Abstract
Here, we present an analysis of 215,634 EST and cDNA sequences of a major vector of human malaria Anopheles gambiae structured into the AnoEST database. The expressed sequences are grouped into clusters using genomic sequence as template and associated with inferred functional annotation, including the following: corresponding Ensembl gene prediction, putative orthologous genes in other species, homology to known proteins, protein domains, associated Gene Ontology terms, and corresponding classification into broad GO-slim functional groups. AnoEST is a vital resource for interpretation of expression profiles derived using recently developed A. gambiae cDNA microarrays. Using these cDNA microarrays, we have experimentally confirmed the expression of 7961 clusters during mosquito development. Of these, 3100 are not associated with currently predicted genes. Moreover, we found that clusters with confirmed expression are nonbiased with respect to the current gene annotation or homology to known proteins. Consequently, we expect that many as yet unconfirmed clusters are likely to be actual A. gambiae genes. [AnoEST is publicly available at http://komar.embl.de, and is also accessible as a Distributed Annotation Service (DAS).]
Blood-feeding anopheline mosquitoes are obligatory vectors for the transmission of the malaria parasites of the genus Plasmodium. The parasites undergo asexual development within mammalian hosts and produce gametocytes which, when ingested by the mosquito, initiate the sexual cycle that culminates with production of sporozoites. In turn, an infected mosquito takes another bloodmeal and sporozoites are released into the circulation of a naive host, thus completing the transmission cycle. Human malaria causes over 1 million deaths every year in the developing world. Recently, in recognition of the great importance of Anopheles gambiae in global health, its genome has been sequenced by an international scientific consortium (Holt et al. 2002), and transcriptomic approaches were initiated with the sequencing of Expressed Sequence Tags (ESTs) prepared from cultured cells (Dimopoulos et al. 2000). Four thousand ESTs were used to construct the first mosquito cDNA microarray, the 4K microarray platform (Dimopoulos et al. 2002). These arrays were used to detect genes that are up-regulated in the mosquito, specifically during infection with parasites and bacteria (Dimopoulos et al. 2002) and to identify differences between parasite-susceptible and refractory mosquitoes (Kumar et al. 2003). However, insufficient annotation of the EST sequences hindered such studies and greatly limited the capacity of researchers to derive appropriate interpretations. In the context of the Anopheles genome project, nearly 83,000 ESTs from naive and blood-fed adult mosquitoes were sequenced (Holt et al. 2002), and in silico analysis of these data detected genes up-regulated in the mosquitoes after a blood meal (Ribeiro et al. 2004). Furthermore, nearly 63,000 single reads from a full-length cDNA library were recently deposited in nucleotide databases by Genoscope (http://www.genoscope.org/). Two other EST libraries were constructed from pooled developmental stages of A. gambiae (NAP1) or adult heads (NAH), and clones from these libraries are currently being sequenced (G.K. Christophides, unpubl.; F. Collins, unpubl.). Twenty thousand of these ESTs were used to build a new cDNA microarray platform (20K or MMC1), which is currently used in various experimental approaches to identify genes that are temporally and spatially regulated in mosquitoes during development, parasite and viral infection, and insecticide treatment (G.K. Christophides, unpubl.). The increasing amount of information obtained from such studies necessitated the development of computational approaches to provide functional annotation and interpretation of the derived data.
Here, we report a large-scale study of malaria mosquito A. gambiae EST and cDNA sequences structured into the newly developed AnoEST database. Using these cDNA microarray data in conjunction with AnoEST, we have experimentally confirmed expression of 7961 clusters during mosquito development. Of these, 3100 are not associated with currently predicted genes (Holt et al. 2002; Birney et al. 2004). Moreover, we found that clusters with confirmed expression are nonbiased with respect to the current gene annotation or homology to known proteins, and consequently, we might expect that many of the unconfirmed clusters are likely to be actual A. gambiae genes. The AnoEST resource is a vital resource for the interpretation of expression profiles derived using the A. gambiae cDNA microarrays, providing inferred functional annotation of the expressed genomic loci, including similarities to known proteins, protein domains, and Gene Ontology (GO) (Ashburner et al. 2000) functional categories.
Results and Discussion
A. gambiae EST classification
We collected from public sequence databases (Benson et al. 2004; Kulikova et al. 2004; Miyazaki et al. 2004) 215,634 A. gambiae expressed sequences (178,618 from 5′-sequences and 37,015 from 3′-sequences) originating from 179,955 clones. Of these sequences, 211,468 were aligned to 593,349 regions on the nuclear or mitochondrial genome. For 203,812 expressed sequences, a unique genomic origin could be recognized. We clustered ESTs (assigned them into groups representing distinct expressed loci) using the genomic sequence as template, as described in the Methods section. This allows for a more specific assignment of ESTs into clusters, as it prevents merging of distinct gene loci due to chimeric ESTs or domains with highly similar sequences. Three types of clusters were distinguished as follows: (1) T-clusters (Transcribed clusters) that have at least one supporting EST from this genomic locus, (2) N-clusters (with No uniquely matched ESTs) that share regions of high sequence identity to EST/cDNA sequences, but cannot be confidently identified as expressed (this fraction also includes recent duplications when the corresponding EST/cDNAs could have been derived from any one of the duplicated regions), and (3) U-clusters (Unaligned) of ESTs that failed to align to the genome. The derived EST clusters were further identified with current Ensembl (Birney et al. 2004) gene predictions, annotated with orthology/homology to known proteins and protein domains using sequence-analysis techniques, and tentatively associated with GO (Gene Ontology) and GO-slim functional categories (see Methods section).
The descriptive statistics of the AnoEST data is provided in Table 1, which includes the numbers of different types of clusters and their annotation with respect to the Ensembl, UniProt/SWISS-PROT (Apweiler et al. 2004), and InterPro (Mulder et al. 2003) databases. Of the derived T-clusters, 13,173 (61%) are supported by more than one EST each; 9944 (75%) of these have a statistically significant hit to known proteins in SWISS-PROT. Although a single EST is commonly considered unreliable as evidence of expression (Okazaki et al. 2002), between one-fourth and one-half of 8305 EST singletons are supported by various sequence features indicative of protein-coding genes; they accommodate a correct gene model according to Ensembl gene predictions, encode known protein domains, or have significant homologs in SWISS-PROT. As discussed below, we have also used transcriptomic data to verify expression of a substantial fraction of T-clusters during mosquito development.
Table 1.
Descriptive statistics of A. gambiae EST clusters
| Clusters (with ≥2 ESTs) | Ensembl | SWISS-PROT | InterPro | |
|---|---|---|---|---|
| TCLAG | 21,478 (13,173) | 11,608 (8048) | 14,131 (9944) | 8015 (6368) |
| NCLAG | 46,560 (20,219)a | 2079 (1105) | 6971 (4247) | 2456 (1485) |
| UCLAG | 3881 (82) | n.a. | 1236 (52) | 199 (25) |
Numbers refer to overlaps with the current Ensembl gene set (14,364 genes in total), homology to known proteins in the SWISS-PROT Knowledgebase (matching 9791 sequences), and hits with protein domains in the InterPro database (matching 2144 distinct domains). Numbers referring to clusters supported by at least two sequences are marked in bold. The numbers of distinct Ensembl genes overlapping with T-clusters and N-clusters are 9639 (8020) and 1821 (1076), respectively, as some Ensembl genes overlap with more than one EST cluster.
35,660 (17,143) of N-clusters, i.e. 77 percent (85%) respectively, are contributed by only 863 ESTs, which are aligned with 50 to 191 distinct genomic loci.
In total, 11,608 T-clusters overlap with 10,726 Ensembl gene models (of 14,364 Ensembl predictions as of Aug. 10, 2004, v23.2b.1), indicating that, despite very strict clustering criteria, the analysis probably engendered only a minor number of fragmentation artifacts. On average, the derived EST clusters overlap with Ensembl gene models by about 920 nt, corresponding to 70% of the shorter loci; 2695 clusters overlap Ensembl gene models by >90%. Only 452 EST clusters have shorter than 20% overlaps; these probably derive from UTRs. Interestingly, 9870 T-clusters (4789 of which are supported by two or more EST/cDNAs) have no associated Ensembl gene predictions.
N-type clusters are quite different; they are twice as numerous, but have only one-sixth as many Ensembl overlaps as do the T-type clusters (Table 1). Moreover, 35,660 (77%) of the N-type clusters are formed by only 863 ESTs, each of which is aligned to at least 50 distinct genomic loci. These likely represent transposable elements in A. gambiae, as 24,984 N-type clusters show significant homology to known transposable elements in RepBase9.12 (Jurka 2000; http://www.girinst.org). In contrast, only 1312 T-clusters (61 with confirmed expression, see below) are homologous to repetitive elements. A total of 2079 N-type clusters are currently annotated as genes. However, only 860 N-clusters correspond to recently duplicated genes, of which 220 have a corresponding gene model. The portion of ESTs failing to align to the nuclear or mitochondrial genome of A. gambiae (U-type clusters) constitutes <3% of all sequences (Table 1). Some U-type clusters may correspond to as yet unsequenced regions of the genome, while most of them are likely to be of erroneous origin (data not shown).
Analysis of T-clusters
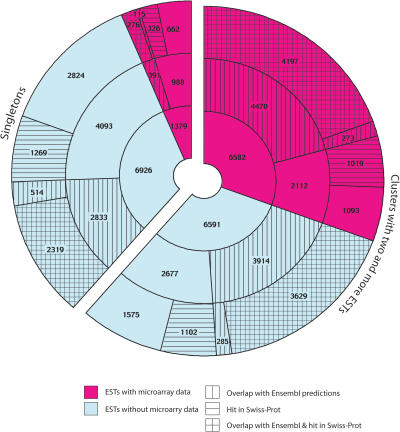
We have used both bioinformatic and transcriptomic methods to analyze in detail the category of T-type clusters, which represent the most prominent fraction of the mosquito genes. This dual analysis is summarized in Figure 1 and Table 2, separately for singletons (left) and for clusters with ≥2 ESTs (right). We categorized clusters as having or lacking corresponding Ensembl gene predictions, homologs in SWISS-PROT, or overlaps of Ensembl and SWISS-PROT hits. Transcriptomic analysis utilized a developmental data set encompassing expression profiles in embryos, larvae, pupae, and adult mosquitoes (G.K. Christophides, in prep.) to highlight the fraction of clusters with experimentally verified expression. These data were collected using 20,000-element cDNA microarrays (MMC1). Of the 8922 corresponding T-clusters, we scored 1379 singletons (17% of the total number of singletons) and 6582 clusters with ≥2 ESTs (50% of such clusters), summing up to 90% of the microarray elements as being significantly expressed in at least two developmental stages (see Methods).
Figure 1.
Analysis of the 21,478 T-clusters. The chart lists numbers of T-clusters, of which expression during mosquito development was confirmed by microarray experiments (pink) and numbers of clusters for which microarray-based expression was not tested or detected (blue). Numbers are provided separately for clusters with two or more ESTs (right) and singletons (clusters with one EST, left). For each category, the numbers of clusters with and without Ensembl gene predictions, as well as the numbers with and without homologs in UniProt/SWISS-PROT are indicated. The inner ring lists the total number of EST clusters with and without microarray data, and the outer two rings partition these clusters according to the associated annotation.
Table 2.
T-clusters with and without supporting microarray expression data
| Singletons
|
Clusters with ≥2 ESTs
|
|||
|---|---|---|---|---|
| Confirmed | No data | Fraction | Confirmed | No data |
| 1379 (100%) | 6926 (100%) | Total | 6582 (100%) | 6591 (100%) |
| 28.4% | 40.9% | With Ensembl gene prediction | 67.9% | 59.4% |
| 43.7% | 51.8% | With homology in SWISS-PROT | 79.2% | 71.8% |
| 20.0% | 33.5% | With Ensembl and SWISS-PROT hit | 63.8% | 55.1% |
| 71.6% | 59.1% | No Ensembl | 32.1% | 40.6% |
| 48.0% | 40.8% | No Ensembl, no SWISS-PROT hit | 16.6% | 23.9% |
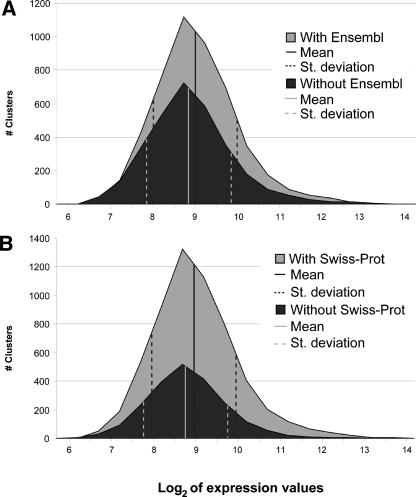
First, we explored the question of whether clusters with verified expression but without annotation represent low-level transcriptional leakage or whether they are expressed at levels comparable to those of recognized genes. For this purpose, we compared the distribution of log2-transformed values of expression for T-clusters with and without Ensembl gene prediction and for the fraction of clusters with and without SWISS-PROT homologs. As shown in Figure 2, in both cases, genes with and without annotation showed rather similar distributions, with only a small shift toward lower expression values in the absence of annotation, which was slightly more pronounced for clusters with SWISS-PROT homologs. Only 61 T-type clusters with confirmed expression, 20 of which have a corresponding gene model, show significant homology to A. gambiae transposable elements. This comparison suggested that most of the 3100 EST clusters that are currently lacking a predicted gene model have detectable expression and are likely to be actual genes.
Figure 2.
(A) Comparison of log2 expression value distributions for T-clusters with and without overlaps with Ensembl gene predictions. The graph also depicts mean and standard deviation values for the corresponding distributions. (B) Comparison of log2 expression value distributions for T-clusters with and without homology hits in the SWISS-PROT knowledgebase; mean and standard deviation values are also shown.
We then compared the T-cluster subsets with verified expression with those lacking microarray data (mostly not represented on the microarrays). These subsets were reasonably similar in terms of presence or absence of corresponding Ensembl predictions, SWISS-PROT homologs, or both (Table 2). As expected, the microarray expressed subset was substantially (fivefold) smaller than the subset lacking microarray data in the case of singletons, whereas the subsets were of equal size for ≥2 ESTs clusters. Surprisingly, the prevalence of Ensembl and SWISS-PROT hits was actually higher among singletons lacking supporting microarray data (Table 2).
Based on the analysis summarized in Figures 1 and 2 and in Table 2, our working hypothesis is that a substantial fraction of EST singletons represents actual genes, as do most of the ≥2 ESTs clusters. These data suggest that the number of genes in the A. gambiae genome may be substantially higher than currently predicted. A similar conclusion has been drawn recently for the Drosophila melanogaster genome using a combined bioinformatics and expression profiling approach (Hild et al. 2003).
Interface to the AnoEST database
The data discussed above have been structured into a relational database, for which we developed a user-friendly Web interface, available at http://komar.embl.de. It allows querying for the EST/cDNA accession number, clone identifier, derived EST cluster identifier, Ensembl gene identifier, SWISS-PROT accession numbers of homologous proteins, and associated GO terms, permitting logical combinations and flexible regular expressions.
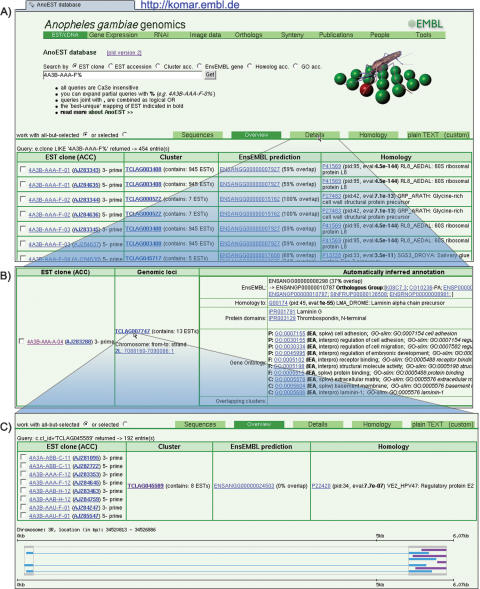
Examples of the available interactive searches are represented in Figure 3. By default, the information on queried sequences is returned in a condensed format showing data corresponding to the best-matching EST cluster (Fig. 3A). The “Sequences” tab at the top of the interface allows retrieval of the sequences in FASTA format and, if required, generates reverse complemented sequences, e.g., for 3′-sequenced clones. The “Details” tab makes available more extensive information on similarity to known proteins and protein domains, orthology, GO, and “GO-slim” categories (Fig. 3B). The annotation available for each corresponding genomic region in Ensembl can also be explored through a direct link to the genome browser. The “Homology” tab refers to the full records of a similarity search of the EST cluster consensus sequence against the UniProt/SWISS-PROT protein database. The records allow manual inspection of the alignments and provide html references to the corresponding entries in the UniProt/SWISS-PROT database.
Figure 3.
Interactive searches available in AnoEST. Searching with an EST clone identifier allows (A) an overview of associated EST sequences, corresponding clusters, overlapping Ensembl genes, and best hit in the SWISS-PROT database and its description. (B) The detailed view gives, in addition, coordinates of EST cluster match to the genome, links to orthologous groups identified on the basis of corresponding Ensembl gene predictions, protein domains collected in the InterPro database, and corresponding GO terms. When examining EST clusters, (C) a graphical representation of overlapping ESTs permits visualization of the underlying exon-intron structure.
When exploring all expressed sequences assigned to one cluster (Fig. 3C), the visualization of EST alignments to genome allows a quick grasp of the gene organization, EST coverage, and quality of the clustering. Sequences derived from 5′- and 3′-ends are colored differently. The scale bar provided indicates the real cluster length over the genomic alignment, sized to fit to the image. The EST cluster image is mapped by html links to EST records for exploring cases of interest.
To make the results more broadly accessible and integrated with the Ensembl genome browser, the data are also available through the DAS protocol (http://komar.embl.de:9000/das). The dump of the data in relational mySQL format is available upon request.
AnoEST utility for microarray analysis
To facilitate functional analysis of transcriptional data derived from cDNA microarrays that have been recently developed (Dimopoulos et al. 2002; G.K. Christophides, unpubl.) and that are already widely used in the mosquito scientific community, the microarray elements are annotated using information from the AnoEST database via the EST identifiers. To allow further exploration of the annotation using specialized software or Excel spreadsheets, the microarray grid annotation (currently the 4K, MMC1/20K, and in the future the full genome array MMC2 currently under development) is provided as tab-delimited text files. The constraints imposed by such representation limit the complexity of included data, e.g., each element is associated with only best-matching EST/cDNA cluster and its functional annotation. In order to draw solid conclusions for the expression of a genomic locus, it is very important in DNA microarray experiments to evaluate all possible cross-hybridizations. That is possible utilizing the “CrossMapping” column included in the files that lists all clusters sharing high-sequence identity for each of the microarray elements. Moreover, as the microarray elements enclose both DNA strands that could potentially contribute to the spot signal, we report all EST clusters that are on opposite strands, but overlap by at least 60 nt. This is summarized in the “Overlapping clusters” column, generated by concatenating the corresponding cluster identifiers ordered by significance of sequence homology to known proteins, which allows users to easily recognize and group them.
Future developments
Together with the Ensembl team, we are planning to use the obtained results for refinement of current gene predictions in the Anopheles genome. This would complement the approach of another Anopheles database, AnoBase (http://www.anobase.org/), which is oriented toward manual refinement of automatically predicted gene models. The functional and expression data available through AnoEST is also being used for the discovery and annotation of alternative splicing events. In the future, we plan to extend AnoEST for use with the previously mentioned new generation of single-exon amplicon microarrays that will permit coupling of transcription profiling of the whole mosquito genome with other high-throughput functional assays, such as the production and use of specific double-stranded RNAs for RNAi gene silencing, and the production of peptides to develop antibody panels. This new microarray platform (MMC2) is designed in the context of an informal Mosquito Microarray Consortium (MMC) that emerged as an initiative to coordinate and standardize global transcriptional studies in A. gambiae. The current AnoEST data on clusters of expressed sequences that are not matched with current Ensembl gene models, as well as on alternatively spliced transcripts, is used to design additional features of MMC2. Although AnoEST was initiated as an independent database, it will be adapted to serve as one of the functional genomics modules of a new integrated genomic data resource for multiple vectors of disease, VectorBase (http://www.vectorbase.org).
Methods
EST clustering
The analysis begins with the collection and processing of all available A. gambiae EST and cDNA sequences, linked with their GenBank/EMBL-Bank/DDBJ accession number, clone name identifier, cDNA strand information, and nucleotide sequence. All sequences are then aligned to the unmasked reference genome using the BLAT algorithm (Kent 2002), considering all matches of 60 or more nucleotides, with at least 96% identity, a level that allows for inaccuracies of EST sequences and polymorphisms and which captures all possible cross-hybridizations on a DNA microarray (Hughes et al. 2001). The accuracy of this step could be further improved at the expense of using orders of magnitude slower algorithms like sim4 (Florea et al. 1998).
ESTs are then clustered (assigned into groups) on the basis of their genomic overlap. For example, two sequences are assigned to the same cluster if their overlap over the aligned regions (exons) is greater than a certain threshold (30 nt in the current version of AnoEST). To avoid CPU-consuming all-against-all EST comparisons, which would be computationally challenging when considering potential alignment of over 200,000 EST sequences with nearly 500,000 genomic loci, we compare ESTs only with the cluster's projection on the genome. DNA strands are considered independently. EST sequences originating from the 3′-end of a clone are deposited in public repositories as reverse complements; therefore, we alter their alignment strand information prior to clustering.
In many cases, an expressed sequence can be aligned to more than one place in the genome (paralogs, transposable elements), making it difficult to identify reliably which genomic locus is actually represented by the EST. To address this, we rank EST to genome alignments using a number-of-matches minus number-of-mismatches scoring scheme, similar to BLAT. The matches with the highest score are then marked as “best”, or as “unique best” when the second-best score is significantly lower (e.g., by more than 15, to reflect the EST sequence error rate and weak support from the data distribution). Clusters including at least one “unique best” EST are identified as TCLAG (for Transcribed CLuster of Anopheles Gambiae, also referred to as T-clusters above), whereas those that share regions of high-sequence identity to EST/cDNA sequences, but there is no one sequence aligned to the locus as “unique best” are identified as NCLAG clusters (with No uniquely matched ESTs). The third type of cluster identifiers, UCLAG, corresponds to ESTs that failed to align (Unaligned) to the A. gambiae nuclear or mitochondrial genome.
In the final step of our clustering procedure, we join clusters that contain ESTs originating from the 5′- and 3′-ends of the same clone, provided that they map as “unique best” to the corresponding EST clusters, and they are on the same chromosome, the same strand, and <30 kb apart.
The choice of many of the above-described parameters reflects a conservative approach that attempts to minimize errors of joining independent expressed loci at the expense of allowing some fragmentation errors, e.g., one gene could be represented by two EST clusters if we do not have sufficient information to link these clusters together. The observed representation of 10,726 Ensembl gene models by 11,608 T-clusters suggests only a minor number of fragmentation artifacts. Use of strand-specific clustering avoids the severe problems of erroneous joining of distinct genes (data not shown). However, some sequences inserted into plasmid in the wrong orientation form erroneous clusters on the strand opposite the actual genes. An upper estimate of such errors is about 11%, counting the number of T-clusters overlapping annotated genes with respect to T-clusters on the opposite strand without annotation (counting overlaps over an average 70%).
Automatic annotation
The derived clusters of expressed sequences are identified with gene models predicted by the Ensembl annotation pipeline, noting the fraction of genomic overlap over all predicted exons and allowing ±150 nt to capture EST clusters derived from UTRs.
We showed previously that genes recognized as 1:1 orthologs in the genomes of A. gambiae and D. melanogaster code on average for proteins with 56% sequence identity (Zdobnov et al. 2002). This suggests that many well-characterized proteins of Drosophila and other evolutionarily more-distant organisms, such as human, share only limited identity with Anopheles proteins. This limits the utility of more comprehensive, but automatically derived nonredundant protein collections such as UniRef90 and even UniRef50 (representing sequences merged at 90% and 50% sequence identity, respectively), where best hits are dominated by poorly annotated predictions from genome-sequencing projects. To capture such weak homologies, we used the sensitive Smith-Waterman algorithm (Smith and Waterman 1981) (as implemented by Paracel) to compare all forward translations of the EST cluster sequences with sequences of known proteins from the manually curated UniProt/SWISS-PROT database. We then extract from that database a concise annotation for the best-matching sequence, identified with a E-value cut-off of <0.001. When available, we tentatively assign Gene Ontology (GO) functional annotation terms to the EST clusters, inferred from the best matching protein in UniProt/SWISS-PROT database. The UniProt/SWISS-PROT to GO mapping is provided by the GOA project at EMBL-EBI (Camon et al. 2004). We traverse the GO hierarchy in a “bottom to top” manner to assign the high-level “GO-slim” functional classes, which can be further compared with the patterns of correlated expression as identified in the DNA microarray experiments. We also analyze the EST cluster sequences for characteristic signatures of known protein domains, using state-of-the-art HMM profiles, as defined in PFAM and SMART (Bateman et al. 2004; Letunic et al. 2004) and summarized in InterPro.
We identified groups of orthologous genes between the predicted full proteomes of A. gambiae and D. melanogaster, and broader orthologous groups, including other animal genomes with full genome coverage using an Inparanoid-like (Remm et al. 2001) procedure. Orthologous genes were then used as markers to identify the conservation of the genomic arrangement (synteny) as described before (Zdobnov et al. 2002) using SyntQL tool (E.M. Zdobnov, unpubl.).
Implementation
AnoEST is implemented as a relational database using mySQL (http://www.mysql.com/). An interactive Web interface to the data is provided using PHP (http://www.php.net/). The data is also accessible through the DAS (http://www.biodas.org) protocol using a Perl-based ProDAS severe (http://www.sanger.ac.uk/Software/analysis/proserver/). EST clustering is implemented in Perl using a DBI interface to the mySQL backend, to allow scaling to higher numbers of sequences without additional computer memory requirements.
Microarray assessment of EST cluster expression
We experimentally assessed the expression of AnoEST-derived clusters utilizing a developmental data set that was recently produced in our laboratory using the MMC1 spotted cDNA microarrays (G.K. Christophides, unpubl.). Briefly, the experimental design interrogated nine different time points of the entire A. gambiae life cycle, from embryos to adults. Hybridizations were performed against an artificially constructed standard reference, containing all spots of the array. Four replicates (three biological and one technical—dye swap) for each time point were performed. Manual inspection and statistical measurements were used to assess spot quality based on signal intensity versus local background levels and spot diameter. Negative spiked-in controls were used to calculate global background levels, and only data above three standard deviations of background intensity levels were considered for further analysis. Data were loaded into Gene-Spring v7.0 (Agilent Technologies), and normalized with the intensity-dependent (lowess) normalization algorithm. After replicate averaging, we selected ESTs that had reliable measurements in at least 33 of the 37 hybridizations and exhibited t-test P-value <0.05 in at least two of the nine time points. These criteria led us to consider the expression of 15,135 ESTs as confirmed during mosquito development. To quantify the level of expression of these ESTs, we considered the maximum intensity signal from the nine time points analyzed.
Acknowledgments
This work was partially supported by NIAID/NIH VectorBase contract (NIAID-DMID-04-34 coordinated by FH Collins), NIAID/NIH U01 AI48846 and EMBL. We acknowledge annotation support from Ensembl (a joint Sanger Institute/EMBL-EBI project funded by the Wellcome Trust) and DNA sequence information from Celera Genomics, Genoscope, Pasteur Institute, EMBL, and the University of Notre-Dame (the major contributors to A. gambiae genomic, EST, and cDNA sequence data). We are also grateful to S. Meister and other members of the F.C. Kafatos group, and to members of the P. Bork group for helpful discussions.
Footnotes
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.3756405. Article published online ahead of print in May 2005. Freely available online through the Genome Research Immediate Open Access option.
References
- Apweiler, R., Bairoch, A., Wu, C.H., Barker, W.C., Boeckmann, B., Ferro, S., Gasteiger, E., Huang, H., Lopez, R., Magrane, M., et al. 2004. UniProt: The Universal Protein knowledgebase. Nucleic Acids Res. 32: D115-D119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner, M., Ball, C.A., Blake, J.A., Botstein, D., Butler, H., Cherry, J.M., Davis, A.P., Dolinski, K., Dwight, S.S., Eppig, J.T., et al. 2000. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 25: 25-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman, A., Coin, L., Durbin, R., Finn, R.D., Hollich, V., Griffiths-Jones, S., Khanna, A., Marshall, M., Moxon, S., Sonnhammer, E.L., et al. 2004. The Pfam protein families database. Nucleic Acids Res. 32: D138-D141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson, D.A., Karsch-Mizrachi, I., Lipman, D.J., Ostell, J., and Wheeler, D.L. 2004. GenBank: Update. Nucleic Acids Res. 32: D23-D26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birney, E., Andrews, T.D., Bevan, P., Caccamo, M., Chen, Y., Clarke, L., Coates, G., Cuff, J., Curwen, V., Cutts, T., et al. 2004. An overview of Ensembl. Genome Res. 14: 925-928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camon, E., Magrane, M., Barrell, D., Lee, V., Dimmer, E., Maslen, J., Binns, D., Harte, N., Lopez, R., and Apweiler, R. 2004. The Gene Ontology Annotation (GOA) Database: Sharing knowledge in Uniprot with Gene Ontology. Nucleic Acids Res. 32: D262-D266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimopoulos, G., Casavant, T.L., Chang, S., Scheetz, T., Roberts, C., Donohue, M., Schultz, J., Benes, V., Bork, P., Ansorge, W., et al. 2000. Anopheles gambiae pilot gene discovery project: Identification of mosquito innate immunity genes from expressed sequence tags generated from immune-competent cell lines. Proc. Natl. Acad. Sci. 97: 6619-6624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimopoulos, G., Christophides, G.K., Meister, S., Schultz, J., White, K.P., Barillas-Mury, C., and Kafatos, F.C. 2002. Genome expression analysis of Anopheles gambiae: Responses to injury, bacterial challenge, and malaria infection. Proc. Natl. Acad. Sci. 99: 8814-8819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florea, L., Hartzell, G., Zhang, Z., Rubin, G.M., and Miller, W. 1998. A computer program for aligning a cDNA sequence with a genomic DNA sequence. Genome Res. 8: 967-974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hild, M., Beckmann, B., Haas, S.A., Koch, B., Solovyev, V., Busold, C., Fellenberg, K., Boutros, M., Vingron, M., Sauer, F., et al. 2003. An integrated gene annotation and transcriptional profiling approach towards the full gene content of the Drosophila genome. Genome Biol. 5: R3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt, R.A., Subramanian, G.M., Halpern, A., Sutton, G.G., Charlab, R., Nusskern, D.R., Wincker, P., Clark, A.G., Ribeiro, J.M., Wides, R., et al. 2002. The genome sequence of the malaria mosquito Anopheles gambiae. Science 298: 129-149. [DOI] [PubMed] [Google Scholar]
- Hughes, T.R., Mao, M., Jones, A.R., Burchard, J., Marton, M.J., Shannon, K.W., Lefkowitz, S.M., Ziman, M., Schelter, J.M., Meyer, M.R., et al. 2001. Expression profiling using microarrays fabricated by an ink-jet oligonucleotide synthesizer. Nat. Biotechnol. 19: 342-347. [DOI] [PubMed] [Google Scholar]
- Jurka, J. 2000. Repbase Update: A database and an electronic journal of repetitive elements. Trends Genet. 9: 418-420. [DOI] [PubMed] [Google Scholar]
- Kent, W.J. 2002. BLAT—the BLAST-like alignment tool. Genome Res. 12: 656-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulikova, T., Aldebert, P., Althorpe, N., Baker, W., Bates, K., Browne, P., van den Broek, A., Cochrane, G., Duggan, K., Eberhardt, R., et al. 2004. The EMBL Nucleotide Sequence Database. Nucleic Acids Res. 32: D27-D30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, S., Christophides, G.K., Cantera, R., Charles, B., Han, Y.S., Meister, S., Dimopoulos, G., Kafatos, F.C., and Barillas-Mury, C. 2003. The role of reactive oxygen species on Plasmodium melanotic encapsulation in Anopheles gambiae. Proc. Natl. Acad. Sci. 100: 14139-14144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic, I., Copley, R.R., Schmidt, S., Ciccarelli, F.D., Doerks, T., Schultz, J., Ponting, C.P., and Bork, P. 2004. SMART 4.0: Towards genomic data integration. Nucleic Acids Res. 32: D142-D144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazaki, S., Sugawara, H., Ikeo, K., Gojobori, T., and Tateno, Y. 2004. DDBJ in the stream of various biological data. Nucleic Acids Res. 32: D31-D34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulder, N.J., Apweiler, R., Attwood, T.K., Bairoch, A., Barrell, D., Bateman, A., Binns, D., Biswas, M., Bradley, P., Bork, P., et al. 2003. The InterPro Database, 2003 brings increased coverage and new features. Nucleic Acids Res. 31: 315-318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okazaki, Y., Furuno, M., Kasukawa, T., Adachi, J., Bono, H., Kondo, S., Nikaido, I., Osato, N., Saito, R., Suzuki, H., et al. 2002. Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature 420: 563-573. [DOI] [PubMed] [Google Scholar]
- Remm, M., Storm, C.E., and Sonnhammer, E.L. 2001. Automatic clustering of orthologs and in-paralogs from pairwise species comparisons. J. Mol. Biol. 314: 1041-1052. [DOI] [PubMed] [Google Scholar]
- Ribeiro, J.M., Topalis, P., and Louis, C. 2004. AnoXcel: An Anopheles gambiae protein database. Insect Mol. Biol. 13: 449-457. [DOI] [PubMed] [Google Scholar]
- Smith, T.F. and Waterman, M.S. 1981. Identification of common molecular subsequences. J. Mol. Biol. 147: 195-197. [DOI] [PubMed] [Google Scholar]
- Zdobnov, E.M., von Mering, C., Letunic, I., Torrents, D., Suyama, M., Copley, R.R., Christophides, G.K., Thomasova, D., Holt, R.A., Subramanian, G.M., et al. 2002. Comparative genome and proteome analysis of Anopheles gambiae and Drosophila melanogaster. Science 298: 149-159. [DOI] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- http://komar.embl.de; AnoEST database.
- http://komar.embl.de:9000/das; AnoEST DAS server.
- http://www.genoscope.org/; Genoscope—Centre National de Séquençage.
- http://www.girinst.org/; Genetic Information Research Institute.
- http://www.anobase.org/; AnoBase database.
- http://www.vectorbase.org; VectorBase database.
- http://www.mysql.com/; MySQL relational database engine.
- http://www.php.net/; PHP scripting language.
- http://www.biodas.org; Distributed Annotation System (DAS).
- http://www.sanger.ac.uk/Software/analysis/proserver/; Perl-based DAS server.