Abstract
Imprinted genes are epigenetically modified genes whose expression is determined according to their parent of origin. They are involved in embryonic development, and imprinting dysregulation is linked to cancer, obesity, diabetes, and behavioral disorders such as autism and bipolar disease. Herein, we train a statistical model based on DNA sequence characteristics that not only identifies potentially imprinted genes, but also predicts the parental allele from which they are expressed. Of 23,788 annotated autosomal mouse genes, our model identifies 600 (2.5%) to be potentially imprinted, 64% of which are predicted to exhibit maternal expression. These predictions allowed for the identification of putative candidate genes for complex conditions where parent-of-origin effects are involved, including Alzheimer disease, autism, bipolar disorder, diabetes, male sexual orientation, obesity, and schizophrenia. We observe that the number, type, and relative orientation of repeated elements flanking a gene are particularly important in predicting whether a gene is imprinted.
Imprinted genes represent a small subset of mammalian genes that are monoallelically expressed in a parent-of-origin manner. Experimental evidence suggests that genomic imprinting evolved ∼180 million years ago in a common ancestor to viviparous mammals after divergence from the egg-laying monotremes (Killian et al. 2000, 2001; Murphy and Jirtle 2003). Imprinting is postulated to provide a means by which the paternal and maternal genomes exert counteracting growth effects during development (Haig and Graham 1991). This is exemplified by the paternally expressed Igf2 and the maternally expressed Igf2r stimulating and inhibiting embryonic growth, respectively.
Prader-Willi syndrome (PWS) and Angelman syndrome (AS) are examples of neurodevelopmental disorders linked to imprinted gene dysregulation. Evidence is also mounting that imprinted genes play a significant role in the genesis of cancer, obesity, and diabetes (Murphy and Jirtle 2003; Waterland and Jirtle 2003; Feinberg and Tycko 2004). Moreover, imprinted genes are targets through which environmental factors can influence gene expression (Waterland and Jirtle 2004). For these reasons, it is critically important to identify imprinted genes, as well as the cis-acting regulatory elements involved in the establishment and maintenance of imprinting.
To date, most efforts to identify imprinted genes have been experimental, focusing on small regions of a chromosome. High-throughput screens based on differential expression have been performed by using RIKEN cDNA microarrays, and they have led to the discovery of several novel imprinted genes (Mizuno et al. 2002; Nikaido et al. 2003). Although it has been proposed that the concentration of certain types of repeated elements and other sequence characteristics might be useful in distinguishing between monoallelically and biallelically expressed genes (Greally 2002; Ke et al. 2002; Allen et al. 2003), no large-scale predictions have previously been performed based on DNA sequence characteristics alone.
This study describes a machine learning approach across the entire mouse genome for both identifying imprinted gene candidates and predicting their parental expression preference. We collected a series of DNA sequence features within and flanking each locus, such as statistics on repetitive elements, transcription factor binding sites, and CpG islands. Based on these features, we subsequently trained a classifier employing a two-tier prediction strategy. Each gene in the mouse genome was first predicted to be either imprinted or nonimprinted, and then the parental allele preferentially expressed was predicted for all candidate imprinted genes.
Results
Genome-wide prediction of candidate imprinted genes
In applying this classifier to the entire mouse genome, the algorithm predicted 600 genes out of a total of 23,788 annotated autosomal genes to be imprinted (2.5%); 384 (64%) of these candidate imprinted genes were predicted to exhibit maternal expression. The entire set of predictions is listed in the Supplemental Table 6.
The frequency of imprinted gene candidates did not vary significantly either between or within the chromosomes (Supplemental material; Fig. 1). Nevertheless, the frequency of imprinted gene candidates was significantly higher within six autosomal bands than in the rest of the autosome: 12d1 (4/16 were predicted to be imprinted), 7b5 (5/25), 18b1 (4/20), 6a1 (8/45), and 7f5 (16/193). Chromosomal bands 6a1, 7b5, and 7f5 contain known imprinted genes. The candidates on 18b1 are located 10 Mb distal to the imprinted gene Impact, while the candidates on 12d1 are 35 Mb proximal from the imprinted Dlk1/Meg3 cluster.
Figure 1.
Subset of mouse chromosome 7f5 that is homologous to the imprinted human BWS region at 11p15.5. Red, blue, and black coloring define genes predicted to be maternally, paternally, or biallelically expressed, respectively. Solid arrowheads represent genes used for training, while genes not used for training are denoted by empty or shaded arrowheads and rectangles. Vertically hatched arrowheads represent weakly imprinted genes. Empty arrowheads identify genes for which there is conflicting data. Horizontal hatching denotes biallelic test genes. Genes for which no gene name was available are represented by their Ensembl ID. Coordinates are based on Ensembl annotation; units are kilobases.
Assessment of prediction accuracy
We assessed the prediction accuracy of our classifier by using both cross-validation and an independent set of test genes. We randomly partitioned the training data into 44 groups for cross-validation, each containing one imprinted gene and 12-13 control genes. We subsequently withheld one group from the model fitting process and then used that model to make predictions on the withheld group. We repeated this process for all groups and at each iteration chose significant features de novo among all 6831 available features. In this cross-validation assessment, we obtained a specificity of 100% (44/44 imprinted genes correctly identified) and a sensitivity of 93% (495/530 presumably nonimprinted genes correctly identified). There were 30 randomly chosen control genes whose imprinting status is unknown among the 35 false-positive predictions. Since we predicted ∼2.5% of the autosomal genes to be potentially imprinted, we would expect up to 13 imprinted genes within a random sample of 500 genes. Thus, almost half of the 30 control genes predicted to be imprinted might actually be imprinted and not be misclassifications.
Interestingly, even microimprinted genes such as Nnat and U2af1-rs1 were correctly classified, when left out of the training process. Furthermore, nonimprinted genes in whose introns these two imprinted genes are located, Bc10 and Murr1, were correctly predicted not to be imprinted. Bc10 is biallelically expressed in both human (Evans et al. 2001) and mouse (John et al. 2001). Murr1 is biallelically expressed in neonatal mouse (Nabetani et al. 1997), but a recent study found predominant maternal expression in the adult mouse brain, an effect that is proposed to stem from transcriptional interference by U2af1-rs1 (Y. Wang et al. 2004). No human homolog of U2af1-rs1 is known. Misclassification will mostly lead to biallelically expressed genes being erroneously labeled as imprinted, rather than truly imprinted genes being overlooked. Thus, the results of this cross-validation indicated that we could identify a large portion of the imprinted protein coding genes residing in the mouse genome.
To further validate our predictions, we excluded an independent set of experimentally validated genes (Supplemental Table 2) from the model learning process. This test set consisted of 18 genes with random monoallelic expression and 82 genes with biallelic expression or synchronous replication. Our algorithm correctly classified all 18 genes with random monoallelic expression as nonimprinted. It also correctly classified 81 of the 82 genes presumed to have biallelic expression as nonimprinted (sensitivity of 98.7%). The single misclassified gene, Ctsd, is located ∼200 kb proximal from imprinted H19 and was predicted to be paternally expressed (Fig. 1). The nonimprinted status of human CTSD is only inferred from its expression in hydatidiform mole, mature teratoma, and normal placenta (Rachmilewitz et al. 1993). Hydatidiform mole contains exclusively paternal chromosomes (Szulman 1987), whereas mature teratoma arises from parthenogenic origin and contains maternal chromosomes only (Linder et al. 1975). Based on our prediction that Ctsd may be imprinted and paternally expressed, it could also be speculated that a loss of imprinting might contribute to the tumorous growth in mature teratoma. We are unaware of any reports on its imprinting status in any species, including mouse and human, in either adult or embryonic tissue.
In addition, our test set contained four genes that are imprinted in embryonic or adult tissues in human or mouse (ATP10A, WT1, ALDH1B1, and CPA4). Our algorithm predicted Atp10a, Wt1, and Aldh1b1 to be imprinted. Murine Atp10a is imprinted in some mouse strains (Kashiwagi et al. 2003); its human homolog is imprinted in the brain and in lymphoblasts (Meguro et al. 2001). The human genes, WT1 (Mitsuya et al. 1997; Malik et al. 2000; Dallosso et al. 2004) and ALDH1B1 (http://lpg.nci.nih.gov/LPG/lee/proj2), are reported to be imprinted; however, the imprinting status of their murine homologs remains unknown. Our algorithm also predicted Cpa4 to be nonimprinted. Although human CPA4 is imprinted in several human tissues (Bentley et al. 2003; Kayashima et al. 2003), it is unknown whether Cpa4 is imprinted in mouse. Interestingly, the homeobox gene DLX5 is imprinted and maternally expressed in human (Okita et al. 2003); however, our algorithm classified it as nonimprinted. While this manuscript was under review, Dlx5 was identified as being biallelically expressed in mouse (Kimura et al. 2004).
Despite the fact that the data on which we based our predictions included characteristics of flanking sequence as far as 100 kb from a gene, it appeared that our algorithm operated on a “single-gene level” and was not misled by long-range phenomena. This is illustrated in Figure 1, which shows the gene cluster at human chromosome 11p15.5 that is involved in Beckwith-Wiedemann syndrome (BWS), an organ overgrowth syndrome. Prediction accuracy is clearly demonstrated by the correct classification of the nonimprinted genes Tnnt3 and Cars, despite their close location to imprinted genes.
Significant genomic features for predicting imprinting status
Previous efforts to determine the sequence characteristics of imprinted genes have demonstrated that imprinted loci are deficient in short interspersed transposable elements (SINEs), particularly in the more ancient MIRs (Greally 2002; Ke et al. 2002). We similarly found that imprinted genes contain a low concentration of SINEs in their flanking regions. In addition, we determined that the orientation of these repetitive elements is of even greater discriminatory value. We denoted repeats oriented in the same direction relative to the gene by a plus sign (+), whereas oppositely oriented repeats are denoted by a minus sign (-).
The most important feature used in the classifier was the ratio of ± counts of repetitive elements (Fig. 2B). Repetitive elements within the introns were of greatest importance, followed by those in the region 10 kb upstream of the gene, whereas those 1 kb upstream of the gene were least important (Fig. 2C). Endogenous retrovirus (ERV) elements, ERV1 and ERVL, were the repeat features of greatest average importance, followed by LINE (long interspersed elements) L1s (Fig. 2D).
Figure 2.
Box plots of the absolute weights of the features used in the imprinted vs. nonimprinted classifier. (A) The largest number of features was based on repetitive elements, followed by transcription factor binding sites, CpG islands, and other miscellaneous features. (B) On average, the ratios of ± counts of repetitive elements carried the greatest absolute weight (P = 3 × 10-12). (C) Data on repetitive elements within the introns were the most important (P = 4 × 10-4), followed by the 10-kb upstream region (P = 6 × 10-3), while the 1-kb upstream window was of least importance (P = 5 × 10-10). (D) Among the repetitive elements, ERV1 (P = 2 × 10-5) and ERVL (P = 5 × 10-3) were of greatest average importance, followed by LINE L1 elements (P = 4 × 10-15). (E) SRF (P = 1 × 10-4), NFuE1 (P = 2 × 10-3), and AP2 (P = 1 × 10-2) were the most important transcription factor binding sites. The dotted line represents the overall mean.
Of the transcription factor binding sites investigated, serum response factor (SRF), NFuE1, and AP2 were most important in predicting imprinted genes (Fig. 2E). SRF is involved in the activation of “immediate early” genes (Schratt et al. 2001), in muscle differentiation (Vandromme et al. 1992; Soulez et al. 1996), and in mesoderm formation (Arsenian et al. 1998). The four-member family of AP2 transcription factors is essential for development and morphogenesis (Schorle et al. 1996; Zhang et al. 1996), and is involved in apoptosis (Moser et al. 1997), cell growth, and differentiation (Byrne et al. 1994). We are unaware of any role of NFuE1 in development. A complete list of the features used, along with their weights and average values, can be found in Supplemental Table 4.
Prediction of parental preference
We trained a separate classifier in order to predict whether an imprinted gene is expressed from the maternal or paternal allele. This model is based on 23 maternally expressed genes and 20 paternally expressed genes. Gnas was omitted from this part of the analysis due to the complex parental expression patterns at this locus (Beaudet 2004). We predicted maternal expression for 384 (64%) of the 600 potentially imprinted gene candidates.
We again assessed the accuracy of the model by using both cross-validation and an independent test set. We randomly split the training data into 20 groups, each containing one paternally and one to two maternally expressed imprinted genes, since there were slightly more maternally expressed genes in our training set. We again chose the features included in the model de novo among 2841 features. We achieved a sensitivity of 95% (19/20 paternally expressed genes correctly identified) and a specificity of 100% (23/23 maternally expressed genes correctly identified) in this cross-validation experiment.
The test set for assessing the prediction of parental preference consisted of three genes: one showing maternal expression (Atp10a) and two showing paternal expression (Wt1, Xist). All three genes were correctly classified. Xist is a particularly challenging example because it is solely expressed from the paternal allele in rodent extraembryonic tissue (Tagaki and Sasaki 1975), but is randomly expressed in rodent embryonic and human tissues (Plath et al. 2002). Appropriate sequence data for its oppositely imprinted antisense transcript, Tsix, were unavailable at the time of this study.
Among the features of greatest significance for the prediction of parental preference was the relative orientation of LINE L1 elements upstream versus the LINE L1ME elements downstream, along with the presence of Oct1, PU1, and CEBP binding sites upstream, and TGTCTGCAG consensus enhancer sites downstream. In paternally expressed genes the upstream LINE L1 elements tended to be oriented in the same relative way as L1ME elements downstream. In contrast, these two classes of repeats tended to be oppositely oriented in maternally expressed genes (P =9 × 10-5). Similarly, we found the MaLR LTRs upstream of paternally expressed genes to be predominantly oriented in the same direction as the gene, whereas those upstream of maternally expressed genes oppositely oriented (P =4 × 10-3). The discriminatory features employed in the parental preference classifier are summarized in the Supplemental Table 5.
Discussion
In this article, we presented a machine learning algorithm to predict novel imprinted genes in the mouse. We determined a number of DNA characteristics to be particularly significant for this task, including data on the presence and orientation of repetitive elements such as LINE L1s and ERVs. In addition to identifying imprinted gene candidates, our algorithm also predicted from which allele these genes are likely to be expressed.
There is mounting evidence of a parent-of-origin effect in complex conditions such as Alzheimer disease, autism, bipolar disorder, diabetes, male sexual orientation, obesity, and schizophrenia. This suggests the involvement of imprinted genes in their etiology. In Table 1 we present a set of human genes whose mouse homologs are predicted to be imprinted and which map to regions in the human genome that are linked to complex conditions with parent-of-origin-dependent inheritance.
Table 1.
Homologs of imprinted gene candidates showing linkage to human conditions
| Condition | Chromosome | Locus | Coordinate | Expressiona,b | Linkage | Reference |
|---|---|---|---|---|---|---|
| Alzheimer | 10p11 | D10S1208 | 35.3 | m | linkage to late-onset Alzheimer disease (AD)b | Bassett et al. (2002) |
| Epc1 | 32.6 | M | ||||
| C1 orf9 | 35.7 | M | ||||
| Atopy | 13q14 | D13S161 | 46.7 | p | linkage to atopyb | Bhattacharyya et al. (2000) |
| LCP1 | 45.6 | P | ||||
| Autism | 9p22 | D9S157 | 17.6 | p | linkage to autismb | Lamb et al. (2005) |
| C9orf39 | 17.1 | P | ||||
| Bipolar | 13q13 | D13S1493 | 32.9 | m | linkage to bipolar disorderb | McInnis et al. (2003) |
| Q5TBK1 | 31.9 | M | ||||
| 18q22 | D18S878 | 61.6 | p | linkage to bipolar disorderb | McInnis et al. (2003) | |
| SERPINB2 | 59.7 | M | ||||
| Diabetes | 11q13-14 | D11S2371-D11S2002 | 73.2-79.6 | p | linkage to birth weight (indirectly to diabetes)b | Lindsay et al. (2002) |
| NEU3 | 74.4 | M | mice overexpressing Neu3 develop diabetic phenotype | Sasaki et al. (2003) | ||
| OR2AT4 | 74.5 | M | ||||
| CAPN5 | 76.5 | M | ||||
| Homosexuality | 10q26 | D10S217 | 129.4 | m | linkage to male sexual orientationb | Mustanski et al. (2005) |
| C10orf90 | 128.1 | P | ||||
| STK32C | 133.9 | M | ||||
| NKX6-2 | 134.4 | M | ||||
| Obesity | 10p12 | D10S197 | 26.5 | m | linkage to obesityb | Dong et al. (2005) |
| GAD2 | 26.5 | M | candidate gene for obesity | Boutin et al. (2003) | ||
| Paternal UPD | 14q12 | D14S608 | 27.9 | paternal UPD results in fetal malformationsb | Kurosawa et al. (2002) | |
| NOVA1 | 26.0 | M | ||||
| FOXG13 | 28.3 | P | ||||
| Schizophrenia | 2p12 | D2S139 | 76.7 | p | linkage to relative hand skillb, Tourette syndrome and schizophrenia | Francks et al. (2003); Simonic et al. (2001); DeLisi et al. (2002) |
| LOXL3 | 74.7 | M | ||||
| DOK1 | 74.7 | P | ||||
| HK2 | 75.0 | M | ||||
| TACR1 | 75.2 | P | ||||
| 22q12 | D22S283 | 35.1 | m | linkage to schizophreniab | DeLisi et al. (2002) | |
| Q96PY3 | 36.1 | P | ||||
| C22orf23 | 36.7 | M | ||||
| Thrombosis | 5q13 | D5S2003 | 74.6 | m | linkage to high FVIII levelsb | Berger et al. (2005) |
| Q9P109 | 74.4 | M | ||||
| 9q22 | D9S910 | 98.6 | p | linkage to high FVIII levelsb | Berger et al. (2005) | |
| SAMD6 | 98.6 | P | ||||
| ALG2 | 99.1 | M | ||||
| NR4A3 | 99.7 | P | ||||
| TXNDC4 | 99.8 | P | ||||
| 11q21 | D11S4176 | 93.7 | m | linkage to high FVIII levelsb | Berger et al. (2005) | |
| JMJD2D | 94.4 | M |
Assignment of cytogenetic band and coordinates (in megabases) is based on Ensembl version 29.35b.
Genes predicted to be expressed from the maternal allele are denoted by M, paternally expressed genes by P.
Parent-of-origin effect was observed (m and p denote maternal and paternal effects, respectively).
For example, the homeobox gene NKX6-2 is located on human chromosome 10q26 near the marker D10S217 that Mustanski et al. (2005) found was maternally linked to male sexual orientation. Its expression is tightly controlled in a tissue-specific way with highest expression in the brain (Lee et al. 2001). Moreover, we predicted that its murine homolog is imprinted and maternally expressed. Increased maternal transmission of male homosexuality has previously been observed (Hamer et al. 1993), and it is proposed to result in part from imprinted genes (Bocklandt and Hamer 2003). A genome-wide screen for methylated CpG islands found a germline differentially methylated region (DMR) at this location (Strichman-Almashanu et al. 2002), lending additional support to the prediction of imprinted genes residing in this genome region. Mustanski et al. (2005) also found linkage to male sexual orientation in bands 7q36 and 8p12, but without any indications of a parental effect. Interestingly, there were no predicted imprinted gene candidates mapping to these two regions.
Another gene predicted to be imprinted and maternally expressed is Gad2. Its human homolog, located at chromosomal location 10p12, is identified as a candidate gene for obesity (Boutin et al. 2003). A recent parent-of-origin linkage study on obesity identified a maternal effect at 10p12 (Dong et al. 2005). This study found maximal linkage on chromosome 10 at marker D10S197, which resides within an intron of GAD2. These findings support the prediction that Gad2 is imprinted, at least in some tissues.
Prediction of parental expression preference is of particular interest in the light of the conflict theory for imprinting evolution that was originally postulated by Haig and Graham (1991). It predicts that paternally expressed genes promote prenatal and postnatal growth, while maternally expressed genes exhibit a growth suppressor role. This pattern holds true for at least seven maternally and seven paternally expressed genes involved in prenatal or postnatal growth. Similarly, at least five maternally and five paternally expressed genes can act as tumor suppressors and oncogenes, respectively. Interestingly, this parental expression pattern is followed by the two highest ranking imprinted gene candidates in our set. Ngfb, located on chromosome 13f3, was predicted to be paternally expressed. It is involved in neuronal development (Misko et al. 1987) and acts as an oncogene in numerous malignancies (Tokusashi et al. 2004). Second-ranked Cables1, located ∼1.2 Mb proximal to the imprinted gene, Impact, was predicted to be maternally expressed. It inhibits cell growth and suppresses tumor formation in nude mice (Tan et al. 2003).
Other promising candidates for experimental investigation include two clusters on mouse chromosome 12, whose homologs are located on human chromosome 14. Both maternal and paternal uniparental disomy (UPD) for chromosome 14 indicate three separate imprinted regions on this chromosome (Kotzot 2001). Meta-analysis also predicts three chromosomal bands 14q11-q13, 14q22-q24, and 14q31-q32 to contain imprinted genes (Sutton and Shaffer 2000). Frequent loss of heterozygosity (LOH) in human malignant mesothelioma in the same chromosomal bands further supports the prediction of several tumor suppressor genes residing within these regions (De Rienzo et al. 2000).
Chromosome region 14q32 contains the imprinted genes DLK1 and GTL2 (Wylie et al. 2000), and a number of other imprinted genes have been identified in orthologous regions in sheep and mouse (Miyoshi et al. 2000; Schmidt et al. 2000; Charlier et al. 2001); however, no imprinted genes have yet been found in the other two heterodisomic areas.
Syt14l and Kcnh5, on mouse chromosome 12d1, were predicted to be paternally and maternally expressed, respectively. The human homologs of these imprinted gene candidates, SYT14L and KCNH5, are located at chromosome location 14q23. These two genes are flanked by markers D14S592 and D14S277, for which frequent LOH is observed in tumors (De Rienzo et al. 2000).
Foxg1 (Bf-1) and Nova1, at mouse chromosome location 12c1, were also predicted to be reciprocally imprinted. Mice that are homozygous for a null mutation in either of these genes are not viable after birth (Xuan et al. 1995; Jensen et al. 2000). Foxg1 (Bf-1) was predicted to be paternally expressed and is the homolog of avian Qin, which has growth enhancer (Ahlgren et al. 2003) and oncogenic effects (Li and Vogt 1993). Human paternal UPD involving the homologous region at 14q12 results in fetal malformations (Towner et al. 2001), and LOH at 14q11-13 is present in numerous malignancies (Lee et al. 1997; Abujiang et al. 1998; Mutirangura et al. 1998). One region of frequent LOH in malignant mesothelioma, delimited by markers D14S1003 and D14S297 (De Rienzo et al. 2000), also contains FOXG1B and NOVA1. Located in between these two genes is marker D14S608, and paternal UPD at this marker has been shown to result in distinctive human malformations (Kurosawa et al. 2002).
The most important features our algorithm identified to distinguish imprinted genes were the ratio of the number of ± oriented SINE Alu and LINE L1 elements, the relative percentage of ± oriented B2 elements, and the number of ERV1 LTRs (long terminal repeats) 10-100 kb downstream of the gene. The relative orientation of repetitive elements in flanking sequences may contribute to physical chromosomal interactions that are important in controlling genomic imprinting. Physical chromosomal pairing has been observed in the vicinity of the imprinted PW and AS loci (LaSalle and Lalande 1996).
Many ERV families are predominantly expressed in placenta, germ cells, and embryonic tissues (Lower 1999). Intercisternal A-type particles (IAPs), also referred to as murine ERVKs, form one family of ERVs. Insertion of IAP sequences at the Agouti (Duhl et al. 1994; Michaud et al. 1994; Argeson et al. 1996) and AxinFu (Vasicek et al. 1997) loci results in epigenetically regulated phenotypes in the mouse that correlate with the extent of DNA methylation at the associated IAP (Michaud et al. 1994; Argeson et al. 1996; Morgan et al. 1999; Rakyan et al. 2003; Waterland and Jirtle 2003). Parent-of-origin effects also exist at these loci (Reed 1937; Wolff 1978; Belyaev et al. 1981; Morgan et al. 1999; Rakyan et al. 2003) that are likely due to differing degrees of IAP resistance to epigenetic reprogramming in the male and female genomes during gametogenesis and after fertilization (Lane et al. 2003). Moreover, IAPs, particularly ERVLs, are highly expressed at the two-cell stage (Evsikov et al. 2004), which coincides with the transcriptional activation of the embryonic genome (Wang et al. 2001).
An involvement of a LINE L1 element in imprinting control is documented, for example, in the paternally expressed Snrpn. This gene is associated with two DMRs. The region at the 5′ end is maternally methylated, while the region at the 3′ end is paternally methylated (Shemer et al. 1997). This second DMR consists of a LINE L1 element (Hajkova et al. 2002).
The timing of the remethylation of LINE L1 and IAPs in the male germ cells is tightly regulated (Lees-Murdock et al. 2003), but these elements adhere to a slightly different time course for methylation than the DMRs of imprinted genes (Li et al. 2004). Furthermore, IAPs are mostly resistant to demethylation, in particular during preimplantation, whereas LINE L1 elements are significantly demethylated during both preimplantation development and in primordial germ cells (Lane et al. 2003). A unifying role for these repeats in imprinting control remains to be elucidated.
Conclusion
The total number of imprinted murine genes is an open question. An initial estimate of ∼100 imprinted genes was derived from restriction landmark genome scanning (Hayashizaki et al. 1994); however, this original estimate was subsequently discounted as overly conservative (Reik and Walter 2001). A recent study in humans also indicates that variation of gene expression between alleles is more common than previously believed (Lo et al. 2003). Our prediction of 600 imprinted autosomal genes is somewhat higher than the early estimate of ∼100 but is still lower than the 2114 candidate imprinted genes reported by using expression profiling of the FANTOM2 set of full-length murine cDNAs (Nikaido et al. 2003).
A possible explanation for our lower estimate for the number of imprinted genes in the mouse than that obtained by using the FANTOM2 cDNA set is that our study is largely restricted to the protein-coding genes annotated in Ensembl, whereas the FANTOM2 set contains numerous noncoding and antisense transcripts. More importantly, expression profiling is based only on differences in parthenogenote and androgenote mRNA levels. This approach can lead to a false-positive rate as high as 42% because the transcription of nonimprinted genes may be regulated by the products of imprinted control genes (Mizuno et al. 2002). Given the high false-positive rate associated with expression profiling, our estimate seems realistic.
Computational prediction of imprinted genes cannot substitute for experimental validation, but it certainly represents a valuable supplement. It may also be the only way to identify imprinted domains where experimental disruption leads to offspring lethality. Moreover, in silico predictions can assist in the laborious process of experimental determination of imprinted genes by predicting candidate genes with the highest probability of being imprinted. The sequence characteristics we have identified as good predictors of imprint status may also have the added benefit of furthering our understanding of the mechanism by which monoallelic expression of imprinted genes is established and/or maintained.
Methods
Mouse genome data
We compiled the positive training set of 44 genes from a list of imprinted mouse genes at the Imprinted Gene Catalog (http://cancer.otago.ac.nz/IGC/Web/home.html). To compile the negative training set of 530 genes, we merged 30 genes located within or near imprinted regions that experimental evidence has shown to be biallelically expressed and 500 genes presumed to be nonimprinted. The latter group of genes was chosen at random from autosomal chromosomal bands known or not suspected to contain imprinted genes, and were intended to represent the overall characteristics of biallelically expressed genes. The entire set of training and testing genes is presented in the Supplemental Tables 1 through 3. We retrieved DNA sequence for all annotated murine genes from Ensembl (http://www.ensembl.org; version 16.30) in the following regions: the concatenated exon sequences, the concatenated intron sequences, and the regions 100 kb, 10 kb, 5 kb, 2 kb, and 1 kb upstream and downstream of the gene.
Feature measurements
We determined the presence of repeated elements by using RepeatMasker (http://repeatmasker.genome.washington.edu/RM/RepeatMasker.html). Subsequently, we calculated the following statistics for different repeat classes: the total count per sequence window, the percentage of the window covered, the ratio
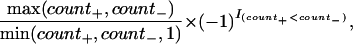 |
where count+ denotes the count of repeated elements in the same orientation as the gene, count- denotes the count of repeated elements in the opposite orientation as the gene, and I(count+ < count-) represents an indicator function, which is equal to one if the condition count+ < count- is met, and zero otherwise. Additionally, we computed the ratio of the window covered as a function of the orientation as
 |
The statistics of repeated elements were determined in nonoverlapping windows 100-10 kb and 10-5 kb upstream and downstream, while the windows 5 kb, 2 kb, and 1 kb were considered in an overlapping fashion.
We used CpG Island Searcher (Takai and Jones 2002) to count the number of CpG islands. CpG island information was recorded as counts of the number of islands found and the percentage of the sequence window consisting of a CpG island (percentage
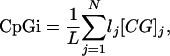 |
where N is the total number of CpG islands found within that sequence window, L is the total length of the sequence window, lj the length of the j-th CpG island, and [CG]j the CG content of the j-th CpG island). We incorporated the two-island rule formulated by Onyango et al. (2000) as an indicator function, whose value was one if the number of CpG islands was two or greater, and zero otherwise. Following the method described by Feltus et al. (2003), we classified each CpG island in the upstream window as either methylation prone or methylation resistant. This model was trained by using the DNA pattern frequencies given in Supplemental Table 4 and implemented in Equbits Foresight (http://www.equbits.com). We subsequently calculated total counts, mean, and running mean of these predictions over 10-kb windows.
By using FASTA (Pearson and Lipman 1988) and custom Perl scripts, we investigated the presence of consensus-binding sites in the upstream and downstream regions. We searched for the enhancer motifs, TGTTTGCAG, TGTCTGCAG, the CTCF binding motif, CCGC**GG*GGC (Wylie et al. 2000), and motifs 3, 7, and 11 described by Z. Wang et al. (2004), i.e., AGAATAAATGAAAAAAAAAATAAAAG, ATATTATGTTTTTTTTCATTTTCAAT, and ATTTTTTTATTTTTATTTTATTTTTTTTTTTTAAAA, respectively.
By using Etandem (http://www.hgmp.mrc.ac.uk/Software/EMBOSS/Apps/etandem.html), we counted the number of dinucleotide (nucleotide repeats of length 2) and tetranucleotide repeats (length 4), as well as simple repeats (nucleotide repeats with minimal length of 5 and maximal length of 100) within the 10 kb upstream region.
It was previously shown that one CpG island is frequently located within a 2-kb region upstream of the gene and occasionally it overlaps the first exon (Onyango et al. 2000). To incorporate these findings in a systematic manner, we encoded the answers to the following questions by means of indicator variables. First, does the first exon at least partially contain a CpG island? Second, does the last exon at least partially contain a CpG island? Third, is there a downstream enhancer consensus binding site (TGTTTGCAG) within either 2 kb upstream or the first 2.6 kb downstream of the first exon? Fourth, is there a downstream enhancer consensus binding site (TGTCTGCAG) within either 2 kb upstream or the first 3.5 kb downstream of the first exon? Fifth, is there an upstream CTCF consensus binding site (CCGC**GG*GGC) in the region 2 kb upstream of the gene to the end of the first exon. A sixth indicator variable was set to one if rules 1 and 2 were both met. We also determined putative transcription factor binding sites within 10 kb upstream by using the RGSiteScan program (http://www.mgs.bionet.nsc.ru/mgs/programs/yura/rgscan1.html). We summarized the transcription factor binding data for non-overlapping windows of 1 kb size to limit the size of the feature vector.
To characterize the interdependence between sets of features, we computed all pairwise interactions between two features within the training set, performed a t-test, and ranked them by P-value. In the imprinted versus nonimprinted model, we retained the top 4000 pairwise interactions. Since all of the interactions had a P-value below a Bonferroni-corrected cutoff at the α = 0.05 level, we added them to the original feature matrix. In the parental preference model, we retained the top 10 pairwise-interactions (P-values ≤ 0.0003).
Classification method
Equbits Foresight was used for classification. The classifier is based on support vector machines and was originally developed for QSAR data. The feature data were scaled to unit length when predicting the imprinting status of a gene, and a linear kernel was used to rank the features by weight. All features that had a non-zero weight were retained for use in an RBF (radial basis function) kernel. In the final imprinted versus nonimprinted classifier, we employed 722 features. The number of features employed ranged from 704-738 during the cross-validation stage of analysis. Only the top 30 features were retained for the prediction of parental expression preference.
Acknowledgments
We thank Hamilton Hitchings, Balaji Krishnapuram, and Jason Stajich for helpful discussions. Computation of the feature data, in particular acquisition of repeat data spanning the entire mouse genome, was made possible by the Duke University Computer Science cluster facility supported by IBM and the National Science Foundation (EIA-9972879). P.P.L. gratefully acknowledges financial support from the Duke University Program for Bioinformatics and Genome Technology. A.J.H. is thankful for support from an Alfred P. Sloan Fellowship and a National Science Foundation CAREER Award. R.L.J. was supported by NIH grants CA25951, ES08823, and ES13053.
Footnotes
[Supplemental material is available online at www.genome.org.]
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.3303505.
References
- Abujiang, P., Mori, T.J., Takahashi, T., Tanaka, F., Kasyu, I., Hitomi, S., and Hiai, H. 1998. Loss of heterozygosity (LOH) at 17q and 14q in human lung cancers. Oncogene 17: 3029-3033. [DOI] [PubMed] [Google Scholar]
- Ahlgren, S., Vogt, P., and Bronner-Fraser, M. 2003. Excess FoxG1 causes overgrowth of the neural tube. J. Neurobiol. 57: 337-349. [DOI] [PubMed] [Google Scholar]
- Allen, E., Horvath, S., Tong, F., Kraft, P., Spiteri, E., Riggs, A.D., and Marahrens, Y. 2003. High concentrations of long interspersed nuclear element sequence distinguish monoallelically expressed genes. Proc. Natl. Acad. Sci. 100: 9940-9945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argeson, A.C., Nelson, K.K., and Siracusa, L.D. 1996. Molecular basis of the pleiotropic phenotype of mice carrying the hypervariable yellow (Ahvy) mutation at the agouti locus. Genetics 142: 557-567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arsenian, S., Weinhold, B., Oelgeschlager, M., Ruther, U., and Nordheim, A. 1998. Serum response factor is essential for mesoderm formation during mouse embryogenesis. EMBO J. 17: 6289-6299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassett, S.S., Avramopoulos, D., and Fallin, D. 2002. Evidence for parent of origin effect in late-onset Alzheimer disease. Am. J. Med. Genet. 114: 679-686. [DOI] [PubMed] [Google Scholar]
- Beaudet, A.L. 2004. Complex imprinting. Nat. Genet. 36: 793-795. [DOI] [PubMed] [Google Scholar]
- Belyaev, D.K., Ruvinsky, A.O., and Borodin, P.M. 1981. Inheritance of alternative states of the fused gene in mice. J. Hered. 72: 107-112. [DOI] [PubMed] [Google Scholar]
- Bentley, L., Nakabayashi, K., Monk, D., Beechey, C., Peters, J., Birjandi, Z., Khayat, F.E., Patel, M., Preece, M.A., Stanier, P., et al. 2003. The imprinted region on human chromosome 7q32 extends to the carboxypeptidase A gene cluster: An imprinted candidate for Silver-Russell syndrome. J. Med. Genet. 40: 249-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger, M., Mattheisen, M., Kulle, B., Schmidt, H., Oldenburg, J., Bickeboller, H., Walter, U., Lindner, T.H., Strauch, K., and Schambeck, C.M. 2005. High factor VIII levels in venous thromboembolism show linkage to imprinted loci on chromosomes 5 and 11. Blood 105: 638-644. [DOI] [PubMed] [Google Scholar]
- Bhattacharyya, S., Leaves, N.I., Wiltshire, S., Cox, R., and Cookson, W.O. 2000. A high-density genetic map of the chromosome 13q14 atopy locus. Genomics 70: 286-291. [DOI] [PubMed] [Google Scholar]
- Bocklandt, S. and Hamer, D.H. 2003. Beyond hormones: A novel hypothesis for the biological basis of male sexual orientation. J. Endocrinol. Invest. 26: 8-12. [PubMed] [Google Scholar]
- Boutin, P., Dina, C., Vasseur, F., Dubois, S., Corset, L., Seron, K., Bekris, L., Cabellon, J., Neve, B., Vasseur-Delannoy, V., et al. 2003. GAD2 on chromosome 10p12 is a candidate gene for human obesity. PLoS. Biol. 1: E68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne, C., Tainsky, M., and Fuchs, E. 1994. Programming gene expression in developing epidermis. Development 120: 2369-2383. [DOI] [PubMed] [Google Scholar]
- Charlier, C., Segers, K., Wagenaar, D., Karim, L., Berghmans, S., Jaillon, O., Shay, T., Weissenbach, J., Cockett, N., Gyapay, G., et al. 2001. Human-ovine comparative sequencing of a 250-kb imprinted domain encompassing the callipyge (clpg) locus and identification of six imprinted transcripts: DLK1, DAT, GTL2, PEG11, antiPEG11, and MEG8. Genome Res. 11: 850-862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dallosso, A.R., Hancock, A.L., Brown, K.W., Williams, A.C., Jackson, S., and Malik, K. 2004. Genomic imprinting at the WT1 gene involves a novel coding transcript (AWT1) that shows deregulation in Wilms' tumours. Hum. Mol. Genet. 13: 405-415. [DOI] [PubMed] [Google Scholar]
- DeLisi, L.E., Shaw, S.H., Crow, T.J., Shields, G., Smith, A.B., Larach, V.W., Wellman, N., Loftus, J., Nanthakumar, B., Razi, K., et al. 2002. A genome-wide scan for linkage to chromosomal regions in 382 sibling pairs with schizophrenia or schizoaffective disorder. Am. J. Psychiatry 159: 803-812. [DOI] [PubMed] [Google Scholar]
- De Rienzo, A., Jhanwar, S.C., and Testa, J.R. 2000. Loss of heterozygosity analysis of 13q and 14q in human malignant mesothelioma. Genes Chromosomes Cancer 28: 337-341. [PubMed] [Google Scholar]
- Dong, C., Li, W.D., Geller, F., Lei, L., Li, D., Gorlova, O.Y., Hebebrand, J., Amos, C.I., Nicholls, R.D., and Price, R.A. 2005. Possible genomic imprinting of three human obesity-related genetic loci. Am. J. Hum. Genet. 76: 427-437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duhl, D.M., Vrieling, H., Miller, K.A., Wolff, G.L., and Barsh, G.S. 1994. Neomorphic agouti mutations in obese yellow mice. Nat. Genet. 8: 59-65. [DOI] [PubMed] [Google Scholar]
- Evans, H.K., Wylie, A.A., Murphy, S.K., and Jirtle, R.L. 2001. The neuronatin gene resides in a “micro-imprinted domain” on human chromosome 20q11.2. Genomics 77: 99-104. [DOI] [PubMed] [Google Scholar]
- Evsikov, A.V., de Vries, W.N., Peaston, A.E., Radford, E.E., Fancher, K.S., Chen, F.H., Blake, J.A., Bult, C.J., Latham, K.E., Solter, D., et al. 2004. Systems biology of the 2-cell mouse embryo. Cytogenet. Genome Res. 105: 240-250. [DOI] [PubMed] [Google Scholar]
- Feinberg, A.P. and Tycko, B. 2004. The history of cancer epigenetics. Nat. Rev. Cancer 4: 1-11. [DOI] [PubMed] [Google Scholar]
- Feltus, F.A., Lee, E.K., Costello, J.F., Plass, C., and Vertino, P.M. 2003. Predicting aberrant CpG island methylation. Proc. Natl. Acad. Sci. 100: 12253-12258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francks, C., DeLisi, L.E., Shaw, S.H., Fisher, S.E., Richardson, A.J., Stein, J.F., and Monaco, A.P. 2003. Parent-of-origin effects on handedness and schizophrenia susceptibility on chromosome 2p12-q11. Hum. Mol. Genet. 12: 3225-3230. [DOI] [PubMed] [Google Scholar]
- Greally, J.M. 2002. Short interspersed transposable elements (SINEs) are excluded from imprinted regions in the human genome. Proc. Natl. Acad. Sci. 99: 327-332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haig, D. and Graham, C. 1991. Genomic imprinting and the strange case of the insulin-like growth factor II receptor. Cell 64: 1045-1046. [DOI] [PubMed] [Google Scholar]
- Hajkova, P., Erhardt, S., Lane, N., Haaf, T., El-Maarri, O., Reik, W., Walter, J., and Surani, M.A. 2002. Epigenetic reprogramming in mouse primordial germ cells. Mech. Dev. 117: 15-23. [DOI] [PubMed] [Google Scholar]
- Hamer, D.H., Hu, S., Magnuson, V.L., Hu, N., and Pattatucci, A.M. 1993. A linkage between DNA markers on the X chromosome and male sexual orientation. Science 261: 321-327. [DOI] [PubMed] [Google Scholar]
- Hayashizaki, Y., Shibata, H., Hirotsune, S., Sugino, H., Okazaki, Y., Sasaki, N., Hirose, K., Imoto, H., Okuizumi, H., and Muramatsu, M. 1994. Identification of an imprinted U2af binding protein related sequence on mouse chromosome 11 using the RLGS method. Nat. Genet. 6: 33-40. [DOI] [PubMed] [Google Scholar]
- Jensen, K.B., Dredge, B.K., Stefani, G., Zhong, R., Buckanovich, R.J., Okano, H.J., Yang, Y.Y., and Darnell, R.B. 2000. Nova-1 regulates neuron-specific alternative splicing and is essential for neuronal viability. Neuron 25: 359-371. [DOI] [PubMed] [Google Scholar]
- John, R.M., Aparicio, S.A., Ainscough, J.F., Arney, K.L., Khosla, S., Hawker, K., Hilton, K.J., Barton, S.C., and Surani, M.A. 2001. Imprinted expression of neuronatin from modified BAC transgenes reveals regulation by distinct and distant enhancers. Dev. Biol. 236: 387-399. [DOI] [PubMed] [Google Scholar]
- Kashiwagi, A., Meguro, M., Hoshiya, H., Haruta, M., Ishino, F., Shibahara, T., and Oshimura, M. 2003. Predominant maternal expression of the mouse Atp10c in hippocampus and olfactory bulb. J. Hum. Genet. 48: 194-198. [DOI] [PubMed] [Google Scholar]
- Kayashima, T., Yamasaki, K., Yamada, T., Sakai, H., Miwa, N., Ohta, T., Yoshiura, K., Matsumoto, N., Nakane, Y., Kanetake, H., et al. 2003. The novel imprinted carboxypeptidase A4 gene (CPA4) in the 7q32 imprinting domain. Hum. Genet. 112: 220-226. [DOI] [PubMed] [Google Scholar]
- Ke, X., Thomas, S.N., Robinson, D.O., and Collins, A. 2002. The distinguishing sequence characteristics of mouse imprinted genes. Mamm. Genome 13: 639-645. [DOI] [PubMed] [Google Scholar]
- Killian, J.K., Byrd, J.C., Jirtle, J.V., Munday, B.L., Stoskopf, M.K., MacDonald, R.G., and Jirtle, R.L. 2000. M6P/IGF2R imprinting evolution in mammals. Mol. Cell 5: 707-716. [DOI] [PubMed] [Google Scholar]
- Killian, J.K., Nolan, C.M., Wylie, A.A., Li, T., Vu, T.H., Hoffman, A.R., and Jirtle, R.L. 2001. Divergent evolution in M6P/IGF2R imprinting from the Jurassic to the Quaternary. Hum. Mol. Genet. 10: 1721-1728. [DOI] [PubMed] [Google Scholar]
- Kimura, M.I., Kazuki, Y., Kashiwagi, A., Kai, Y., Abe, S., Barbieri, O., Levi, G., and Oshimura, M. 2004. Dlx5, the mouse homologue of the human-imprinted DLX5 gene, is biallelically expressed in the mouse brain. J. Hum. Genet. 49: 273-237. [DOI] [PubMed] [Google Scholar]
- Kotzot, D. 2001. Comparative analysis of isodisomic and heterodisomic segments in cases with maternal uniparental disomy 14 suggests more than one imprinted region. Clin. Genet. 60: 226-231. [DOI] [PubMed] [Google Scholar]
- Kurosawa, K., Sasaki, H., Sato, Y., Yamanaka, M., Shimizu, M., Ito, Y., Okuyama, T., Matsuo, M., Imaizumi, K., Kuroki, Y., et al. 2002. Paternal UPD14 is responsible for a distinctive malformation complex. Am. J. Med. Genet. 110: 268-272. [DOI] [PubMed] [Google Scholar]
- Lamb, J.A., Barnby, G., Bonora, E., Sykes, N., Bacchelli, E., Blasi, F., Maestrini, E., Broxholme, J., Tzenova, J., Weeks, D., et al. 2005. Analysis of IMGSAC autism susceptibility loci: Evidence for sex limited and parent of origin specific effects. J. Med. Genet. 42: 132-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane, N., Dean, W., Erhardt, S., Hajkova, P., Surani, A., Walter, J., and Reik, W. 2003. Resistance of IAPs to methylation reprogramming may provide a mechanism for epigenetic inheritance in the mouse. Genesis 35: 88-93. [DOI] [PubMed] [Google Scholar]
- LaSalle, J. and Lalande, M. 1996. Homologous association of oppositely imprinted chromosomal domains. Science 272: 725-728. [DOI] [PubMed] [Google Scholar]
- Lee, D.J., Koch, W.M., Yoo, G., Lango, M., Reed, A., Califano, J., Brennan, J.A., Westra, W.H., Zahurak, M., and Sidransky, D. 1997. Impact of chromosome 14q loss on survival in primary head and neck squamous cell carcinoma. Clin. Cancer Res. 3: 501-505. [PubMed] [Google Scholar]
- Lee, S.H., Davison, J.A., Vidal, S.M., and Belouchi, A. 2001. Cloning, expression and chromosomal location of NKX6B TO 10Q26, a region frequently deleted in brain tumors. Mamm. Genome 12: 157-162. [DOI] [PubMed] [Google Scholar]
- Lees-Murdock, D.J., De, F.E., and Walsh, C.P. 2003. Methylation dynamics of repetitive DNA elements in the mouse germ cell lineage. Genomics 82: 230-237. [DOI] [PubMed] [Google Scholar]
- Li, J. and Vogt, P.K. 1993. The retroviral oncogene qin belongs to the transcription factor family that includes the homeotic gene fork head. Proc. Natl. Acad. Sci. 90: 4490-4494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J.Y., Lees-Murdock, D.J., Xu, G.L., and Walsh, C.P. 2004. Timing of establishment of paternal methylation imprints. Genomics 84: 952-960. [DOI] [PubMed] [Google Scholar]
- Linder, D., McCaw, B.K., and Hecht, F. 1975. Parthenogenic origin of benign ovarian teratomas. N. Engl. J. Med. 292: 63-66. [DOI] [PubMed] [Google Scholar]
- Lindsay, R.S., Kobes, S., Knowler, W.C., and Hanson, R.L. 2002. Genome-wide linkage analysis assessing parent-of-origin effects in the inheritance of birth weight. Hum. Genet. 110: 503-509. [DOI] [PubMed] [Google Scholar]
- Lo, H.S., Wang, Z., Hu, Y., Yang, H.H., Gere, S., Buetow, K.H., and Lee, M.P. 2003. Allelic variation in gene expression is common in the human genome. Genome Res. 13: 1855-1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lower, R. 1999. The pathogenic potential of endogenous retroviruses: Facts and fantasies. Trends Microbiol. 7: 350-356. [DOI] [PubMed] [Google Scholar]
- Malik, K., Salpekar, A., Hancock, A., Moorwood, K., Jackson, S., Charles, A., and Brown, K.W. 2000. Identification of differential methylation of the WT1 antisense regulatory region and relaxation of imprinting in Wilms' tumor. Cancer Res. 60: 2356-2360. [PubMed] [Google Scholar]
- McInnis, M.G., Lan, T.H., Willour, V.L., McMahon, F.J., Simpson, S.G., Addington, A.M., MacKinnon, D.F., Potash, J.B., Mahoney, A.T., Chellis, J., et al. 2003. Genome-wide scan of bipolar disorder in 65 pedigrees: Supportive evidence for linkage at 8q24, 18q22, 4q32, 2p12, and 13q12. Mol. Psychiatry 8: 288-298. [DOI] [PubMed] [Google Scholar]
- Meguro, M., Kashiwagi, A., Mitsuya, K., Nakao, M., Kondo, I., Saitoh, S., and Oshimura, M. 2001. A novel maternally expressed gene, ATP10C, encodes a putative aminophospholipid translocase associated with Angelman syndrome. Nat. Genet. 28: 19-20. [DOI] [PubMed] [Google Scholar]
- Michaud, E.J., van Vugt, M.J., Bultman, S.J., Sweet, H.O., Davisson, M.T., and Woychik, R.P. 1994. Differential expression of a new dominant agouti allele (Aiapy) is correlated with methylation state and is influenced by parental lineage. Genes & Dev. 8: 1463-1472. [DOI] [PubMed] [Google Scholar]
- Misko, T.P., Radeke, M.J., and Shooter, E.M. 1987. Nerve growth factor in neuronal development and maintenance. J. Exp. Biol. 132: 177-190. [DOI] [PubMed] [Google Scholar]
- Mitsuya, K., Sui, H., Meguro, M., Kugoh, H., Jinno, Y., Niikawa, N., and Oshimura, M. 1997. Paternal expression of WT1 in human fibroblasts and lymphocytes. Hum. Mol. Genet. 6: 2243-2246. [DOI] [PubMed] [Google Scholar]
- Miyoshi, N., Wagatsuma, H., Wakana, S., Shiroishi, T., Nomura, M., Aisaka, K., Kohda, T., Surani, M.A., Kaneko-Ishino, T., and Ishino, F. 2000. Identification of an imprinted gene, Meg3/Gtl2 and its human homologue MEG3, first mapped on mouse distal chromosome 12 and human chromosome 14q. Genes Cells 5: 211-220. [DOI] [PubMed] [Google Scholar]
- Mizuno, Y., Sotomaru, Y., Katsuzawa, Y., Kono, T., Meguro, M., Oshimura, M., Kawai, J., Tomaru, Y., Kiyosawa, H., Nikaido, I., et al. 2002. Asb4, Ata3, and Dcn are novel imprinted genes identified by high-throughput screening using RIKEN cDNA microarray. Biochem. Biophys. Res. Commun. 290: 1499-1505. [DOI] [PubMed] [Google Scholar]
- Morgan, H.D., Sutherland, H.G., Martin, D.I., and Whitelaw, E. 1999. Epigenetic inheritance at the agouti locus in the mouse. Nat. Genet. 23: 314-318. [DOI] [PubMed] [Google Scholar]
- Moser, M., Pscherer, A., Roth, C., Becker, J., Mucher, G., Zerres, K., Dixkens, C., Weis, J., Guay-Woodford, L., Buettner, R., et al. 1997. Enhanced apoptotic cell death of renal epithelial cells in mice lacking transcription factor AP-2β. Genes & Dev. 11: 1938-1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy, S.K. and Jirtle, R.L. 2003. Imprinting evolution and the price of silence. Bioessays 25: 577-588. [DOI] [PubMed] [Google Scholar]
- Mustanski, B.S., Dupree, M.G., Nievergelt, C.M., Bocklandt, S., Schork, N.J., and Hamer, D.H. 2005. A genomewide scan of male sexual orientation. Hum. Genet. 116: 272-278. [DOI] [PubMed] [Google Scholar]
- Mutirangura, A., Pornthanakasem, W., Sriuranpong, V., Supiyaphun, P., and Voravud, N. 1998. Loss of heterozygosity on chromosome 14 in nasopharyngeal carcinoma. Int. J. Cancer 78: 153-156. [DOI] [PubMed] [Google Scholar]
- Nabetani, A., Hatada, I., Morisaki, H., Oshimura, M., and Mukai, T. 1997. Mouse U2af1-rs1 is a neomorphic imprinted gene. Mol. Cell. Biol. 17: 789-798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikaido, I., Saito, C., Mizuno, Y., Meguro, M., Bono, H., Kadomura, M., Kono, T., Morris, G.A., Lyons, P.A., Oshimura, M., et al. 2003. Discovery of imprinted transcripts in the mouse transcriptome using large-scale expression profiling. Genome Res. 13: 1402-1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okita, C., Meguro, M., Hoshiya, H., Haruta, M., Sakamoto, Y., and Oshimura, M. 2003. A new imprinted cluster on the human chromosome 7q21-q31, identified by human-mouse monochromosomal hybrids. Genomics 81: 556-559. [DOI] [PubMed] [Google Scholar]
- Onyango, P., Miller, W., Lehoczky, J., Leung, C.T., Birren, B., Wheelan, S., Dewar, K., and Feinberg, A.P. 2000. Sequence and comparative analysis of the mouse 1-megabase region orthologous to the human 11p15 imprinted domain. Genome Res. 10: 1697-1710. [DOI] [PubMed] [Google Scholar]
- Pearson, W.R. and Lipman, D.J. 1988. Improved tools for biological sequence comparison. Proc. Natl. Acad. Sci. 85: 2444-2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plath, K., Mlynarczyk-Evans, S., Nusinow, D.A., and Planning, B. 2002. Xist RNA and the mechanism of X chromosome inactivation. Annu. Rev. Genet. 36: 233-278. [DOI] [PubMed] [Google Scholar]
- Rachmilewitz, J., Gonik, B., Goshen, R., Ariel, I., Schneider, T., de Groot, N., and Hochberg, A. 1993. Use of a novel system for defining a gene imprinting region. Biochem. Biophys. Res. Commun. 196: 659-664. [DOI] [PubMed] [Google Scholar]
- Rakyan, V.K., Chong, S., Champ, M.E., Cuthbert, P.C., Morgan, H.D., Luu, K.V., and Whitelaw, E. 2003. Transgenerational inheritance of epigenetic states at the murine Axin(Fu) allele occurs after maternal and paternal transmission. Proc. Natl. Acad. Sci. 100: 2538-2543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed, S.C. 1937. The inheritance and expression of fused: A new mutation in the house mouse. Genetics 22: 1-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reik, W. and Walter, J. 2001. Genomic imprinting: Parental influence on the genome. Nat. Rev. Genet. 2: 21-32. [DOI] [PubMed] [Google Scholar]
- Sasaki, A., Hata, K., Suzuki, S., Sawada, M., Wada, T., Yamaguchi, K., Obinata, M., Tateno, H., Suzuki, H., and Miyagi, T. 2003. Overexpression of plasma membrane-associated sialidase attenuates insulin signaling in transgenic mice. J. Biol. Chem. 278: 27896-27902. [DOI] [PubMed] [Google Scholar]
- Schmidt, J.V., Matteson, P.G., Jones, B.K., Guan, X.J., and Tilghman, S.M. 2000. The Dlk1 and Gtl2 genes are linked and reciprocally imprinted. Genes & Dev. 14: 1997-2002. [PMC free article] [PubMed] [Google Scholar]
- Schorle, H., Meier, P., Buchert, M., Jaenisch, R., and Mitchell, P.J. 1996. Transcription factor AP-2 essential for cranial closure and craniofacial development. Nature 381: 235-238. [DOI] [PubMed] [Google Scholar]
- Schratt, G., Weinhold, B., Lundberg, A.S., Schuck, S., Berger, J., Schwarz, H., Weinberg, R.A., Ruther, U., and Nordheim, A. 2001. Serum response factor is required for immediate-early gene activation yet is dispensable for proliferation of embryonic stem cells. Mol. Cell Biol. 21: 2933-2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shemer, R., Birger, Y., Riggs, A.D., and Razin, A. 1997. Structure of the imprinted mouse Snrpn gene and establishment of its parental-specific methylation pattern. Proc. Natl. Acad. Sci. 94: 10267-10272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonic, I., Nyholt, D.R., Gericke, G.S., Gordon, D., Matsumoto, N., Ledbetter, D.H., Ott, J., and Weber, J.L. 2001. Further evidence for linkage of Gilles de la Tourette syndrome (GTS) susceptibility loci on chromosomes 2p11, 8q22 and 11q23-24 in South African Afrikaners. Am. J. Med. Genet. 105: 163-167. [DOI] [PubMed] [Google Scholar]
- Soulez, M., Rouviere, C.G., Chafey, P., Hentzen, D., Vandromme, M., Lautredou, N., Lamb, N., Kahn, A., and Tuil, D. 1996. Growth and differentiation of C2 myogenic cells are dependent on serum response factor. Mol. Cell. Biol. 16: 6065-6074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strichman-Almashanu, L.Z., Lee, R.S., Onyango, P.O., Perlman, E., Flam, F., Frieman, M.B., and Feinberg, A.P. 2002. A genome-wide screen for normally methylated human CpG islands that can identify novel imprinted genes. Genome Res. 12: 543-554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton, V.R. and Shaffer, L.G. 2000. Search for Imprinted regions on chromosome 14: Comparison of maternal and paternal UPD cases with cases of chromosome 14 deletion. Am. J. Med. Genet. 93: 381-387. [DOI] [PubMed] [Google Scholar]
- Szulman, A.E. 1987. In Gestational trophoblastic disease (eds. A.E. Szulman and H.J. Bucksbaum), pp. 27-44. Springer-Verlag, New York.
- Tagaki, N. and Sasaki, M. 1975. Preferential inactivation of the paternally derived X chromosome in the extraembryonic membranes of the mouse. Nature 256: 640-642. [DOI] [PubMed] [Google Scholar]
- Takai, D. and Jones, P.A. 2002. Comprehensive analysis of CpG islands in human chromosomes 21 and 22. Proc. Natl. Acad. Sci. 99: 3740-3745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan, D., Kirley, S., Li, Q., Ramnath, N., Slocum, H.K., Brooks, J.S., Wu, C.L., and Zukerberg, L.R. 2003. Loss of cables protein expression in human non-small cell lung cancer: A tissue microarray study. Hum. Pathol. 34: 143-149. [DOI] [PubMed] [Google Scholar]
- Tokusashi, Y., Asai, K., Tamakawa, S., Yamamoto, M., Yoshie, M., Yaginuma, Y., Miyokawa, N., Aoki, T., Kino, S., and Kasai, S., et al. 2004. Expression of NGF in hepatocellular carcinoma cells with its receptors in non-tumor cell components. Int. J. Cancer. 114: 39-45. [DOI] [PubMed] [Google Scholar]
- Towner, D., Yang, S.P., and Shaffer, L.G. 2001. Prenatal ultrasound findings in a fetus with paternal uniparental disomy 14q12-qter. Ultrasound Obstet. Gynecol. 18: 268-271. [DOI] [PubMed] [Google Scholar]
- Vandromme, M., Gauthier-Rouviere, C., Carnac, G., Lamb, N., and Fernandez, A. 1992. Serum response factor p67SRF is expressed and required during myogenic differentiation of both mouse C2 and rat L6 muscle cell lines. J. Cell. Biol. 118: 1489-1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasicek, T.J., Zeng, L., Guan, X.J., Zhang, T., Costantini, F., and Tilghman, S.M. 1997. Two dominant mutations in the mouse fused gene are the result of transposon insertions. Genetics 147: 777-786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Q., Chung, Y.G., deVries, W.N., Struwe, M., and Latham, K.E. 2001. Role of protein synthesis in the development of a transcriptionally permissive state in one-cell stage mouse embryos. Biol. Reprod. 65: 748-754. [DOI] [PubMed] [Google Scholar]
- Wang, Y., Joh, K., Masuko, S., Yatsuki, H., Soejima, H., Nabetani, A., Beechey, C.V., Okinami, S., and Mukai, T. 2004. The mouse Murr1 gene is imprinted in the adult brain, presumably due to transcriptional interference by the antisense-oriented U2af1-rs1 gene. Mol. Cell. Biol. 24: 270-279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Z., Fan, H., Yang, H.H., Hu, Y., Buetow, K.H., and Lee, M.P. 2004. Comparative sequence analysis of imprinted genes between human and mouse to reveal imprinting signatures. Genomics 83: 395-401. [DOI] [PubMed] [Google Scholar]
- Waterland, R.A. and Jirtle, R.L. 2003. Transposable elements: Targets for early nutritional effects on epigenetic gene regulation. Mol. Cell. Biol. 23: 5293-5300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ____. 2004. Early nutrition, epigenetic changes at transposons and imprinted genes, and enhanced susceptibility to adult chronic diseases. Nutrition 20: 63-68. [DOI] [PubMed] [Google Scholar]
- Wolff, G.L. 1978. Influence of maternal phenotype on metabolic differentiation of agouti locus mutants in the mouse. Genetics 88: 529-539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wylie, A.A., Murphy, S.K., Orton, T.C., and Jirtle, R.L. 2000. Novel imprinted DLK1/GTL2 domain on human chromosome 14 contains motifs that mimic those implicated in IGF2/H19 regulation. Genome Res. 10: 1711-1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xuan, S., Baptista, C.A., Balas, G., Tao, W., Soares, V.C., and Lai, E. 1995. Winged helix transcription factor BF-1 is essential for the development of the cerebral hemispheres. Neuron 14: 1141-1152. [DOI] [PubMed] [Google Scholar]
- Zhang, J., Hagopian-Donaldson, S., Serbedzija, G., Elsemore, J., Plehn-Dujowich, D., McMahon, A.P., Flavell, R.A., and Williams, T. 1996. Neural tube, skeletal and body wall defects in mice lacking transcription factor AP-2. Nature 381: 238-241. [DOI] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- http://lpg.nci.nih.gov/LPG/lee/proj2; M.P. Lee's laboratory.
- http://cancer.otago.ac.nz/IGC/Web/home.html; Imprinted Gene Catalog.
- http://www.ensembl.org; Ensembl.
- http://repeatmasker.genome.washington.edu/RM/RepeatMasker.html; RepeatMasker.
- http://www.equbits.com; Equbits Foresight.
- http://www.mgs.bionet.nsc.ru/mgs/programs/yura/rgscan1.html; RGSiteScan.
- http://www.hgmp.mrc.ac.uk/Software/EMBOSS/Apps/etandem.html; Etandem.




