Abstract
Epstein-Barr virus (EBV)-negative Burkitt lymphomas (BLs) can be infected in vitro with prototype EBV strains to study how the virus may affect the phenotype of tumor cells. Studies thus far have concentrated on the use of transforming B95-8 and nontransforming P3HR1 strains. Immunological and phenotypic differences between the sublines infected with these two strains were reported. The majority of these differences, if not all, can be attributed to the lack of EBNA-2 coding sequences in the P3HR1 strain. The recent development of a selectable Akata strain has opened up new possibilities for infecting epithelial and T cells as well. We infected five EBV-negative BL lines with the recombinant Akata virus. Our results indicate that the infected cell lines BL28, Ramos, and DG75 express EBNA-1, EBNA-2, and LMP1, the viral proteins associated with type III latency, and use both YUK and QUK splices. In contrast, two EBV-negative variants of Akata and Mutu when reinfected displayed restricted type I latency and expressed only EBNA-1. All clones of infected Mutu cells used the QUK splice exclusively. The usage of Qp was observed in a majority of Akata clones. Some Akata clones, however, were found to have double promoter usage (Qp and C/Wp) but at 4 months after infection did not express EBNA-2. The results demonstrate differential regulation of EBV latency in BLs with the same recombinant viral strain and suggest that the choice of latency type may be cell dependent. The restricted latency observed for infected Akata and Mutu cells indicates that a BL may opt for type I latency in the absence of immune pressure as well.
Epstein-Barr virus (EBV) belongs to the gammaherpesviruses and is strongly associated with human tumors like Burkitt lymphoma (BL) and nasopharyngeal carcinoma. It also efficiently transforms B lymphocytes in vitro, giving rise to lymphoblastoid cell lines (11).
Based on viral latent gene expression and the expression of surface activation markers, three main types of EBV latency have been characterized (25). In phenotypically representative BLs, the EBV-encoded latent protein expression is limited to EBNA-1 only, termed latency type I or type I phenotype. Such a phenotype is also exemplified in B lymphocytes of EBV-positive healthy individuals, with additional expression of LMP2A. The EBNA-1 transcript is generated through Q promoter usage (2, 22, 32).
The majority of type I BLs, after several passages in tissue culture, tend to drift to a more blastoid phenotype. The viral genes expressed in this situation include EBNA-1 to EBNA-6, LMP1, LMP2A, and LMP2B. This is termed type III phenotype or latency type III. Such a form of latency is also observed in lymphoblastoid cell lines. At the initial stages of infection, the W promoter is used. This is followed by a switch to the C promoter once a stable line has been established (37). The usage of either the W or the C promoter gives rise to a giant message which is alternatively spliced to encode all EBNAs (6). The intermediate type II latency (EBNA1+, LMP1+, and LMP2A+) occurs in nasopharyngeal carcinomas and Hodgkin's disease and other non-B-cell malignancies in which the virus is present (5, 27, 38).
EBV-negative BLs can be converted to positive status, in vitro, through a cumbersome and repetitive infection with the prototype B95-8 strain (12, 13). Such infected cells express all EBV growth- and transformation-associated proteins (20). The availability of a recombinant Akata-derived EBV with a selectable marker has enormously facilitated studies dealing with the role of EBV in transformation of B- and non-B-cell lines (9, 15, 28).
Previous studies have indicated that reinfection of the EBV-negative form of Akata cells with Akata cell-derived EBV results in type I latency and hence expression of EBNA-1 only (15, 26, 29). This is in sharp contrast to the infected sublines resulting from B95-8 infection of EBV-negative BLs, which exhibit the type III phenotype as long as the virus integrity is maintained (4, 8, 20, 33). This raises the question whether such diversity in viral gene expression in EBV-infected sublines with two different strains is due to differences related to the viral strain or to the cells in which the virus resides. This issue was addressed by infecting five EBV-negative BLs with the recombinant Akata virus. We found that BL28, Ramos, and DG75 display type III latency upon infection. The other two EBV-negative variants, Akata and Mutu, when reinfected with the Akata virus show a type I phenotype, just as their original EBV-positive parental counterparts do.
We chose the following cell lines as targets of infection. BL28, DG75, and Ramos are EBV-negative BL lines. Akata and Mutu were originally described as EBV-positive type I BL lines. The clones used in this study, Akata 2A8 (26), Mutu 9, and Mutu 30 (3, 24, 30), are EBV-negative variants isolated from the EBV-positive parental lines. The EBV-negative status of these lines was previously verified by DNA PCR and in situ hybridization (3, 24, 30). We further confirmed the EBV-negative status of these lines by DNA PCR using primers derived from Y2 and Y3 exons (data not shown). All lines were maintained in RPMI 1640, supplemented with 10% fetal calf serum, 100 IU of penicillin, and 100 μg of streptomycin/ml.
The infection protocol was essentially the same as described previously (28). Briefly, 5 × 106 cells from each line were suspended in 2 ml of diluted virus supernatant (1:1). The cells were incubated at 37°C for 2 h, with intermittent gentle mixing. At the end of the incubation, a further 3 ml of complete medium was added and the cells were kept for an additional 48 h at 37°C. Subsequently, the cells were washed once, cloned by limiting dilution, and placed in a 96-well plate in selection medium containing 1 mg of G418/ml. The medium was replaced every 3 days. The clones were continuously maintained in the selection medium. The G418-resistant infected cells were isolated from all the cell lines with a single infection only. Several clones from each line grew in a 96-well plate and were randomly selected for further experiments. The sensitivity of each line to G418 was tested, and at 1 mg of G418/ml, the uninfected cells died within 10 days.
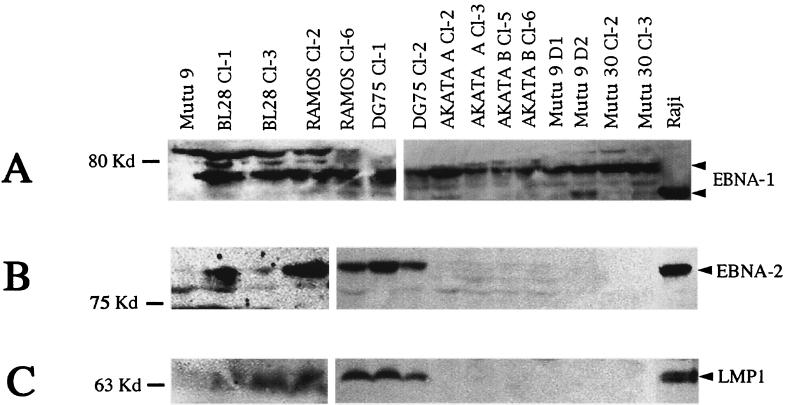
The expression of EBNA-1, EBNA-2, and LMP1 was verified by immunoblotting. The cell lysate was prepared by sonicating 107 cells in 1 ml of Laemmli buffer (16). Cell extract equivalent to 106 cells was loaded in each slot. The proteins were separated by discontinuous gel electrophoresis and blotted on Protran filters (Schleicher and Schuell) (35). EBNA-1 was detected with a polyclonal human serum (diluted 1:5 in milk) by incubating the filters with the primary antibody overnight at 4°C on a rotor. The expression of EBNA-2 and LMP1 was verified by using PE2 and CS1-4 monoclonal antibodies, respectively, diluted as recommended by the manufacturer (Dako). Figure 1A shows EBNA-1 expression in two clones each of BL28, Ramos, DG75, Akata, Mutu 9, and Mutu 30 cells. The infected Akata A and B cells represent two separate infections of the same line. All the clones examined showed high EBNA-1 expression. The EBNA-1 detected in the infected cells was of higher molecular weight than that detected in Raji cells. This strain-dependent variation in the size of EBNA-1 is suggested to be due to differences in the Gly-Ala repeat length in the EBNA-1 sequence. The faster-migrating band in the Mutu 9 D2 clone may represent a degraded species of EBNA-1.
FIG. 1.
EBV latent protein expression in Akata virus-infected BLs. (A) EBNA-1 expression in Akata virus-infected BLs using a polyclonal human serum from a healthy seropositive donor; (B) EBNA-2 expression as seen with monoclonal antibody PE2; (C) LMP1 expression in the infected cells detected with CS1-4 monoclonal antibody.
The expression of EBNA-2 and LMP1 in two clones from each line is shown in Fig. 1B and 1C, respectively. All the clones of the infected cell lines BL28, Ramos, and DG75 examined showed detectable levels of EBNA-2 and LMP1. Among infected Akata cells, a few clones were EBNA-2 positive early after infection (data not shown); however, at 4 months, none of these clones showed detectable EBNA-2 or LMP1 (Fig. 1B and C). None of the infected Mutu 9 and Mutu 30 cells expressed EBNA-2 or LMP1. Tables 2 and 3 summarize EBNA-1, EBNA-2, and LMP1 expression in infected BL28, DG75, Ramos, and Akata and Mutu cells, respectively.
TABLE 2.
EBV latent protein expression and promoter usage in Akata virus-infected clones of BL28, DG75, and Ramos cell linesa
| Cell line | EBNA-1 | EBNA-2 | LMP1 | QUK | Y3UK |
|---|---|---|---|---|---|
| Raelb | + | − | − | + | − |
| Rajib | + | + | + | + | + |
| BL28 | − | − | − | − | − |
| BL28 cl-1 | + | + | + | + | + |
| BL28 cl-2 | + | + | + | + | + |
| BL28 cl-3 | + | + | + | + | + |
| BL28 cl-4 | + | + | + | + | + |
| BL28 cl-5b | + | + | + | + | + |
| BL28 cl-6 | + | + | + | + | + |
| Ramosb | − | − | − | − | − |
| Ramos cl-1 | + | + | + | + | + |
| Ramos cl-2 | + | + | + | + | + |
| Ramos cl-3 | + | + | + | + | + |
| Ramos cl-4 | + | + | + | + | + |
| Ramos cl-5 | + | + | + | + | + |
| Ramos cl-6b | + | + | + | + | + |
| DG75 | − | − | − | − | − |
| DG75 cl-1 | + | + | + | + | + |
| DG75 cl-2b | + | + | + | + | + |
EBV protein expression was verified by immunoblotting, and promoter usage was verified by RT-PCR. Immunoblotting and RT-PCR were repeated at least twice for each sample. PCR was performed 3 to 4 months after infection.
These lines were also examined for either Qp or W/Cp usage by using additional sets of nested primers that distinguish the two transcripts.
TABLE 3.
EBV latent gene expression and promoter usage in Akata and Mutu cells infected with Akata virusa
| Cell line | EBNA-1 | EBNA-2 | LMP1 | QUK | Y3UK |
|---|---|---|---|---|---|
| Akata 2A8b | − | − | − | − | − |
| Akata Ac cl-2e | + | − | − | + | − |
| Akata A cl-3 | + | − | − | + | + |
| Akata A cl-4 | + | − | − | NDf | ND |
| Akata A cl-5 | + | − | − | + | − |
| Akata A cl-6 | + | − | − | + | − |
| Akata A cl-7e | + | − | − | + | + |
| Akata Bc cl-1 | + | − | − | ND | ND |
| Akata B cl-2 | + | − | − | ND | ND |
| Akata B cl-3e | + | − | − | + | − |
| Akata B cl-4 | + | − | − | + | + |
| Akata B cl-5 | + | − | − | + | − |
| Akata B cl-6 | + | − | − | + | + |
| Akata B cl-7e | + | − | − | + | + |
| Mutu 9de | − | − | − | − | − |
| Mutu 9 C1e | + | − | − | + | − |
| Mutu 9 D1e | + | − | − | + | − |
| Mutu 9 D2 | + | − | − | ND | ND |
| Mutu 9 D3 | + | − | − | + | − |
| Mutu 30d | − | − | − | − | − |
| Mutu 30 cl-1e | + | − | − | + | − |
| Mutu 30 cl-2 | + | − | − | + | − |
| Mutu 30 cl-3 | + | − | − | ND | ND |
See Table 2, footnote a.
Akata 2A8 is an EBV-negative variant of the Akata cell line.
Akata A- and Akata B-infected cells represent two sets generated from different infections. Some Akata clones were EBNA-2 positive 6 weeks after infection but at 4 months did not express the protein.
Mutu 9 and Mutu 30 are two EBV-negative clones derived from Mutu cells.
These lines were also examined for either Qp or W/Cp usage by using additional sets of nested primers that distinguish the two transcripts.
ND, not determined.
The promoter usage in Akata virus-infected cells was verified by reverse transcription-PCR (RT-PCR). Total RNA was extracted by the Trizol method as recommended by the manufacturer (Life Technologies). The first-strand cDNA was generated from 2 μg of RNA by using avian myeloblastosis virus reverse transcriptase. The RNA was incubated with 25 μl of a reaction mixture containing 50 mM Tris HCl (pH 8.5), 145 mM KCl, 10 mM MgCl2, 1 mM deoxynucleoside triphosphate, 4 μM random hexamers, 30 U of DNase-free RNase inhibitor, and 15 U of avian myeloblastosis virus reverse transcriptase (Promega) for 60 min at 42°C and then for 5 min at 95°C to inactivate the reverse transcriptase. The reactions were performed in an automated thermal cycler (Techne PHC-2).
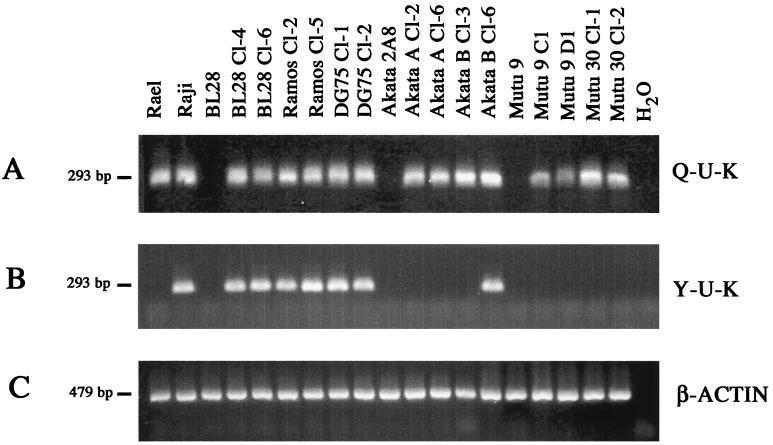
Three microliters of cDNA was used as template for PCR amplifications. The reaction was carried out in a 50-μl volume containing 20 mM Tris (pH 8.5); 50 mM KCl; MgCl2; 1 μM primers; and 200 mM (each) dATP, dGTP, dCTP, and dTTP. One unit of Taq polymerase (Promega) was added per 50-μl reaction mixture. For the nested PCR, 2 μl of amplification product of the first-round PCR was carried over. The primer (Life Technologies) sequences and PCR conditions are described in Table 1. The primers for verification of β-actin expression were purchased from Clontech, and the RT-PCR conditions were according to the manufacturers' specifications. Figure 2 shows a schematic representation of the transcripts, promoters, primer location, and EBV genome coordinates. The clones BL28 cl-4 and cl-6, Ramos cl-2 and cl-5, and DG75 cl-1 and cl-2 were found positive for both QUK and YUK splicing (Fig. 3A and B). The promoter usage in additional clones is shown in Table 2. Among infected Akata cells, three patterns of promoter usage were found. First, Akata A cl-2 and cl-6 and Akata B cl-3 showed only QUK splicing. Second, in Akata B cl-4, cl-6, and cl-7 and some Akata A clones, double QUK and YUK splices were detected (Fig. 3A and B; Table 3). Third, we also observed that Akata A cl-5 and Akata B cl-5 were both QUK and YUK positive and expressed EBNA-2 at 6 weeks after infection (data not shown) but that at 4 months postinfection they no longer used YUK and did not express EBNA-2 (Table 3). It is important to point out that none of the Akata clones were positive for EBNA-2 and LMP1 expression by immunoblotting at 4 months after infection (Fig. 1B and C; Table 3). The exclusive QUK usage was found in all infected Mutu 9 and Mutu 30 cell lines. None of these clones showed YUK usage (Fig. 3A and B; Table 3). These results were further confirmed by using nested PCR primers that distinguish transcripts originating from either Qp or W/Cp (Tables 1 to 3). The results of the control PCR for β-actin expression are shown in Fig. 3C. Rael and Raji cells were used as positive controls for QUK and YUK usage, respectively. Raji cells were found to use the Q promoter as well (Fig. 3A). The Q usage in Raji cells indicates a heterogeneity in this line and suggests that there may exist a subpopulation that displays the restricted latency. A previous study described similar Q usage in Raji cells (31).
TABLE 1.
PCR primers and conditionsa
| Promoter and splice | Sequence | Primer pair(s) | Mg2+ concn (mM) | Annealing temp (°C) |
|---|---|---|---|---|
| Q promoter (QUK splice) | ||||
| 5′ Q1 | 5′-GTGCGCTACCGGATGGCG-3′ | Q1-K1 | 1 | 63 |
| 3′ K1 | 5′-GGTCTCCGGACACCATCTCT-3′ | |||
| 5′ U1 | 5′-AGCTTCCCTGGGATGAGCGT-3′ | U1-K2 | 1.5 | 66 |
| 3′ K2 | 5′-TCTTCCCCGTCCTCGTCCAT-3′ | |||
| 5′ Q1A | 5′-AGCGTGCGCTACCGGAT-3′ | Q1A-K1 | 1.5 | 60 |
| Q1-K2 | 1.5 | 63 | ||
| W or C promoter (YUK splice) | ||||
| 5′ Y3 | 5′-CGTGTGACGTGGTGTAAAGT-3′ | Y3-K1 | 2 | 63 |
| 3′ K1 | 5′-GGTCTCCGGACACCATCTCT-3′ | |||
| 5′ U1 | 5′-AGCTTCCCTGGGATGAGCGT-3′ | U1-K2 | 1.5 | 66 |
| 3′ K2 | 5′-TCTTCCCCGTCCTCGTCCAT-3′ | |||
| 5′ Y3A | 5′-TGGCGTGTGACGTGGTGTAA-3′ | Y3A-K1 | 1.5 | 63 |
| Y3-K2 | 1.5 | 62 |
Denaturing was done at 94°C for 2 min, extension was done at 72°C for 2 min, and annealing was done for 1 min. First-round PCR used 30 cycles, and second-round PCR used 35 cycles.
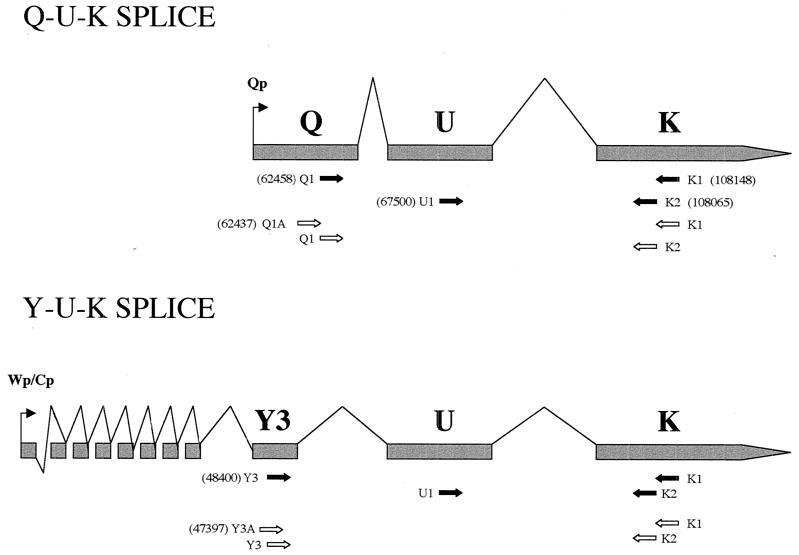
FIG. 2.
Qp- and W/Cp-derived transcripts and their splice patterns. The numbers in parentheses are EBV genome coordinates. The arrows indicate the primer locations. The solid arrows indicate a set of primers which detect the common U and K exons. The open arrows show a set in which the nested primers distinguish splice patterns derived from either Qp or W/Cp.
FIG. 3.
Promoter usage in the infected cells. (A) Usage of the Q promoter as revealed by RT-PCR using primers that detect the QUK splice; (B) YUK splice usage; (C) expression of β-actin. Detailed PCR conditions are described in Table 1.
EBV infection of originally negative BLs provides a valuable tool to study the effects of the virus on the phenotype of the tumor cell. Generally, this is a tedious and often unsuccessful process. The use of recombinant technology permitted the generation of EBV derived from Akata cells with a selection marker incorporated into the viral genome (28). We have utilized this recombinant viral strain to address the question whether the choice of latency is virus or cell dependent. Previous reports indicated that EBV-negative Akata cells, when reinfected with Akata cell-derived EBV, demonstrate type I latency and thus expression of EBNA-1 only. This is intriguing since, almost invariably, the infection of EBV-negative BLs with B95-8 virus results in a lymphoblastoid phenotype and type III latency. This prompted us to ask if such a difference is a characteristic of the Akata cells or the viral strain and if this could be applicable to other BL lines. We chose three standard EBV-negative BL lines and two BL lines that had lost the endogenous viral genome but, before losing it, could sustain type I latency and EBNA-1 expression, even after several years in tissue culture.
We observed that all Akata and Mutu clones expressed only EBNA-1. In contrast, infected BL28, Ramos, and DG75 cells expressed EBNA-1, EBNA-2, and LMP1, an indication of the type III phenotype. The choice of different types of latency in various BL lines demonstrates that these lines can vary with regard to the restrictive control that they can exert on the same viral strain. It would be interesting to see if different viral strains would vary in their sensitivity to restrictions of the same host cell. Such an issue could be addressed by infecting EBV-negative Mutu and Akata cells with the B95-8 virus. If the resultant infected cells expressed the type I phenotype and hence only EBNA-1, this would speak for cellular control regardless of the viral strain. However, if the B95-8-infected Akata or Mutu cells displayed a type III phenotype, this would indicate that latency is dependent on the viral strain. However, infection of Akata and Mutu cells with the B95-8 virus may prove extremely difficult technically, as we have failed to get such infected cells after repeated attempts.
Previous studies have shown variations in tumorigenicity, agarose clonability, and recognition by cytotoxic T lymphocytes when BL lines were infected with either the B95-8 or the P3HR1 strain (13, 20, 33, 34). However, such differences could be attributable not only to clonal variations and differences between cell lines but also to the fact that the P3HR1 genome carries a deletion in the EBNA-2 coding region (23). Using the present series of infected cell lines, the individual role of viral gene effect on the phenotype of the infected cells can be studied, since the use of the same viral strain results in different types of latency. Furthermore, since there is no clear phenotypic distinction between EBV-positive and EBV-negative type I BL lines, the significance of the viral presence is under constant scrutiny. The series of infected Mutu and Akata cell lines which retain the type I phenotype seems to be an ideal setting to compare the parental lines (EBV positive and type I), EBV-negative variants, and variants infected with Akata cell-derived EBV (type I) to see if a true difference could be correlated with the presence of EBV in the above-mentioned BL lines.
Marchini et al. (18) have previously shown that, when four EBV-negative BL lines were infected with a recombinant P3HR1 virus, the latent EBV gene expression was limited to EBNA-1 only. The apparent discrepancy between that study and our findings that standard EBV-negative BL lines when reinfected with Akata virus resulted in type III latency could be due to differences in the viral strains. It is known that P3HR1-infected cells generally do not express LMP1, since EBNA-2, which induces LMP1 expression, is missing in this strain.
The double YUK and QUK splices and promoter usage seen in infected BL28, Ramos, and DG75 cell lines suggest either that the same cell may use both types of programs or that cells having individual types of latency separately may coexist. Interestingly, some of the Akata clones which were positive for EBNA-2 by Western blotting and for YUK splicing at about 6 weeks after infection neither used YUK splicing nor expressed EBNA-2 at 4 months postinfection. The infected Akata cells maintaining the YUK usage at 4 months postinfection but negative for EBNA-2 expression may suggest that in a fraction of cells EBNA-2 may be expressed below the limit of detection. It is also conceivable that the Akata clones with double promoter usage may still be in the process of an inverse shift, as seen for those clones which initially had QUK and YUK splices but became exclusive QUK users. All this suggests that this particular virus strain in its original environment may sustain type I latency only. The Mutu 9 and Mutu 30 clones, however, had type I latency from the initial stages and maintained it even 4 months after infection.
The BL cell may originate in the germinal center from a centrocyte or a centroblast (14). The c-myc translocation keeps the cell cycling even if its phenotype represents that of a resting cell. It has been speculated that the translocation-carrying cell may then become infected with EBV, which may provide an additional event toward malignant transformation (17). The finding that in vitro-infected Akata and Mutu clones display type I latency just like phenotypically representative BL lines favors such a scenario. Moreover, since phenotypically representative BL lines express nonimmunogenic EBNA-1 only (19, 36), it is assumed that the lack of expression of immunogenic EBNA-3 to EBNA-6 and LMPs could be due to pressure exercised by the functional immune system of the host (7, 10, 21). Our data showing that type I latency can be achieved in more than one BL line, in vitro, in the absence of immune pressure, suggest that the choice of restricted type I latency may be regulated by cellular factors as well.
DNA methylation is suggested to be one of the mechanisms by which type III latency is suppressed. The virus thus may avert immune surveillance, as the potentially immunogenic EBNAs are not expressed. The pharmacological demethylation of the C promoter and consequently the induction of EBNA-2 to EBNA-6 may provide target antigens for EBV-specific T-cell surveillance of the tumor cells. However, in a pilot study, the therapeutic use of demethylating agent 5-Aza C in patients with EBV-associated tumors did not lead to induction of EBNA-2 to EBNA-6 (1). This may have been due to pharmacokinetics and inadequate 5-Aza C dosage. On the basis of our findings, however, it could also be surmised that additional parameters which control types of viral latency may operate in EBV-infected cells, and further studies will be required to understand such mechanisms in order to improve the treatment of EBV-associated tumors.
Acknowledgments
This work was supported by grants from the MURST, Ministero della Sanità, Progetto AIDS, Associazione Italiana di Ricerca sul Cancro (AIRC), and Istituto Pasteur-Cenci-Bolognetti foundation. P.T. is supported by AIRC.
We thank J. Sixbey for EBV-negative Akata and Mutu cells and Sandro Valia for help with the photographic work.
REFERENCES
- 1.Ambinder R F, Robertson K D, Tao Q. DNA methylation and the Epstein-Barr virus. Semin Cancer Biol. 1999;9:369–375. doi: 10.1006/scbi.1999.0137. [DOI] [PubMed] [Google Scholar]
- 2.Chen F, Zou J Z, di Renzo L, Winberg G, Hu L F, Klein E, Klein G, Ernberg I. A subpopulation of normal B cells latently infected with Epstein-Barr virus resembles Burkitt lymphoma cells in expressing EBNA-1 but not EBNA-2 or LMP1. J Virol. 1995;69:3752–3758. doi: 10.1128/jvi.69.6.3752-3758.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chodosh J, Holder V P, Gan Y J, Belgaumi A, Sample J, Sixbey J W. Eradication of latent Epstein-Barr virus by hydroxyurea alters the growth-transformed cell phenotype. J Infect Dis. 1998;177:1194–1201. doi: 10.1086/515290. [DOI] [PubMed] [Google Scholar]
- 4.Ehlin-Henriksson B, Manneborg-Sandlund A, Klein G. Expression of B-cell-specific markers in different Burkitt lymphoma subgroups. Int J Cancer. 1987;39:211–218. doi: 10.1002/ijc.2910390215. [DOI] [PubMed] [Google Scholar]
- 5.Fahraeus R, Hu L F, Ernberg I, Finke J, Rowe M, Klein G, Falk K, Nilsson E, Yadav M, Busson P, Kallin B. Expression of Epstein-Barr virus-encoded proteins in nasopharyngeal carcinoma. Int J Cancer. 1988;42:329–338. doi: 10.1002/ijc.2910420305. [DOI] [PubMed] [Google Scholar]
- 6.Farrell P J. Epstein-Barr virus immortalizing genes. Trends Microbiol. 1995;3:105–109. doi: 10.1016/s0966-842x(00)88891-5. [DOI] [PubMed] [Google Scholar]
- 7.Gavioli R, De Campos-Lima P O, Kurilla M G, Kieff E, Klein G, Masucci M G. Recognition of the Epstein-Barr virus-encoded nuclear antigens EBNA-4 and EBNA-6 by HLA-A11-restricted cytotoxic T lymphocytes: implications for down-regulation of HLA-A11 in Burkitt lymphoma. Proc Natl Acad Sci USA. 1992;89:5862–5866. doi: 10.1073/pnas.89.13.5862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hurley E A, Agger S, McNeil J A, Lawrence J B, Calendar A, Lenoir G, Thorley-Lawson D A. When Epstein-Barr virus persistently infects B-cell lines, it frequently integrates. J Virol. 1991;65:1245–1254. doi: 10.1128/jvi.65.3.1245-1254.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Imai S, Nishikawa J, Takada K. Cell-to-cell contact as an efficient mode of Epstein-Barr virus infection of diverse human epithelial cells. J Virol. 1998;72:4371–4378. doi: 10.1128/jvi.72.5.4371-4378.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Khanna R, Burrows S R, Kurilla M G, Jacob C A, Misko I S, Sculley T B, Kieff E, Moss D J. Localization of Epstein-Barr virus cytotoxic T cell epitopes using recombinant vaccinia: implications for vaccine development. J Exp Med. 1992;176:169–176. doi: 10.1084/jem.176.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kieff E, Liebowitz D. The Epstein-Barr virus. In: Fields B, Knipe D, editors. Virology. New York, N.Y: Raven Press; 1989. pp. 1889–1929. [Google Scholar]
- 12.Klein G, Giovannela B, Westman A, Stehlin J S, Mumford D. An EBV-genome-negative cell line established from an American Burkitt lymphoma; receptor characteristics. EBV and permanent conversion into EBV-positive sublines by in vitro infection. Intervirology. 1975;5:319–334. doi: 10.1159/000149930. [DOI] [PubMed] [Google Scholar]
- 13.Klein G, Zeuthen J, Terasaki P, Billing R, Honig R, Jondal M, Westman A, Clements G. Inducibility of the Epstein-Barr virus (EBV) cycle and surface marker properties of EBV-negative lymphoma lines and their in vitro EBV-converted sublines. Int J Cancer. 1976;18:639–652. doi: 10.1002/ijc.2910180513. [DOI] [PubMed] [Google Scholar]
- 14.Klein G. Dysregulation of lymphocyte proliferation by chromosomal translocations and sequential genetic changes. Bioessays. 2000;22:414–422. doi: 10.1002/(SICI)1521-1878(200005)22:5<414::AID-BIES3>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 15.Komano J, Maruo S, Kurozumi K, Oda T, Takada K. Oncogenic role of Epstein-Barr virus-encoded RNAs in Burkitt's lymphoma cell line Akata. J Virol. 1999;73:9827–9831. doi: 10.1128/jvi.73.12.9827-9831.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 17.Lenoir G, Bornkamm G W. Burkitt lymphoma, a human cancer model for the study of multistep development of cancer: proposal of a new scenario. Adv Viral Oncol. 1987;7:173–202. [Google Scholar]
- 18.Marchini A, Longnecker R, Kieff E. Epstein-Barr virus (EBV)-negative B-lymphoma cell lines for clonal isolation and replication of EBV recombinants. J Virol. 1992;66:4972–4981. doi: 10.1128/jvi.66.8.4972-4981.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mukherjee S, Trivedi P, Dorfman D M, Klein G, Townsend A. Murine cytotoxic T lymphocytes recognize an epitope in an EBNA-1 fragment, but fail to lyse EBNA-1-expressing mouse cells. J Exp Med. 1998;187:445–450. doi: 10.1084/jem.187.3.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Murray R J, Young L S, Calender A, Gregory C D, Rowe M, Lenoir G M, Rickinson A B. Different patterns of Epstein-Barr virus gene expression and of cytotoxic T-cell recognition in B-cell lines infected with transforming (B95.8) or nontransforming (P3HR1) virus strains. J Virol. 1988;62:894–901. doi: 10.1128/jvi.62.3.894-901.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Murray R J, Kurilla M G, Brooks J M, Thomas W A, Rowe M, Kieff E, Rickinson A B. Identification of target antigens for the human cytotoxic T cell response to Epstein-Barr virus (EBV): implications for the immune control of EBV-positive malignancies. J Exp Med. 1992;176:157–168. doi: 10.1084/jem.176.1.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qu L, Rowe D T. Epstein-Barr virus latent gene expression in uncultured peripheral blood lymphocytes. J Virol. 1992;66:3715–3724. doi: 10.1128/jvi.66.6.3715-3724.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rabson M, Gradoville L, Heston L, Miller G. Non-immortalizing P3J-HR-1 Epstein-Barr virus: a deletion mutant of its transforming parent, Jijoye. J Virol. 1982;44:834–844. doi: 10.1128/jvi.44.3.834-844.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Razzouk B I, Srinivas S, Sample C E, Singh V, Sixbey J W. Epstein-Barr virus DNA recombination and loss in sporadic Burkitt's lymphoma. J Infect Dis. 1996;173:529–535. doi: 10.1093/infdis/173.3.529. [DOI] [PubMed] [Google Scholar]
- 25.Rowe M, Rowe D T, Gregory C D, Young L S, Farrell P J, Rupani H, Rickinson A B. Differences in B cell growth phenotype reflect novel patterns of Epstein-Barr virus latent gene expression in Burkitt's lymphoma cells. EMBO J. 1987;6:2743–2751. doi: 10.1002/j.1460-2075.1987.tb02568.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ruf I K, Rhyne P W, Yang H, Borza C M, Hutt-Fletcher L M, Cleveland J L, Sample J T. Epstein-Barr virus regulates c-MYC, apoptosis, and tumorigenicity in Burkitt lymphoma. Mol Cell Biol. 1999;19:1651–1660. doi: 10.1128/mcb.19.3.1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shibata D K. Epstein-Barr virus and gastric carcinoma. Am J Pathol. 1992;140:769–774. [PMC free article] [PubMed] [Google Scholar]
- 28.Shimizu N, Yoshiyama H, Takada K. Clonal propagation of Epstein-Barr virus (EBV) recombinants in EBV-negative Akata cells. J Virol. 1996;70:7260–7263. doi: 10.1128/jvi.70.10.7260-7263.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shimizu N, Tanabe-Tochikura A, Kuroiwa Y, Takada K. Isolation of Epstein-Barr virus (EBV)-negative cell clones from the EBV-positive Burkitt's lymphoma (BL) line Akata: malignant phenotypes of BL cells are dependent on EBV. J Virol. 1994;68:6069–6073. doi: 10.1128/jvi.68.9.6069-6073.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Srinivas S K, Sample J T, Sixbey J W. Spontaneous loss of viral episomes accompanying Epstein-Barr virus reactivation in a Burkitt's lymphoma cell line. J Infect Dis. 1998;177:1705–1709. doi: 10.1086/517427. [DOI] [PubMed] [Google Scholar]
- 31.Tao Q, Robertson K D, Manns A, Hildesheim A, Ambinder R F. Epstein-Barr virus (EBV) in endemic Burkitt lymphoma: molecular analysis of primary tumor tissue. Blood. 1998;91:1373–1381. [PubMed] [Google Scholar]
- 32.Tierney R J, Steven N, Young L S, Rickinson A B. Epstein-Barr virus latency in blood mononuclear cells: analysis of viral gene transcription during primary infection and in the carrier state. J Virol. 1994;68:7374–7385. doi: 10.1128/jvi.68.11.7374-7385.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Torsteinsdottir S, Masucci M G, Ehlin-Henriksson B, Brautbar C, Ben Bassat H, Klein G, Klein E. Differentiation-dependent sensitivity of human B-cell-derived lines to major histocompatibility complex-restricted T-cell cytotoxicity. Proc Natl Acad Sci USA. 1986;83:5620–5624. doi: 10.1073/pnas.83.15.5620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Torsteinsdottir S, Andersson M L, Avila-Carino J, Ehlin-Henriksson B, Masucci M G, Klein G, Klein E. Reversion of tumorigenicity and decreased agarose clonability after EBV conversion of an IgH/myc translocation-carrying BL line. Int J Cancer. 1989;43:273–278. doi: 10.1002/ijc.2910430219. [DOI] [PubMed] [Google Scholar]
- 35.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Trivedi P, Masucci M G, Winberg G, Klein G. The Epstein-Barr-virus-encoded membrane protein LMP but not the nuclear antigen EBNA-1 induces rejection of transfected murine mammary carcinoma cells. Int J Cancer. 1991;48:794–800. doi: 10.1002/ijc.2910480527. [DOI] [PubMed] [Google Scholar]
- 37.Woisetschlaeger M, Yandava C N, Furmanski L A, Strominger J L, Speck S H. Promoter switching in Epstein-Barr virus during the initial stages of infection of B lymphocytes. Proc Natl Acad Sci USA. 1990;87:1725–1729. doi: 10.1073/pnas.87.5.1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Young L S, Dawson C W, Clark D, Rupani H, Busson P, Tursz T, Johnson A, Rickinson A B. Epstein-Barr virus gene expression in nasopharyngeal carcinoma. J Gen Virol. 1988;69:1051–1065. doi: 10.1099/0022-1317-69-5-1051. [DOI] [PubMed] [Google Scholar]