Abstract
Loss-of-function mutations in the NF1 tumor suppressor gene underlie the familial cancer syndrome neurofibromatosis type I (NF1). The NF1-encoded protein, neurofibromin, functions as a Ras-GTPase activating protein (RasGAP). Accordingly, deregulation of Ras is thought to contribute to NF1 development. However, the critical effector pathways involved in disease pathogenesis are still unknown. We show here that the mTOR pathway is tightly regulated by neurofibromin. mTOR is constitutively activated in both NF1-deficient primary cells and human tumors in the absence of growth factors. This aberrant activation depends on Ras and PI3 kinase, and is mediated by the phosphorylation and inactivation of the TSC2-encoded protein tuberin by AKT. Importantly, tumor cell lines derived from NF1 patients, and a genetically engineered cell system that requires Nf1-deficiency for transformation, are highly sensitive to the mTOR inhibitor rapamycin. Furthermore, while we show that the activation of endogenous Ras leads to constitutive mTOR signaling in this disease state, we also demonstrate that in normal cells Ras is differentially required for mTOR signaling in response to various growth factors. Thus, these findings identify the NF1 tumor suppressor as an indispensable regulator of TSC2 and mTOR. Furthermore, our results also demonstrate that Ras plays a critical role in the activation of mTOR in both normal and tumorigenic settings. Finally, these data suggest that rapamycin, or its derivatives, may represent a viable therapy for NF1.
Keywords: neurofibromin, Ras
NF1 is a prevalent familial cancer syndrome affecting 1 in 3,500 individuals worldwide (1). The hallmark feature of the disease is the development of benign and malignant peripheral nerve sheath tumors (neurofibromas and MPNSTs, respectively). However, NF1 patients can also exhibit cognitive deficits, hamartomatous lesions of the iris, and bone deformations and are predisposed to developing myeloid malignancies, gliomas, and pheochromocytomas (1, 2).
Although loss-of-function mutations in the NF1 gene result in the deregulation of Ras (3–7), the specific effector pathways responsible for disease pathogenesis have not been defined. Interestingly, NF1 belongs to a group of disorders known as the phakomatoses (8). This family of diseases exhibits several common features, including the development of hamartomas [lesions not classically defined as benign tumors but rather extraneous masses of well differentiated tissue indigenous to the site of their development (9)]. Recently, many of the genes responsible for these diseases, such as PTEN (Cowden disease), LKB1 (Peutz–Jeghers syndrome), and TSC1 and TSC2 (tuberous sclerosis complex), have been linked in a common biochemical pathway (10, 11). Inactivation of all of these genes in primary cells results in a deregulation of TOR (target of rapamycin) signaling (12–17). Furthermore, in some cases the cognate human tumor cells appear to be highly sensitive to TOR inhibitors (rapamycin or rapamycin derivatives) (12, 13). We therefore sought to determine whether the TOR pathway might also be critically deregulated in NF1, in part, to identify a potential therapeutic target for this devastating disorder.
TOR is an evolutionarily conserved serine/threonine protein kinase that regulates cell growth and proliferation in yeast, flies, and mammals (11). Mammalian TOR (mTOR) has been shown to be a critical sensor of nutrients, growth factors, and cellular energy state and integrates signals from each of these pathways to control a variety of cellular processes (11, 18, 19). Growth factors activate mTOR via a PI3 kinase-dependent mechanism, whereas nutrients (amino acids) affect this pathway further downstream, at the level of tuberin and/or mTOR itself (11). Because neurofibromin can negatively regulate the Ras/PI3 kinase pathway, we hypothesized that the mTOR pathway might also be deregulated in NF1-deficient cells.
In this article, we show that mTOR is constitutively activated in Nf1-deficient primary cells. This aberrant activation depends on Ras and PI3 kinase, which inactivate the TSC2 gene product tuberin via AKT. Cells derived from NF1 patient tumors similarly exhibit a constitutive phosphorylation of tuberin and an aberrant activation of mTOR, whereas patient-matched NF1+/- cells do not. Importantly, NF1-deficient malignant human tumor cell lines are exquisitely sensitive to rapamycin. We further explored the requirement for Ras activation in the normal regulation of mTOR and found that Ras differentially contributes to mTOR activation in response to growth factors. Specifically, suppression of Ras activity significantly attenuated mTOR activation in response to l-α-lysophosphatidic acid (LPA) and insulin but had little or no effect on mTOR activation in response to platelet-derived growth factor (PDGF). Thus, these findings identify the NF1 tumor suppressor as a previously uncharacterized component of the mTOR pathway. Furthermore, they demonstrate that physiological levels of Ras-GTP critically modulate mTOR activation in both normal and tumorigenic settings. Moreover, they suggest the potential utility of rapamycin derivatives in treating NF1 tumors.
Materials and Methods
Cell Culture and Immunoblots. MEFs and NIH 3T3 cells were grown in DMEM supplemented with FCS or calf serum, respectively. Littermate matched MEFs or NIH 3T3 cells were plated in serum-free DMEM at a density of 1.0 × 106 cells per 10-cm plate. After 18 h, 200 nM insulin, 6 μM LPA, or 20 ng/ml PDGF was added. Where indicated, cells were pretreated with DMSO, 20 nM rapamycin (Calbiochem), and 200 nM Wortmannin (Sigma-Aldrich) for 30 min. For amino acid withdrawal studies, cells were washed and media was replaced with Dulbecco's PBS for the indicated times. After stimulation, cells were lysed in 1% SDS boiling lysis buffer. Neurofibroma-derived patient-matched NF1+/- and NF1-/- Schwann cells were isolated and cultured as previously described in DMEM supplemented with 10% FCS, 0.5 μM forskolin (NF1+/- cells only), 2.5 μg/ml insulin, 0.5 mM IBMX, and 10 nM heregulin β1 (20, 21). Schwann cells were seeded at a concentration of 2.5 × 105 cells per 6-cm plate in DMEM containing 0.1% serum without forskolin, insulin, or heregulin and lysed as described above. Identical results were obtained in Schwann cell experiments that were acutely performed in the presence or absence of forskolin. Human MPNST cell lines were generated from NF1 patients as described in ref. 22. Clarified lysates were normalized for protein levels and analyzed by Western blotting with the following antibodies: phospho-p70S6K (T-389), phospho-tuberin (S-939), phosphotuberin (T-1462), phospho-Akt (S-473), tuberin (C-20) from Santa Cruz Biotechnology, and protein kinase Bα/AKT1, actin, and β-tubulin from Sigma-Aldrich.
Retroviruses, Lentiviruses, and Transfections. Primary MEFs or NIH3T3 cells were infected with retrovirus expressing empty vector, human NF1 GAP-related domain (GRD) (MSCV-GRD-pac), a dominant-negative p53 (pBabe-hygro-p53DD), or pLPC-E1A12S and selected with the appropriate antibiotic. For lentiviral production, HEK293T cells were transfected with the Δ8.2 lentiviral construct (encoding gag, pol, rev), VSVG, and either empty pKO vector or the pKO vector containing the following sequence transcribing short-hairpin RNAs specific to NF1: 5′-TTATAAATAGCCTGGAAAAGG-3′. Virus-containing medium was then used to infect HEK293 cells. Approximately 8–10 h later, infected cells were transiently transfected (FuGENE, Roche) with 0.5 μg of pCDNA3-Flag vector or 0.5 μg of pCDNA-Flag encoding C-terminally tagged wild-type tuberin, the AKT-phosphorylation site mutant tuberin S939A/T1462A (both generous gifts from Brendan Manning, Harvard School of Public Health, Boston), or pCDNA-Flag-NF1-GRD (7). All infected cells were cotransfected with 0.5 μg of rat pRK7-HA-p70S6K (also from Brendan Manning).
Ras Activation Analysis. NIH3T3 cells were plated in DMEM containing 3% serum at a density of 1.0 × 106 cells per 10-cm plate. After 18 h, cell lysates were normalized, and Ras-GTP was detected by using a Ras-activation assay per the manufacturer's protocol (Upstate Biotechnology).
Immunoprecipitations. HEK293 lysates were prepared in 1.0 ml of lysis buffer [20 mM Tris, pH 7.5/150 mM NaCl/1 mM MgCl2/1% Nonidet P-40/10% glycerol/50 mM NaF/30 mM Na4O7P2/Complete protease inhibitor mixture (Roche)/1 μM Microcystin-LR (Calbiochem)/100 nM Calyculin-A (Calbiochem)]. Immunoprecipitated proteins were resolved along with total cell extracts (10% of immunoprecipitation volume) by SDS/PAGE, and immunoblots were performed with the indicated antibodies.
MPNST Proliferation Studies. MPNSTs were seeded at 6.5 × 104 cells per 6-cm well in normal growth medium containing rapamycin (0.01, 0.1, 1.0, 10, or 100 nM) or an equal volume of vector (DMSO). After 7 days, cells were trypsinized, and live cells were counted on triplicate plates by using trypan blue exclusion.
Soft Agar Assays. Nf1-/- and Nf1+/+ MEFs were infected with pLPC-E1A12S-puro and pBabe-p53DD-Hygro. Then, 1.0 × 104 selected MEFs were suspended in DMEM containing 0.34% agarose containing either rapamycin (0.1, 1.0, 10, 20, or 50 nM) or equal volumes of DMSO. Cells were seeded onto a 0.5% agar base. Four to 6 weeks after the initial seed, colony growth was assayed by photographing and counting 10 random fields of view per sample, in triplicate plates or wells. Colony size was assessed by using imagej v.1.32j software (http://rsb.info.nih.gov/ij).
Results
Because neurofibromin can negatively regulate the Ras/PI3 kinase pathway, we hypothesized that the mTOR pathway might also be deregulated in NF1-deficient cells.
The most commonly used in vivo readout of mTOR activation is the phosphorylation of a well characterized substrate, S6 kinase (S6K1), at T-389. Phosphorylation at this site depends on mTOR and is required for maximal S6K1 activation. Serum-starved wild-type mouse embryonic fibroblasts (MEFs) exhibited little activation of AKT or S6K1 (Fig. 1A). However, AKT was aberrantly activated in serum-deprived Nf1-null MEFs. Notably, S6K1 was also hyperphosphorylated in these cells in the absence of any growth factors. The aberrant activation of S6K1 was inhibited by rapamycin, demonstrating its dependence on mTOR. We also examined whether amino acid deprivation would inhibit mTOR activation in Nf1-deficient cells. Consistent with the observation that nutrients integrate into the mTOR pathway downstream of PI3 kinase activation, amino acid withdrawal resulted in the appropriate termination of the mTOR signal in both wild-type and Nf1-deficient cells (Fig. 1B). Thus, neurofibromin deficiency specifically results in the deregulation of the mTOR pathway in response to growth factor deprivation.
Fig. 1.
The mTOR pathway is aberrantly regulated in Nf1-/- MEFs. (A) Western blot analysis of serum-starved primary Nf1+/+ and Nf1-/- MEFs. Abbreviations are as follows: NF1, neurofibromin; pS6K, p70S6K phosphorylated at T389; pAKT, Akt phosphorylated at S473; RAPA, rapamycin. Cells were serum-starved and treated with rapamycin (20 nM) for 30 min where indicated. (B) Western blot analysis of primary Nf1+/+ and Nf1-/- MEFs deprived of amino acids (AA).
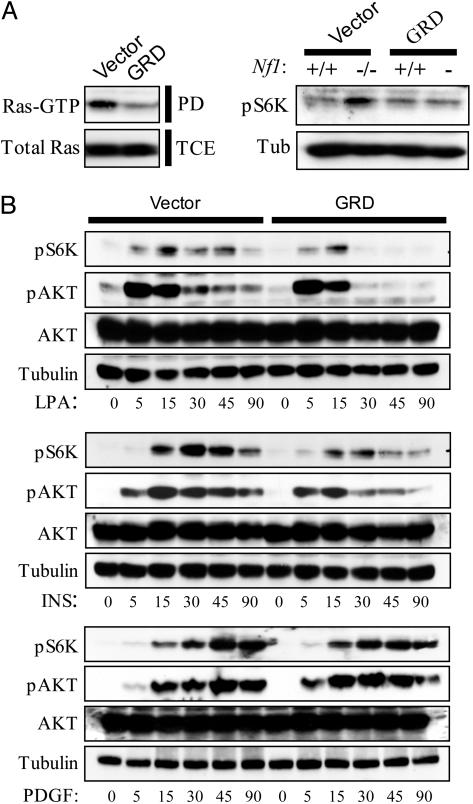
Because neurofibromin is a RasGAP, we investigated whether the aberrant activation of the mTOR pathway in Nf1-deficient cells depended on the inappropriate activation of Ras. Notably, we and others have previously shown that Ras-GTP levels are deregulated in Nf1-deficient cells (5–7, 23). We therefore sought to determine whether expression of only the catalytic GRD of neurofibromin would be sufficient to suppress the observed defect in S6K1 activation. This fragment has previously been reported to rescue specific defects associated with Nf1-deficiency and dramatically suppressed Ras activation (Fig. 2A Left) (23). Moreover, it restored the level of phospho-S6K1 in Nf1-deficient cells to levels equivalent to those observed in wild-type cells under the same conditions (Fig. 2 A Right). These results indicate that the hyperactivation of this pathway in Nf1-deficient cells depends on the absence of its RasGAP activity, rather than an additional uncharacterized function of neurofibromin.
Fig. 2.
mTOR activation in normal and Nf1-/- fibroblasts requires active Ras. (A) Ras GTP levels were quantitated by using a pull-down assay in cells expressing either the NF1-GRD or empty vector (Left). GTP-bound Ras, isolated from the pull-down assay (PD), and total Ras protein, present in the total cell extract (TCE), are shown. Western blot analysis of Nf1+/+ and Nf1-/- MEFs expressing the NF1-GRD or empty vector (Right). (B) Western analysis of serum-starved 3T3 fibroblasts expressing either the NF1-GRD or empty vector in response to 200 nM insulin (Top), 6 μM LPA (Middle), or 20 ng/ml PDGF (Bottom) for the indicated times.
Although the overexpression of an oncogenic Ras allele can induce S6K1 activation (24), the extent to which Ras normally participates in regulating mTOR in the context of growth factor signaling has been debated. Notably, growth factor receptors can activate PI3 kinase via Ras-dependent and independent mechanisms, the redundancy of which has been a conundrum in the field. Nevertheless, the Ras-independent PI3 kinase pathway is assumed to be the primary mTOR activation signal, and Ras is generally excluded from discussions of this pathway entirely (10, 11). This exclusion may partially be attributed to earlier studies suggesting that Ras was not required for mTOR activation in response to PDGF (25). However, it is plausible that different growth factors might preferentially engage one pathway versus the other. Furthermore, in vivo where growth factors are likely to be limiting, both pathways may be required to elicit maximal mTOR activation and a subsequent biological response. In any case, the requirement for physiological levels of Ras activation in regulating the mTOR pathway has not been thoroughly elucidated.
The observation that neurofibromin-deficiency results in constitutive mTOR activation indicates that the activation of endogenous Ras can stimulate this pathway. Because expression of the GRD of neurofibromin dramatically inhibited Ras-GTP levels in wild-type cells, this reagent was also used to determine whether Ras activation is required for maximal activation of the mTOR pathway in response to various growth factors. Importantly, the GRD significantly attenuated AKT activation and blunted S6K1 activation in response to LPA in wild-type cells (Fig. 2B). It also inhibited S6K1 phosphorylation in response to insulin. Finally, the GRD had no effect on PDGF-induced mTOR activation, consistent with previous reports (25). Interestingly, in response to LPA and insulin the GRD appeared to have the most dramatic effect on AKT and S6K1 at later time points, rather than immediately after growth factor treatment. This finding may suggest that Ras-independent mechanisms of PI3 kinase activation are important for the initial activation of this pathway, whereas Ras is required for a more sustained activation. This model is consistent with the fact that Ras-independent activation of PI3 kinase occurs more proximal to receptor activation than Ras-dependent activation of its effectors, in both receptor tyrosine kinase and G protein-coupled receptor signaling. Regardless, these data indicate that Ras activation is differentially required for maximal activation of the mTOR pathway in response to growth factors.
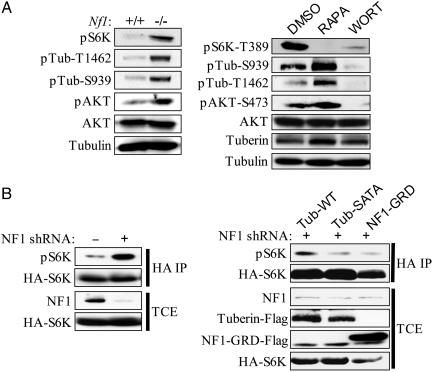
To determine whether the inappropriate activation of mTOR in Nf1-deficient cells depended on the Ras/PI3 kinase effector pathway, we examined S6K1 phosphorylation in Nf1-deficient cells in the presence of the PI3 kinase inhibitor wortmannin. We found that wortmannin dramatically reduced AKT activation and S6K1 phosphorylation in serum-deprived Nf1-deficient cells (Fig. 3A). The primary target of AKT in this pathway is thought to be tuberin, which in response to insulin is phosphorylated at two distinct sites, T-1462 and S-939, by this kinase (26–28). AKT phosphorylation of tuberin has been shown to inactivate the TSC1/TSC2 complex, resulting in the subsequent activation of Rheb and mTOR, through an unknown mechanism (29–32). Importantly, in Nf1-deficient serum-starved cells tuberin was constitutively phosphorylated at T-1462 and S-939, in contrast to wild-type cells where phosphorylation was minimal under these conditions (Fig. 3A). Phosphorylation at these sites was also suppressed in the presence of wortmannin (Fig. 3A). Finally, a mutant TSC2 gene, encoding alanine substitutions at T-1462 and S-939, suppressed mTOR activation in Nf1-deficient cells (Fig. 3B).
Fig. 3.
Hyperactivation of the PI3 kinase pathway in Nf1-/- primary MEFs results in tuberin inactivation. (A Left) Western blot analysis of serum-starved Nf1+/+ and Nf1-/- MEFs. Abbreviations are as follows: pS6K, p70S6K phosphorylated at T389; pAKT, AKT phosphorylated at S473; pTub-T1462, tuberin phosphorylated at T1462; pTub-S939, tuberin phosphorylated at S939. (A Right) Western blot analysis of tuberin phosphorylation in serum-starved Nf1-/- MEFs after treatment with DMSO, rapamycin, or wortmannin. (B Left) HEK293 cells expressing either pLKO lentiviral vector or a short hairpin RNA (shRNA) against NF1 were transfected with an hemagglutinin-tagged p70S6K reporter. (B Right) HEK293 cells expressing shRNA against NF1 were transfected with the indicated tuberin or NF1-GRD constructs along with a hemagglutinin-tagged p70S6K reporter. Abbreviations are as follows: Tub-WT, wild-type tuberin; Tub-SATA, tuberin with alanine mutations at the AKT phosphorylation sites S939/T1462. Hemagglutinin immunoprecipitates (HA IP) and total cell extracts (TCE) were immunoblotted with the indicated antibodies.
To determine whether NF1-deficient tumors from NF1 patients similarly exhibit an aberrant activation of the mTOR pathway, we examined this pathway in NF1+/- and NF1-/- Schwann cells derived from human neurofibromas. Neurofibromas are known to be extremely heterogenous lesions (1). Based on genetic studies of human tumors as well as mouse modeling data, it is believed that neurofibromas develop in NF1 patients as a result of “second hit” mutations in Schwann cells or Schwann cell precursors, which then act as a seed population to recruit other cells (NF1+/- Schwann cells, fibroblasts, mast cells, and perineurial cells) into a developing lesion (20, 33–37). Methods for isolating Schwann cells and then further separating NF1+/- Schwann cells from NF1-/- Schwann cells have been well described (20, 21). We obtained matched NF1+/- and NF1-/- Schwann cells from one cutaneous and one plexiform neurofibroma. In both cases S6K1, AKT, and tuberin phosphorylation was significantly higher in the NF1-null cells as compared with the matched NF1+/- cells (Fig. 4A). Furthermore, S6K1 phosphorylation was blocked by wortmannin, indicating that the aberrant activation of the PI3 kinase pathway underlies the activation of mTOR in these tumor cells (Fig. 4B).
Fig. 4.
NF1-/- tumor cells exhibit mTOR hyperactivation and are highly sensitive to rapamycin. (A) Western blot analysis of two independently derived patient-matched NF1+/- and NF1-/- Schwann cells grown in 0.1% serum devoid of exogenous growth factors. (B) Western blot analysis of an NF1-/- tumor cell line treated for 30 min with the following concentrations of inhibitors: DMSO, 0.1%; rapamycin, 20 nM; wortmannin, 200 nM. (C) NF1-/- tumor cell lines were grown for 7 days in the presence of the indicated concentrations of rapamycin and counted in triplicate.
It is commonly believed that tumors become dependent on the dysregulation of a specific signaling pathway and are therefore hypersensitive to its down-regulation (38, 39). To determine whether NF1-associated peripheral nerve sheath tumors have become dependent on hyperactivation of the mTOR pathway, we tested the effects of rapamycin on tumors from NF1 patients. For this experiment we used malignant tumors (MPNSTs), which arise from benign neurofibromas. We found that the proliferation of two independently derived MPNST cell lines from two NF1 patients was dramatically suppressed at low concentrations of rapamycin (Fig. 4C, both lines). The IC50 for these experiments was between 1 and 10 nM. Notably, these IC50 values are comparable to or lower than the IC50 values of rapamycin derivatives that were effective on PTEN-deficient tumors, in which activation of the mTOR pathway has been well characterized (12, 13). In addition, in one of the two cell lines, rapamycin induced cell death at the higher indicated concentrations (Fig. 4C and data not shown).
As an independent means of assessing the effects of rapamycin on the tumorigenic properties of NF1-deficient cells, we established a genetically engineered cell system. Cellular transformation is known to require multiple genetic events affecting the Rb, p53, and Ras pathways (40). In MEFs, the combined expression of a dominant-negative p53 gene and the E1A oncogene, which binds and inactivates Rb family members, is not sufficient to promote growth in soft agar. However, we found that in the absence of Nf1, cells expressing these genes did form colonies in soft agar (Fig. 5A). Importantly, rapamycin dramatically suppressed colony growth at low concentrations in this system as well (Fig. 5B). Taken together, these results suggest that NF1-deficient tumor cell lines are exquisitely sensitive to rapamycin and suggest the potential therapeutic utility of rapamycin or its derivatives in treating PNS tumors in NF1 patients.
Fig. 5.
Rapamycin inhibits colony growth in soft agar of Nf1-deficient transformed cells. (A) Nf1+/+ and Nf1-/- MEFs retrovirally transduced with dominant negative p53 (pBabe-hygro-p53DD) and adenoviral E1A (pLPC-E1A12S) were selected and plated in soft agar. (B) Nf1-/- MEFs (with the additional genetic alterations described in A) were grown in soft agar with the indicated concentration of rapamycin.
Discussion
In this article we have shown that the mTOR pathway is critically deregulated in NF1-deficient primary cells and human tumors. Importantly, PNS tumors derived from NF1 patients are highly sensitive to the mTOR inhibitor rapamycin, suggesting that it or its derivatives might be useful therapeutically. To date there is no effective treatment or cure for NF1. The hallmark neurofibromas, even when benign, can be painful, debilitating, and can grow large enough to encompass an entire body region (1). In addition to a high tumor burden encumbered by some patients, many cannot be surgically resected because of underlying nerve involvement. Furthermore, lesions that are surgically reduced typically regrow. Therefore, the evidence that mTOR deregulation participates in NF1-related tumorigenesis may represent a therapeutic breakthrough for this disease. Notably, NF1 is quite prevalent, >10 times more prevalent than most other phakomatotic disorders, and even more common than TSC, a disease for which the effects of rapamycin are currently being assessed in clinical trials.
We also found that tuberin mediates the effects of NF1-deficiency on mTOR. Although AKT has been shown to be activated in Nf1-deficient cells, the critical pathogenic targets of this kinase in NF1-associated tumors were unknown (41). We have demonstrated that tuberin is a target of AKT in human tumors derived from NF1 patients. Furthermore, inactivation of tuberin by AKT is absolutely required for mTOR activation in Nf1-deficient cells. These data identify NF1 as a new player in the cascade of tumor suppressors and oncogenes involved in mTOR activation. We have previously shown that neurofibromin is rapidly degraded in response to growth factors, and that its reappearance is required to appropriately terminate the Ras signal in wild-type cells (7). Therefore, it is likely that neurofibromin plays an active role in appropriately regulating this signal in response to growth factors. In any case, these data demonstrate that neurofibromin is indispensable for the appropriate suppression of the mTOR signal in the absence of mitogenic stimuli.
In addition, although the activation of mTOR in response to growth factors was known to be PI3 kinase-dependent, the extent to which Ras participates in this process was unknown. By using Nf1-deficient cells we were able to uniquely determine that the activation of endogenous Ras, via the loss of neurofibromin, results in pathogenic levels of mTOR activation. Furthermore, we also showed that in wild-type cells, Ras activation is required for maximal mTOR activation in response to LPA and insulin. Although this may be a subtle point, the amplitude and duration of Ras effector pathways has been shown to be critical for specifying biological responses (42). Therefore, it is possible that threshold levels of mTOR activation are not achieved in the absence of a Ras signal, which may have important biological consequences in certain settings. Interestingly, Ras activation does not appear to be required in response to PDGF, indicating a differential role for this pathway in response to different growth factors.
Finally, rapamycin derivatives have been suggested as potential therapeutic agents for a variety of cancers (43, 44). In particular, it has been suggested that these drugs may be useful in combination with other small-molecule inhibitors (43). The basis for this suggestion is that multiple genes are mutated in tumors; therefore, inhibition of more than one pathway may be required for a therapeutic effect. On this basis neurofibromatosis type I may represent a uniquely treatable disease. Notably, benign symptoms, such as neurofibromas, can be quite severe. However, they are likely to occur from the mutation of a single gene: NF1. Therefore it is possible that rapamycin derivatives may represent a single-hit therapy for these lesions, either by preventing tumor development and/or promoting their regression. Thus, these agents may represent one of the first viable therapies for this untreatable and devastating tumor-type. However, this benefit is likely to extend beyond peripheral nerve sheath tumors to include other NF1 symptoms. Recently, the mTOR pathway has been shown to be activated in a high percentage of acute myeloid leukemias (in patients that do not have NF1) (45). Furthermore, these patients appear to respond to rapamycin, suggesting that the aberrant activation of mTOR is also critical for the development of this tumor type. These observations, in conjunction with our data demonstrating a direct connection between NF1 and mTOR, support the possibility that these agents will also be useful in treating NF1-associated myeloid malignancies.
Acknowledgments
We thank D. W. Clapp (School of Medicine, Indiana University, Indianapolis) for the MSCV-GRD-pac; O. Martens (University of Ghent, Ghent, Belgium) for NF1 sequence analysis; B. Manning, R. J. Shaw, J. Blenis, P. Roux, and members of the Cichowski laboratory for helpful discussions. M.F.J. is funded through National Institute of Neurological Disorders and Stroke Grant NS24279 and a National Neurofibromatosis Foundation Young Investigator Award. This work was supported by Department of Defense Grant NF020033 (to K.C.), American Cancer Society Grant RSG-02-047-01-DDC (to K.C.), Fonds voor Wetenschappelijk Onderzoek Vlaanderen Grant G.0096.02 (to E.L.), and the Interuniversity Attraction Poles grant from the Federal Office for Scientific, Technical, and Cultural Affairs of Belgium (to E.L.; 2002, 2006; P5/25).
Author contributions: C.M.J., E.E.R., and K.C. designed research; C.M.J. and E.E.R. performed research; M.F.J., H.B., and E.L. contributed new reagents/analytic tools; C.M.J., E.E.R., and K.C. analyzed data; and K.C. wrote the paper.
Abbreviations: GRD, GAP-related domain; LPA, l-α-lysophosphatidic acid; PDGF, platelet-derived growth factor.
References
- 1.Riccardi, V. M. (1992) Neurofibromatosis: Phenotype, Natural History, and Pathogenesis (Johns Hopkins Univ. Press, Baltimore).
- 2.Bader, J. L. (1986) Ann. N.Y. Acad. Sci. 486, 57-65. [DOI] [PubMed] [Google Scholar]
- 3.Basu, T. N., Gutmann, D. H., Fletcher, J. A., Glover, T. W., Collins, F. S. & Downward, J. (1992) Nature 356, 713-715. [DOI] [PubMed] [Google Scholar]
- 4.DeClue, J. E., Papageorge, A. G., Fletcher, J. A., Diehl, S. R., Ratner, N., Vass, W. C. & Lowy, D. R. (1992) Cell 69, 265-273. [DOI] [PubMed] [Google Scholar]
- 5.Kim, H. A., Rosenbaum, T., Marchionni, M. A., Ratner, N. & DeClue, J. E. (1995) Oncogene 11, 325-335. [PubMed] [Google Scholar]
- 6.Bollag, G., Clapp, D. W., Shih, S., Adler, F., Zhang, Y. Y., Thompson, P., Lange, B. J., Freedman, M. H., McCormick, F., Jacks, T. & Shannon, K. (1996) Nat. Genet. 12, 144-148. [DOI] [PubMed] [Google Scholar]
- 7.Cichowski, K., Santiago, S., Jardim, M., Johnson, B. W. & Jacks, T. (2003) Genes Dev. 17, 449-454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Korf, B. R. (2004) Neuroimaging Clin. N. Am. 14, 139-148, vii. [DOI] [PubMed] [Google Scholar]
- 9.Kumar, V., Fausto, N. & Abbas, A. K. (2004) Robbins and Cotran Pathologic Basis of Disease (Elsevier Science, Amsterdam).
- 10.Harris, T. E. & Lawrence, J. C., Jr. (2003) Sci. STKE 212, 9. [DOI] [PubMed] [Google Scholar]
- 11.Hay, N. & Sonenberg, N. (2004) Genes Dev. 18, 1926-1945. [DOI] [PubMed] [Google Scholar]
- 12.Podsypanina, K., Lee, R. T., Politis, C., Hennessy, I., Crane, A., Puc, J., Neshat, M., Wang, H., Yang, L., Gibbons, J., et al. (2001) Proc. Natl. Acad. Sci. USA 98, 10320-10325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Neshat, M. S., Mellinghoff, I. K., Tran, C., Stiles, B., Thomas, G., Petersen, R., Frost, P., Gibbons, J. J., Wu, H. & Sawyers, C. L. (2001) Proc. Natl. Acad. Sci. USA 98, 10314-10319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shaw, R. J., Bardeesy, N., Manning, B. D., Lopez, L., Kosmatka, M., DePinho, R. A. & Cantley, L. C. (2004) Cancer Cell 6, 91-99. [DOI] [PubMed] [Google Scholar]
- 15.Corradetti, M. N., Inoki, K., Bardeesy, N., DePinho, R. A. & Guan, K. L. (2004) Genes Dev. 18, 1533-1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kwiatkowski, D. J., Zhang, H., Bandura, J. L., Heiberger, K. M., Glogauer, M., el-Hashemite, N. & Onda, H. (2002) Hum. Mol. Genet. 11, 525-534. [DOI] [PubMed] [Google Scholar]
- 17.Zhang, H., Cicchetti, G., Onda, H., Koon, H. B., Asrican, K., Bajraszewski, N., Vazquez, F., Carpenter, C. L. & Kwiatkowski, D. J. (2003) J. Clin. Invest. 112, 1223-1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fingar, D. C. & Blenis, J. (2004) Oncogene 23, 3151-3171. [DOI] [PubMed] [Google Scholar]
- 19.Raught, B., Gingras, A. C. & Sonenberg, N. (2001) Proc. Natl. Acad. Sci. USA 98, 7037-7044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Serra, E., Rosenbaum, T., Winner, U., Aledo, R., Ars, E., Estivill, X., Lenard, H. G. & Lazaro, C. (2000) Hum. Mol. Genet. 9, 3055-3064. [DOI] [PubMed] [Google Scholar]
- 21.Rosenbaum, T., Rosenbaum, C., Winner, U., Muller, H. W., Lenard, H. G. & Hanemann, C. O. (2000) J. Neurosci. Res. 61, 524-532. [DOI] [PubMed] [Google Scholar]
- 22.Frahm, S., Mautner, V. F., Brems, H., Legius, E., Debiec-Rychter, M., Friedrich, R. E., Knofel, W. T., Peiper, M. & Kluwe, L. (2004) Neurobiol. Dis. 16, 85-91. [DOI] [PubMed] [Google Scholar]
- 23.Hiatt, K. K., Ingram, D. A., Zhang, Y., Bollag, G. & Clapp, D. W. (2000) J. Biol. Chem. 276, 7240-7245. [DOI] [PubMed] [Google Scholar]
- 24.Roux, P. P., Ballif, B. A., Anjum, R., Gygi, S. P. & Blenis, J. (2004) Proc. Natl. Acad. Sci. USA 101, 13489-13494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ming, X. F., Burgering, B. M., Wennstrom, S., Claesson-Welsh, L., Heldin, C. H., Bos, J. L., Kozma, S. C. & Thomas, G. (1994) Nature 371, 426-429. [DOI] [PubMed] [Google Scholar]
- 26.Manning, B. D., Tee, A. R., Logsdon, M. N., Blenis, J. & Cantley, L. C. (2002) Mol. Cell. 10, 151-162. [DOI] [PubMed] [Google Scholar]
- 27.Inoki, K., Li, Y., Zhu, T., Wu, J. & Guan, K. L. (2002) Nat. Cell Biol. 4, 648-657. [DOI] [PubMed] [Google Scholar]
- 28.Potter, C. J., Pedraza, L. G. & Xu, T. (2002) Nat. Cell Biol. 4, 658-665. [DOI] [PubMed] [Google Scholar]
- 29.Garami, A., Zwartkruis, F. J., Nobukuni, T., Joaquin, M., Roccio, M., Stocker, H., Kozma, S. C., Hafen, E., Bos, J. L. & Thomas, G. (2003) Mol. Cell 11, 1457-1466. [DOI] [PubMed] [Google Scholar]
- 30.Tee, A. R., Manning, B. D., Roux, P. P., Cantley, L. C. & Blenis, J. (2003) Curr. Biol. 13, 1259-1268. [DOI] [PubMed] [Google Scholar]
- 31.Zhang, Y., Gao, X., Saucedo, L. J., Ru, B., Edgar, B. A. & Pan, D. (2003) Nat. Cell Biol. 5, 578-581. [DOI] [PubMed] [Google Scholar]
- 32.Inoki, K., Li, Y., Xu, T. & Guan, K. L. (2003) Genes Dev. 17, 1829-1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kluwe, L., Friedrich, R. & Mautner, V. F. (1999) Genes Chromosomes Cancer 24, 283-285. [DOI] [PubMed] [Google Scholar]
- 34.Serra, E., Puig, S., Otero, D., Gaona, A., Kruyer, H., Ars, E., Estivill, X. & Lazaro, C. (1997) Am. J. Hum. Genet. 61, 512-519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cichowski, K., Shih, T. S., Schmitt, E., Santiago, S., Reilly, K., McLaughlin, M. E., Bronson, R. T. & Jacks, T. (1999) Science 286, 2172-2176. [DOI] [PubMed] [Google Scholar]
- 36.Zhu, Y., Ghosh, P., Charnay, P., Burns, D. K. & Parada, L. P. (2002) Science 296, 920-922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cichowski, K. & Jacks, T. (2001) Cell 104, 593-604. [DOI] [PubMed] [Google Scholar]
- 38.Weinstein, I. B. (2002) Science 297, 63-64. [DOI] [PubMed] [Google Scholar]
- 39.Shamji, A. F., Nghiem, P. & Schreiber, S. L. (2003) Mol. Cell 12, 271-280. [DOI] [PubMed] [Google Scholar]
- 40.Hahn, W. C. & Weinberg, R. A. (2002) Nat. Rev. Cancer 2, 331-341. [DOI] [PubMed] [Google Scholar]
- 41.Donovan, S., See, W., Bonifas, J., Stokoe, D. & Shannon, K. M. (2002) Cancer Cell 2, 507-514. [DOI] [PubMed] [Google Scholar]
- 42.Marshall, C. J. (1995) Cell 80, 179-185. [DOI] [PubMed] [Google Scholar]
- 43.Sawyers, C. L. (2003) Cancer Cell 4, 343-348. [DOI] [PubMed] [Google Scholar]
- 44.Luo, J., Manning, B. D. & Cantley, L. C. (2003) Cancer Cell 4, 257-262. [DOI] [PubMed] [Google Scholar]
- 45.Recher, C., Beyne-Rauzy, O., Demur, C., Chicanne, G., Dos Santos, C., Mansat-De Mas, V., Benzaquen, D., Laurent, G., Huguet, F. & Payrastre, B. (2004) Blood 18, 18. [DOI] [PubMed] [Google Scholar]