 structural variants per accession compared with the TA1675 reference (Supplementary Table 9).
Gene discovery
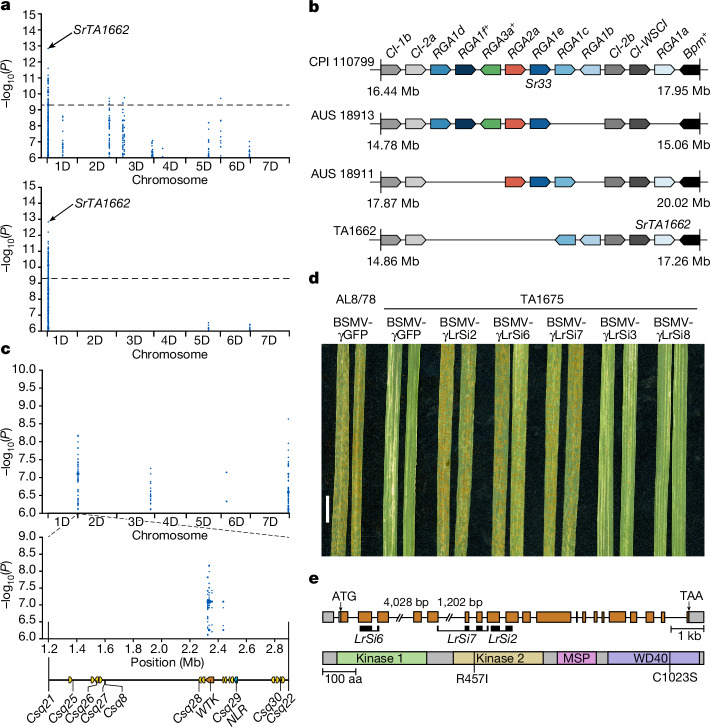
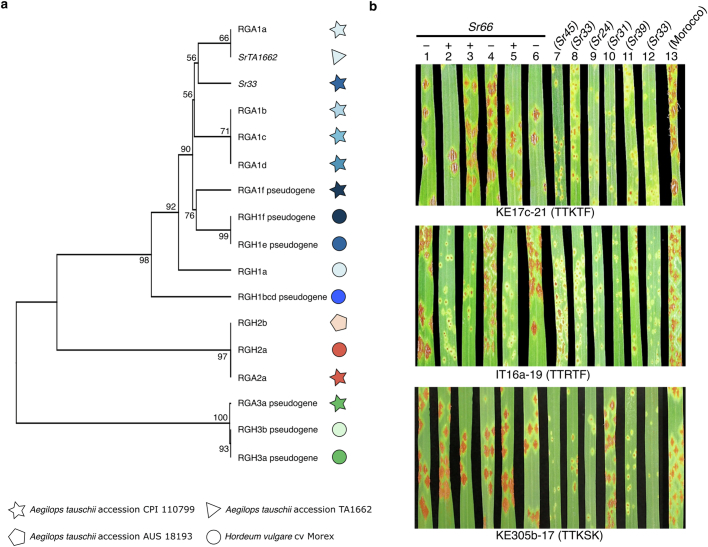
The highly contiguous Ae. tauschii assemblies generated here present an opportunity for gene discovery and characterization by comparative haplotype analyses. Here, we assessed the value of the Ae. tauschii genomic resources with a focus on rust resistance genes. The three fungal wheat rust diseases, leaf rust (caused by Puccinia triticina (Pt)), stripe rust (P. striiformis f. sp. tritici) and stem rust (P. graminis f. sp. tritici (Pgt)), are among the most devastating and most ubiquitous wheat diseases, causing considerable yield losses22. The stem rust resistance gene SrTA1662 was introgressed into bread wheat from Ae. tauschii accession TA1662 and genetically mapped to the stem rust resistance locus SR33 on chromosome arm 1DS23,24. Because the original mapping could not establish whether the stem rust resistance gene from TA1662 was a new gene or was allelic to Sr33, the gene was given the temporary designation SrTA166223. Sr33 and SrTA1662 encode intracellular nucleotide-binding leucine-rich repeat (NLR) immune receptors belonging to the Mla family14,24,25. Here, we repeated the k-mer-based association mapping that led to the initial discovery of the SrTA1662 candidate gene14. Compared with the short-read based Ae. tauschii assemblies14, mapping the k-mers against our high-quality Ae. tauschii genome strongly decreased the noise in the k-mer-based association approach (Fig. 2a). A detailed haplotype analysis revealed that SrTA1662 is a paralogue rather than an allele of Sr33 (Fig. 2b, Extended Data Fig. 4a and Supplementary Table 10). When we compared the stem rust infection phenotypes of Ae. tauschii lines predicted to carry only Sr33 or SrTA1662, we observed that the two genes appeared to have different specificities (Supplementary Table 11). We confirmed this notion by inoculating SrTA1662 transgenic wheat lines14 and Sr33 introgression lines with five Pgt isolates (Extended Data Fig. 4b and Supplementary Table 12). Our analysis so far showed that SrTA1662 confers resistance to a subset of the Pgt isolates avirulent on Sr33. In line with the nomenclature standards for wheat gene designation26, we therefore renamed SrTA1662 to Sr66.
Fig. 2. Haplotype analyses and cloning of a disease resistance gene.
a, Effect of assembly quality on association genetics. Significantly associated k-mers for resistance to Pgt race QTHJC mapped to two Ae. tauschii TA1662 assemblies (top, low-quality, contig N50 = 196 kb (ref. 14); bottom, high-quality, contig N50 = 58.21 Mb (this study)). The chromosome arm 1DS disequilibrium block contains the stem rust resistance gene SrTA1662 (renamed Sr66). b, Different Mla haplotypes24 reflected by analysis of resistance gene analogues (RGA) in Ae. tauschii CPI 110799 (Sr33 donor), AUS 18913, AUS 18911 and TA1662 (SrTA1662 donor). Boxes indicates genes; + indicates pseudogenes. Alleles are indicated by matching colour and position. The locus is flanked by subtilisin-chymotrypsin inhibitor (CI, grey) and pumilio (Bpm, black) genes. Unrelated genes present in this region are omitted. Locus length and gene distribution are not drawn to scale. c, k-mer-based genome-wide association study (GWAS) with Pt race BBBDB mapped to the Ae. tauschii TA1675 assembly. The chromosome arm 2D peak corresponds to leaf rust resistance locus LR39. The diagram shows the LR39 interval delimited by bi-parental mapping, flanked by markers Csq21 and Csq22, and markers Csq8 and Csq25-Csq30 co-segregating with LR39. Arrows indicate candidate genes. d, Effects of VIGS on susceptibility to leaf rust. AL8/78, susceptible control; BSMV-γGFP, barley stripe mosaic virus (BSMV) expressing a GFP silencing construct (control); BSMV-γLrSi2, BSMV-γLrSi6 and BSMV-γLrSi7 are silencing constructs specific for the WTK gene. BSMV-γLrSi3 and BSMV-γLrSi8 are silencing construct specific for the NLR gene. Probe specificities were evaluated using the TA1675 assembly. Chlorosis in BSMV-γGFP controls represent virus symptoms. Scale bar, 1 cm. e, Gene structure of Lr39 (top) and domain architecture of the Lr39 protein (bottom). Grey boxes represent untranslated regions, orange boxes are exons and lines are introns. VIGS probes are indicated. R457I and C1023S indicate the two Lr39 amino acid changes between the TA1675 (resistant) and AL8/78 (susceptible) lines. MSP, major sperm protein domain.
Extended Data Fig. 4. Haplotype analysis leads to the designation of stem rust resistance gene Sr66.
a, Phylogeny showing the relationship across Mla genes from Ae. tauschii and barley. Resistance Gene Analogs (RGA) represent Ae. tauschii and Resistance Gene Homologs (RGH) represent barley cultivar Morex. The Ae. tauschii RGA gene sequences were derived from different accessions (Supplementary Table 10). RGA/RGH families 1, 2 and 3 are indicated in blue, red and green, respectively. The tree was constructed using the unweighted UPGMA algorithm. Bootstrap support values are shown based on 5,000 replicates. b, SrTA1662 (Sr66) and Sr33 display different race specificities. Reactions to Puccinia graminis f. sp. tritici isolates KE17c-21 (race TTKTF), IT16a-19 (TTRTF), and KE305b-17 (TTKSK) of transgenic SrTA1662 (Sr66) wheat lines and non-transgenic nulls (1 to 6) and wheat Sr gene introgression lines and controls (7 to 13). 1, Fielder null (DPRM0050); 2, Sr66 (DPRM0051); 3, Sr66 (DPRM0059); 4, Fielder null (DPRM0062); 5, Sr66 (DPRM0071); 6, Fielder null (DPRM0072); 7. Sr45 (RL5406); 8. Sr33 (RL5405); 9. Sr24 (LcSr24Ag); 10. Sr31 (Little Club/Agent (CI 13523)); 11. Sr39 (RL5711); 12. Sr33 (Chinese Spring); 13. cv. Morocco.
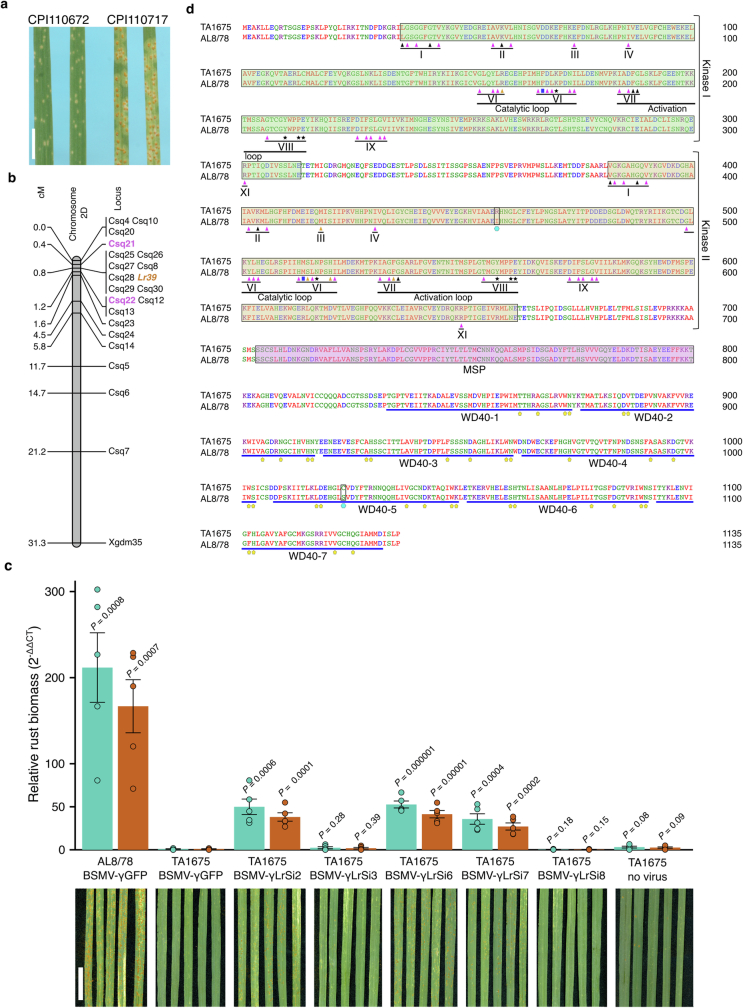
Ae. tauschii accession TA1675, for which a chromosome-scale reference assembly was generated in this study, carries the leaf rust resistance gene Lr39, which was mapped to the short arm of chromosome 2D27,28. k-mer-based association mapping with the Pt isolate BBBDB (avirulent against Lr39)29 revealed a peak at the telomeric end of chromosome arm 2DS, corresponding to the 2.33–2.45 Mb region in the TA1675 assembly (Fig. 2c). This location overlapped with markers flanking LR39 (positions 1.20–2.84 Mb) that were identified based on bi-parental genetic mapping30 (Fig. 2c and Extended Data Fig. 5a,b). The genomic region underlying the association peak contained 3 candidate genes in TA1675 (1 wheat tandem kinase (WTK) and 2 genes of unknown function), and the interval identified through bi-parental mapping harboured 16 genes (Supplementary Table 13). On the basis of functional annotations and polymorphisms compared with the susceptible Ae. tauschii accession AL8/78, the most promising candidate was AeT.TA1675.r1.2D000150, which encodes a WTK, a protein family that has a prominent role in disease resistance in wheat31–33. Virus-induced gene silencing (VIGS) of the WTK candidate gene in TA1675 resulted in greater susceptibility to leaf rust (Fig. 2d and Extended Data Fig. 5c). Silencing of an NLR-encoding gene (AeT.TA1675.r1.2D000200) located just outside the peak region did not result in increased susceptibility (Fig. 2c,d), indicating that the WTK gene is Lr39. The predicted genomic sequence of the Lr39 candidate gene is 11,699 bp in length with 21 exons. The corresponding 3,408-bp coding sequence encodes an 1,135-amino acid protein with two N-terminal kinase domains of the LRR_8B subfamily, followed by a major sperm protein domain and a WD40 repeat-containing domain (WD40) at the C terminus (Fig. 2e and Extended Data Fig. 5d). Compared with the susceptible Ae. tauschii accession AL8/78, Lr39 from TA1675 carried two amino acid changes located in the kinase 2 and WD40 domains, respectively (Fig. 2e and Extended Data Fig. 5d).
Extended Data Fig. 5. Bi-parental genetic mapping of LR39 and analysis of key conserved domains in Lr39.
a, Phenotypes of Ae. tauschii parents inoculated with the Puccinia triticina race Pt 26-1,3 (accession 316). CPI110672 (synonymous TA1675) carries Lr39. CPI110717 is the susceptible parent. Scale bar = 1 cm. b, Fine mapping of LR39 in chromosome arm 2DS. Markers Csq21 and Csq22 are flanking the LR39 locus whereas Csq8, Csq25, Csq26, Csq27, Csq28, Csq29 and Csq30 are co-segregating. c, Fungal biomass quantification using qPCR after virus-induced gene silencing (VIGS). Cereal rust specific primers amplifying the 28 S large subunit region (LSU - blue) or the internal transcribed spacer 1 (ITS1 - red) were used. Values represent means and error bars standard errors. Statistical analyses were done using a two tailed t-test against the TA1675 γGFP control. BSMV-γLrSi2, BSMV-γLrSi6, and BSMV-γLrSi7 are silencing constructs specific for the WTK gene. BSMV-γLrSi3 and BSMV-γLrSi8 are silencing construct specific for the NLR gene. N = 5 independent biological replicates. Scale bar = 5 cm. d, Analysis of key conserved domains of the Lr39 protein. The kinase 1 domain is highlighted by a green box, kinase 2 by a yellow box, the major sperm protein (MSP) domain by a pink box, and the seven WD40-repeats are underlined by blue lines. Roman numerals represent conserved kinase subdomains. Black triangles = ATP binding site predicted by InterPro; magenta triangles = key conserved residues; black asterisks = putative substrate binding site; blue squares = residue determining RD and non-RD kinases; brown triangles = polymorphism in the key conserved residues. In kinase 1, a key residue histidine is replaced by arginine in subdomain VI. In kinase 2, substitutions of residues glutamic acid to methionine in subdomain III, aspartic acid to serine and asparagine to histidine in subdomain VI form a catalytic loop, and aspartic acid to glycine in subdomain VII in the activation loop. Yellow pentagons = key conserved residues of WD40 repeats predicted by InterPro. Cyan hexagon = two polymorphic residues of TA1675 compared to AL8/78.
Several recent studies have highlighted the importance of using genome assemblies of resistance gene-containing reference accessions for gene cloning and designation11,34. The analyses we present here would have been difficult, if not impossible, using previous Ae. tauschii assemblies, because none of them are donors of Sr33, SrTA1662 (Sr66) or Lr39.
Lineage-specific haplotype blocks
Bread wheat has become one of the most successful and widely cultivated crop species, and is adapted to a wide range of climatic conditions1. Continuous gene flow by natural and artificial introgressions from wild wheat relatives has increased the genetic diversity of bread wheat following a domestication bottleneck9–13. For example, around 1% of the extant bread wheat D genome originated from Ae. tauschii L3, indicating multiple hybridization events that gave rise to the extant bread wheat D genome14. The genetic distinctness and geographical restriction of Ae. tauschii L3 (Fig. 1a,b) makes this lineage an ideal example to study the spatial dynamics of introgressions. We hypothesize that bread wheat landraces with higher L3 genome content, possibly representing a more ancestral state of the L3 introgression(s), have been preserved in ex situ collections but are rare and geographically restricted.
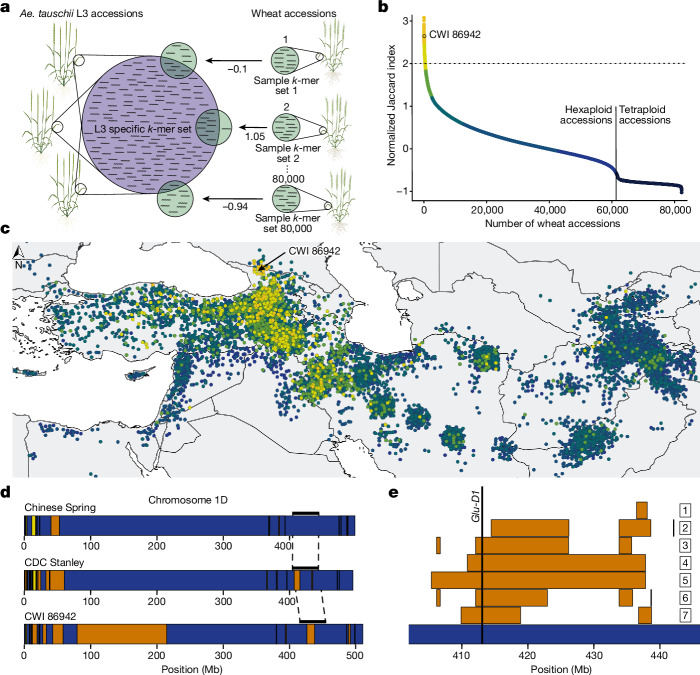
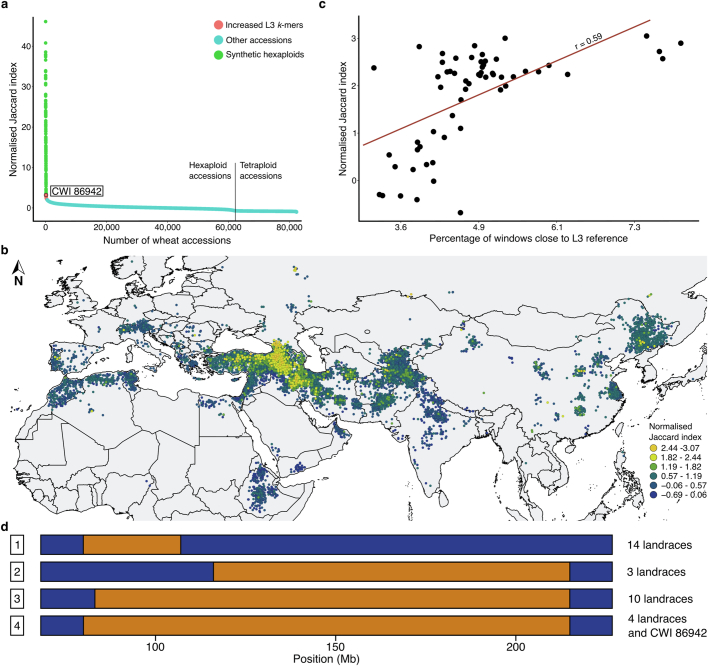
To identify bread wheat accessions with above-average proportions of L3 genome, we developed the ‘Missing Link Finder’ pipeline (Fig. 3a). Missing Link Finder estimates the similarity between a species- or lineage-specific reference k-mer set and sample k-mer sets generated from genotyping data of individual wheat accessions, computing the result as Jaccard similarity coefficients. To deploy Missing Link Finder, we used a reference k-mer set consisting of 769 million Ae. tauschii L3-specific k-mers14 and compared it to individual sample k-mer sets from 82,293 genotyped wheat accessions (6.16 million k-mers per accession on average)35. We identified 503 bread wheat accessions with an above-average (more than twofold) normalized Jaccard index (a value of 0 indicates an average number of L3 k-mers), indicative of increased Ae. tauschii L3 content (Fig. 3b and Extended Data Fig. 6a). The 139 accessions with the highest Jaccard indices are synthetic hexaploid wheats (Extended Data Fig. 6a), most of which (122 accessions) were produced using an Ae. tauschii accession collected in Georgia (CWI 94855), the only country where Ae. tauschii L3 has been found in the present day14. We also identified 364 bread wheat landraces with putatively increased proportions of L3 introgressions (Fig. 3b and Extended Data Fig. 6a). One of the bread wheat landraces with the highest Jaccard indices, CWI 86942 (PI 572674), was collected in the Samegrelo-Zemo Svaneti region of Georgia36. We observed a gradient of decreasing L3 proportions (as revealed by Jaccard indices) with increasing geographical distance from Georgia (Fig. 3c and Extended Data Fig. 6b).
Fig. 3. Ae. tauschii L3 introgressions in bread wheat.
a, Diagram of the Missing Link Finder pipeline. An Ae. tauschii L3-specific k-mer set (769 million L3-specific k-mers; blue circle) was compared to individual sample k-mer sets generated from more than 80,000 genotyped wheat accessions (green circles). The result is indicated as normalized Jaccard indices. b, Distribution of normalized Jaccard scores across 82,154 wheat accessions. The horizontal dotted line indicates the two-fold threshold. The 139 synthetic hexaploid wheat lines with increased Jaccard indices have been removed and are shown in Extended Data Fig. 6a. c, The Jaccard indices show a gradual decline with increasing geographical distance from Georgia. Dots represent individual bread wheat accessions for which exact coordinates were available. Colours represent different normalized Jaccard indices corresponding to b. A full map is shown in Extended Data Fig. 6b. Eastern bread wheat accessions from Tajikistan with high Jaccard indices carry the same L3 introgression segments as bread wheat landraces from Georgia, indicating a common origin of the L3 haplotype blocks (Supplementary Table 18). d, Diagram of chromosome 1D in the wheat lines Chinese Spring, CDC Stanley and CWI 86942. Haplotype blocks corresponding to Ae. tauschii L1 are indicated in yellow, L2 is indicated in blue, and L3 is in orange. The black bars above the chromosome indicate the region shown in e. e, Diagram of a portion of the long arm of bread wheat chromosome 1D. Shown are different lengths of the L3 introgression segment in various bread wheat lines. The numbers correspond to the following accessions chosen for their diverse recombination patterns in this locus: (1) CWI 86929; (2) CWI 30140; (3) CWI 57175; (4) CWI 84686; (5) CWI 84704; (6) CWI 86481; (7) CDC Stanley.
Extended Data Fig. 6. Tracing lineage-specific Ae. tauschii haplotype blocks in bread wheat.
a, Normalized Jaccard scores across 82,293 wheat accessions (including the 139 synthetic hexaploid wheats). Green indicates 139 synthetic hexaploid wheat accessions with k-mer enrichments of up to 40-fold. Red indicates bread wheat landraces with increased (2 to 3-fold) normalized Jaccard index. b, The Jaccard indices show a gradual decline with increasing geographic distance from Georgia. Dots represent individual bread wheat accessions for which exact coordinates were available. Colors represent different normalized Jaccard indices. c, Correlation between normalized Jaccard indices and the percentage of L3 genome based on whole-genome sequencing data. d, Diagram of a portion of chromosome arm 1DS. The chromosome positions indicated in Mb are according to the CWI 86942 assembly. Haplotype blocks corresponding to Ae. tauschii L2 are indicated in blue, and L3 in orange. Shown are different lengths of the L3 haplotype segment in various bread wheat lines. 1, CWI 84680, CWI 84694, CWI 84704, CWI 84686, CWI 14537, GEO-L1, WATDE0105, WATDE0944, WATDE0957, WATDE1005, WATDE1018, WATDE1017, WATDE0113, WATDE1010; 2, C33, WATDE1031, WATDE1032; 3, BW 50849, CWI 14244, CWI 28055, WATDE0026, WATDE0749, WATDE0047, WATDE0739, WATDE0999, WATDE1003, WATDE0993; 4, CWI 86929, CWI 86942, WATDE0975, WATDE0973, WATDE0974. The IBSpy variation values for the Watkins lines (WATDE) were extracted from Cheng et al.38.
To further quantify and explore the L3 contents of CWI 86942 and other landraces, we generated an annotated chromosome-scale assembly of CWI 86942 using PacBio circular consensus sequencing16 and chromosome conformation capture18 (Extended Data Table 1 and Extended Data Fig. 3d). In addition, we produced WGS data (tenfold coverage) of 36 hexaploid wheat landraces with higher (greater than 2) Jaccard indices using short-read Illumina-based sequencing. For comparison, we also sequenced 23 wheat landraces with Jaccard indices of less than 2 (Supplementary Table 14). Our analysis focused on landraces to avoid detection of L3 haplotype blocks that might be the result of artificial introgressions. We observed a good correlation between the Jaccard indices and the Ae. tauschii L3 content estimated based on WGS data (Extended Data Fig. 6c), supporting the idea that Missing Link Finder is a suitable pipeline to identify rare wheat accessions with above-average introgressions. CWI 86942 contained approximately 7.0% of L3 introgressions, compared with the 0.5 to 1.9% in other bread wheat assemblies14. Most notable was a 135-Mb L3 segment in the pericentromeric region of chromosome 1D (Fig. 3d), which represents the largest Ae. tauschii L3 haplotype block reported in bread wheat so far. This segment contains 587 predicted genes, of which 112 showed presence–absence variation or a disruptive mutation compared to the corresponding L2 segment in wheat cultivar Kariega (Supplementary Table 15). In addition to CWI 86942, this L3 haplotype segment, or parts thereof, were found in multiple bread wheat landraces collected between the 1920s and the 1930s (Extended Data Fig. 6d), indicating that this segment is not the result of synthetic hexaploid wheat breeding37,38. A second notable L3 segment was found on the long arm of chromosome 1D in multiple bread wheat landraces (Fig. 3e). This segment carries a superior wheat quality allele at the Glu-D1 locus that originated from Ae. tauschii L339. In modern bread wheat (for example, wheat cultivars CDC Landmark, CDC Stanley and Jagger), this L3 segment is around 8.5 Mb in size. We identified a group of bread wheat landraces originating from Azerbaijan where the corresponding L3 segment was up to 36.35 Mb in size (Fig. 3e). This L3 introgression showed various lengths in different bread wheat landraces, reflecting extensive recombination. We further estimated the cumulative proportion of L3 introgression across a comprehensive set of 126 hexaploid wheat landraces, including the WGS data from the 59 landraces generated in this study and publicly available sequencing data (Supplementary Table 16). Using identity-by-state, we determined that 16.6% of the wheat D genome, corresponding to 666.0 Mb and containing 8,779 high-confidence genes (25.6%), can be covered with Ae. tauschii L3 haplotype blocks across these landraces (Supplementary Table 17). Although the proportion of Ae. tauschii L3 introgressions in most modern bread wheat cultivars is marginal (approximately 1% relative to the entire D genome), the cumulative size of L3 introgressions across multiple bread wheat landraces is considerable.
Origin and evolution of the wheat D genome
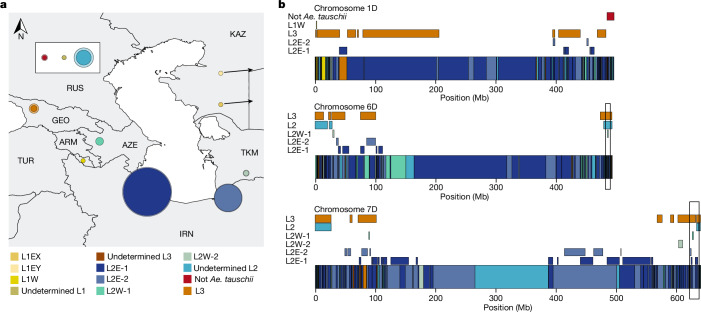
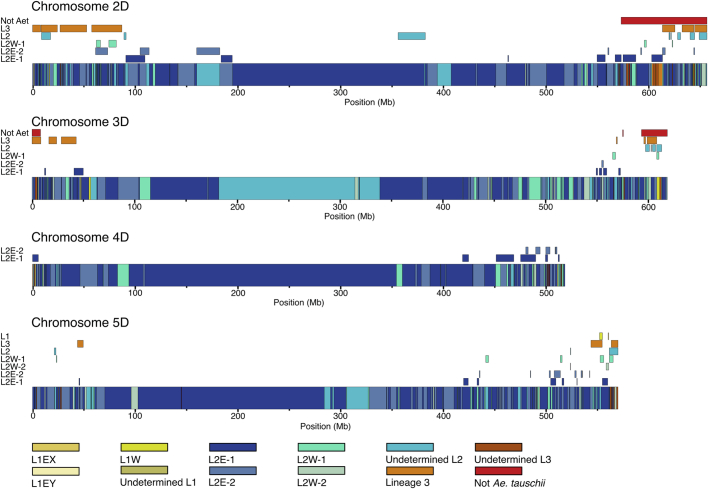
We determined the complexity and origin of the D genome across 17 hexaploid wheat lines, for which chromosome-scale assemblies are available11,34,40,41,20,42,43. We divided the wheat genomes into 50-kb windows and assigned each window to an Ae. tauschii subpopulation based on identity-by-state13. We observed that all four Ae. tauschii L2 subpopulations contributed genomic segments to the bread wheat D genome (Supplementary Table 19). Consistent with previous reports5, the largest proportion of the wheat D genome (45.6–51.3%) originated from subpopulation L2E-1, which is mainly found in the southwestern Caspian Sea region. Subpopulation L2E-2 (southeastern Caspian Sea) contributed 24.7–27.3% to the wheat D genome (Fig. 4a). Up to 6.9% of the wheat D genome was identical (based on identity-by-state analysis) to Ae. tauschii subpopulation L2W, with 4.1-5.0% coming from subpopulation L2W-1 and subpopulation L2W-2 contributing 1.7–2.2% (Fig. 4a,b and Extended Data Fig. 7). We could assign another 10.7–19.5% of the wheat D genome to L2, but without being able to infer the exact subpopulation (Supplementary Note 3), indicating that these segments originated from Ae. tauschii L2 haplotypes that were not captured in our diversity panel (Fig. 4a,b and Extended Data Fig. 7). The contributions from Ae. tauschii L1 and L3 ranged from 0.7% to 1.1% and 1.6% to 7.0%, respectively. Genomic windows representing 0.1–2.4% of the hexaploid wheat D genome had a different origin than Ae. tauschii. These windows include previously described introgressions, such as the tall wheatgrass (Thinopyrum ponticum) introgression on chromosome 3D in bread wheat cultivar LongReach Lancer or the putative Aegilops markgrafii/Aegilops umbellulata introgression on chromosome 2D of cultivars Julius, ArinaLrFor, SY Mattis and Jagger11,12 (Supplementary Table 19). A list of predicted genes and their corresponding subpopulation origin is provided in Supplementary Table 20. The number of Ae. tauschii subpopulations that contributed to the hexaploid wheat D genome does not necessarily reflect the number of independent hybridization events, because the Ae. tauschii line that contributed the D genome may have already been admixed. To infer the minimal number of hybridizations that gave rise to the extant wheat D genome, we assessed the number of haplotypes present at any given position in the wheat D genome. We used Chinese Spring as a reference and identified 50-kb windows showing no identity-by-state across the 126 hexaploid wheat landraces for which WGS data were available. Such windows indicate the presence of at least two haplotypes in the hexaploid wheat gene pool. Consecutive 50-kb windows with no identity-by-state were concatenated into alternative haplotype blocks. The origins of alternative haplotype blocks were then assigned to one of the Ae. tauschii subpopulations using identity-by-state (Supplementary Table 21). In total, 71.4% of the wheat D genome was covered by a single haplotype across the analysed hexaploid wheat lines (59.7% of genes). The remaining 28.6% of the wheat D genome showed multiple haplotypes (21.0% of the wheat D genome had two haplotypes (26.2% of genes), 6.7% had three haplotypes (12.2% of genes) and 0.9% had four haplotypes (1.9% of genes)) (Fig. 4b, Extended Data Fig. 7 and Supplementary Table 22). The maximum number of haplotypes corresponding to different Ae. tauschii subpopulations for any given window was four, indicating that the bread wheat D genome evolved through at least four hybridizations.
Fig. 4. Different Ae. tauschii subpopulations contributed to the hexaploid wheat D genome.
a, Proportions of Ae. tauschii subpopulations that make up the wheat D genome. Inner circles in solid colours represent the average proportions across 17 hexaploid wheat assemblies. The outer lighter circles represent the maximum proportion found across the 17 wheat genomes. The geographical location for each subpopulation was assigned on the basis of representative accessions. RUS, Russian Federation. b, Minimal number of hybridization events that gave rise to the extant bread wheat D genome. Diagrams show chromosomes 1D, 6D and 7D in Chinese Spring. The coloured boxes along the chromosomes represent the haplotypes present in Chinese Spring. Coloured rectangles above the chromosomes represent alternative haplotype blocks identified across 126 hexaploid wheat landraces (cumulative length of alternative haplotype blocks across all 126 landraces). Colours refer to the Ae. tauschii subpopulations. The maximum number of haplotype blocks was four. Black boxes highlight the regions on chromosome 3D and 7D in which four overlapping haplotypes are found.
Extended Data Fig. 7. Minimal number of hybridizations that gave rise to the extant bread wheat D genome.
Shown are graphical representations of Chinese Spring chromosomes 2D, 3D, 4D and 5D. The colored boxes in the chromosomes represent the haplotypes found in Chinese Spring. Colored rectangles above the chromosomes represent alternative haplotype blocks identified across 126 hexaploid wheat landraces (cumulative length of alternative haplotype blocks across all 126 landraces). Colors refer to the Ae. tauschii subpopulations following the legend. The maximum number of haplotype blocks is four.
Discussion
The comprehensive genomic resources generated in this study enabled haplotype analysis and cloning of rust resistance genes and they offered a detailed insight into the composition and origin of the bread wheat D genome. Crop domestication has often been considered as a relatively simple linear progression44. Our analyses support a model of protracted domestication that is more complex, involving recurring episodes of hybridization and gene flow that resulted in patchwork-like haplotype patterns across the bread wheat D genome. We largely confirm that an Ae. tauschii L2 population from the southwestern Caspian Sea region was the major donor of the bread wheat D genome5 (Fig. 4a), with smaller genomic segments originating from different Ae. tauschii lineages5,14. In contrast to previous reports, however, our work revealed a much more complex patterning of the bread wheat D genome. We determined that all four L2 subpopulations, as well as L1 and L3, contributed segments to the extant bread wheat D genome. Compared to the AB subgenomes, the bread wheat D genome shows a lower genetic diversity, indicative of a much lower rate of introgression from wild progenitors2,6. The patchwork pattern seen in the bread wheat D genome is somewhat surprising given that most Ae. tauschii L2 accessions in our diversity panel showed a low degree of admixture, with a well-defined population structure following their geographical distribution (Extended Data Fig. 2). A possible explanation for this observation is that the Ae. tauschii accession that gave rise to the bread wheat D genome was admixed, carrying genomic segments from different subpopulations. Remnants of Aegilops species have been identified at several pre-agricultural settlements in the Fertile Crescent45, indicating that Aegilops species were used as food source or persisted as weeds in pre-agricultural cultivation of other wild cereals. The gathering and possible management of Ae. tauschii for food, or its co-cultivation as a weed over an extended period might have resulted in mixing of Ae. tauschii populations with different geographical origins, leading to an increase of admixed accessions close to human settlements. Such an admixed Ae. tauschii population might have later given rise to the bread wheat D genome. This scenario would also explain why the bread wheat D genome forms a separate clade from Ae. tauschii in many phylogenetic and population structure analyses5,14,15,46 (Fig. 1b). Alternatively, the Ae. tauschii accession that gave rise to the bread wheat D genome was non-admixed, and recurrent hybridizations resulted in the observed mosaic-like haplotype pattern.
Another important finding of this study is the large cumulative size of alternative haplotype (non-L2E) blocks in the bread wheat D genome. Following hexaploidization, genetic material from the other Ae. tauschii lineages (L1 and L3) became incorporated into the bread wheat D genome and were subsequently broken into smaller fragments via recombination. Although the proportions of alternative haplotype blocks are low in individual elite wheat cultivars, the different segments accumulate to considerable lengths across various genotypes. This notion is evidenced by the cumulative size of L3 segments that span a total of 666.0 Mb. We assessed 126 hexaploid wheat landraces, and although we selected for accessions with increased L3 genome content, it is likely that the proportion of remnant L3 segments in the bread wheat gene pool is even higher. This finding raises important questions about the adaptive potential of alternative haplotype blocks for wheat breeding.
Methods
Establishing Ae. tauschii genomic resources
Plant material
We compiled a database comprising 1,124 Ae. tauschii accessions with associated passport data in Supplementary Table 1 (Supplementary Note 1). Duplicate germplasm bank IDs were identified and passport data collated using the Plant Genetic Resources for Food and Agriculture (PGRFA) database (https://www.genesys-pgr.org/) or other sources as indicated in Supplementary Table 1. From this database, seed of 228 non-redundant accessions were obtained from the Open Wild Wheat Consortium Ae. tauschii Diversity Panel collection deposited at the Germplasm Resource Unit (GRU) of the John Innes Centre; 48 accessions from the Cereal Crop Wild Relatives (Triticeae) collection of the GRU; 19 accessions from the Designing Future Wheat (DFW) Wheat Academic Toolkit collection of the GRU that have been used as synthetic hexaploid wheat D genome donors; 223 accessions from the Wheat Genetics Resource Center (WGRC) of Kansas State University; 34 accessions from the Plant Gene Resources of Canada (PGRC); 84 accessions donated by the Institute of Botany, Plant Physiology and Genetics of the Tajikistan National Academy of Sciences; 20 accessions donated by the Azerbaijan National Academy of Sciences; and 37 accessions donated by Quaid-i-Azam University. Accession P-99.95-1.1 was obtained from the Deposited Published Research Material collection of the GRU.
We also resequenced and analysed 60 hexaploid wheat landraces. The list can be found in Supplementary Table 14. Out of the 60 wheat landraces, 57 were received from the International Maize and Wheat Improvement Center (CIMMYT) and 3 were from the International Center for Agricultural Research in the Dry Areas (ICARDA).
Re-sequencing of Ae. tauschii and hexaploid wheat accessions
In this study, we generated short-read WGS data for 350 Ae. tauschii accessions (Supplementary Table 1) and 59 hexaploid wheat accessions (Supplementary Table 14). We isolated DNA following the CTAB protocol described by Abrouk et al.47 from leaf tissue of 2-week-old seedlings under prior dark treatment for 48 h. DNA was quantified using the Qubit dsDNA HS Assay (Thermo Fisher Scientific) and purity was determined according to 260/280 and 260/230 absorbance ratios using a Nanodrop spectrophotometer. PCR-free paired-end libraries were constructed and sequenced on an Illumina Novaseq 6000 instrument, yielding a median 8.3-fold coverage per sample (ranging from 5.87- to 16.86-fold) for the Ae. tauschii samples and a minimum tenfold coverage for the bread wheat samples (Supplementary Tables 1 and 14). Library preparation and sequencing was performed as a service by Novogene.
Library construction and RNA sequencing
Seedlings of Ae. tauschii accessions TA10171, TA1675 and TA2576 were raised as 5–6 seeds per pot (6 × 6 × 10 cm) in a growth chamber at 22–24 °C under long-day photoperiods of 16/8 h day/night cycle with high-output white-fluorescent tubes until the third leaf stage (about 2–3 weeks old), and then transferred to a 4 °C growth chamber with a long-day photoperiod for vernalization. After a nine-week vernalization period, all the plants were moved back to the original growth chamber under the controlled conditions mentioned above. In total, 45 tissue samples were collected: From each of the three accessions, three biological replicates were taken from each of: young leaf, root, stem, flag leaf and inflorescence. Samples were collected at the same time of day at approximately 5–6 h after daylight. The seedling leaves and roots were harvested after two weeks of recovery in the original growth chamber and rinsed with water to remove soil particles. When the plants had 4–5 tillers, the stems, flag leaves and inflorescences were harvested together. The green inflorescences were collected immediately after pollination. The 5-cm-long stem sections and youngest flag leaves were measured from the top of the same inflorescences. Samples were placed in liquid nitrogen after harvest and stored at −80 °C.
The samples were ground into a fine powder in liquid nitrogen in a ceramic mortar and pestle to isolate RNA using the Qiagen RNeasy Mini Kit following the manufacturer’s protocol. The quality of RNA was determined on a 1% agarose gel, and RNA concentration was measured using a NanoPhotometer (Implen) at 260 nm and 280 nm. Sample collection time and relative details are listed in Supplementary Table 23. High-quality RNA samples were delivered for RNA integrity test, poly-A mRNA enrichment, library construction and PE100 sequencing using the Illumina NovaSeq system (Génome Québec, Canada).
PacBio HiFi genome sequencing; primary assembly of the Ae. tauschii genomes and CWI 86942
We selected 46 Ae. tauschii accessions, including 11 L1 accessions, 34 L2 accessions and 1 L3 accession. These 46 accessions were selected to span the geographical range of the species (Fig. 1a) and provide a collection of phenotypes related to disease and pest resistance, abiotic tolerance and agromorphological traits of strategic interest to the Open Wild Wheat Consortium for bread wheat improvement (Supplementary Table 4). We included a higher proportion of accessions from L2 relative to L1 based on reported phylogenies showing that L2 is more genetically diverse than L15,14,21. A single L3 accession was selected based on low genetic diversity observed among five non-redundant L3 accessions in the phylogeny reported by Gaurav et al.14. Several accessions were selected to maximize the genetic diversity based on a core subset sampling analysis using Core Hunter (v3)48, using the ‘average entry-to-nearest-entry’ distance measure, aiming to maximally represent the diversity of the panel of 242 non-redundant accessions published by Gaurav et al.14. The bread wheat landrace CWI 86942 was selected based on a high L3 k-mer content.
For the Ae. tauschii accessions, ‘high molecular weight’ genomic DNA was isolated from leaf tissue of three to four-week-old dark-treated seedlings. We followed the high molecular weight DNA isolation protocol optimized by Driguez et al.49 for long-read sequencing. DNA integrity was confirmed using the FemtoPulse system (Agilent). DNA was quantified using the Qubit dsDNA HS Assay (Thermo Fisher Scientific) and purity was determined according to 260/280 and 260/230 ratios using a Nanodrop spectrophotometer. For the bread wheat accession CWI 86942, leaves from two-week old seedlings were collected from two different plants and high molecular weight DNA extraction was performed as mentioned above49. All the library preparation and Circular Consensus Sequencing (CCS) was performed on a PacBio Sequel II instrument, as a service by Novogene.
For Ae. tauschii, HiFi reads were assembled using hifiasm (v0.16.1)50 with parameters “-l0 -u -f38” optimized for homozygous and large genomes (-l0 -f38) and to minimize misassemblies by disabling the post-join contigs step (-u), favouring accuracy over contiguity. Sequencing coverage ranged from 18 to 47-fold depending on the accession, except for the three Ae. tauschii lineage reference accessions (TA10171, TA1675 and TA2576) for which the coverage was increased to 67 - 97-fold. For assembly validation and quality control, we used QUAST (v5.0.2)51 to calculate the assembly metrics, Merqury (v1.3)52 to estimate the base-call accuracy and k-mer completeness based on 21-mer produced from the short-read WGS data14 and BUSCO (v5.3.1)17 with the embryophyta_odb10 database to determine the completeness of each genome assembly. The number of homozygous SNPs and short insertion–deletion mutations (indels) was determined comparing the HiFi assemblies against the respective WGS data (Supplementary Table 24). They are in the range of 3,416–40,885 homozygous SNPs or indels per accession.
For CWI 86942, we performed the primary contig-level assembly with 484.33 Gb of HiFi reads (32-fold coverage) using the LJA assembler (v0.2)53 with default parameters. Assembly metrics and QC were performed with QUAST (v5.0.2)51 and BUSCO (v5.3.1)17 with the embryophyta_odb10 database.
Chromosome conformation capture sequencing and chromosome-scale scaffolding
In situ Hi-C libraries were prepared for TA1675 and TA10171 from two-week-old Ae. tauschii plants according to the previously published protocol54. Libraries were quantified and sequenced (paired-end, 2 ×111 cycles) using the Illumina NovaSeq 6000 device (Illumina) at IPK Gatersleben55, yielding 316 million paired-end reads (150 bp) for TA1675 and 215 million paired-end reads for TA10171.
For TA2576, two-week-old, dark-treated leaf tissue samples were harvested and cross-linked with formaldehyde for library preparation and Hi-C sequencing by Phase Genomics, yielding 543 million paired-end reads (150 bp). For CWI 86942, two Omni-C libraries were generated and sequenced from two-week-old, dark-treated leaf tissue samples as a service by Dovetail Genomics. The total yield was 715 million paired-end reads (150 bp).
Scaffolding into pseudomolecules for TA10171, TA1675, TA2576 and CWI 86942 was performed from their primary assemblies and their specific Hi-C and Omni-C data, respectively. Hi-C and Omni-C reads were processed with Juicer (v1.6)56 (for the Hi-C reads, parameter: -s DpnII) to convert raw fastq reads to chromatin contacts and remove duplicates. The chromatin contacts were used to scaffold the contig-level assemblies using 3D-DNA (v190716)57 (using run-asm-pipeline.sh with -r 0 parameter). Scaffolds were visualized, manually oriented and ordered using Juicebox (v2.20.00)58.
RagTag assembly of 43 Ae. tauschii accessions
The remaining 43 contig-level assemblies were scaffolded into chromosome-scale assemblies using RagTag (v2.1.0)59 and the three high-quality genomes (TA10171, TA1675 and TA2576) as anchors. In brief, the primary contig-level assemblies were scaffolded using RagTag scaffold against the respective chromosome-scale reference assemblies generated in this study. After running RagTag scaffold, the placed contigs had the exact same lengths as the primary contigs before running RagTag scaffold. Also, the number of gaps in each RagTag assembly corresponds to the number of placed contigs minus seven (number of chromosomes) (Supplementary Table 6). This indicates that RagTag scaffold did not introduce misassemblies or duplicated contig ends. The scaffolded assemblies were validated with dot-plots generated using MashMap (v3.0.6)60 against the corresponding reference assembly. While being great resources for gene discovery and comparative analyses, reference-guided assemblies are limited in their ability to study large structural rearrangements.
Repeat and gene annotation
Paired-end RNA-seq reads for TA10171, TA1675 and TA2576 were first cleaned using Trimmomatic (v0.40)61 with the following settings “ILLUMINACLIP:TruSeq3-PE.fa:2:30:10:2:True LEADING:30 TRAILING:30 MINLEN:36”. Trimmed paired-end reads were aligned to the corresponding genome assembly using STAR (v2.7.10b)62 with the parameters “--twopassMode basic --outFilterMismatchNMax 5 --outFilterMatchNminOverLread 0.80 --alignMatesGapMax 100000 --outSAMstrandField intronMotif --runMode alignReads” and the results were filtered and sorted using SAMtools (v1.10)63. Then, the BRAKER (v3.0.3)64–66 pipeline was used to predict de novo gene models using RNA-Seq and protein data mode with the Viridiplantae protein models provided by OrthoDB (v11). Predicted gene annotations obtained from BRAKER were processed using a combination of NCBI BLAST+ (v2.9.0-2)67, AGAT (v1.2.1) (https://github.com/NBISweden/AGAT), InterProScan (v5.64-96.0)68,69, and R (v4.2.0). Outputs from BRAKER3 were first converted to gff3 and CDS and protein sequences were extracted using “agat_sp_extract_sequences.pl” from AGAT package. BLASTn was used to perform a reciprocal BLAST of the predicted CDS against themselves, and a unidirectional BLAST against the Ensembl nrTEplantsJune2020.fa repetitive elements database, using default search parameters. The putative functions for each annotated gene model were predicted using InterProScan with default parameters for the following databases: FunFam, SFLD, PANTHER, Gene3D, PRINTS, Coils, SUPERFAMILY, SMART, CDD, PIRSR, ProSitePatterns, AntiFam, Pfam, MobiDBLite, PIRSF, NCBIfam. R (v4.2.0) (in R studio) was used to visualize and filter these results. Predicted transcripts with fewer than 50 exact and fewer than 150 inexact self-BLAST results were retained. Predicted transcripts were retained from the final de novo annotation if there were (1) no exact matches to the transposable elements database, and (2) at least one domain predicted by any of: FunFam, PANTHER, Gene3D, SUPERFAMILY, ProSitePatterns, Pfam, CDD, InterPro. Predicted genes were considered as ‘low confidence’ if there were no exact matches to the database of original transcript predictions. The remaining annotated genes were considered as ‘high confidence’. Validation and annotation completeness was performed using agat_sp_statistics.pl and BUSCO (v5.4.7)17 running in transcriptome mode with the poales_odb10 database. We used OrthoFinder70 (v2.5.4) with default parameters to perform gene family analysis.
Repeat annotation was performed using RepeatMasker (v4.1.2-p1)71 and the Ensembl nrTEplantsJune2020 repetitive elements database72 using the RMBlast engine.
For bread wheat accession CWI 86942, gene model prediction was performed using a lifting approach similarly to the one described in Abrouk et al.73 with a combination of liftoff (v1.6.3)74, AGAT and gffread (v0.11.7)75. In brief, gene model annotations of hexaploid wheat line Chinese Spring, Kariega, Fielder, ArinaLrFor, Julius and Norin61 were independently transferred using liftoff (parameters: -a 0.9 -s 0.9 -copies -exclude_partial -polish) and all the output gff files were merged into a single file using the Perl script agat_sp_merge_annotations.pl. The merged file was then post-processed using gffread tools (parameters: --keep-genes -N -J) to retain transcripts with start and stop codons, and to discard transcripts with (1) premature stop codons, and/or (2) having introns with non-canonical splice sites. In total, 147,646 gene models were predicted for which the putative functional annotations were assigned using a protein comparison against the UniProt database (2021_03) using DIAMOND (v2.1.8)76 (parameters: -f 6 -k1 -e 1e-6). PFAM domain signatures and GO were assigned using InterproScan (v5.55-88.09)68,69. The BUSCO score showed a completeness of 99.2% (96.4% duplicated) with the poales_odb10 database17.
k-mer matrix generation, redundancy and diversity analyses
k-mer matrix generation
We developed an optimized k-mer matrix workflow to generate a presence/absence k-mer matrix for large diversity panels (Supplementary Note 2) (https://github.com/githubcbrc/KGWASMatrix). We counted k-mers (k = 51) in raw sequencing data for 350 accessions generated in this study, 306 accessions published by Gaurav et al.14, 275 accessions published by Zhou et al.15 and 24 accessions by Zhao et al.6. The 35 accessions with less than fivefold sequencing coverage were discarded to avoid affecting the k-mer count. k-mers with a count of one were discarded prior to generating the k-mer matrix. k-mers were retained in the k-mer matrix by a minimum occurrence of 6 across accessions and a maximum occurrence of (N − 6), where N is the total number of accessions.
Redundancy analysis
A redundancy analysis was performed using a subset of 100,000 random k-mers sampled from the k-mer matrix of 920 Ae. tauschii accessions. The complete matrix contained 10,078,115,665 k-mers. The pairwise comparisons between accessions were performed by computing the sum of the presence–absence values (0 and 1) per k-mer between 2 accessions of the matrix. To determine the divergence, we computed the total number of 1 present in the summed string, each one corresponding to a difference in the presence/absence of the k-mer in the 2 compared accessions. In the sum, the presence of a k-mer in two accessions would result in a 2 and the absence in both accessions in a 0. A threshold of 96% shared k-mers was used to call redundancy based on control lines determined by Gaurav et al.14 to be genetically redundant based on a SNP analysis.
Estimation of the genetic diversity in the 46 Ae. tauschii accessions selected for high-quality genome assemblies
We computed the k-mer accumulation across the 46 Ae. tauschii accessions by analysing their k-mer presence or absence in the k-mer matrix. First, we extracted a k-mer sub-matrix for the 46 accessions and removed k-mers that were absent from all accessions. The k-mer accumulation was computed by counting the number of k-mers present in the first accession, then adding new k-mers from the second accession (that is, not present in the previous accession) and sequentially adding new k-mers until accession 46. This computation was iterated 100 times using randomly shuffled sub-matrices, and the mean and standard deviation were calculated. The mean cumulative k-mer counts were fitted to a logarithmic function (y = a + b × log(x)] using the Python function optimize.curve_fit from SciPy library (v1.8.0)77. The fitted data were plotted using the Python seaborn library (v0.11.2)78 to visualize the k-mer-based accumulation curve. We calculated the k-mer frequency across the full panel of 920 Ae. tauschii accessions in comparison to the genetic diversity in the 46 accessions. The k-mers were divided into two groups: k-mers present and absent in the 46 accessions. We extracted k-mer sub-matrices for each group and computed the occurrence of the k-mers across the 920 accessions (Extended Data Fig. 1c). We plotted the square root transformation of the k-mer frequency using the Python seaborn library (v0.11.2)78.
SNP calling
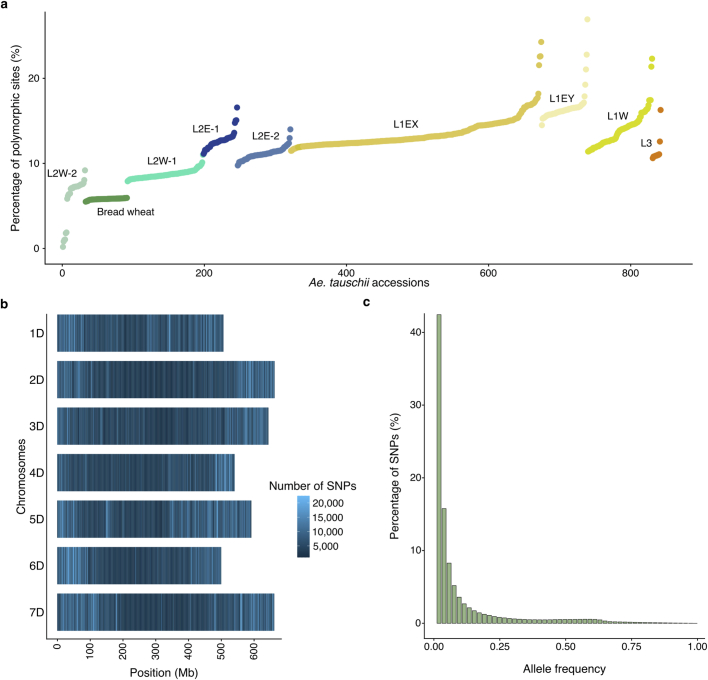
Fastq raw reads were trimmed using Trimmomatic (v0.38)61 with the following settings “ILLUMINACLIP:adapters.fa:2:30:10 LEADING:3 TRAILING:3 SLIDINGWINDOW:4:15 MINLEN:36”. Cleaned reads were mapped on the TA1675 assembly using BWA mem (v0.7.17)79 and sorted with SAMtools (v1.8)63. Variants were called using BCFtools mpileup (v1.9)63 with the setting “-C 60 -q 5 -Q 20”, and only SNPs were retained as variants. The filtering was performed using BCFtools, retaining only sites with a maximum depth of 40, a quality higher than 100 and an allele count higher than 1. For quality check, we counted the percentage of divergent sites using re-sequencing data from TA1675 against the chromosome-scale TA1675 reference assembly, revealing an error rate of 0.18%. For the assessment of assembly quality (Supplementary Table 24), homozygous indels were also retained (maximum number of raw reads supporting an indel (IDV) = 3, maximum fraction of raw reads supporting an indel (IMF) = 0.3, depth between 5 and 40, a quality higher than 30 and an allele count higher than one). We further computed the SNP density across the chromosomes and calculated allele frequency (Extended Data Fig. 8a–c and Supplementary Tables 25–27). In total, 957 Ae. tauschii and 59 wheat landraces accessions reached the quality threshold of a coverage higher than fivefold after trimming and were used for SNP calling. The SNP data set was used for phylogenetic, ancestry, and nucleotide diversity analyses.
Extended Data Fig. 8. SNP data statistics.
a, The percentage of polymorphic sites for each Ae. tauschii accession compared to the TA1675 (L2) reference accession. Each color represents an Ae. tauschii or bread wheat group. b, SNP density in windows of 1 Mb computed across the 7 chromosomes of TA1675. c, Allele frequency distribution.
The phylogenetic tree was built from the filtered SNPs using vcfkit (v0.1.6)80 with the UPGMA algorithm. Ancestry analysis was performed using the sNMF (Fast and Efficient Estimation of Individual Ancestry Coefficients) approach available in the LEA R package (v3.10.2)81. For each run, we performed 20 repetitions using the following parameters “alpha = 10, tolerance = 0.00001, iterations = 300” up to K = 28. Supplementary Table 28 shows the minimum cross-entropy values for 20 sNMF runs across different values of K.
We estimated the nucleotide diversity (π) in the 493 non-redundant accession of Ae. tauschii using the filtered SNP calls against the TA1675 reference assembly. We calculated π over 10-kb windows of the genome using VCFtools (v0.1.16)82 (parameter --window-pi 10000).
Structural variant calling
We determined the structural variation across the 46 high-quality assemblies with reference to the chromosome-scale assembly of TA1675. Structural variants of >50 bp in length and up to 100 kb were called using the PacBio structural variant calling and analysis suite (pbsv) (v2.9.0) and following the pipeline described at (https://github.com/PacificBiosciences/pbsv). In brief, HiFi sequencing reads in bam format were aligned to the reference genome using pbmm2 aligner (v1.10.0) (https://github.com/PacificBiosciences/pbmm2). The bam file was indexed as CSI suitable for larger genomes. Signatures of structural variation were detected and structural variants were called per accession in vcf format, then concatenated into a single bed file per lineage.
The Ae. tauschii genomes facilitate gene discovery
k-mer-based genome-wide association in Ae. tauschii
We followed the k-mer GWAS (kGWAS) pipeline described by Gaurav et al.14 using the Python scripts available at (https://github.com/wheatgenetics/owwc/tree/master/kGWAS) and the phenotype data for stem rust and leaf rust available for this panel to specifically run the association mapping and plotting using default parameters. The association mapping analyses showing the effect of assembly quality in Ae. tauschii accession TA1662 were performed using previously published phenotype data for reaction to Pgt race QTHJC14. The kGWAS for leaf rust to identify Lr39 in Ae. tauschii accession TA1675 was performed using phenotype data for reaction to Pt race BBBDB (Supplementary Table 29).
SrTA1662 haplotype analysis
To identify the SrTA1662 locus in the contig-level assembly of Ae. tauschii accession TA1662, we performed a BLASTn (v2.12.0)67 search of the SrTA1662 gene sequence (GenBank ID MW526949.1) published by Gaurav et al.14. To identify the SR33 locus in the contig-level assembly of Ae. tauschii accession CPI 110799 (the original source of Sr33), we searched for the RGA1e (also known as Sr33) gene sequence (GenBank ID KF031291.1) published by Periyannan et al.24. RGA1e gene sequences were also searched against the contig-level assemblies of accessions AUS 18911 (KF031299.1) and AUS 18913 (KF031284.1)24. For all accessions, the genes were found within a single contig that located to the chromosome arm 1DS based on the scaffolding against the TA1675 reference assembly. In the four accessions, we performed BLASTn searches for additional Ae. tauschii resistance gene analogues (RGA1a-d, RGA2a-b, RGA3a) reported by Periyannan et al.24 (GenBank ID KF031285.1–KF031299.1). To confirm that this region is orthologous to the Mla locus in barley, we searched for the presence of the pumilio (Bpm) gene homologue and subtilisin-chymotrypsin inhibitor (CI) genes in gene-lifting annotations for AUS 18911, AUS 18913, CPI 110799 and TA1662. Bpm and CI genes were previously reported flanking Resistance Gene Homologues (RGH) of the Mla locus83. The gene-lifting annotations were generated using liftoff v1.6.174 with default parameters based on the TA1675 genome annotation.
Phylogenetic analysis of RGAs in Ae. tauschii
To provide further evidence for the homology of the SrTA1662 (SR66) and SR33 loci in Ae. tauschii and the Mla locus in barley, we performed phylogenetic analyses of RGA and RGH gene sequences. Clustal algorithm with default parameters was used for the DNA sequence multiple alignment. We used the unweighted UPGMA algorithm with bootstrap testing to support the tree topology with 5,000 replicates. The phylogenetic analyses were performed using MEGA (v11)84,85.
Leaf rust inoculations and association studies
The evaluation of resistance and susceptibility in 149 Ae. tauschii L2 accessions was conducted against the North American Pt race isolate BBBDB 1-186 using seedlings organized in cone racks. Every cone rack housed 98 cones, and each cone was sown with three seeds. The primary leaves of seedlings, aged 8–9 days, were subjected to inoculation by distributing 1 ml of inoculum per cone rack, which consisted of 15 mg of spores in 1 ml of Soltrol 170 (Chevron Phillips Chemical Company). The delivery to each plant was 0.05 mg of urediniospores. Post-inoculation, the phytotoxicity from the oil carrier, Soltrol 170, was mitigated by mildly fanning the leaves for 2 h under the illumination of 400-Watt HPS bulbs to expedite the evaporation of the carrier oil. The seedlings, once inoculated, were placed in mist chambers maintained at 22 °C, where 100% humidity was sustained using a domestic ultrasonic humidifier for a period of 16–18 h in the absence of light. Subsequently, the seedlings were transferred to a greenhouse with a 16-h day cycle, maintaining nocturnal and diurnal temperatures at 15 °C and 20 °C, respectively. The phenotypic assessment of disease was undertaken at 10 and 12 dpi using an infection type scoring range of 0 to 3 + , as standardized by Long and Kolmer87, and depicted as the mean of three individual replicates per accession (Supplementary Table 29).
For use in GWAS, the qualitative scores were converted to a quantitative score by assigning numerical values to the infection types. This was achieved by the kGWAS pipeline (https://github.com/wheatgenetics/owwc/tree/master/kGWAS) that performs Stakman IT to numeric scale 1 conversion (RunAssociation_GLM.py with -st parameter).
Bi-parental mapping of LR39 and candidate gene identification
An Ae. tauschii bi-parental mapping population (n = 123) was generated by crossing the leaf rust resistant Ae. tauschii accession CPI 110672 (synonymous TA1675) with the leaf rust susceptible accession CPI 110717. The mapping population was segregating for a single dominant leaf rust resistance gene (P = 0.606) when inoculated with the Australian Pt isolate 26-1,3 (PBI culture no 316) and phenotyped at the Plant Breeding Institute, Cobbitty30. Bulk segregant analysis of selected homozygous resistant and susceptible F2 progenies with the 90 K SNP array88 placed the leaf rust resistance locus to chromosome 2DS. The mapping population was further genotyped with markers derived from the 90 K iSelect SNP array, the TA1675 genomic sequence, and a marker closely linked to LR39 (Xgdm35) (Supplementary Table 30 and 31)28,89. Linkage analysis was performed using MapDisto (v2.0)90 with default parameters such as LOD (logarithm of the odds) threshold of 3.0, maximum recombination frequency of 0.3 and removal of loci with 10% missing data. Genetic distances were calculated using the Kosambi mapping function, and the map was created using MapChart (v2.32)91. Markers flanking LR39 were anchored to the TA1675 reference assembly. Annotated high-confidence genes at the delimited physical interval were screened for protein homology using BLASTp to identify diversity between TA1675 and AL8/78 (Supplementary Table 13). The conserved domains and critical residues of WTK and NLR were identified using the amino acid sequences in the NCBI Conserved Domain search (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) and InterPro (https://www.ebi.ac.uk/interpro/search/sequence/) databases. The polymorphic SNP corresponding to R457I in WTK was converted to a KASP marker diagnostic for Lr39 (Supplementary Table 31).
Virus-induced gene silencing
To develop candidate gene-specific VIGS probes, the predicted coding sequences of candidate genes were searched against the TA1675 transcriptome database using siRNA-Finder (siFi21) software (v1.2.3-0008)92. Based on the RNA interference (RNAi) design plot, regions predicted to have a higher number of efficient probes and fewer off-targets were used for designing silencing probes for the WTK (LrSi2:258 bp, LrSi6:254 bp and LrSi7:248 bp) and the NLR (LrSi3:234 bp and LrSi8:257 bp) candidate genes. The silencing probe sequences were verified for specificity using a BLAST search against the TA1675 reference assembly (<80% sequence identity for hits other than the target candidate). Designed probes were flanked by XbaI and ApaI and synthesized at GenScript Biotech followed by cloning into the BSMV-γ vector in an antisense direction. The resulting constructs were transformed into Agrobacterium tumefaciens strain GV3101. The Agrobacterium clones were grown overnight at 28 °C in lysogeny broth with appropriate antibiotics. Cells were collected by centrifugation at 3,500g for 10 min, then re-suspended using infiltration buffer (10 mM MgCl2, 10 mM MES pH 6.5 with KOH buffer and 150 mM acetosyringone) and adjusted to an OD600 of 1.0 followed by incubation at 28 °C for 3 h. Equal volumes of BSMV-α and BSMV-β were mixed with respective BSMV-γ silencing probes or BSMV-γGFP and infiltrated into Nicotiana benthamiana leaves. Infiltrated leaves were collected 5 days after infiltration and homogenized with virus inoculation buffer (10 mM monopotassium phosphate containing 1% Celite (Thermo Fisher Scientific, 68855-54-9)). The homogenate containing viral particles was rub inoculated onto five to ten seedlings of TA1675 at the three leaf stage. After two weeks of recovery and viral symptoms appearing, the seedlings were inoculated with Pt isolate B9414. Prior to inoculating TA1675, Pt isolate B9414 was propagated on seedlings of the susceptible wheat cultivar Thatcher. Freshly collected urediniospores were suspended in Isopar L and sprayed onto plants using a high-pressure air sprayer. After inoculation, plants were placed in the dark overnight in an incubation box equipped with a humidifier and then transferred to a growth chamber with a 16/8 h day/night cycle, with 21 °C/18 °C growth conditions. Leaf rust phenotypes were recorded at 12 days after inoculation by scanning the leaves at 600 dots per inch on an Epson Perfection V850 Pro scanner. For leaf rust biomass quantification, DNA was extracted from Pt-inoculated leaves using the CTAB method47. DNA concentrations were measured using a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific). A 20 µl qPCR reaction containing Power SYBR Green PCR Mix (Applied Biosystems 4367659), ~25 ng of DNA, and primers specific to the Puccinia 28 S large subunit or the internal transcribed spacer region93 and Triticeae elongation factor-specific primers94 was run using the ABI QuantStudio 6 Flex Real-Time PCR machine. The 2−ΔΔCT method was used to normalize rust gene amplification values relative to the Ae. tauschii elongation factor endogenous control.
PCR conditions
A 20 μl PCR containing 100 ng of genomic DNA, 1X GoTaq Flexi green buffer, 1.5 mM MgCl2, 200 μM dNTP, 200 nM primers and 1 unit of Taq polymerase (M829B, Promega) was used for various fragment amplifications. Primer sequences are shown in Supplementary Table 31. A touchdown PCR protocol was used as follows: initial denaturation at 94 °C for 30 s; annealing at 65 °C for 30 s, decreasing by 1 °C per cycle; and extension at 72 °C for 60 s, followed by repeating these steps for 14 cycles. After enrichment, the program continued for 29 cycles as follows: 94 °C for 30 s, 58 °C for 30 s, and 72 °C for 60 s. PCR products of cleaved amplified polymorphic sequence (CAPS) markers were digested with appropriate restriction enzymes by following the manufacturer’s instructions. A 5 μl reaction (2.5 μl of KASP Master Mix (Low ROX KBS-1016-016), 0.07 μl of assay mix and 2.5 μl (25 ng) of DNA) was used for KASP markers. PCR cycling was performed in an ABI QuantStudio 6 Flex Real-Time PCR machine as follows: preread at 30 °C for 60 s; hold stage at 94 °C for 15 min; and then ten touchdown cycles (94 °C for 20 s; touchdown at 61 °C, decreasing by 0.6 °C per cycle for 60 s), followed by 29 additional cycles (94 °C for 20 s; 55 °C for 60 s). The plates were then read at 30 °C for endpoint fluorescent measurement.
Tracing lineage-specific Ae. tauschii haplotype blocks in the bread wheat genome
Missing link finder pipeline
We generated canonical 51-mers for each of the 82,293 genotyped wheat accessions from Sansaloni et al.35 using their respective DArTseq markers and Jellyfish (v 2.3.0)95. For each accession, the k-mers were sorted and stored as text files. From the k-mer matrix available from Gaurav et al.14, k-mers present only in Ae. tauschii L3 were extracted, sorted, and stored as text files. Pairwise comparisons of the sample-specific k-mers from the 82,293 wheat accessions and the L3-specific k-mers were performed using the comm bash command. Jaccard indices were computed with the following formula, where A is the set of k-mers from a single accession and L is the L3-specific k-mer set.
The script is available on Github (https://github.com/emilecg/wheat_evolution).
Determining the extent of L3 in wheat lines using whole-genome re-sequencing data of 59 hexaploid wheat landraces
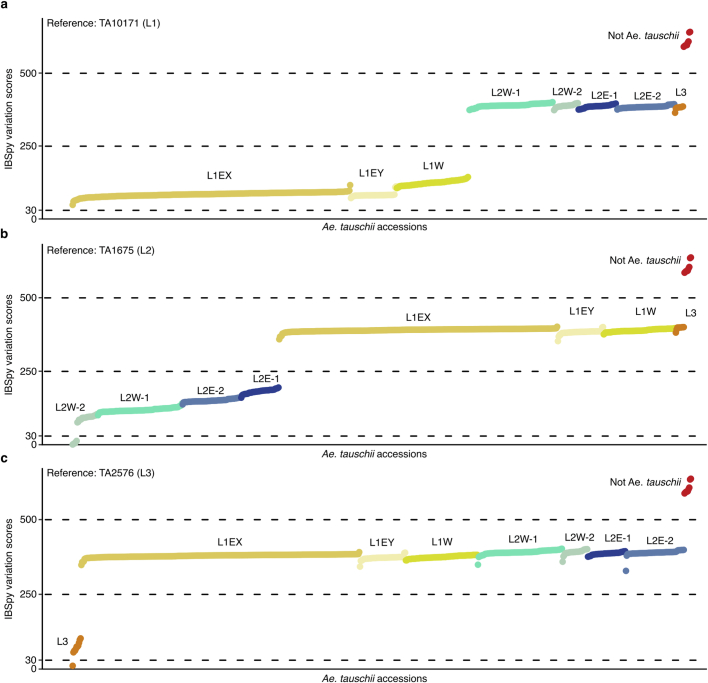
Raw reads were trimmed using Trimmomatic (v0.38)61 with the following settings “ILLUMINACLIP:adapters.fa:2:30:10 LEADING:3 TRAILING:3 SLIDINGWINDOW:4:15 MINLEN:36”. KMC (v3.1.2)96 was used to generate 31-mer sets for the 59 resequenced wheat landraces (Supplementary Table 14). IBSpy (v0.4.6)13 was run with TA2576 as a reference and the bread wheat landraces as queries with a k-mer size of 31 and a window size of 50,000 bp as parameters. A variation score threshold of ≤150 was used to determine how many windows were in common between the L3 reference and the wheat landraces. IBSpy variation values of ≤150 were determined to be optimal to account for the relatively low intra-lineage variation present in L3 (Extended Data Fig. 9 and Supplementary Table 32). The percentage of matching 50-kb windows was used as a proxy to determine the extent of introgression in the landraces.
Extended Data Fig. 9. IBSpy variation score distribution.
Shown are the average variation scores for each Ae. tauschii accession (represented as a dot) against TA10171 (L1) (a), TA1675 (L2) (b), and TA2576 (L3) (c) (Supplementary Table 32). Based on the distribution, we defined IBSpy values ≤ 30 as identical by state, values > 30 ≤ 250 as being the same Ae. tauschii lineage as the reference, values > 250 ≤ 500 as being a different Ae. tauschii lineage, and values > 500 as not being Ae. tauschii.
Differences in genes in the 135-Mb L3 introgression block on chromosome 1D
The protein sequences of genes annotated in the interval of the introgression on the Kariega genome and on the CWI 86942 genome were compared using DIAMOND and visualized with the Persephone genome browser. In case genes were annotated in both the genomes, their amino acid sequences were aligned using the Needleman–Wunsch algorithm to determine the percentage of identity. The absence of genes in one of the two annotations was investigated manually with the BLAST algorithm integrated into Persephone. Annotated genes found to be part of transposable elements were excluded from the analysis.
Presence of the 135-Mb L3 haplotype block on chromosome 1D in wheat landraces
The presence of the 135-Mb L3 haplotype block was manually confirmed in 12 out of the 126 wheat landraces (Supplementary Table 16). CWI 86942 and another Georgian landrace (CWI 86929) had the largest block size (Extended Data Fig. 6d).
To further determine how widespread the presence of the chromosome 1D L3 segment was, we downloaded the IBSpy variation file from 1,035 hexaploid wheat accessions (827 landraces and 208 modern cultivars) and L3 line BW_01028 (https://opendata.earlham.ac.uk/wheat/under_license/toronto/WatSeq_2023-09-15_landrace_modern_Variation_Data/IBSpy_variations_10WheatGenomes/) against the Chinese Spring RefSeq v1.0 assembly38. We found an additional 20 wheat accessions that carry at least parts of this segment. We defined the start and end of the L3 segments in these 20 accessions by determining the difference between the variation value of BW_01028 (L3) and the corresponding variation value of the twenty accessions. If the difference was ≤150, we defined the accession to carry the L3 segment.
Bread wheat D genome subpopulations contribution
The approach used for a quantitative estimation of the contributions of the different subpopulations to the D genomes and the estimation of technical artifacts are described in Supplementary Note 3 and Supplementary Table 33. The manual curation process that allowed counting the minimal number of hybridizations required to explain the presence of different haplotypes is described in Supplementary Note 4.
Data visualization
We used the R package karyoploteR (v1.20.3)97 for the haplotype representation of the chromosomes in Figs. 3d,e and 4b and Extended Data Fig. 7. The remaining plots were produced with ggplot2 (v3.4.2)98 and the Python seaborn library (v0.11.2)78. Maps in Figs. 1a, 3c and 4a and Extended Data Fig. 6b were generated using QGIS (v3.32.3).
Germplasm availability
All the 60 wheat landraces analysed in this study listed in Supplementary Table 14 are available upon request from the CIMMYT (https://www.cimmyt.org/) and ICARDA (https://www.icarda.org/) gene banks. Seed of accessions from the Open Wild Wheat Consortium Ae. tauschii Diversity Panel collection, Cereal Crop Wild Relatives (Triticeae) collection, DFW Wheat Academic Toolkit collection and Deposited Published Research Material collection can be obtained from the Germplasm Resource Unit (GRU) of the John Innes Center; seed from accessions with WGRC bank ID as the only primary ID (Supplementary Table 1) can be obtained from the Wheat Genetics Resource Center (WGRC) of Kansas State University; 34 accessions can be obtained from the Plant Gene Resources of Canada (PGRC); 84 accessions donated by the Institute of Botany, Plant Physiology and Genetics of the Tajikistan National Academy of Sciences were deposited in the Wheat Genetics Resource Center (WGRC) as were 37 accessions donated by Quaid-i-Azam University; 20 accessions donated by the Azerbaijan National Academy of Sciences can be made available upon request.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Online content
Any methods, additional references, Nature Portfolio reporting summaries, source data, extended data, supplementary information, acknowledgements, peer review information; details of author contributions and competing interests; and statements of data and code availability are available at 10.1038/s41586-024-07808-z.
Supplementary information
This file contains Supplementary Notes 1–4.
Supplementary Tables 1–33.
Acknowledgements
This research used the Shaheen supercomputer and the Ibex cluster managed by the Supercomputing Core Laboratory at King Abdullah University of Science and Technology (KAUST). We thank the systems administrators and computational scientists for help with debugging and overall support. The authors thank B. Steuernagel, D. Keyes, L. Fabbian and K. G. Kise for bioinformatics advice; E. P. Faig for greenhouse assistance; I. Walde for technical assistance with Hi-C library preparation and sequencing; H. Guo for administering the OWWC sequencing; J. Poland and L. Gao for advice on PacBio sequencing; A. Bentley, B. Keller, H. Bürstmayr, J. Faris, M. Maccaferri, M. Bozzoli, R. Horsnell, D. Seung, J. Balk, R. McNelly and T. O’Hara for nominating Ae. tauschii lines of strategic interest; R. McIntosh for critical reading of the manuscript; E. Waller for assistance with the OWWC website; GrainGenes resources for hosting the online database; USDA for infrastructure support in terms of providing computational resources at SCINet high performance cluster; and University of Maryland supercomputing resources (http://hpcc.umd.edu) for computational work for the database. This publication is based on work supported by KAUST awards ORFS-CRG10-2021-4735 to B.B.H.W., ORFS-CRG11-2022-5076 to S.G.K. and URF/1/4352-01-01, FCC/1/1976-44-01, FCC/1/1976-45-01, REI/1/5234-01-01 and REI/1/5414-01-01 to X.G.; Australian Government Research Training Program and the University of Queensland Centennial Scholarships to N.A. during genetic mapping of Lr39; Academy of Scientific Research and Technology (ASRT) project ID 19385 and Climate Change Adaptation and Nature Conservation (GREEN FUND-ASRT) to A.F.E.; National Major Agricultural Science and Technology project NK2022060101 and National Key Research and Development Program of China 2021YFF1000204 to L.M.; a Genebank3.0 project from the German Federal Ministry of Education and Research (grant no. FKZ 031B1300A) to J.C.R.; grants from the Next‐Generation BioGreen 21 Linked Program (PJ015786) of the Rural Development Administration (RDA), South Korea to J.-Y.L.; RDA/USDA-ARS cooperation agreement no. 58-0210-9-226-F to J.-Y.L. and Y.Q.G.; ARS project 2030-21430-015-000 to S.S.X. and Y.Q.G.; the UK Biotechnology and Biological Sciences Research Council (BBSRC) Institute Strategic Programme Designing Future Wheat (BB/P016855/1) and a European Research Council grant (ERC-2019-COG-866328) to C.U.; the Mexican Consejo Nacional de Ciencia y Tecnología (CONACYT; 2018-000009-01EXTF-00306) to J.Q.-C.; funding from Department of Biotechnology, Government of India to P.C. and S.K.; USDA-NIFA Capacity Fund via South Dakota Agricultural Experiment Station to W.L.; a Bayer (previously Monsanto) Beachell Borlaug International Scholar’s Program to S. Ghosh; National Science Foundation of United States grant number 2102953 to J.D.; ARS Project 2030-21000-056-00D to G.R.L.; an USDA-NIFA Grant (2022-67013-36362) to V.K.T.; a NERC Independent Research Fellowship (NE/T011025/1) to L.T.D.; and 4D Wheat: Diversity, Domestication, Discovery and Delivery project (C.J.P.), funded by Genome Canada, Agriculture and Agri-Food Canada, Western Grains Research Foundation, Saskatchewan Ministry of Agriculture, Saskatchewan Wheat Development Commission, Alberta Wheat Commission, Manitoba Crop Alliance, Ontario Research Fund, and the Canadian Agricultural Partnership.
Extended data figures and tables
Author contributions
A.G.-M., C.G., E.C.-G., S.M.R., A.R., N.C., A.B.P., C.-Y.H., K.D.L., W.J.R., F.Y.N., S. Grewal, G.S.B. and B.B.H.W. configured Ae. tauschii diversity panel and distributed seed. A.G.-M., A.P., A.B.P. and C.-Y.H. extracted Ae. tauschii DNA. S.H., M. Abbasi, M.L., N.K. and G.S.B. sampled tissue and extracted RNA from Ae. tauschii. A.G.-M., S.H., A.F.E., L.M., A. Sharon, A.H., J.C.R., M.K., M.M., N.S., D.P., S.L., J.-Y.L., P.C., S.K., P.Z., R.F.P., D.-C.L., W.L., J.D., V.K.T., S. Grewal, C.J.P., H.S.C., J.E., L.T.D., S.S.X., Y.Q.G., X.X., G.S.B. and B.B.H.W. outlined Ae. tauschii sequencing strategy and acquired DNA and RNA sequences. E.C.-G. and N.A. extracted DNA for bread wheat landraces. A.G.-M., E.C.-G., S.H., L.F.R., H.S.C. and M. Abrouk undertook sequence data quality control, curation, back-up and/or distribution. A. Salhi, X.G. and S.B. assembled k-mer matrix. E.C.-G. performed variant calling and filtering. A.G.-M. and E.C.-G. performed redundancy analysis. E.C.-G. and A.G.-M. performed genome-wide phylogenetic analysis and E.C.-G. performed structure analysis. A.G.-M. and H.S.C. assembled Ae. tauschii genomes and A.G.-M. assessed their quality. S.H. and M. Abrouk performed genome annotations. A.G.-M. performed structural variant analysis. A.G.-M. performed k-mer GWAS. N.A., M.P., S. Ghosh, R.C., Y.W., E.L.O., P.Z., R.F.P., W.B.M., W.H.P.B., B.J.S., J.A.K. and O.M. performed rust phenotyping. A.G.-M. and E.C.-G. performed Sr33 and Sr66 comparative genomics analyses. N.A., E.S.L., Y.D. and S.K.P. generated bi-parental population and mapped Lr39. N.A. and Y.W. identified Lr39 candidate gene, performed VIGS and annotated Lr39. E.C.-G. developed the ‘Missing Link Finder’ pipeline, identified L3-enriched bread wheat lines, and assembled CWI 86942 genome. C.U. and J.Q.-C. provided prepublication access to and helped implement IBSpy pipeline, and E.C.-G., C.U. and J.Q.-C. identified L3 haploblocks in wheat and calculated their accumulative size. E.C.-G. assigned extant wheat genome segments to ancestral Ae. tauschii populations and determined the minimum number of hybridizations. G.Y. and H.I.A. provided scientific advice. P.K.S., G.R.L. and V.K.T. implemented the Ae. tauschii genome database. E.C.-G., A.G.-M., N.A., S.H., A. Salhi, M. Abrouk, S.B., G.S.B., B.B.H.W. and S.G.K. conceived and designed experiments. E.C.-G., A.G.-M., N.A., C.G., B.B.H.W. and S.G.K. designed figures. S.G.K., E.C.-G., A.G.-M. and B.B.H.W. drafted first version of the manuscript with inputs and edits from N.A., S.H., A. Salhi, C.G., G.S.B. and S. Ghosh. A.G.-M. and B.B.H.W. managed the Open Wild Wheat Consortium (www.openwildwheat.org). All authors read, commented on and approved the final version of the manuscript. Authors are grouped by institution acronym and then by first name in the author list and author contribution statement, except for the first seven and last six authors in the list.
Peer review
Peer review information
Nature thanks Anthony Hall, John Lovell and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer review reports are available.
Data availability
The sequencing data and genome assemblies generated in this study were submitted to NCBI under BioProject number PRJNA956839, including the raw Illumina reads for 350 Ae. tauschii accessions, the raw PacBio reads, the Hi-C data, the raw RNA-seq reads from 5 tissues of Ae. tauschii accessions TA10171, TA1675 and TA2576, the raw Illumina reads for 59 wheat landraces and the raw PacBio reads and Omni-C data of the wheat landrace CWI 86942. The genome assemblies of the 46 Ae. tauschii accessions, the assemblies and annotations for CWI 86942, TA10171, TA1675 and TA2576, the variant call (SNP) file, the k-mer matrix for 920 Ae. tauschii accessions, the phylogenetic tree for 493 non-redundant Ae. tauschii accessions, the structural variant call (SV) files, the IBSpy variation tables, the predictions of the subpopulations contributing to the 17 hexaploid wheat assemblies, an excel file containing the RagTag scaffold output agp files and the dot-plots produced by MashMap used to validate the RagTag scaffolding are available at Dryad (10.5061/dryad.vmcvdnd0d (ref. 99); 10.5061/dryad.wm37pvmvd (ref. 100); 10.5061/dryad.wpzgmsbvm (ref. 101); 10.5061/dryad.p5hqbzkvx (ref. 102)). The Lr39 genomic sequence was deposited in NCBI Genbank under accession number OR567850. The TA10171 (L1), TA1675 (L2) and TA2576 (L3) genomes are available for online BLAST, Jbrowse visualization and synteny analysis with the currently available Triticinae genomes at (https://wheat.pw.usda.gov/GG3pangenome/wheat/D/taus_home.php). The Ensembl nrTEplants repetitive element database (June 2020) was used for repeat content prediction. Viridiplantae protein models from OrthoDB v.11 were used to predict de novo gene models for the annotated Ae. tauschii genomes. The predicted translated proteins were annotated using the following databases: FunFam, SFLD, PANTHER, Gene3D, PRINTS, Coils, SUPERFAMILY, SMART, CDD, PIRSR, ProSitePatterns, AntiFam, Pfam, MobiDBLite, PIRSF and NCBIfam. We downloaded sequencing data for 306 accessions from NCBI BioProject number PRJNA685125, 275 accessions from NCBI BioProject number PRJNA705859, and 24 accessions from the China National Center for Bioinformation–National Genomics Data Center under accession number PRJCA005979.
Code availability
The k-mer matrix generation pipeline for large diversity panels is available at GitHub (https://github.com/githubcbrc/KGWASMatrix). The custom script for estimating the cumulative k-mer content is available at GitHub (https://github.com/andreagonzam/tauschii_pangenome). The custom scripts for Missing Link Finder pipeline and haplotype analysis are available at GitHub (https://github.com/emilecg/wheat_evolution).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Emile Cavalet-Giorsa, Andrea González-Muñoz, Naveenkumar Athiyannan
Contributor Information
Brande B. H. Wulff, Email: brande.wulff@kaust.edu.sa
Simon G. Krattinger, Email: simon.krattinger@kaust.edu.sa
Extended data
is available for this paper at 10.1038/s41586-024-07808-z.
Supplementary information
The online version contains supplementary material available at 10.1038/s41586-024-07808-z.
References
- 1.Dubcovsky, J. & Dvorak, J. Genome plasticity a key factor in the success of polyploid wheat under domestication. Science316, 1862–1866 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhou, Y. et al. Triticum population sequencing provides insights into wheat adaptation. Nat. Genet.52, 1412–1422 (2020). [DOI] [PubMed] [Google Scholar]
- 3.Tadesse, W. et al. Genetic gains in wheat breeding and its role in feeding the world. Crop Breed. Genet. Genom.1, e190005 (2019). [Google Scholar]
- 4.Marcussen, T. et al. Ancient hybridizations among the ancestral genomes of bread wheat. Science345, 1250092 (2014). [DOI] [PubMed] [Google Scholar]
- 5.Wang, J. et al. Aegilops tauschii single nucleotide polymorphisms shed light on the origins of wheat D‐genome genetic diversity and pinpoint the geographic origin of hexaploid wheat. New Phytol.198, 925–937 (2013). [DOI] [PubMed] [Google Scholar]
- 6.Zhao, X. et al. Population genomics unravels the Holocene history of bread wheat and its relatives. Nat. Plants9, 403–419 (2023). [DOI] [PubMed] [Google Scholar]
- 7.Gaut, B. S., Seymour, D. K., Liu, Q. & Zhou, Y. Demography and its effects on genomic variation in crop domestication. Nat. Plants4, 512–520 (2018). [DOI] [PubMed] [Google Scholar]
- 8.Leal‐Bertioli, S. C. et al. Segmental allopolyploidy in action: increasing diversity through polyploid hybridization and homoeologous recombination. Am. J. Bot.105, 1053–1066 (2018). [DOI] [PubMed] [Google Scholar]
- 9.Cheng, H. et al. Frequent intra- and inter-species introgression shapes the landscape of genetic variation in bread wheat. Genome Biol.20, 136 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.He, F. et al. Exome sequencing highlights the role of wild-relative introgression in shaping the adaptive landscape of the wheat genome. Nat. Genet.51, 1194–1194 (2019). [DOI] [PubMed] [Google Scholar]
- 11.Walkowiak, S. et al. Multiple wheat genomes reveal global variation in modern breeding. Nature588, 277–283 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Keilwagen, J. et al. Detecting major introgressions in wheat and their putative origins using coverage analysis. Sci. Rep.12, 1908 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ahmed, H. I. et al. Einkorn genomics sheds light on history of the oldest domesticated wheat. Nature620, 830–838 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gaurav, K. et al. Population genomic analysis of Aegilops tauschii identifies targets for bread wheat improvement. Nat. Biotechnol.40, 422–431 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou, Y. et al. Introgressing the Aegilops tauschii genome into wheat as a basis for cereal improvement. Nat. Plants7, 774–786 (2021). [DOI] [PubMed] [Google Scholar]
- 16.Wenger, A. M. et al. Accurate circular consensus long-read sequencing improves variant detection and assembly of a human genome. Nat. Biotechnol.37, 1155–1162 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Manni, M., Berkeley, M. R., Seppey, M., Simão, F. A. & Zdobnov, E. M. BUSCO update: novel and streamlined workflows along with broader and deeper phylogenetic coverage for scoring of eukaryotic, prokaryotic, and viral genomes. Mol. Biol. Evol.38, 4647–4654 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lieberman-Aiden, E. et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science326, 289–293 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang, L. et al. Aegilops tauschii genome assembly Aet v5.0 features greater sequence contiguity and improved annotation. G311, jkab325 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhu, T. et al. Optical maps refine the bread wheat Triticum aestivum cv. Chinese Spring genome assembly. Plant J.107, 303–314 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mizuno, N., Yamasaki, M., Matsuoka, Y., Kawahara, T. & Takumi, S. Population structure of wild wheat D‐genome progenitor Aegilops tauschii Coss.: implications for intraspecific lineage diversification and evolution of common wheat. Mol. Ecol.19, 999–1013 (2010). [DOI] [PubMed] [Google Scholar]
- 22.Savary, S. et al. The global burden of pathogens and pests on major food crops. Nat. Ecol. Evol.3, 430–439 (2019). [DOI] [PubMed] [Google Scholar]
- 23.Olson, E. L. et al. Simultaneous transfer, introgression, and genomic localization of genes for resistance to stem rust race TTKSK (Ug99) from Aegilops tauschii to wheat. Theor. Appl. Genet.126, 1179–1188 (2013). [DOI] [PubMed] [Google Scholar]
- 24.Periyannan, S. et al. The gene Sr33, an ortholog of barley Mla genes, encodes resistance to wheat stem rust race Ug99. Science341, 786–788 (2013). [DOI] [PubMed] [Google Scholar]
- 25.Arora, S. et al. Resistance gene cloning from a wild crop relative by sequence capture and association genetics. Nat. Biotechnol.37, 139–143 (2019). [DOI] [PubMed] [Google Scholar]
- 26.Boden, S. et al. Updated guidelines for gene nomenclature in wheat. Theor. Appl. Genet.136, 72 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Raupp, W., Brown-Guedira, G. & Gill, B. Cytogenetic and molecular mapping of the leaf rust resistance gene Lr39 in wheat. Theor. Appl. Genet.102, 347–352 (2001). [Google Scholar]
- 28.Singh, S. et al. Lr41, Lr39, and a leaf rust resistance gene from Aegilops cylindrica may be allelic and are located on wheat chromosome 2DS. Theor. Appl. Genet.108, 586–591 (2004). [DOI] [PubMed] [Google Scholar]
- 29.Kolmer, J. A. & Fajolu, O. Virulence phenotypes of the wheat leaf rust pathogen, Puccinia triticina, in the United States from 2018 to 2020. Plant Dis.106, 1723–1729 (2022). [DOI] [PubMed] [Google Scholar]
- 30.Athiyannan, N. Molecular Genetic Characterisation of Triple Rust Resistance in Aegilops tauschii. PhD thesis, Univ. of Queensland (2018).
- 31.Wang, Y. et al. An unusual tandem kinase fusion protein confers leaf rust resistance in wheat. Nat. Genet.55, 914–920 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yu, G. et al. The wheat stem rust resistance gene Sr43 encodes an unusual protein kinase. Nat. Genet.55, 921–926 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Klymiuk, V., Coaker, G., Fahima, T. & Pozniak, C. J. Tandem protein kinases emerge as new regulators of plant immunity. Mol. Plant Microbe Interact.34, 1094–1102 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Athiyannan, N. et al. Long-read genome sequencing of bread wheat facilitates disease resistance gene cloning. Nat. Genet.54, 227–231 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sansaloni, C. et al. Diversity analysis of 80,000 wheat accessions reveals consequences and opportunities of selection footprints. Nat. Commun.11, 4572 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Beridze, R. K., Hanelt, P., Kandelaki, V. N., Sakvarelidze, I. M. & Schultze-Motel, J. A further mission to the Georgian SSR 1984 for collecting indigenous material of cultivated plants. Kulturpflanze33, 199–212 (1985). [Google Scholar]
- 37.Rosyara, U. et al. Genetic contribution of synthetic hexaploid wheat to CIMMYT’s spring bread wheat breeding germplasm. Sci. Rep.9, 12355 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cheng, S. et al. Harnessing landrace diversity empowers wheat breeding. Nature10.1038/s41586-024-07682-9 (2024). [DOI] [PMC free article] [PubMed]
- 39.Delorean, E. et al. High molecular weight glutenin gene diversity in Aegilops tauschii demonstrates unique origin of superior wheat quality. Commun. Biol.4, 1242 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sato, K. et al. Chromosome-scale genome assembly of the transformation-amenable common wheat cultivar ‘Fielder’. DNA Res.28, dsab008 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kale, S. M. et al. A catalogue of resistance gene homologs and a chromosome-scale reference sequence support resistance gene mapping in winter wheat. Plant Biotechnol. J.20, 1730–1742 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang, Z. Y. et al. Helitron and CACTA DNA transposons actively reshape the common wheat-AK58 genome. Genomics114, 110288 (2022). [DOI] [PubMed] [Google Scholar]
- 43.Guo, W. et al. Origin and adaptation to high altitude of Tibetan semi-wild wheat. Nat. Commun.11, 5085 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gonçalves-Dias, J., Singh, A., Graf, C. & Stetter, M. G. Genetic incompatibilities and evolutionary rescue by wild relatives shaped grain amaranth domestication. Mol. Biol. Evol.40, msad177 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Weide, A., Riehl, S., Zeidi, M. & Conard, N. J. A systematic review of wild grass exploitation in relation to emerging cereal cultivation throughout the Epipalaeolithic and aceramic Neolithic of the Fertile Crescent. PLoS ONE13, e0189811 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Singh, N. et al. Genomic analysis confirms population structure and identifies inter-lineage hybrids in Aegilops tauschii. Front. Plant Sci.10, 9 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Abrouk, M. et al. Fonio millet genome unlocks African orphan crop diversity for agriculture in a changing climate. Nat. Commun.11, 4488 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thachuk, C. et al. Core Hunter: an algorithm for sampling genetic resources based on multiple genetic measures. BMC Bioinformatics10, 243 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Driguez, P. et al. LeafGo: Leaf to Genome, a quick workflow to produce high-quality de novo plant genomes using long-read sequencing technology. Genome Biol.22, 256 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cheng, H. Y., Concepcion, G. T., Feng, X. W., Zhang, H. W. & Li, H. Haplotype-resolved de novo assembly using phased assembly graphs with hifiasm. Nat. Methods18, 170–175 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gurevich, A., Saveliev, V., Vyahhi, N. & Tesler, G. QUAST: quality assessment tool for genome assemblies. Bioinformatics29, 1072–1075 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rhie, A., Walenz, B. P., Koren, S. & Phillippy, A. M. Merqury: reference-free quality, completeness, and phasing assessment for genome assemblies. Genome Biol.21, 245 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bankevich, A., Bzikadze, A. V., Kolmogorov, M., Antipov, D. & Pevzner, P. A. Multiplex de Bruijn graphs enable genome assembly from long, high-fidelity reads. Nat. Biotechnol.40, 1075–1081 (2022). [DOI] [PubMed] [Google Scholar]
- 54.Padmarasu, S., Himmelbach, A., Mascher, M. & Stein, N. in Plant Long Non-Coding RNAs: Methods and Protocols, Vol. 1933 (eds Chekanova, J. A. & Wang, H.-L. V.) 441–472 (Springer, 2019).
- 55.Himmelbach, A., Walde, I., Mascher, M. & Stein, N. Tethered chromosome conformation capture sequencing in Triticeae: a valuable tool for genome assembly. Bio Protoc.8, e2955 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Durand, N. C. et al. Juicer provides a one-click system for analyzing loop-resolution Hi-C experiments. Cell Syst.3, 95–98 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dudchenko, O. et al. De novo assembly of the Aedes aegypti genome using Hi-C yields chromosome-length scaffolds. Science356, 92–95 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Durand, N. C. et al. Juicebox provides a visualization system for Hi-C contact maps with unlimited zoom. Cell Syst.3, 99–101 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Alonge, M. et al. Automated assembly scaffolding using RagTag elevates a new tomato system for high-throughput genome editing. Genome Biol.23, 258 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jain, C., Koren, S., Dilthey, A., Phillippy, A. M. & Aluru, S. A fast adaptive algorithm for computing whole-genome homology maps. Bioinformatics34, i748–i756 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics30, 2114–2120 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Danecek, P. et al. Twelve years of SAMtools and BCFtools. Gigascience10, giab008 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Brůna, T., Hoff, K. J., Lomsadze, A., Stanke, M. & Borodovsky, M. BRAKER2: automatic eukaryotic genome annotation with GeneMark-EP+ and AUGUSTUS supported by a protein database. NAR Genom. Bioinformatics3, lqaa108 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hoff, K. J., Lange, S., Lomsadze, A., Borodovsky, M. & Stanke, M. BRAKER1: unsupervised RNA-Seq-based genome annotation with GeneMark-ET and AUGUSTUS. Bioinformatics32, 767–769 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hoff, K. J., Lomsadze, A., Borodovsky, M. & Stanke, M. in Gene Prediction: Methods and Protocols, Vol. 1962 (ed. Kollmar, M.) 65–95 (Springer, 2019).
- 67.Camacho, C. et al. BLAST+: architecture and applications. BMC Bioinformatics10, 421 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Blum, M. et al. The InterPro protein families and domains database: 20 years on. Nucleic Acids Res.49, D344–D354 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jones, P. et al. InterProScan 5: genome-scale protein function classification. Bioinformatics30, 1236–1240 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Emms, D. M. & Kelly, S. OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biol.20, 238 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Smit, A., Hubley, R. & Green, P. RepeatMasker Open-4.0 http://www.repeatmasker.org (2015).
- 72.Contreras‐Moreira, B. et al. K‐mer counting and curated libraries drive efficient annotation of repeats in plant genomes. Plant Genome14, e20143 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Abrouk, M. et al. Chromosome-scale assembly of the wild wheat relative Aegilops umbellulata. Sci. Data10, 739 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Shumate, A. & Salzberg, S. L. Liftoff: accurate mapping of gene annotations. Bioinformatics37, 1639–1643 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pertea, G. & Pertea, M. GFF utilities: GffRead and GffCompare. F1000Research9, 304 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Buchfink, B., Reuter, K. & Drost, H.-G. Sensitive protein alignments at tree-of-life scale using DIAMOND. Nat. Methods18, 366–368 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Virtanen, P. et al. SciPy 1.0: fundamental algorithms for scientific computing in Python. Nat. Methods17, 261–272 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Waskom, M. L. Seaborn: statistical data visualization. J. Op. Source Softw.6, 3021 (2021). [Google Scholar]
- 79.Li, H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. Preprint at arXiv10.48550/arXiv.1303.3997 (2013).
- 80.Cook, D. E. & Andersen, E. C. VCF-kit: assorted utilities for the variant call format. Bioinformatics33, 1581–1582 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gain, C. & François, O. LEA 3: Factor models in population genetics and ecological genomics with R. Mol. Ecol. Resour.21, 2738–2748 (2021). [DOI] [PubMed] [Google Scholar]
- 82.Danecek, P. et al. The variant call format and VCFtools. Bioinformatics27, 2156–2158 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wei, F., Wing, R. A. & Wise, R. P. Genome dynamics and evolution of the Mla (powdery mildew) resistance locus in barley. Plant Cell14, 1903–1917 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Stecher, G., Tamura, K. & Kumar, S. Molecular Evolutionary Genetics Analysis (MEGA) for macOS. Mol. Biol. Evol.37, 1237–1239 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tamura, K., Stecher, G. & Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis version 11. Mol. Biol. Evol.38, 3022–3027 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kolmer, J., Bajgain, P., Rouse, M., Li, J. & Zhang, P. Mapping and characterization of the recessive leaf rust resistance gene Lr83 on wheat chromosome arm 1DS. Theor. Appl. Genet.136, 115 (2023). [DOI] [PubMed] [Google Scholar]
- 87.Long, D. & Kolmer, J. A North American system of nomenclature for Puccinia recondita f. sp. tritici. Phytopathology79, 525–529 (1989). [Google Scholar]
- 88.Wang, S. et al. Characterization of polyploid wheat genomic diversity using a high‐density 90,000 single nucleotide polymorphism array. Plant Biotechnol. J.12, 787–796 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Pestsova, E., Ganal, M. & Röder, M. Isolation and mapping of microsatellite markers specific for the D genome of bread wheat. Genome43, 689–697 (2000). [PubMed] [Google Scholar]
- 90.Heffelfinger, C., Fragoso, C. A. & Lorieux, M. Constructing linkage maps in the genomics era with MapDisto 2.0. Bioinformatics33, 2224–2225 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Voorrips, R. E. MapChart: software for the graphical presentation of linkage maps and QTLs. J. Heredity93, 77–78 (2002). [DOI] [PubMed] [Google Scholar]
- 92.Lück, S. et al. siRNA-Finder (si-Fi) software for RNAi-target design and off-target prediction. Front. Plant Sci.10, 1023 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Barnes, C. W. & Szabo, L. J. Detection and identification of four common rust pathogens of cereals and grasses using real-time polymerase chain reaction. Phytopathology97, 717–727 (2007). [DOI] [PubMed] [Google Scholar]
- 94.Chang, Q. et al. A unique invertase is important for sugar absorption of an obligate biotrophic pathogen during infection. New Phytol.215, 1548–1561 (2017). [DOI] [PubMed] [Google Scholar]
- 95.Marçais, G. & Kingsford, C. A fast, lock-free approach for efficient parallel counting of occurrences of k-mers. Bioinformatics27, 764–770 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kokot, M., Długosz, M. & Deorowicz, S. KMC 3: counting and manipulating k-mer statistics. Bioinformatics33, 2759–2761 (2017). [DOI] [PubMed] [Google Scholar]
- 97.Gel, B. & Serra, E. karyoploteR: an R/Bioconductor package to plot customizable genomes displaying arbitrary data. Bioinformatics33, 3088–3090 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wickham, H. ggplot2: Elegant Graphics for Data Analysis (Springer-Verlag, 2016).
- 99.Cavalet-Giorsa, E. et al. Population genomics of the wild wheat Aegilops tauschii (Open wild wheat consortium phase II) [Dataset]. Dryad10.5061/dryad.vmcvdnd0d (2024).
- 100.Cavalet-Giorsa, E. et al. k-mer matrix Aegilops tauschii diversity panel (Open wild wheat consortium phase II) Part 3/3 [Dataset]. Dryad10.5061/dryad.wm37pvmvd (2024).
- 101.Cavalet-Giorsa, E. et al. k-mer matrix Aegilops tauschii diversity panel (Open wild wheat consortium phase II) Part 2/3 [Dataset]. Dryad10.5061/dryad.wpzgmsbvm (2024).
- 102.Cavalet-Giorsa, E. et al. k-mer matrix Aegilops tauschii diversity panel (Open wild wheat consortium phase II) Part 1/3 [Dataset]. Dryad10.5061/dryad.p5hqbzkvx (2024).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
This file contains Supplementary Notes 1–4.
Supplementary Tables 1–33.
Data Availability Statement
The sequencing data and genome assemblies generated in this study were submitted to NCBI under BioProject number PRJNA956839, including the raw Illumina reads for 350 Ae. tauschii accessions, the raw PacBio reads, the Hi-C data, the raw RNA-seq reads from 5 tissues of Ae. tauschii accessions TA10171, TA1675 and TA2576, the raw Illumina reads for 59 wheat landraces and the raw PacBio reads and Omni-C data of the wheat landrace CWI 86942. The genome assemblies of the 46 Ae. tauschii accessions, the assemblies and annotations for CWI 86942, TA10171, TA1675 and TA2576, the variant call (SNP) file, the k-mer matrix for 920 Ae. tauschii accessions, the phylogenetic tree for 493 non-redundant Ae. tauschii accessions, the structural variant call (SV) files, the IBSpy variation tables, the predictions of the subpopulations contributing to the 17 hexaploid wheat assemblies, an excel file containing the RagTag scaffold output agp files and the dot-plots produced by MashMap used to validate the RagTag scaffolding are available at Dryad (10.5061/dryad.vmcvdnd0d (ref. 99); 10.5061/dryad.wm37pvmvd (ref. 100); 10.5061/dryad.wpzgmsbvm (ref. 101); 10.5061/dryad.p5hqbzkvx (ref. 102)). The Lr39 genomic sequence was deposited in NCBI Genbank under accession number OR567850. The TA10171 (L1), TA1675 (L2) and TA2576 (L3) genomes are available for online BLAST, Jbrowse visualization and synteny analysis with the currently available Triticinae genomes at (https://wheat.pw.usda.gov/GG3pangenome/wheat/D/taus_home.php). The Ensembl nrTEplants repetitive element database (June 2020) was used for repeat content prediction. Viridiplantae protein models from OrthoDB v.11 were used to predict de novo gene models for the annotated Ae. tauschii genomes. The predicted translated proteins were annotated using the following databases: FunFam, SFLD, PANTHER, Gene3D, PRINTS, Coils, SUPERFAMILY, SMART, CDD, PIRSR, ProSitePatterns, AntiFam, Pfam, MobiDBLite, PIRSF and NCBIfam. We downloaded sequencing data for 306 accessions from NCBI BioProject number PRJNA685125, 275 accessions from NCBI BioProject number PRJNA705859, and 24 accessions from the China National Center for Bioinformation–National Genomics Data Center under accession number PRJCA005979.
The k-mer matrix generation pipeline for large diversity panels is available at GitHub (https://github.com/githubcbrc/KGWASMatrix). The custom script for estimating the cumulative k-mer content is available at GitHub (https://github.com/andreagonzam/tauschii_pangenome). The custom scripts for Missing Link Finder pipeline and haplotype analysis are available at GitHub (https://github.com/emilecg/wheat_evolution).