Abstract
Recently, we identified a large number of ultraconserved (uc) sequences in noncoding regions of human, mouse, and rat genomes that appear to be essential for vertebrate and amniote ontogeny. Here, we used similar methods to identify ultraconserved genomic regions between the insect species Drosophila melanogaster and Drosophila pseudoobscura, as well as the more distantly related Anopheles gambiae. As with vertebrates, ultraconserved sequences in insects appear to occur primarily in intergenic and intronic sequences, and at intron-exon junctions. The sequences are significantly associated with genes encoding developmental regulators and transcription factors, but are less frequent and are smaller in size than in vertebrates. The longest identical, nongapped orthologous match between the three genomes was found within the homothorax (hth) gene. This sequence spans an internal exon-intron junction, with the majority located within the intron, and is predicted to form a highly stable stem-loop RNA structure. Real-time quantitative PCR analysis of different hth splice isoforms and Northern blotting showed that the conserved element is associated with a high incidence of intron retention in hth pre-mRNA, suggesting that the conserved intronic element is critically important in the post-transcriptional regulation of hth expression in Diptera.
Evolutionary conservation of DNA sequences between related or distant biological species is usually believed to reflect conservation of protein structure and function or of important cis-acting regulatory elements. Indeed, many homologous proteins carrying out evolutionarily conserved functions display a high degree of identity of their protein-coding nucleotide sequences. However, in such cases, nucleotide sequence identity very rarely reaches 100%, due to the accumulation of synonymous and other types of substitutions.
Recently, we identified almost 500 sequences equal to or longer than 200 bp that showed 100% identity between orthologous regions of the human, rat, and mouse genomes (Bejerano et al. 2004; Woolfe et al. 2005). These ultraconserved elements (uc-elements) are widely distributed in the genome, and are highly conserved among mammals and birds, and in many cases, fish. Shorter ultraconserved elements are even more common (around 5000 ≥ 100 bp, and around 50,000 ≥ 50 bp), as are elements with slightly less than 100% identity (M. Pheasant and J.S. Mattick, unpubl.). Most uc-elements are located in intergenic sequences, often distant from known protein-coding sequences, or in introns, often overlapping splice junctions, and are significantly associated with genes encoding developmental regulators, transcription factors, and RNA-binding proteins (Bejerano et al. 2004). These associations suggest a regulatory role that is fiercely conserved and therefore presumably essential to vertebrate and amniotic ontogeny. While there is some experimental support for this possibility (Woolfe et al. 2005), the precise biological functions and mechanism of action of these uc-elements remain to be determined.
In this study, we examined whether uc-elements can also be identified in invertebrate metazoan species by comparing the sequenced genomes of two fruitfly species, Drosophila melanogaster and Drosophila pseudoobscura, and the mosquito Anopheles gambiae. All three species belong to the order of Diptera, which diverged from their nearest common ancestor around 330 million years ago (Gaunt and Miles 2002). The divergence time between modern Drosophila and Anopheles species is estimated to be around 250 million years (Fig. 1; Gaunt and Miles 2002), comparable to the divergence time between human and birds (Kumar and Hedges 1998). The two Drosophila species are estimated to have diverged around 25-30 million years ago (Powell 1996), allowing significant changes in genomic DNA sequences to occur (71% identity in alignable regions, covering ∼60% of the genome) (Richards et al. 2005), comparable to those between humans and rodents (69% identity in alignable regions, covering 40% of the genome) (Waterston et al. 2002), which diverged earlier, but have longer generation times than insects.
Figure 1.
Summary of phylogenetic relationships of insect species addressed in this study. The tree structure and estimated divergence times are derived from data presented in M.W. Gaunt (Gaunt and Miles 2002) and J.R. Powell (Powell 1996).
We identified all identical nongapped matches longer than 50 bp between the genomes of the mosquito and two fruitflies. Excluding one snRNA sequence (snRNA:U6), the longest match (92 bp) between all three insect genomes was found at an exon-intron junction within the homothorax gene (Table 1). The hth gene (Pai et al. 1998) encodes a homeodomain-containing protein, which belongs to the MEIS protein family (Steeman et al. 1997), members of which have been shown to be involved in various developmental processes and neoplastic transformations (Moskow et al. 1995; Kurant et al. 1998; Mercader et al. 1999; Maeda et al. 2001; Bessa et al. 2002; Van Auken et al. 2002; Wernet et al. 2003). Analysis of the structure and expression of the ultraconserved element suggests that it is involved in regulation of hth expression level via formation of putative RNA secondary structure and intron retention.
Table 1.
Ultraconserved genomic sequences among insects
| Number of uc-elements
|
||||||||
|---|---|---|---|---|---|---|---|---|
| Splice site
|
||||||||
| Category | Total size (bp) | Total | ncRNA | Intergenic | Exonic | % ibp | Intronic | |
| D. mel./D. ps. | ||||||||
| ≥ 50 bp | 1,524,657 | 23,699 | 291 | 14,487 | 2,070 | 1037 | 31% | 5,814 |
| ≥ 80 bp | 293,630 | 3,076 | 46 | 1,854 | 250 | 287 | 32% | 639 |
| ≥ 100 bp | 98,190 | 841 | 8 | 477 | 83 | 133 | 31% | 140 |
| ≥ 150 bp | 9,234 | 52 | 8 | 11 | 32 | 32% | 1 | |
| ≥ 200 bp | 2,362 | 11 | 4 | 7 | 26% | |||
| D. mel./A. gam. | ||||||||
| ≥ 50 bp | 8,297 | 126 | 90 | 1 | 30 | 3 | 53% | 2 |
| ≥ 80 bp | 1,238 | 14 | 12 | 1 | 1 | 87% | ||
| D. mel./D. ps./A. gam. | ||||||||
| ≥ 50 bp | 5,944 | 87 | 76 | 1 | 7 | 2 | 69% | 1 |
| ≥ 80 bp | 1,022 | 12 | 11 | 1 | 87% | |||
The numbers of sequences that are conserved between D. melanogaster (D. mel.) and D. pseudoobscura (D. ps.), and between these species and A. gambiae (A. gam.) are shown. ncRNA sequences include tRNAs, snRNAs, snoRNAs, and some unknown noncoding RNAs. Sequences that overlapped an exon were classified as exonic if they overlapped by at least 95%, and otherwise as “splice site.” % ibp indicates an average percentage of base pairs located within intron. Intronic sequences do not overlap any annotated exon.
Results
Identification of ultraconserved sequences in insects
Table 1 shows the numbers of sequences that are conserved by length categories between D. melanogaster and D. pseudoobscura, and between these species and A. gambiae, as well as their position relative to annotated noncoding RNAs, known or predicted exons of protein-coding genes, introns, and intergenic regions.
We found over 23,000 sequences of 50 bp or more that are 100% conserved between D. melanogaster and D. pseudoobscura, covering over 1.5 Mb of genomic sequence. These sequences appear to be mainly located in intergenic sequences and introns, and although we cannot rule out the possibility that some of them may overlap unrecognized exons, this distribution is similar to that observed for uc-elements in vertebrates (Bejerano et al. 2004). The number of exact matches of 50 bp between these genomes (based on 76% identity over their alignable sequences) that would be expected by random chance is 105, and the number over 80 bp is 0.02, indicating that the vast majority of these matches have been preserved by purifying selection. In addition, the distribution of the matches changes with increasing length, with the majority of the longest matches located at splice sites.
We examined the Gene Ontology (GO) molecular function annotations for the genes harboring exonic, splice site, and intronic uc-elements, as well as the nearest genes, left and right, within 5 kb, for the intergenic uc-elements. Intronic and intergenic sequences are enriched for genes with GO terms related to transcription factors (4.2-fold enrichment, P < 2 χ 10-12 and 3.2-fold enrichment, P <1 χ 10-22, respectively), which is similar to earlier findings in vertebrate genomes (Bejerano et al. 2004; Woolfe et al. 2005; Table 2). The transcription factor enrichment is up to 7.7-fold for genes with intronic uc-elements ≥80 bp. By comparison, genes with exonic or splice site uc-elements longer than 80 bp are enriched for ion channel/transporter activity GO annotations (6.8-fold enrichment, P <3 χ 10-13); of the 10 genes associated with the longest of these uc-elements, seven encoded neurological ion channels/transporter proteins affecting neurological function (Table 3). Interestingly, six of these genes have been shown to have extensive RNA editing at multiple sites (Hanrahan et al. 2000; Hoopengardner et al. 2003), suggesting a possible link between these phenomena. Not surprisingly, uc-elements are also significantly associated with the GO biological process terms morphogenesis, organogenesis, and behavior.
Table 2.
The 10 known genes harboring the largest intronic ultraconserved sequences between D. melanogaster and D. pseudoobscura
| Gene | Molecular function | Size of uc-element (bp) |
|---|---|---|
| RhoGAPp 190 | Rho GTPase activator activity | 160 |
| Awh (Arrowhead) | Homeobox transcription factor | 144 |
| acj6 (abnormal chemosensory jump 6) | Homeobox transcription factor | 144 |
| CG 16791 | Unknown function | 143 |
| caup (caupolican) | Homeobox transcription factor | 139 |
| sif (still life) | Rho guanyl-nucleotide exchange factor activity | 138 |
| CG14521 | Immunoglobulin-like protein | 134 |
| svp (seven up) | COUP transcription factor | 134 |
| Ubx (Ultrabithorax) | Homeobox transcription factor | 132 |
| nAcRalpha-34E (nicotinic Acetylcholine Receptor_34E) | Ion-channel | 131 |
| hh (hedgehog) | Endopeptidase activity | 131 |
The full list of uc-elements can be found at http://www.imb.uq.edu.au/groups/mattick/drosophila_uc/.
Table 3.
The 10 genes harboring the largest ultraconserved sequences between D. melanogaster and D. pseudoobscura overlapping with exons and splice sites
| Gene | Molecular function | Location | Size of ucelement (bp) |
|---|---|---|---|
| para (paralytic) | Ion channel | Splice site (12 ibp) | 246 |
| Splice site (92 ibp) | 199 | ||
| Rd1 (Resistant to dieldrin) | Ion channel | Exonic | 233 |
| eag (ether-a-go-go) | Ion channel | Exonic | 224 |
| Sh (Shaker) | Ion channel | Splice site (44 ibp) | 216 |
| Splice site (13 ibp) | 204 | ||
| Cf2 (Chorion factor 2) | Transcription factor | Splice site (26 ibp) | 211 |
| nAcRalpha-34E (nicotinic Acetylcholine Receptor_34E) | Ion channel | Exonic (3′ UTR) | 209 |
| Sk (small conductance calcium-activated potassium channel) | Ion channel | Exonic | 203 |
| cpo (couch potato) | RNA binding | Splice site (43 ibp) | 202 |
| slo (slowpoke) | Ion channel | Splice site (25 ibp) | 200 |
| hth (homothorax) | Transcription factor | Splice site (149 ibp) | 197 |
ibp indicates the number of base pairs located within the intron. The full list of uc-elements can be found at http://www.imb.uq.edu.au/groups/mattick/drosophila_uc/.
The genes containing the highest density of uc-elements include genes encoding RNA-binding proteins, homeotic developmental regulators (homothorax, Ultrabithorax, fruitless, and cut), and genes involved in neural development and function (Supplemental Table 1). The gene with the most uc-elements enclosed within its borders encodes the Drosophila bruno paralog Bru-3. It is remarkable that the total length of the 168 uc-elements within the Bru-3 gene is 11.2 kb, which is over five times longer than its protein-coding sequence (2.1 kb). Additionally, there is a cluster of 102 uc-elements within the 96 kb upstream of the transcription start site of Bru-3, totaling nearly 18 kb of ultraconserved sequence in the ∼220-kb region encompassing this gene.
Although many of the genes with the highest number of uc-elements are also amongst the longest genes in the Drosophila genome, there was no general correlation between the length of the transcription unit and number of associated uc-elements. For example, the gene encoding SNF4A-γ (72 kb) has no uc-elements, whereas the relatively short gene encoding Glu-RI (11 kb) has 14 uc-elements.
Several recent studies have demonstrated that a significant portion of the cis-regulatory modules in Drosophila genomes reside within conserved regions (Bergman et al. 2002; Emberly et al. 2003; Berman et al. 2004). To assess the extent to which transcription-factor binding sites impact on conservation of the sequences identified in this study, we compared the manually curated set of D. melanogaster transcription-factor binding sites available as the “FlyReg” track on the UCSC genome browser (Bergman et al. 2004) with our set of uc-elements. We found that only 19 of the 23,699 uc-sequences longer than 50 bp contain any “FlyReg” annotation. The small proportion (∼0.06%) of overlap of the two data sets suggests that conservation of these transcription-factor binding sites only makes a minor contribution to the conservation of longer sequences present in the uc-elements data set.
MicroRNAs (miRNAs) play critical roles in Drosophila development and are known to be conserved over large evolutionary distances (Aravin et al. 2003; Brennecke et al. 2003; Lai et al. 2003). We found that 17 of the total of 78 miRNAs in the D. melanogaster miRNA registry (Griffiths-Jones 2004) are present in our set of uc-elements ≥50 bp (Table 4). Two of these miRNA genes are also detectable in the A. gambiae genome. The presence of such a large portion of the known miRNA genes in our ultraconserved data set suggests that this approach may also help to identify novel miRNA genes.
Table 4.
The list of miRNA genes present within the set of ultraconserved sequences between D. melanogaster and D. pseudoobscura longer than 50 bp
| MicroRNA | Genomic location | Length of the uc-element | Reference |
|---|---|---|---|
| dme-mir-125 | Downstream of CG10283 | 113 | 1 |
| dme-mir-307 | Intron of Mmp2 (CG1794) | 92 | 1 |
| dme-mir-9a | Intergenic | 89 | 1 |
| dme-mir-100 | Downstream of CG10283 | 86 | 1 |
| dme-mir-2b-2 | Intron of Spi (CG10334) | 86 | 1 |
| dme-mir-2a-2 | Intron of Spi (CG10334) | 83 | 1 |
| dme-mir-279 | Upstream of CG31044 | 75 | 1 |
| dme-mir-280 | Intergenic | 69 | 2 |
| dme-mir-iab-4* | Within iab-4 RNA | 63 | 1 |
| dme-bantam | Intergenic | 59 | 1 |
| dme-mir-219 | Intergenic | 59 | 2 |
| dme-mir-9c* | grp intron (CG17161) | 58 | 1 |
| dme-mir-287 | Intergenic | 55 | 2 |
| dme-let-7 | Intergenic | 54 | 1 |
| dme-mir-277 | Downstream of Fmr1 (CG6203) | 54 | 1 |
| dme-mir-289 | Intron of bru-3 (CG12478) | 53 | 2 |
| dme-mir-288 | Intergenic | 50 | 2 |
microRNAs marked with an asterisk were also found in the Anopheles genome. References are (1) Aravin et al. 2003, (2) Lai et al. 2003. The full list of uc-elements can be found at http://www.imb.uq.edu.au/groups/mattick/drosophila_uc/.
We then examined the sequences that are exactly conserved between Drosophila and the mosquito A. gambiae (Table 1). The longest sequences that are conserved in all three genomes are located within a snRNA:U6 gene cluster (85, 88, and 93 bp), and at the intron-exon junction of the homothorax (hth) gene (92 bp). The hth gene encodes for a homeodomain containing protein (Pai et al. 1998), which belongs to the MEIS protein family (Steeman et al. 1997). The Meis1 gene was originally identified as a protooncogene involved in neoplastic transformation of hematopoietic cells (Moskow et al. 1995). hth orthologs have been also identified in worms (Van Auken et al. 2002), amphibians (Steeman et al. 1997), and mammals (Moskow et al. 1995; Steeman et al. 1997).
HTH/MEIS proteins have been shown to be involved in several key developmental processes during embryonic development, including the regulation of nervous system differentiation in Drosophila (Kurant et al. 1998) and Xenopus (Maeda et al. 2001). Drosophila hth has also been reported to be involved in eye development (Bessa et al. 2002) and photoreceptor cell-fate determination (Wernet et al. 2003), and the role of HTH/MEIS proteins in limb development has been shown to be evolutionarily conserved in fruit fly, chicken, and mouse (Mercader et al. 1999).
Analysis of the hth ultraconserved sequence
Alignment of the conserved sequence from the D. melanogaser hth gene to the orthologous genomic regions from D. pseudoobscura and A. gambiae revealed that the 92-bp identical match between the three insect genomes extends to 186 bp between the two Drosophila species (Fig. 2A). To gain more information about the nature and the extent of the observed evolutionary conservation, we searched NCBI Genome Trace Archive for sequence similarities with the ultraconserved sequence of the D. melanogaster hth exon-intron junction against the genomes of other Drosophila species, and the honey bee (Apis mellifera). We found that the orthologous sequences from D. simulans and D. yakuba, which diverged from D. melanogaster ∼2.5 and 5 million years ago, respectively (Powell 1996), have 100% identity with D. melanogaster sequence over a 312 bp genomic fragment (data not shown).
Figure 2.
(A) Alignment of five orthologous fragments of the hth gene from various insects. The species are Drosophila melanogaster (D. mel.), Drosophila pseudoobscura (D. ps.), Drosophila virilis (D. vir.), Anopheles gambiae (A. gam.), and Apis melifera (A. mel.). The conserved intronic sequence is shown in lowercase; uppercase represents the adjacent constitutive exon 6. The donor splice-site consensus is underlined. Black arrowheads show the start and end of the predicted hairpin fold region in the first four species.(B) RNA hairpin structures of the highly conserved intronic sequence predicted by MFOLD 3.1. The donor splice-site consensus is marked by the bracket. Splice-site positions are indicated by black arrowheads. Open arrows show substituted nucleotides in the A. gambiae sequence. (C) Splicing diagram of the hth gene (not shown to scale). Gray and white boxes represent protein-coding and noncoding exons, respectively. Straight lines connect constitutively spliced exons. Angled lines show sites of alternative splicing. The transcript BT010238 terminates at the exon 7′. Positions of primers used in real-time PCR are indicated by arrows. The fragment amplified by tF/tR (HTH-t) primer pair is present in all hth RNA transcripts, and thus, detects total transcriptional output of the hth gene. The fragment amplified by sF/sR (HTH-s) primer pair detects the presence of spliced hth mRNA (GenBank accession BT010238). The fragment amplified by sF/iR (HTH-i) primer pair spans the border of the constitutive exon and the adjacent intron, and thus detects the presence of intron-retained hth RNA isoforms. A fragment amplified by cF/cR (HTH-c) primer pair is present in all hth RNA transcripts except BT010238, and thus, detects abundance of the remaining hth transcripts. The enlarged portion of the diagram shows the position of the ultraconserved element (thick black line) and the location of DNA probes used in RNA-blot hybridization.
D. virilis, which diverged from D. melanogaster around 40-60 million years ago (Gaunt and Miles 2002) shows a similar pattern of conservation to that of D. pseudoobscura, which is separated from D. melanogaster by ∼25-30 million years (Fig. 2A).
Interestingly, both D. virilis and D. pseudoobscura show some synonymous nucleotide substitutions in the adjacent protein-coding exon sequence, but do not have any mismatches in the conserved intronic fragment (Fig. 2A). In A. gambiae, which diverged from the Drosophila lineage ∼250 million years ago, there are only three nucleotide substitutions over the 138-bp sequence within the intron. These data clearly indicate that this intronic sequence is under strong selective constraint, which is greater than that in the adjacent protein-coding sequence. On the other hand, despite the fact that we successfully identified a sequence with a high degree of similarity to the Drosophila hth protein-coding sequence in the A. mellifera genome, we were unable to find any significant similarity in the adjacent intron (Fig. 2A). This observation suggests a tight link between primary sequence conservation and the function of this intronic element, and that this is relevant to the Diptera, but not to the Hymenoptera.
One possibility is that the conservation of the intronic sequence might be due to formation of a pre-mRNA secondary structure, which we assessed using the MFOLD 3.1 nucleic acids structure prediction program (Zuker 2003). The structures predicted by MFOLD using sequences from D. melanogaster and A. gambiae are shown in Figure 2B. Interestingly, we found that nucleotide substitutions present in A. gambiae did not affect the overall structure prediction, suggesting a compensatory nature of these mutations. An important observation is that the predicted RNA hairpin structure includes the donor splice site of the intron and a small portion of the adjacent exon (Fig. 2B). Extension of the folding sequence up to 800 nucleotides in either the 5′ or 3′ direction along the gene did not alter the prediction of the putative hairpin structure, indicating its local thermodynamic stability. In contrast, no similar or strong secondary structure is predicted by MFOLD for the orthologous A. mellifera sequence (data not shown).
Analysis of hth RNA expression
To determine the effect that the putative hairpin structure might have on hth pre-mRNA splicing at this splice site, we used real-time PCR to examine different hth RNA splice isoforms (Fig. 3). To quantify the total transcriptional output from the hth locus, we designed a pair of primers (tF/tR) spanning two constitutive coding exons that are present in all known hth RNA transcripts (Fig. 2C). Another pair of primers (sF/iR) were directed at sequences flanking the ultraconserved splice site. The position of the sF forward primer within the constitutive coding exon ensured that sF/iR pair would detect the presence of the conserved intronic element. As a control for the splicing of the hth mRNAs, two pairs of primers (sF/sR and cF/cR) were designed to detect the spliced form of one of the known hth transcripts (Fig. 2C). sF/sR pair detects a homothorax isoform that terminates at the alternative exon 7′ (GenBank accession BT010238), and thus, lacks the sequence encoding a homeodomain present in other transcripts. cF/cR pair spans across the constitutive exons present in all other known transcripts (GenBank accessions AF026788, AF036584, AF035825, and AF032865) thus providing data of relative abundance of these transcripts to the total transcriptional output and the BT010238 transcript.
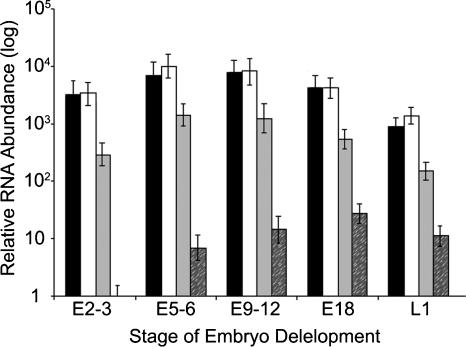
Figure 3.
Relative quantification of hth splice variants by real-time PCR. The diagram shows the relative quantification of hth mRNA splice variants. Black bars represent relative abundance of the HTH-t amplicon, reflecting total transcriptional output from the hth gene. Relative abundance of the intron-retained and spliced BT010238 transcripts is shown as white and dark-gray bars, respectively. Light-gray bars demonstrate the relative abundance of the other known RNA isoforms detected by the cF/cR primer pair. Mean and standard deviation were calculated based on six independent values obtained in two experiments performed in triplicate. Note, the abscissa is a log scale.
Previous studies (Rieckhof et al. 1997; Kurant et al. 1998), have shown that hth RNAs are present in relatively high abundance at early stages of Drosophila development, so we decided to examine the splicing pattern of the hth transcripts during these stages. Surprisingly, we found that hth transcripts that retained the conserved intronic element constitute the majority of the total hth RNA pool (Fig. 3). Intron-retained hth RNAs were at least 10 times more abundant than any of the previously recorded hth transcripts, which were difficult to detect in Northerns, but were quantifiable by PCR.
These data led us to conclude that the putative hairpin structure present in the hth pre-mRNA transcripts could be involved in the regulation of hth splicing by blocking the donor splice site and causing intron retention. To explore this hypothesis further, we performed Northern blot hybridization using exon- and intron-specific DNA probes (Fig. 2C). A membrane hybridized with 32P-labeled intron-specific probe (probe II) is shown in the Figure 4A. Two major bands of ∼4.5 and 7.5 kb were detected using this probe. The same hybridization pattern was observed after stripping the membrane and reprobing it with the exon-specific probe I (Fig. 4B). Remarkably, both of the detected bands were longer than any of the known spliced hth transcripts (Supplemental Table 2). Together, these results strongly indicate that the retained intron is present in the majority of hth steady-state RNAs.
Figure 4.
Expression analysis hth transcripts. (A,B) RNA-blot hybridizations of 10 μg of total RNA prepared from several stages of Drosophila development. The stages of embryo development are indicated as follows: 2-3 h post-laying (E2-E3), 5-6 h post-laying (E5-E6), 9-12 h postlaying (E9-E12), 18 h post-laying (E18), first instar larvae (L1), and third instar larvae (L3). (A) A membrane hybridized with the intron-specific probe II. (B) The same membrane hybridized with the exon-specific probe I after stripping. The positions of 0.24-9.5 kb RNA Ladder bands are indicated on the right. (C) shows a photograph of the ethidium bromide-stained gel with Drosophila ribosomal RNAs.
Discussion
The results shown here indicate that highly conserved noncoding sequences also occur in insect genomes, although they are smaller and fewer in number than those observed in vertebrates (Bejerano et al. 2004). It should be noted however, that while the divergence time between D. melanogaster and D. pseudoobscura is less than that between humans and rodents, and the divergence time between Drosophila/Anopheles is similar to that between mammals and birds, analysis of the Drosophila and Anopheles genomes has revealed that these two insects have diverged significantly faster than vertebrates, presumably due to a greater number of generations and differential substitution rates (Waterston et al. 2002; Zdobnov et al. 2002). This may account, at least in part, for the existence of fewer sequences conserved between them. Nevertheless, as with the mammalian genomes (Bejerano et al. 2004), we found biases between different classes of the uc-elements and their GO annotation. Similarly to mammalian uc-elements, the Drosophila intronic and intergenic uc-elements are enriched for genes with GO terms related to transcription factor activity, and thus, we suggest that they are likely to be a general feature of the genetic programming of the ontogeny of complex organisms via as-yet-unknown mechanisms. Given this prediction, these elements should be less abundant in the simpler metazoa. Indeed, this is the case in nematodes where conserved sequences occur mainly in exons, with no significant bias toward regulatory proteins (M. Pheasant and J.S. Mattick, unpubl.).
A functional bias in the GO annotation is also present in the uc-elements overlapping exons and splice sites, although it is different between vertebrate and insect genomes. In mammalian genomes, this class is enriched for RNA-binding GO terms (Bejerano et al. 2004), whereas in insects, it has significant over-representation of ion channel/transporter activity annotations. Moreover, this class includes the longest uc-elements in insects. As noted above, we found that seven of the 10 genes associated with the longest uc-elements encode ion channels/transporter proteins (Table 3), six of which undergo extensive RNA editing (Hanrahan et al. 2000; Hoopengardner et al. 2003). The latter may partly explain the high level of conservation in this class of uc-elements, as RNA adenosine deaminases (ADARs) require the presence of double-stranded RNA as a substrate (Bass 2002). Thus, a strong selection constraint may apply to the primary RNA sequence involved in intramolecular duplex formation that can be appropriately recognized by ADARs. Conservation of secondary structure is also important for miRNA precursor processing (Denli et al. 2004).
Similarly, the high level of conservation of the intronic sequence in the homothorax gene is likely due to selective pressure to maintain putative RNA secondary structure around the donor splice site, in this case, apparently to regulate splicing rather than RNA editing. However, since the processes of alternative splice-site selection and RNA editing can be closely related (Bass 2002), we cannot rule out the possibility of RNA editing in the homothorax gene, although we did not detect any A-to-G substitution around the conserved splice site in the sequenced hth cDNAs prepared from mixed stage embryos (data not shown). In the honey bee, the function of the RNA hairpin structure appears to have been lost (or was not present in the common ancestor of the Diptera and the Hymenoptera), since the nucleotide sequence of the orthologous intronic fragment has diverged, except for a few nucleotides representing the donor splice site consensus (Weir and Rice 2004; Fig. 2A).
A role for secondary structure in pre-mRNA splicing was first proposed almost 20 years ago (Solnick 1985). However, artificially introduced secondary structures were examined in that study, and despite the success of these proof-of-concept experiments, their biological relevance has remained uncertain. Due to the fact that systematic identification of RNA secondary structures was and still is a challenging task, there have been relatively few studies addressing this issue. Goguel et al. (1993) have shown that in yeast, even a 6-bp long hairpin structure inserted at the 5′ splice site could reduce splicing efficiency, while insertion of a 15-bp long hairpin structure led to almost complete inhibition of splicing. These authors showed that sequestration of splicing occurs at the early stage of U1 ribonucleoprotein complex assembly. The first example of an effect of a naturally occurring intramolecular RNA duplex on pre-mRNA splicing efficiency was demonstrated by Blanchette and Chabot (1997), who showed that an 84-nucleotide duplex-forming region covering the 5′ splice site decreases splicing efficiency and is associated with a reduction in the assembly of U1-dependent spliceosomal complex.
By comparison, the putative hairpin structure identified in our study covers 135 nucleotides, 96 of which are paired, suggesting that a similar mechanism is used in the homothorax transcripts, in that the thermodynamically stable hairpin structure predicted to be formed around the donor splice site would block access of the U1 RNP complex assembly, resulting in intron retention. The intron-retained hth transcripts constitute the majority of the total hth steady-state RNA pool during Drosophila embryogenesis.
Considering the abundance of the intron-retained transcripts, we were surprised that their existence has not been reported previously. Analysis of the published studies on hth expression has led us to the following observations. The hth mRNA expression data are largely derived from in situ hybridization experiments, which utilized various nucleic acid probes (Rieckhof et al. 1997; Kurant et al. 1998; Inoue et al. 2002; Prpic and Tautz 2003), usually directed against the coding part of the hth transcript. This means that while providing valuable information about the pattern of expression of the homothorax gene, the in situ hybridization experiments did not distinguish between alternatively spliced or/and intron-retained transcripts. Interestingly, Kurant et al. (1998) have published an RNA-blot hybridization similar to the one presented in the Figure 4. As in our experiment, two major hth transcripts of ∼4.0-4.5 kb and 6.0-6.5 kb were detected (Kurant et al. 1998). However, the authors did not comment on why the detected transcripts appeared to be substantially longer than expected from the length of the known hth cDNAs. Based on our RNA-blot hybridization data, we suggest that the transcripts detected in the work of Kurant et al. (1998) are the same as those shown in Figure 4. The discrepancy in length between known hth cDNA clones and the observed length of the hybridized bands can be explained by the presence of an 1869-bp long retained intron in these transcripts. Thus, we conclude that intron retention is a key component of the regulation hth splicing in vivo.
A similar role of pre-mRNA secondary structure affecting splicing has been found for the 3′ acceptor spice site in other genes (Coleman and Roesser 1998; Hefferon et al. 2004). Blocking the acceptor splice site via intramolecular duplex formation caused skipping of the subsequent exon in rat Calcitonin/CGRP (Coleman and Roesser 1998) and human CFTR RNA transcripts (Hefferon et al. 2004). Interestingly, in the latter case, a degree of secondary structure seemed to be needed for proper splice-site selection, as removal of the whole duplex-forming region also caused exon skipping. Duplex and/or secondary structure formation inside of an intron has been shown to be important for mutually exclusive exon selection in fibroblast growth factor receptor 2 (Muh et al. 2002; Baraniak et al. 2003), as well as for exon inclusion in mouse and human fibronectin (Buratti et al. 2004) and YL8A yeast transcripts (Howe and Ares 1997). In addition, two recently published studies on a genome-wide scale have demonstrated that intron retention is common (Galante et al. 2004; Ner-Gaon et al. 2004). Another comparative genomic study has shown that alternative splicing is often associated with a high level of nucleotide sequence conservation (Philipps et al. 2004). Pre-mRNA secondary structure formation seems to play a vital role in regulation of gene expression by influencing the exon composition of the spliced mRNA via splice-site selection and by controlling the level of functional mRNA via intron retention. Although some generality in these mechanisms is becoming evident the molecular nature of the involved components, and how these secondary structures may be resolved to allow productive splicing for protein synthesis, remains to be determined.
Methods
Identification of ultraconserved elements
A mirror of the UCSC genome browser and database (Kent et al. 2002; Karolchik et al. 2003; http://genome.ucsc.edu/) was created with the D. melanogaster Release 3.2 genome sequence and annotations (dm1, March 2004) (Celniker and Rubin 2003; Gelbart et al. 2003), the D. pseudoobscura genome assembly (dp2, August 2003) (http://www.hgsc.bcm.tmc.edu/projects/drosophila/) (Richards et al. 2005), and the Anopheles gambiae MOZ2 draft genome sequence (anoGam1, February 2003) (Holt et al. 2002). Pairwise “AXT” (Kent et al. 2003) alignments for dm1/dp2 and dm1/anoGam1 were scanned to identify 100% conserved regions. The fraction of uc-elements annotated as coding, intronic, and intergenic were determined using the UCSC “BDGP Genes” and “RefSeq” data (Pruitt and Maglott 2001; Karolchik et al. 2003).
Percent identity of alignable regions between genomes was calculated using the “axtAndBed” and “axtCalcMatrix” (Karolchik et al. 2003) programs.
The calculation of expected number of the uc-elements was performed as follows. First, we calculated the fractional identity (Base Id) of the alignable sequence between D. melanogaster and D. pseudoobscura genomes as the total number of aligned identical bases/total number of aligned identical bases + total number of aligned substituted bases. Then, the expected number of uc-elements of length L = NL = Genome Length χ (Base Id)L.
Accession numbers
The FlyBase accession number of the Drosophila homothorax gene is CG17117. The Ensembl (http://www.ensembl.org/) accession number of the homothorax othologous gene in Anopheles gambiae is ENSANGG00000022368. The Ensembl accession number of the orthologous homothorax gene in Apis mellifera is ENSAPMG00000006561. The NCBI Trace Archive (http://www.ncbi.nlm.nih.gov/Traces/trace.cgi?) accession number of the Drosophila virilis trace used in sequence alignment is ti_471898326.
Gene Ontology enrichment and P-values
Gene Ontology (GO) annotations were taken from the UCSC GO database (June 2004) (Camon et al. 2004; Harris et al. 2004). FlyBase (Gelbart et al. 2003) gene identifiers were used to make sure a gene was counted only once where there were multiple transcript isoforms. A Perl script and SQL code (M. Pheasant, unpubl.) were created to calculate enrichment of GO terms and “Fisher's Exact” P-values against a background of all GO annotated FlyBase identifiers in the UCSC Known Genes database. For significance, we required at least twofold enrichment, P < 10-10, and at least 10 associated FlyBase genes in the uc-elements sample.
Drosophila embryo collection
D. melanogaster Oregon Red wild-type flies were used in all experiments. Flies were reared on a standard cornmeal diet (Sigma) at 25°C with a 12-h light/12-h dark cycle. Embryos were collected using standard procedures for synchronized development (Rothwell and Sullivan 2000). In brief, flies were transferred to the embryo collection bottles and were allowed to lay eggs on apple juice medium for 40 min. The first two batches of eggs were discarded to ensure that all laid eggs were synchronous. The laid eggs were incubated at 25°C for variable times. After the incubation period, embryos were harvested, snap frozen in the liquid nitrogen, and subsequently stored at -80°C until RNA extraction.
RNA isolation and cDNA synthesis
Total RNA was extracted from several developmental stages of Drosophila embryos using TRIZOL reagent (Invitrogen) following the manufacturer's protocol. The RNA concentration and purity were determined photometrically by measuring absorbance at 260 nm and A260/A280 ratio (Smart-Spec 3000, BioRad). For cDNA synthesis, 1 μg of total RNA was treated with DNase I (Invitrogen) and then utilized as a template for randomly primed reverse transcription using Omniscript Reverse Transcriptase Kit (QIAGEN), according to the manufacturer's instructions. The resulting cDNA was diluted 1:25 (v/v) with nuclease-free water. A total of 5 μL of the cDNA was used as a template for quantitative real-time PCR.
Quantitative real-time PCR
Relative abundance of different splice isoforms of the hth RNA transcripts was determined by real-time PCR using SYBR Green RCR Master Mix (Applied Biosystems) according to manufacture's protocol. The PCR amplification was performed on the ABI PRISM 7700 sequence-detection system (Applied Biosystems) in a final volume of 25 μL using standard cycling parameters (2 min, 50°C; 10 min, 95°C; 30 sec, 95°C; 1 min 60°C, with the latter two steps repeated for 45 times). Primers for the real-time PCR reaction were designed using Vector NTI 9.0 software package (Invitrogen), following primer design guidelines given in the SYBR green PCR Master Mix and RT-PCR protocol (Applied Biosystems). The melting temperatures of all primers were between 58 and 62°C. All primers were purchased from Proligo (Proligo Australia Pty Ltd).
The primers used for detection of total transcriptional output from the hth locus were as follows:
tF 5′-TCATGTATCGCCGGTCGGTAATCA-3′;
tR 5′-CAAAAGCGGGAATAGCGGATGTT-3′.
The primers used for detection of the spliced hth transcript BT010238 were as follows:
sF 5′-GCAGCAACGCCGACTCCACA-3′;
sR 5′-GCATAGGTCTCCTCCAGCGGTGA-3′.
The reverse primer used in combination with the sF to detect intron-retained hth transcripts was as follows:
iR 5′-GGGTGCACCACCGCATTGTA-3′.
The primers used for detection of transcriptional output from the hth locus excluding BT010238 transcript were as follows:
cF-5′-CCGGCGATTGTATTCCTCAGTCTT-3′;
cR-5′-CCTCTCCACTTCCTATGCTGGCATT-3′.
Primers for the 18S ribosomal RNA were as follows:
18S-F 5′-CTTATGGGACATGTGCTTTTATTAGGCTAA-3′;
18S-R 5′-AAAGTTGATAGGGCAGACATTTGAAAGA-3′.
The final optimized concentration of primers was 100 nM for hth amplicons and 300 nM for 18S ribosomal RNA amplicon, which was used as an endogenous control and normalization standard. The absence of inter- and/or intramolecular duplex formation between primers was confirmed in a control real-time PCR reaction lacking template. Relative quantification was performed as described in ABI Prism 7700 Sequence Detection System User Bulletin #2 (Applied Biosystems) according to Comparative CT Method. In brief, threshold cycle (CT) values of experimental samples were normalized to corresponding CT values of the 18S ribosomal RNA control, and then quantified relative to the sample with the maximal CT value (calibrator). All real-time PCR reactions were done in triplicates. The results were confirmed in two independent experiments.
Northern blot hybridization
For RNA-blot hybridization, 10 μg of total RNA was loaded onto an 0.8% (w/v) denaturing agarose gel containing 50% (v/v) of formaldehyde. After electrophoretic separation, RNA was transferred on a Hybond+ membrane (Amersham Life Sciences). Blotted RNA was UV cross-linked to the membrane using a Stratalinker 1800 apparatus (Stratagene). Random primed 32P-labeled DNA probes were prepared as follows. DNA fragments were labeled with [α-32P]dCTP (3000 Ci/mmol, Perkin Elmer) using a rediprime-II random prime labeling system (Amersham Pharmacia). Unincorporated nucleotides were removed using a Micro-Spin G-50 Column (Amersham Pharmacia) according to the manufacturer's instructions. Hybridization was carried out using ULTRAhyb hybridization buffer (Ambion) according to manufacture's recommendations.
Primers used for amplification of DNA probe fragments were as follows:
Probe I: PI-F 5′-GCAGGTACACGAGTTATGCGATAA-3′;
PI-R 5′-GTCTGGTGTACTAGCTCCATCGGT-3′;
Probe II: PII-F 5′-GTGAGTACCCCCTTCGCGGGAGCAC-3′;
PII-R 5′-ACAGTGTGCAGCAACATTCATTTC-3′.
After post-hybridization washes, membranes were exposed to a PhosphorImager screen (Amersham Pharmacia). The probe was removed by pouring boiling 0.5% (w/v) SDS onto the membrane and letting it cool to room temperature.
Acknowledgments
This work was supported by the Australian Research Council and the Queensland State Government. We thank Igor Makunin and Cas Simons for advice and helpful discussion during the preparation of this study. We thank Kelin Ru for technical assistance with the DNA sequencing. We also thank the members of the D. pseudoobscura genome sequencing consortium for making their data and assemblies available in advance of formal publication.
Footnotes
[Supplemental material is available online at www.genome.org and http://www.imb.uq.edu.au/groups/mattick/drosophila_uc/.]
Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.3545105. Article published online before print in May 2005.
References
- Aravin, A.A., Lagos-Quintana, M., Yalcin, A., Zavolan, M., Marks, D., Snyder, B., Gaasterland, T., Meyer, J., and Tuschl, T. 2003. The small RNA profile during Drosophila melanogaster development. Dev. Cell 5: 337-350. [DOI] [PubMed] [Google Scholar]
- Baraniak, A.P., Lasda, E.L., Wagner, E.J., and Garcia-Blanco, M.A. 2003. A stem structure in fibroblast growth factor receptor 2 transcripts mediates cell-type-specific splicing by approximating intronic control elements. Mol. Cell Biol. 23: 9327-9337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bass, B.L. 2002. RNA editing by adenosine deaminases that act on RNA. Annu. Rev. Biochem. 71: 817-846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bejerano, G., Pheasant, M., Makunin, I., Stephen, S., Kent, W.J., Mattick, J.S., and Haussler, D. 2004. Ultraconserved elements in the human genome. Science 304: 1321-1325. [DOI] [PubMed] [Google Scholar]
- Bergman, C.M., Pfeiffer, B.D., Rincon-Limas, D.E., Hoskins, R.A., Gnirke, A., Mungall, C.J., Wang, A.M., Kronmiller, B., Pacleb, J., Park, S., et al. 2002. Assessing the impact of comparative genomic sequence data on the functional annotation of the Drosophila genome. Genome Biol. 3: R86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergman, C.M., Carlson, J.W., and Celniker, S.E. 2004. Drosophila DNase I footprint database: A systematic genome annotation of transcription factor binding sites in the fruitfly, D. melanogaster. Bioinformatics doi: 10.1093/bioinformatics/bti173. [DOI] [PubMed]
- Berman, B.P., Pfeiffer, B.D., Laverty, T.R., Salzberg, S.L., Rubin, G.M., Eisen, M.B., and Celniker, S.E. 2004. Computational identification of developmental enhancers: Conservation and function of transcription factor binding-site clusters in Drosophila melanogaster and Drosophila pseudoobscura. Genome Biol. 5: R61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessa, J., Gebelein, B., Pichaud, F., Casares, F., and Mann, R.S. 2002. Combinatorial control of Drosophila eye development by eyeless, homothorax, and teashirt. Genes & Dev. 16: 2415-2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchette, M. and Chabot, B. 1997. A highly stable duplex structure sequesters the 5′ splice site region of hnRNP A1 alternative exon 7B. RNA 3: 405-419. [PMC free article] [PubMed] [Google Scholar]
- Brennecke, J., Hipfner, D.R., Stark, A., Russell, R.B., and Cohen, S.M. 2003. bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell 113: 25-36. [DOI] [PubMed] [Google Scholar]
- Buratti, E., Muro, A.F., Giombi, M., Gherbassi, D., Iaconcig, A., and Baralle, F.E. 2004. RNA folding affects the recruitment of SR proteins by mouse and human polypurinic enhancer elements in the fibronectin EDA exon. Mol. Cell Biol. 24: 1387-1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camon, E., Magrane, M., Barrell, D., Lee, V., Dimmer, E., Maslen, J., Binns, D., Harte, N., Lopez, R., and Apweiler, R. 2004. The Gene Ontology Annotation (GOA) Database: Sharing knowledge in Uniprot with Gene Ontology. Nucleic Acids Res. 32: D262-D266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celniker, S.E. and Rubin, G.M. 2003. The Drosophila melanogaster genome. Annu. Rev. Genomics Hum. Genet. 4: 89-117. [DOI] [PubMed] [Google Scholar]
- Coleman, T.P. and Roesser, J.R. 1998. RNA secondary structure: An important cis-element in rat calcitonin/CGRP pre-messenger RNA splicing. Biochemistry 37: 15941-15950. [DOI] [PubMed] [Google Scholar]
- Denli, A.M., Tops, B.B., Plasterk, R.H., Ketting, R.F., and Hannon, G.J. 2004. Processing of primary microRNAs by the Microprocessor complex. Nature 432: 231-235. [DOI] [PubMed] [Google Scholar]
- Emberly, E., Rajewsky, N., and Siggia, E.D. 2003. Conservation of regulatory elements between two species of Drosophila. BMC Bioinformatics 4: 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galante, P.A., Sakabe, N.J., Kirschbaum-Slager, N., and de Souza, S.J. 2004. Detection and evaluation of intron retention events in the human transcriptome. RNA 10: 757-765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaunt, M.W. and Miles, M.A. 2002. An insect molecular clock dates the origin of the insects and accords with palaeontological and biogeographic landmarks. Mol. Biol. Evol. 19: 748-761. [DOI] [PubMed] [Google Scholar]
- Gelbart, W., Bayraktaroglu, L., Bettencourt, B., Campbell, K., Crosby, M., Emmert, D., Hradecky, P., Huang, Y., Letovsky, S., Matthews, B., et al. 2003. The FlyBase database of the Drosophila genome projects and community literature. Nucleic Acids Res. 31: 172-175.12519974 [Google Scholar]
- Goguel, V., Wang, Y., and Rosbash, M. 1993. Short artificial hairpins sequester splicing signals and inhibit yeast premessenger RNA splicing. Mol. Cell Biol. 13: 6841-6848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones, S. 2004. The microRNA Registry. Nucleic Acids Res. 32: D109-D111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanrahan, C.J., Palladino, M.J., Ganetzky, B., and Reenan, R.A. 2000. RNA editing of the Drosophila para Na+ channel transcript: Evolutionary conservation and developmental regulation. Genetics 155: 1149-1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris, M.A., Clark, J., Ireland, A., Lomax, J., Ashburner, M., Foulger, R., Eilbeck, K., Lewis, S., Marshall, B., Mungall, C., et al. 2004. The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res. 32: D258-D261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hefferon, T.W., Groman, J.D., Yurk, C.E., and Cutting, G.R. 2004. A variable dinucleotide repeat in the CFTR gene contributes to phenotype diversity by forming RNA secondary structures that alter splicing. Proc. Natl. Acad. Sci. 101: 3504-3509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt, R.A., Subramanian, G.M., Halpern, A., Sutton, G.G., Charlab, R., Nusskern, D.R., Wincker, P., Clark, A.G., Ribeiro, J.M.C., Wides, R., et al. 2002. The genome sequence of the malaria mosquito Anopheles gambiae. Science 298: 129-149. [DOI] [PubMed] [Google Scholar]
- Hoopengardner, B., Bhalla, T., Staber, C., and Reenan, R. 2003. Nervous system targets of RNA editing identified by comparative genomics. Science 301: 832-836. [DOI] [PubMed] [Google Scholar]
- Howe, K.J. and Ares, M. 1997. Intron self-complementarity enforces exon inclusion in a yeast pre-mRNA. Proc. Natl. Acad. Sci. 94: 12467-12472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue, Y., Mito, T., Miyawaki, K., Matsushima, K., Shinmyo, Y., Heanue, T.A., Mardon, G., Ohuchi, H., and Noji, S. 2002. Correlation of expression patterns of homothorax, dachshund, and Distal-less with the proximodistal segmentation of the cricket leg bud. Mech. Dev. 113: 141-148. [DOI] [PubMed] [Google Scholar]
- Karolchik, D., Baertsch, R., Diekhans, M., Furey, T.S., Hinrichs, A., Lu, Y.T., Roskin, K.M., Schwartz, M., Sugnet, C.W., Thomas, D.J., et al. 2003. The UCSC Genome Browser Database. Nucleic Acids Res. 31: 51-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent, W.J., Sugnet, C.W., Furey, T.S., Roskin, K.M., Pringle, T.H., Zahler, A.M., and Haussler, D. 2002. The human genome browser at UCSC. Genome Res. 12: 996-1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent, W.J., Baertsch, R., Hinrichs, A., Miller, W., and Haussler, D. 2003. Evolution's cauldron: Duplication, deletion, and rearrangement in the mouse and human genomes. Proc. Natl. Acad. Sci. 100: 11484-11489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, S. and Hedges, S.B. 1998. A molecular timescale for vertebrate evolution. Nature 392: 917-920. [DOI] [PubMed] [Google Scholar]
- Kurant, E., Pai, C.Y., Sharf, R., Halachmi, N., Sun, Y.H., and Salzberg, A. 1998. dorsofonals/homothorax, the Drosophila homologue of meis1, interacts with extradenticle in patterning of the embryonic PNS. Development 125: 1037-1048. [DOI] [PubMed] [Google Scholar]
- Lai, E.C., Tomancak, P., Williams, R.W., and Rubin, G.M. 2003. Computational identification of Drosophila microRNA genes. Genome Biol. 4: R42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda, R., Mood, K., Jones, T.L., Aruga, J., Buchberg, A.M., and Daar, I.O. 2001. Xmeis1, a protooncogene involved in specifying neural crest cell fate in Xenopus embryos. Oncogene 20: 1329-1342. [DOI] [PubMed] [Google Scholar]
- Mercader, N., Leonardo, E., Azpiazu, N., Serrano, A., Morata, G., Martinez, C., and Torres, M. 1999. Conserved regulation of proximodistal limb axis development by Meis1/Hth. Nature 402: 425-429. [DOI] [PubMed] [Google Scholar]
- Moskow, J.J., Bullrich, F., Huebner, K., Daar, I.O., and Buchberg, A.M. 1995. Meis1, a PBX1-related homeobox gene involved in myeloid leukemia in BXH-2 mice. Mol. Cell Biol. 15: 5434-5443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muh, S.J., Hovhannisyan, R.H., and Carstens, R.P. 2002. A non-sequence-specific double-stranded RNA structural element regulates splicing of two mutually exclusive exons of fibroblast growth factor receptor 2 (FGFR2). J. Biol. Chem. 277: 50143-50154. [DOI] [PubMed] [Google Scholar]
- Ner-Gaon, H., Halachmi, R., Savaldi-Goldstein, S., Rubin, E., Ophir, R., and Fluhr, R. 2004. Intron retention is a major phenomenon in alternative splicing in Arabidopsis. Plant J. 39: 877-885. [DOI] [PubMed] [Google Scholar]
- Pai, C.Y., Kuo, T.S., Jaw, T.J., Kurant, E., Chen, C.T., Bessarab, D.A., Salzberg, A., and Sun, Y.H. 1998. The Homothorax homeoprotein activates the nuclear localization of another homeoprotein, Extradenticle, and suppresses eye development in Drosophila. Genes & Dev. 12: 435-446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philipps, D.L., Park, J.W., and Graveley, B.R. 2004. A computational and experimental approach toward a priori identification of alternatively spliced exons. RNA 10: 1838-1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell, J.R. 1996. Progress and prospects in evolutionary biology. Oxford University Press, Oxford, UK.
- Prpic, N.M. and Tautz, D. 2003. The expression of the proximodistal axis patterning genes Distal-less and dachshund in the appendages of Glomeris marginata (Myriapoda: Diplopoda) suggests a special role of these genes in patterning the head appendages. Dev. Biol. 260: 97-112. [DOI] [PubMed] [Google Scholar]
- Pruitt, K.D. and Maglott, D.R. 2001. RefSeq and LocusLink: NCBI gene-centered resources. Nucleic Acids Res. 29: 137-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards, S., Liu, Y., Bettencourt, B.R., Hradecky, P., Letovsky, S., Nielsen, R., Thornton, K., Hubisz, M.J., Chen, R., Meisel, R.P., et al. 2005. Comparative genome sequencing of Drosophila pseudoobscura: Chromosomal, gene, and cis-element evolution. Genome Res. 15: 1-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieckhof, G.E., Casares, F., Ryoo, H.D., AbuShaar, M., and Mann, R.S. 1997. Nuclear translocation of extradenticle requires homothorax, which encodes an extradenticle-related homeodomain protein. Cell 91: 171-183. [DOI] [PubMed] [Google Scholar]
- Rothwell, W.F. and Sullivan, W. 2000. Fluorescent analysis of Drosophila embryos. In Drosophila Protocols (eds. W. Sullivan et al.), pp. 151-157. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York.
- Solnick, D. 1985. Alternative splicing caused by RNA secondary structure. Cell 43: 667-676. [DOI] [PubMed] [Google Scholar]
- Steeman, S., Moskow, J.J., Druck, T., Montgomery, J.C., Huebner, K., Daar, I.O., and Buchberg, A.M. 1997. Identification of a conserved family of Meis1-related homeobox gene. Genome Res. 7: 142-156. [DOI] [PubMed] [Google Scholar]
- Van Auken, K., Weaver, D., Robertson, B., Sundaram, M., Saldi, T., Edgar, L., Elling, U., Lee, M., Boese, Q., and Wood, W.B. 2002. Roles of the Homothorax/Meis/Prep homolog UNC-62 and the Exd/Pbx homologs CEH-20 and CEH-40 in C. elegans embryogenesis. Development 129: 5255-5268. [DOI] [PubMed] [Google Scholar]
- Waterston, R.H., Lindblad-Toh, K., Birney, E., Rogers, J., Abril, J.F., Agarwal, P., Agarwala, R., Ainscough, R., Alexandersson, M., An, P., et al. 2002. Initial sequencing and comparative analysis of the mouse genome. Nature 420: 520-562. [DOI] [PubMed] [Google Scholar]
- Weir, M. and Rice, M. 2004. Ordered partitioning reveals extended splice-site consensus information. Genome Res. 14: 67-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wernet, M.F., Labhart, T., Baumann, F., Mazzoni, E.O., Pichaud, F., and Desplan, C. 2003. Homothorax switches function of Drosophila photoreceptors from color to polarized light sensors. Cell 115: 267-279. [DOI] [PubMed] [Google Scholar]
- Woolfe, A., Goodson, M., Goode, D.K., Snell, P., McEwen, G.K., Vavouri, T., Smith, S.F., North, P., Callaway, H., Kelly, K., et al. 2005. Highly conserved non-coding sequences are associated with vertebrate development. PLoS Biol. 3: e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zdobnov, E.M., von Mering, C., Letunic, I., Torrents, D., Suyama, M., Copley, R.R., Christophides, G.K., Thomasova, D., Holt, R.A., Subramanian, G.M., et al. 2002. Comparative genome and proteome analysis of Anopheles gambiae and Drosophila melanogaster. Science 298: 149-159. [DOI] [PubMed] [Google Scholar]
- Zuker, M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31: 3406-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- http://www.imb.uq.edu.au/groups/mattick/drosophila_uc/; IMB research Web site.
- http://www.hgsc.bcm.tmc.edu/projects/drosophila/; Human Genome Sequencing Center at Baylor College of Medicine. [DOI] [PubMed]
- http://www.ensembl.org/; Ensembl Genome Browser.
- http://www.ncbi.nlm.nih.gov/Traces/trace.cgi?; NCBI Trace Archive.
- http://genome.ucsc.edu/; UCSC Genome Browser.