Abstract
We longitudinally measured T-cell receptor transcript frequencies of human immunodeficiency virus type 1 (HIV-1) specific cytotoxic T lymphocytes (CTL) in an individual with rapidly progressive disease and high levels of viremia. CTL clones elicited during acute HIV-1 infection were present at the time of death, despite absent functional CTL responses, arguing against clonal deletion as a mechanism for the decline of CTL responses observed during HIV-1 infection.
Virus-specific cytotoxic T lymphocytes (CTL) appear in acute human immunodeficiency virus type 1 (HIV-1) infection coincident with control of the initial intense burst of viral replication (4, 13). Evidence that these CTL expansions are oligoclonal is supported by studies demonstrating major oligoclonal or monoclonal expansions of CD8+ T cells having a predominant Vβ usage (17). In HIV-1 infection, some of these Vβ expansions have been shown to contain HIV-specific CTL (17), but the long-term fate of these cells has not been determined. Despite the appearance of these early robust immune responses, in most individuals there is a decline in CTL function as the disease progresses (6, 12). Clonal exhaustion, resulting in physical deletion of CTL clones from chronic exposure to high levels of antigen, has been proposed as a mechanism for the decline in CTL activity seen during HIV-1 disease progression (15, 18).
Moskophidis et al. (15) have shown that the presence of rapidly replicating, noncytopathic lymphocytic choriomeningitis virus throughout the lymphoid system in acute murine infection can initiate large expansions of antiviral CTL followed, after a short period of anergy, by clonal deletion. Pantaleo et al. (18), by using CDR3-specific PCR to demonstrate a rapid disappearance of two expanded clonotypes, suggested that clonal exhaustion occurred during acute HIV-1 infection in two humans. Whether the decline in CTL function seen in late-stage HIV-1 disease is because CTL clones, elicited during early stages of disease, undergo clonal exhaustion in the face of a chronically elevated antigen burden remains to be seen.
We have quantitated the magnitude and duration of individual HIV-1-specific CTL expansions based on the molecular analysis of CTL clones obtained by limiting dilution assays. Using oligonucleotide probes complementary to the unique CDR3 region of CTL clones, we evaluated the T-cell receptor (TCR) transcript frequencies in peripheral blood in the chronic stages of disease in an intermediate progressor and during acute- and late-stage disease in an individual with rapidly progressive infection.
Longitudinal tracking of TCR transcripts in PBMC of a chronic HIV-1-infected subject.
To determine the fate of individual CTL clonal frequencies during chronic stable infection we first longitudinally tracked CTL clones in subject 115, an individual with chronic stable disease and persistent functional CTL responses. Subject 115 was known to have been HIV-1 positive since 1987 and had been started on antiretroviral therapy (zidovudine, lamivudine, and indinavir) in July 1996. CTL clones were isolated and maintained by limiting dilution cloning performed on either fresh or thawed cryopreserved peripheral blood mononuclear cells (PBMC) (21). Table 1 summarizes the HLA restriction, epitope recognition, and TCR Vβ gene usage of four CTL clones isolated from this individual. The method used for sequencing the TCR of CTL clones has been previously described in detail (11).
TABLE 1.
Summary of HLA restriction, epitope recognition, and TCR β gene usage of CTL clones obtained from subject 115
| CTL clone | Sequence | Epitope/position | HLA restriction | TCR; Vβ gene usage
|
||
|---|---|---|---|---|---|---|
| Vβ | CDR3 | Jβ | ||||
| 115 M21a | ERYLKDQQL | gp41/584–592 | B14 | 4 | VKDGA | 1.2 |
| 115 E15a | ERYLKDQQL | gp41/584–592 | B14 | 4 | VEDWGGAS | 2.1 |
| 115 D87 | ERYLQDQQL | gp41/584–592 | B14 | 8 | ALNRVD | 2.1 |
| p175b | SLYNTVATL | p17/77–85 | A2 | 5 | FDS | 2.7 |
Previously described CTL clone sequences (10, 11). CDR3 region-specific biotin-labeled probes designed for each individual clonotype were 27 to 30 nucleotides in length and spanned the Dβ region with some overlap of the adjacent Vβ and Jβ regions. The diversity region-specific probes used for subject 115 were the following: 115 M21 Db (AGC GTG AAG GAC GGG GCT GGC), 115 E15 Db (AGC GTT GAA GAT TGG GGC GGA GCG AGC TCC), 115 D87 Db (GCT AGT GCC CTC AAC AGG GTC GAT GAG CAG), and 115 p175b.33 Db (TGC GCC AGC AGC TTT GAC AGC GAG CAG TAC). Sequence data for CTL clones 115 D87, 115 p175b, 18030 D23, and 15160 ETA49 are available from GenBank under accession numbers AF272739, AF272740, AF272741, and AF272742.
For each longitudinal time point of interest, total RNA was isolated from 5 × 106 cryopreserved PBMC. Two types of cDNA libraries were constructed from the RNA. The first library, representing all rearranged TCR transcripts obtained from the subjects' cDNA, was generated by anchored PCR (14). The second cDNA library of clone-specific Vβ transcripts was generated by using the respective Vβ primer and a TCR β-chain constant region primer (5) for PCR amplification. The Vβ-specific primers used have been described previously (11).
Colonies were picked from each library and transferred to nylon membranes (Colony/Plaque Screen [137 mm], NEF-978Y; NEN Life Science Products, Boston, Mass.). Denatured membranes were hybridized with Vβ- or CDR3-specific biotin-labeled oligonucleotide probes. Hybridization conditions were stringently optimized, and membranes were developed according to the manufacturer's specifications (Tropix, Bedford, Mass.). The CDR3-specific probes had 100% specificity and greater than 99% sensitivity on test membranes gridded with positive and negative controls. Controls for the entire procedure were run in parallel, and positive and negative colonies were sequenced for confirmation.
The library containing total PBMC TCR transcripts by anchored PCR was probed with an oligonucleotide specific for the Vβ region of interest to obtain the frequency of clone-specific Vβ-chain transcripts among all TCR β-chain transcripts. The library containing only TCR transcripts of this particular Vβ was then probed with an oligonucleotide probe specific to the CDR3 region of the relevant CTL clone to obtain the frequency of clone-specific CDR3 sequences among the Vβ amplified transcripts. The final clone-specific frequency among all TCR transcripts was obtained by multiplying these two frequencies. Five hundred to 1,500 colonies of PBMC or of the Vβ of interest were probed.
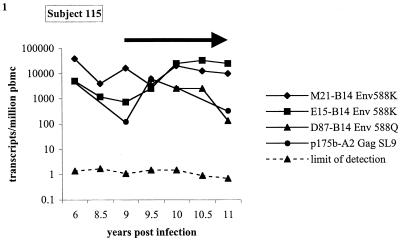
Four individual CTL clones from subject 115, an individual with chronic stable HIV-1 infection, had frequencies ranging from 130 to 40,000 transcripts/million PBMC over 6 years of longitudinal follow-up extending from years 6 to 11 of the disease course (Fig. 1). These translate into clonal frequencies ranging from 0.012% of the total T-cell population for clone 115 p175b at 9 years postinfection to 3.78% of the total T-cell population for clone 115 M21 at 6 years postinfection. Frequencies of individual clonal responses varied by up to 100-fold, but all responses persisted during the time of longitudinal follow-up in the presence of ongoing detectable viremia and during almost 2 years of follow-up on highly active antiretroviral therapy (HAART) during which viral load remained below the limits of detection by commercial RNA PCR (<50 copies/ml). The measured transcript frequencies were detected at frequencies 2 to 5 logs higher than the limit of detection for the assay (Fig. 1).
FIG. 1.
Longitudinal CTL TCR clone transcript frequencies in a chronically infected subject. Longitudinal CTL transcript frequencies of CTL clones M21, E15, D87, and p175b from chronically infected subject 115. CTL clones M21 and E15 recognize the B14 restricted Env epitope 588K (ERYLKDQQL). Clone D87 recognizes the B14 restricted Env variant epitope 588Q (ERYLQDQQL). Clone p175b recognizes the A2 restricted p17 Gag epitope SL9 (SLYNTVATL). The black arrow indicates the time of institution of antiretroviral therapy in subject 115. For each time point the longitudinal limit of detection of the assay is shown for the least prevalent clonotype. The limit of detection at a particular time point for a clonotype is defined by the following formula: (1/total PBMC TCR β-chain transcripts) × (1 CTL clone CDR3 transcript/all Vβ transcripts). The diversity region-specific probes are identified in footnote a of Table 1. CDR3 region-specific biotin-labeled probes designed for each individual clonotype were 27 to 30 nucleotides in length and spanned the Dβ region with some overlap of the adjacent Vβ and Jβ regions.
To validate the reproducibility of our Vβ and CDR3 probing techniques we also did a series of spiking experiments. The HIV-specific clone 115 M21 was mixed with either PBMC from an HIV-1-seronegative subject or with an unrelated CTL clone at frequencies ranging from 0.1 to 10%. The frequencies obtained for this clone with this method were linear over this range (R = 0.95 and P = 0.003). When seronegative PBMC were probed with two CDR3 region probes from two separate HIV-1-specific CTL clones, no positive colonies were detected, despite detectable colonies for the corresponding Vβ.
Comparison of CTL TCR transcript frequencies to persistent functional CTL responses in chronic HIV-1 infection.
In order to address the relationship between transcript frequencies and functional CTL assays, gamma interferon (IFN-γ) enzyme-linked immunospot (ELISPOT) assays and peptide-stimulated limiting dilution precursor frequency assays were performed on the four clones from subject 115.
Precursor frequencies of HIV-1 epitope-specific CTL were estimated by performing limiting dilution on freshly isolated PBMC followed by in vitro stimulation with peptide-pulsed autologous PBMC and irradiated PBMC from an HIV-1 seronegative donor, as previously described (10, 11). Cells secreting IFN-γ in an antigen-specific manner were detected using a standard ELISPOT assay, as previously described (2, 3, 16, 20).
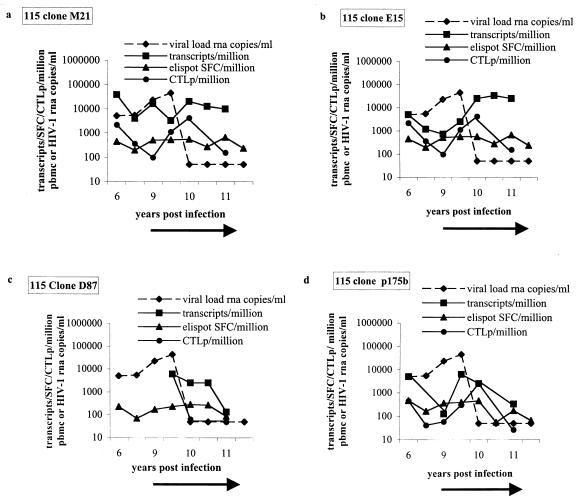
The frequencies of spot-forming cells (SFC) obtained by the IFN-γ ELISPOT assay and peptide-stimulated CTL precursors (CTLp) were comparable and, in general, lower than the corresponding CTL TCR transcript frequencies measured at each time point for the four clones (Fig. 2). The functional responses and the transcript frequencies persist in the absence of therapy and also after nearly 2 years of continuous viral suppression (Fig. 2). These studies indicate that clonal CTL responses, once generated, persist over time. This occurs even when HAART results in a decline in viral load to below the limits of detection and suggests that a new steady state of memory CTL is achieved, as has been described for the chronic phase of other persistent viral infections (1, 16, 22). These comparative studies provide a reference range for the TCR transcript frequencies when CTL functional responses are present in an individual.
FIG. 2.
Comparative analysis of transcript frequency, SFC per million and CTLp per million PBMC, in subject 115 for CTL clones M21, E15, D87, and p175b. Frequencies of CTL transcripts per million PBMC are longitudinally compared to CTL responses measures by IFN-γ ELISPOT assay as SFC per million and compared to CTLp frequencies measured by peptide-stimulated precursor frequency assay for CTL clone M21 (a) and CTL clone E15 (b), clones which recognize the B14 restricted Env epitope ERYLKDQQL, CTL clone D87 (c), which recognizes the variant epitope ERYLQDQQL, and CTL clone p175b (d), which recognizes the A2 restricted Gag epitope SLYNTVATL. The black arrow indicates the time of institution of antiretroviral therapy.
Evaluation of the fate of clonal CTL responses over the course of rapidly progressive infection.
The above data do not address the fate of clonal responses generated in acute infection, nor do they address the fate of such responses over the entire course of infection when functional CTL responses are eventually lost. To address these issues, we evaluated an individual (subject 053i) with rapidly progressive HIV-1 disease who had been identified with acute HIV-1 infection, developed AIDS 13 months after seroconversion, and died within 4 years of infection (8).
Previous studies had shown that this individual had both strong in vivo activated CTL responses and in vitro stimulated memory CTL responses narrowly directed against a dominant B7 restricted epitope in Env, gp41 IPRRIRQGL, and a subdominant B7 restricted epitope in Pol, SPAIFQSSM (8). These functional responses were lost with disease progression. No sequence variation occurred within the targeted epitopes (8). Clones specific for the Env epitope were generated 3, 8, and 39 months after presentation with acute HIV-1 infection. TCR cDNA libraries generated from PBMC obtained at two time points, during seroconversion at 3 months (February 1993) and just prior to death at 45 months (August 1996), were screened with CDR3-and Vβ-specific probes to quantitate the frequency of these clones in the circulation.
At 3 months after presentation, seven IPRRIRQGL-specific CTL clones isolated were analyzed for TCR gene usage (Table 2). The majority population of CTL clones isolated from this time utilized Vβ 6 and Jβ 2.7 in their β-chains and had the TCR β CDR3 sequence WAASS, a clonotype designated CDR3 B1. The minority population had β-chain sequences matching the IPRRIRQGL-specific CTL clone isolated at 8 months with the CDR3 sequence ERSPPGD, a clonotype designated CDR3 B2. The CDR3 sequence for the clone isolated at 39 months is PTAAG, a clonotype designated CDR3 B3.
TABLE 2.
TCR β-chain sequences of B7 restricted Env (gp41/843- 851)-specific CTL clones isolated longitudinally from rapid progressor subject 053ia
| CTL cloneb | TCR β gene usage
|
Date isolatedc | No. of sequences/ no. of colonies screened | ||
|---|---|---|---|---|---|
| Vβ | CDR3 | Jβ | |||
| 53i#2 | 6S1 | WAASS | 2.7 | 2/93 | 12/12 |
| 53i.#9 | 6S1 | WAASS | 2.7 | 2/93 | 10/10 |
| 53i.#14 | 6S1 | WAASS | 2.7 | 2/93 | 10/10 |
| 53i.B2.34 | 6S1 | WAASS | 2.7 | 2/93 | 7/8 |
| 53I.B2.72 | 6S1 | WAASS | 2.7 | 2/93 | 10/10 |
| 53i.B2.89 | 16S1 | ERSPPGD | 2.1 | 2/93 | 9/9 |
| 53i.B2.95 | 16S1 | ERSPPGD | 2.1 | 2/93 | 8/8 |
| 53i.C26 | 16S1 | ERSPPGD | 2.1 | 7/93 | 9/9 |
| 53i.J56 | 14S1 | PTAAG | 2.1 | 2/96 | 13/13 |
The biotin-labeled diversity region-specific probes used for subject 053i were the following: CDR3 B1 clonotype, AGC AGC TTA TGG GCG GCA AGC TCC TAC GAG; CDR3 B2 clonotype, CAA GAA CGT TCC CCA CCG GGG GAT GAG; and CDR3 B3 clonotype; AGC AGT CCC ACG GCC GCC GGT GAG CAG. Sequence data for CTL clones 53i.B2.72, 53i.C26, and 53i.J56 are available from GenBank under accession numbers AF272743, AF272744, and AF272745.
CTL clones recognize the peptide sequence IPRRIRQGL.
2/93, 7/93, and 2/96 represent 3, 8, and 39 months, respectively, after patient presented with acute HIV-1 infection syndrome.
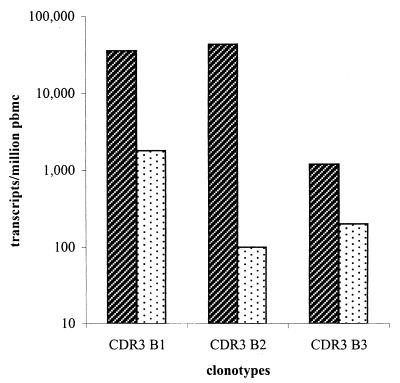
These clonotypes exhibited varying degrees of expansion over the course of the illness (Table 3). With regard to the first clonotype, Vβ 6 constituted 10.99% of all TCR transcripts at seroconversion and the CDR3 B1 clonotype was 32.26% of all Vβ 6 transcripts. The overall clonotype frequency was 3.54% (1 of 28) of all TCR transcripts, indicating an expansion of a single effector clonotype specific for the dominant CTL response during acute HIV-1 infection. At 45 months, the time of death, when no in vivo activated CTL activity was detected, this clonotype was still present at a fairly high frequency of 0.18% (1 of 547) of PBMC transcripts. The second clonotype, CDR3 B2, isolated 8 months after presentation, constituted 87.2% of all Vβ 16 transcripts at seroconversion and represented 4.29% (1 of 23) of all TCR transcripts in PBMC. At the time of death, the CDR3 B2 clonotype was 2.6% of Vβ 16 transcripts, yielding a persistent but lower clonotype frequency of 0.0063% (1 of 15,873) of all TCR transcripts. The third clonotype, CDR3 B3, constituted 1.5% of Vβ 14 and 0.12% (1 of 833) of all TCR transcripts at the time of seroconversion. At the time of death this clonotype made up 2.11% of Vβ 14, with a clonotype frequency of 0.02% (1 of 5,618) of T cells. Figure 3 graphically summarizes the longitudinal CTL transcript frequencies of clonotypes CDR3 B1, CDR3 B2, and CDR3 B3 in subject 053i at seroconversion and at the time of death.
TABLE 3.
Summary of the number of positive colonies obtained out of total number of colonies screened after hybridization of PBMC and Vβ-specific cDNA libraries with biotin-labeled Vβ- and CDR3-specific probes used for calculation of logitudinal TCR frequences in the rapid progressor at seroconversion and at the time of death
| CTL clone (Vβ) | Date PBMC probed (mo/yr)a | Vβ transcripts/total PBMC TCR (β-chain) transcripts (%) | CTL clone CDR3 transcripts/all Vβ transcripts (%) | CTL clone CDR3 transcripts/total PBMC TCR transcripts (%)b | Limit of detection (%)c |
|---|---|---|---|---|---|
| CDR3 B1 (6) | 2/93 | 155/1,400 (10.99) | 271/840 (32.26) | 1/28 (3.54) | 0.00008 |
| 8/96 | 21/828 (2.55) | 78/1,096 (7.2) | 1/547 (0.18) | 0.00011 | |
| CDR3 B2 (16S1) | 2/93 | 41/840 (4.88) | 1,402/1,608 (87.2) | 1/23 (4.29) | 0.00007 |
| 8/96 | 2/828 (0.24) | 21/804 (2.6) | 1/15,873 (0.0063) | 0.00015 | |
| CDR3 B3 (14S1) | 2/93 | 85/1,072 (7.93) | 8/536 (1.5) | 1/833 (0.12) | 0.00017 |
| 8/96 | 7/828 (0.85) | 17/804 (2.11) | 1/5,618 (0.02) | 0.00015 |
2/93 and 8/96 represent seroconversion and time of death, respectively.
Values in this column are the products of the multiplication of the values in the previous two columns.
(1/total PBMC TCR β-chain transcripts) × (1 CTL clone CDR3 transcript/all Vβ transcripts).
FIG. 3.
Longitudinal transcript frequencies of CTL clonotypes of the rapid progressor. Longitudinal transcript frequencies of clonotypes CDR3 B1, CDR3 B2, and CDR3 B3 in the PBMC of the rapid progressor at 3 months ( ) (February 1993, the time of seroconversion) and 45 months (
) (February 1993, the time of seroconversion) and 45 months ( ) (August 1996, the time of death) after acute retrovirus infection.
) (August 1996, the time of death) after acute retrovirus infection.
Our results indicate that once clonal CTL responses are established, they persist in vivo, both in settings of persistent high-level viremia and when HAART lowers viremia to undetectable levels. These studies also extend an earlier study of rapidly progressive infection (8), demonstrating that identical clones generated in acute infection persisted over the course of infection and were still present, albeit at lower levels, at the time of death despite the loss of these epitope-specific CTL functional responses over time. Since no sequence variation occurred within the targeted epitope (8), and yet the CTL response declined, the data suggest both an in vivo defect in the ability to expand to a cognate stimulus and weak immune selection pressure mediated through some CTL responses.
We have demonstrated the cytolytic activity of the Vβ expansions identified in acute HIV-1 infection by isolating functional CTL clones from these expansions in infected persons. In the CTL clones we studied at seroconversion, we found that the frequencies of the expanded CDR3 B1 and CDR3 B2 clonotypes were 3.54 and 4.29% of PBMC and that the frequencies of the respective expanded Vβ 6 and Vβ 16 were 10.99 and 4.88% of PBMC. Our data demonstrate that the composite frequency of three CTL clones specific for the same B7 restricted epitope was 7.95% of PBMC at the time of acute HIV-1 infection. These data indicate that early CTL expansions can be large but still may be insufficient to maintain viral control.
These studies also provide strong evidence that CTL clonal deletion is not the predictable result of persistent high-level viremia in late-stage HIV-1 infection. In murine lymphocytic choriomeningitis virus infection, despite the induction of vigorous CTL responses during initial antigen exposure, exposure to high levels of antigen can lead to clonal exhaustion as a result of excessive stimulation provided by the persistence of elevated viral antigen (15). Additionally, Gallimore et al. and Zajac et al. have shown that in some situations where there is excessive antigen, antigen-specific cells detected by tetramers may have a diminished capacity to produce IFN-γ and lytic activity in vitro (7, 22). Our data support the possibility that clonal deletion of CTL may not predictably occur in late-stage HIV-1 disease, pointing rather to an inability of CTL clones to expand and retain functional activity for the cognate epitope. At least one explanation for this would be a lack of adequate T helper cell function, since there is an association between CTL and T helper cell magnitude (9, 19). Since the method described here discriminates among clones based on the CDR3 region, it can unambiguously define the magnitude of clonal responses and should thus be helpful in defining the relationship between clonal CTL responses and disease progression.
Acknowledgments
We thank Bruce D. Walker for critical review of the manuscript. We thank Debbie J. Ruhl for excellent technical support. We thank M. Gately and Hoffman-LaRoche for the generous gift of interleukin-2.
This research was supported by NIH grants AI-39966 and AI-28568 (S.A.K.). S. A. Islam was supported by a Howard Hughes Medical Institute Postdoctoral Research Fellowship.
REFERENCES
- 1.Ahmed R, Gray D. Immunological memory and protective immunity: understanding their relation. Science. 1996;272:54–60. doi: 10.1126/science.272.5258.54. [DOI] [PubMed] [Google Scholar]
- 2.Altfeld M, Rosenberg E S, Shankarappa R, Mukherjee J S, Hecht F M, Eldridge R L, Addo M M, Poon S H, Phillips M N, Robbins G K, Sax P E, Boswell S, Kahn J O, Brander C, Goulder P J, Levy J A, Mullins J I, Walker B D. Cellular immune responses and viral diversity in individuals treated during acute and early HIV-1 infection. J Exp Med. 2001;193:169–180. doi: 10.1084/jem.193.2.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altfeld M A, Livingston B, Reshamwala N, Nguyen P T, Addo M M, Shea A, Newman M, Fikes J, Sidney J, Wentworth P, Chesnut R, Eldridge R L, Rosenberg E S, Robbins G K, Brander C, Sax P E, Boswell S, Flynn T, Buchbinder S, Goulder P J R, Walker B D, Sette A, Kalams S A. Identification of novel HLA-A2-restricted human immunodeficiency virus type 1-specific cytotoxic T-lymphocyte epitopes predicted by the HLA-A2 supertype peptide-binding motif. J Virol. 2001;75:1301–1311. doi: 10.1128/JVI.75.3.1301-1311.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Borrow P, Lewicki H, Hahn B H, Shaw G M, Oldstone M B A. Virus-specific CD8+ cytotoxic T-lymphocyte activity associated with control of viremia in primary human immunodeficiency virus type 1 infection. J Virol. 1994;68:6103–6110. doi: 10.1128/jvi.68.9.6103-6110.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brander C, Goulder P J, Luzuriaga K, Yang O O, Hartman K E, Jones N G, Walker B D, Kalams S A. Persistent HIV-1-specific CTL clonal expansion despite high viral burden post in utero HIV-1 infection. J Immunol. 1999;162:4796–4800. [PubMed] [Google Scholar]
- 6.Carmichael A, Jin X, Sissons P, Borysiewicz L. Quantitative analysis of the human immunodeficiency virus type 1 (HIV-1)-specific cytotoxic T lymphocyte (CTL) response at different stages of HIV-1 infection: differential CTL responses to HIV-1 and Epstein-Barr virus in late disease. J Exp Med. 1993;177:249–256. doi: 10.1084/jem.177.2.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gallimore A, Glithero A, Godkin A, Tissot A C, Pluckthun A, Elliott T, Hengartner H, Zinkernagel R. Induction and exhaustion of lymphocytic choriomeningitis virus-specific cytotoxic T lymphocytes visualized using soluble tetrameric major histocompatibility complex class I-peptide complexes. J Exp Med. 1998;187:1383–1393. doi: 10.1084/jem.187.9.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hay C M, Ruhl D J, Basgoz N O, Wilson C C, Billingsley J M, DePasquale M P, D'Aquila R T, Wolinsky S M, Crawford J M, Montefiori D C, Walker B D. Lack of viral escape and defective in vivo activation of human immunodeficiency virus type 1-specific cytotoxic T lymphocytes in rapidly progressive infection. J Virol. 1999;73:5509–5519. doi: 10.1128/jvi.73.7.5509-5519.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kalams S A, Buchbinder S P, Rosenberg E S, Billingsley J M, Colbert D S, Jones N G, Shea A K, Trocha A K, Walker B D. Association between virus-specific cytotoxic T-lymphocyte and helper responses in human immunodeficiency virus type 1 infection. J Virol. 1999;73:6715–6720. doi: 10.1128/jvi.73.8.6715-6720.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kalams S A, Johnson R P, Dynan M J, Hartman K E, Harrer T, Harrer E, Trocha A K, Blattner W A, Buchbinder S P, Walker B D. T cell receptor usage and fine specificity of human immunodeficiency virus 1-specific cytotoxic T lymphocyte clones: analysis of quasispecies recognition reveals a dominant response directed against a minor in vivo variant. J Exp Med. 1996;183:1669–1679. doi: 10.1084/jem.183.4.1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kalams S A, Johnson R P, Trocha A K, Dynan M J, Ngo H S, D'Aquila R T, Kurnick J T, Walker B D. Longitudinal analysis of T cell receptor (TCR) gene usage by human immunodeficiency virus 1 envelope-specific cytotoxic T lymphocyte clones reveals a limited TCR repertoire. J Exp Med. 1994;179:1261–1271. doi: 10.1084/jem.179.4.1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Klein M R, van Baalen C A, Holwerda A M, Kerkhof Garde S R, Bende R J, Keet I P, Eeftinck-Schattenkerk J K, Osterhaus A D, Schuitemaker H, Miedema F. Kinetics of Gag-specific cytotoxic T lymphocyte responses during the clinical course of HIV-1 infection: a longitudinal analysis of rapid progressors and long-term asymptomatics. J Exp Med. 1995;181:1365–1372. doi: 10.1084/jem.181.4.1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koup R A, Safrit J T, Cao Y, Andrews C A, McLeod G, Borkowsky W, Farthing C, Ho D D. Temporal association of cellular immune responses with the initial control of viremia in primary human immunodeficiency virus type 1 syndrome. J Virol. 1994;68:4650–4655. doi: 10.1128/jvi.68.7.4650-4655.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Loh E Y, Elliot J F, Cwirla S, Lanier L L, Davis M M. Polymerase chain reaction with single-sided specificity: analysis of T cell receptor d chain. Science. 1989;243:217–220. doi: 10.1126/science.2463672. [DOI] [PubMed] [Google Scholar]
- 15.Moskophidis D, Lechner F, Pircher H, Zinkernagel R M. Virus persistence in acutely infected immunocompetent mice by exhaustion of antiviral cytotoxic effector T cells. Nature. 1993;362:758–761. doi: 10.1038/362758a0. . (Erratum, 364:262.) [DOI] [PubMed] [Google Scholar]
- 16.Murali-Krishna K, Altman J D, Suresh M, Sourdive D J, Zajac A J, Miller J D, Slansky J, Ahmed R. Counting antigen-specific CD8 T cells: a reevaluation of bystander activation during viral infection. Immunity. 1998;8:177–187. doi: 10.1016/s1074-7613(00)80470-7. [DOI] [PubMed] [Google Scholar]
- 17.Pantaleo G, Demarest J F, Soudeyns H, Graziosi C, Denis F, Adelsberger J W, Borrow P, Saag M S, Shaw G M, Sekaly R P, et al. Major expansion of CD8+ T cells with a predominant V beta usage during the primary immune response to HIV. Nature. 1994;370:463–467. doi: 10.1038/370463a0. [DOI] [PubMed] [Google Scholar]
- 18.Pantaleo G, Soudeyns H, Demarest J F, Vaccarezza M, Graziosi C, Paolucci S, Daucher M, Cohen O J, Denis F, Biddison W E, Sekaly R P, Fauci A S. Evidence for rapid disappearance of initially expanded HIV-specific CD8+ T cell clones during primary HIV infection. Proc Natl Acad Sci USA. 1997;94:9848–9853. doi: 10.1073/pnas.94.18.9848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rosenberg E S, Billingsley J M, Caliendo A M, Boswell S L, Sax P E, Kalams S A, Walker B D. Vigorous HIV-1-specific CD4+ T cell responses associated with control of viremia. Science. 1997;278:1447–1450. doi: 10.1126/science.278.5342.1447. [DOI] [PubMed] [Google Scholar]
- 20.Taguchi T, McGhee J R, Coffman R L, Beagley K W, Eldridge J H, Takatsu K, Kiyono H. Detection of individual mouse splenic T cells producing IFN-V and IL-5 using the enzyme-linked immunospot (ELISPOT) assay. J Immunol Methods. 1990;128:65–73. doi: 10.1016/0022-1759(90)90464-7. [DOI] [PubMed] [Google Scholar]
- 21.Walker B D, Chakrabarti S, Moss B, Paradis T J, Flynn T, Durno A G, Blumberg R S, Kaplan J C, Hirsch M S, Schooley R T. HIV-specific cytotoxic T lymphocytes in seropositive individuals. Nature. 1987;328:345–348. doi: 10.1038/328345a0. [DOI] [PubMed] [Google Scholar]
- 22.Zajac A J, Blattman J N, Murali-Krishna K, Sourdive D J, Suresh M, Altman J D, Ahmed R. Viral immune evasion due to persistence of activated T cells without effector function. J Exp Med. 1998;188:2205–2213. doi: 10.1084/jem.188.12.2205. [DOI] [PMC free article] [PubMed] [Google Scholar]