Abstract
Mitophagy plays a vital role in carcinogenesis and tumor progression. However, the research on the mechanism of mitophagy in esophageal cancer metastasis is limited. This study explored the regulatory mechanism of RECQL4 in mitophagy and affects esophageal cancer metastasis. The RECQL4 expression in esophageal cancer tissues and cells was examined by bioinformatics and qRT-PCR. Bioinformatics analysis was used to determine the upstream regulatory factor of RECQL4 and CREB1. Their binding relationship was evaluated by dual luciferase and Chromatin Immunoprecipitation assays. The effects of RECQL4 on esophageal cancer cells viability, metastasis, and mitophagy were examined using CCK-8, Transwell, immunofluorescence, and Western blot assays. The expression of RECQL4 was up-regulated in esophageal cancer tissues and cells. Overexpression of RECQL4 promoted the cells viability, invasion, migration, and epithelial-mesenchymal transition by inhibiting mitophagy. Bioinformatics analysis revealed a positive correlation between RECQL4 and CREB1, their binding relationship was validatied by dual luciferase and ChIP assays. CREB1 knockdown promoted mitophagy and prevented the metastasis of cancer cells, which could be countered by overexpressing RECQL4. In conclusion, CREB1 was found to transcriptionally activate RECQL4 to inhibit mitophagy, thereby promoting esophageal cancer metastasis. Targeting mitophagy could be an effective therapeutic approach for esophageal cancer.
Keywords: RECQL4, CREB1, mitophagy, esophageal cancer, metastasis
Introduction
In the upper digestive tract, esophageal cancer is among the most prevalent malignant tumors. According to statistics, there were about 604,000 new cases globally in 2020, accounting for 5.5% of all malignant tumors. It is predicted that by 2040, about 957,000 new cases will emerge if the current trend persists.(1) Esophageal cancer patients have low five-year survival rates due to high mortality, poor prognosis, and low early detection rates despite recent treatment advancements.(2) Therefore, it is urgent to find the mechanisms of esophageal cancer occurrence and development for improving the current treatment of this disease.
Mitochondria are important energy metabolism organelles and the main site for oxidative phosphorylation to generate adenosine triphosphate (ATP) in cells. They play an irreplaceable role in the normal physiological metabolism of organisms. Mitophagy is triggered in response to harmful stimuli like hypoxia, inflammation, or DNA damage in the cells to eliminate damaged mitochondria and maintain the quality of the mitochondria.(3) Furthermore, mitophagy is crucial in maintaining cellular homeostasis and is often accompanied by dysregulated autophagy during cancer development.(4) Studies have reported that mitophagy plays a dual role in tumor development. By interacting with and stabilizing PINK1, STOML2 in hepatocellular carcinoma can induce mitophagy to promote tumor metastasis.(5) However, there is also emerging evidence demonstrating that the activation of mitophagy can induce cell death and inhibit cancer occurrence and metastasis. For instance, Zhen et al.(6) revealed that flubendazole impairs the permeability and function of the mitochondrial outer membrane in breast cancer and promotes excessive mitophagy by increasing the DRP1 expression, thereby inhibiting the proliferation and migration of breast cancer cells. In addition, the steroidal dimer by001 inhibits the migration of esophageal cancer by inducing mitophagy.(7) These investigations have shown how autophagy plays dynamic roles in either tumor-suppressive or tumor-promoting ways based on the circumstances and stages of cancer development. This may be related to the cellular environment. On the one hand, mitophagy can timely remove dysfunctional mitochondria to inhibit tumor growth. On the other hand, in cancer cells, mitophagy-mediated clearance of proapoptotic mitochondria may be necessary for cell protection.(8) Therefore, to develop effective strategies for controlling cancer metastasis, it is imperative to comprehend how to use mitophagy to exert anti-tumor effects and investigate the mechanism underlying mitophagy.
As a member of the RECQ helicase family, RECQL4 is essential for preserving genome stability because it takes part in several processes including transcription, recombination, replication, and repair of DNA.(9) The human RECQL4 gene, located on chromosome 8, has an N-terminal ATP-dependent helicase domain that causes functional diversity. This characteristic distinguishes the gene from other members of the family.(10) Recent research has shown that mutations in the RECQL4 gene increase the likelihood of tumor occurrence and progression. For example, RECQL4 mutations related to Baller–Gerold syndrome (BGS), Rothmund–Thomson syndrome (RTS), and RAPADILINO (RAPA) syndrome, with RTS and RAPA patients showing a strong predisposition to cancer.(11) Additionally, overexpression of RECQL4 can also lead to the development of tumors. It has been discovered by Guo et al.(12) that ovarian cancer exhibits a high expression of RECQL4. Overexpressing RECQL4 boosts the invasion and proliferation of ovarian cancer cells, whereas RECQL4 silencing had the opposite effect, according to in vitro cell experiments. Previous studies have found that RECQL4 is highly expressed in esophageal squamous cell carcinoma and is positively correlated with the malignant progression of esophageal squamous cell carcinoma cells.(13) Further research is required to fully understand the precise mechanism by which RECQL4 contributes to esophageal cancer metastasis.
In this research, we first discovered that RECQL4 was highly expressed in esophageal cancer through bioinformatics analysis and molecular experiments. In vitro cell experiments showed that overexpression of RECQL4 could inhibit mitophagy and promote the metastasis of esophageal cancer cells. It was also found that there was an upstream transcription factor CREB1 for RECQL4, confirming that CREB1 activated the transcription of RECQL4 and promoted esophageal cancer metastasis by inhibiting mitophagy. In summary, our study confirms the functional mechanism of the CREB1/RECQL4 axis in promoting esophageal cancer metastasis, providing reliable clues for esophageal cancer diagnosis and treatment.
Materials and Methods
Bioinformatics analysis
First, the mRNA expression data for esophageal cancer was downloaded from The Cancer Genome Atlas (TCGA) (https://portal.gdc.cancer.gov/). Next, the ‘edgeR’ package was used to perform a differential analysis of the mRNAs between the tumor group (182) and the normal group (13) (|logFC|>1.0, FDR<0.05) to obtain differentially expressed mRNAs (DEmRNAs). Finally, combining with literature review, the target gene, RECQL4, was identified. Potential transcription factors upstream of RECQL4 in esophageal cancer were predicted using the KnockTF database (http://www.licpathway.net/KnockTF/index.php). Pearson correlation analysis was then performed between the transcription factors and RECQL4, and JASPAR (https://jaspar.genereg.net/) predicted the 2,000 bp motif binding site upstream of RECQL4.
Cell cultivation
Human embryonic kidney cells (293T), human normal esophageal epithelial cells (HEEC), and esophageal cancer cells (OE19, TE-1, EC109) were all purchased from BeNa Culture Collection (China). OE19, TE-1, and EC109 cells were cultured in RPMI-1640 medium (GIBCO, Grand Island, NY) containing 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin. HEEC cells were cultured in Eagle’s minimal essential medium (EMEM) containing 10% FBS and 1% penicillin/streptomycin. 293T cells were cultivated in DMEM containing 10% FBS, 1% penicillin/streptomycin, and 2 mM L-glutamine. All the mentioned cells were cultured at 37°C with 5% CO2 in a cell incubator.
Cell transfection
The pLKO.1-Puro empty vector (sh-NC), pLKO.1-RECQL4 shRNA interference plasmid (sh-RECQL4), pcDNA3.1 empty vector (oe-NC), pcDNA3.1-RECQL4 expression plasmid (oe-RECQL4), and pLKO.1-CREB1 shRNA interference plasmid (sh-CREB1) were all provided by Genechem (China). The above-mentioned plasmids were transfected into esophageal cancer cells and 293T cells by Lipofectamine 2000 (Invitrogen, Carlsbad, CA).
Quantitative real-time polymerase chain reaction (qRT-PCR)
Esophageal cancer cells were treated with Trizol reagent (Invitrogen) to extract total RNA, and cDNA was obtained by reverse transcription using the Hifair®II 1st Strand cDNA Synthesis Kit (Yeasen, Shanghai, China). qRT-PCR was then performed using the UltraSYBR Mixture (Cwbio, Taizhou, China) on an ABI7500 fluorescence quantitative PCR instrument (Invitrogen). With GAPDH serving as an internal reference, the expression of RECQL4 and CREB1 was calculated using the 2−ΔΔCt method. Table 1 lists the necessary primers.
Table 1.
qRT-PCR primer list
| Gene | Sequence (5'→3') |
|---|---|
| RECQL4 | F: TCAACATGAAGCAGAAACACTAC |
| R: CTGCTCGTTCAGGAAACAAGACT | |
| CREB1 | F: TGCAACATCATCTGCTCCCA |
| R: CTGAATAACTGATGGCTGGGC | |
| GAPDH | F: GAGAAGGCTGGGGCTCATTT |
| R: AGTGATGGCATGGACTGTGG |
CCK-8 assay
CCK-8 was used to evaluate cell viability. Esophageal cancer cells were seeded on a 96-well plate with 2,000 cells in each well. After the cells adhered, 10 μl CCK-8 reagent was added to each well at 0, 24, 48, and 72 h (Solarbio, Beijing, China), continue culturing for 2 h, and then a microplate reader (Invitrogen) was used to measure the optical density (OD) value at 450 nm.
Transwell assay
Five thousand cells were seeded in each well of the upper chamber, the chamber with the serum-free medium was added for the migration test, and the chamber coated with Matrigel was prepared for the invasion test. The lower chamber contained 600 μl of complete culture medium per well. After 48 h, the medium was removed from the upper chamber, and a cotton swab was used to wipe off the residual medium, matrix gel, and cells. Once the chamber membrane was fixed with 4% paraformaldehyde, stained with crystal violet, and then cleaned. It was cut out and positioned on a glass slide. The slide was put under an Olympus microscope (Tokyo, Japan) and photographed after added with neutral resin dropwise and covered with a coverslip.
Immunofluorescence
Cells were cultivated over night after being seeded in a 24-well plate. Cells were fixed for 30 min with 4% paraformaldehyde fixative after being washed three times with phosphate-buffered saline (PBS). Fixative was removed, and then cells were permeabilized with 0.1% TritonX-100 in PBS for 15 min. Cells were incubated with primary antibodies against E-cadherin, N-cadherin, MMP-2, and MMP-9 (Abcam, Cambridge, UK) diluted in PBS overnight at 4°C, followed by incubation with secondary antibodies at 37°C in the dark for 1 h. DAPI staining solution was used to observe the cell nucleus, and the fluorescence quencher (Beyotime, Shanghai, China) was sealed and observed under a confocal microscope (Nikon, Tokyo, Japan).
Drug treatment and fluorescence microscopy observation of mitophagy
To study mitophagy, corresponding esophageal cancer cells were treated with 5 μM Olaparib (MedChemExpress, State of New Jersey) for 72 h. The esophageal cancer cells were transfected with the GFP-LC3 plasmid. After being successfully transfected, the cells were cultured for 24 h in a 12-well plate. Using a fluorescence microscope, the DAPI-stained cell nuclei were examined for fluorescence.(14)
Western blot (WB)
Cells were lysed and total proteins were extracted using radioimmunoprecipitation assay (RIPA) buffer (ThermoFisher Scientific, Waltham, MA). The protein concentration was quantified using the bicinchoninic acid (BCA) assay kit (Beyotime). The PAGE gel was prepared and placed in an electrophoresis tank containing electrophoresis solution. Marker and protein samples were then added and the separated proteins were transferred to a polyvinylidene fluoride (PVDF) membrane (Millipore, Billerica, MA). To block the membrane, 5% albumin from bovine serum (BSA) was used at room temperature for 1 h. After incubation with the primary antibodies overnight at 4°C, the membrane was washed three times with Tris-Buffered Saline Tween-20 (TBST) buffer and then incubated with the secondary antibody for 2 h. ECL immunoblot chemiluminescence solution (Millipore) was used to detect the protein bands on a fluorescence and chemiluminescence imaging system (Clinx, Shanghai, China). Primary antibodies included rabbit anti-human LC3B, p62, and GAPDH. The secondary antibody was Goat anti-rabbit IgG H&L (HRP). All antibodies mentioned were purchased from Abcam.
Dual luciferase reporter assay
LipofectamineTM2000 (Invitrogen) was used to co-transfect 293T cells with the luciferase reporter vectors pGL3-RECQL4-promoter-WT and pGL3-RECQL4-promoter-MUT (Promega, Madison, WI), along with sh-NC, and sh-CREB1. The Dual-Luciferase Reporter Assay System (Promega) was used to measure luciferase activity following a 48-h incubation period.
Chromatin immunoprecipitation (ChIP)
Protein-DNA interactions were stabilized by covalent crosslinking with 1% formaldehyde, and cells were lysed to release cellular components. Chromatin was sheared into small fragments using nucleases and incubated overnight with anti-IgG and anti-CREB1 separately. DNA was purified using a DNA purification kit (Beyotime) after crosslinking was reversed. Finally, purified DNA products were quantified by qPCR. The ChIP-qPCR primers are listed in the Table 2.
Table 2.
ChIP-qPCR primer list
| Target site | Sequence (5'→3') |
|---|---|
| Site | F: CAAATGCAGCCACTGCCTC |
| R: GGACAAGCAGCCAATGGGA |
Data statistics and analysis
There were at least 3 independent experiments in each group, and the data were represented as mean ± SD. GraphPad Prism 8.0 was used to perform t test or one-way analysis of variance (ANOVA) analysis on the experimental data. P<0.05 was considered statistically significant.
Results
RECQL4 is highly expressed in esophageal cancer
This study examined the expression level of RECQL4 in esophageal cancer based on the TCGA-esophageal cancer (EC) database. It was discovered that RECQL4 was notably upregulated in esophageal cancer tissues compared to normal tissues (Fig. 1A). Next, qRT-PCR analysis was performed on RECQL4 in HEEC and esophageal cancer cells (OE19, TE-1, EC109), and the results presented that the expression of RECQL4 was upregulated in esophageal cancer cells (Fig. 1B). To summary, RECQL4 was highly expressed in esophageal cancer tissues and cells.
Fig. 1.
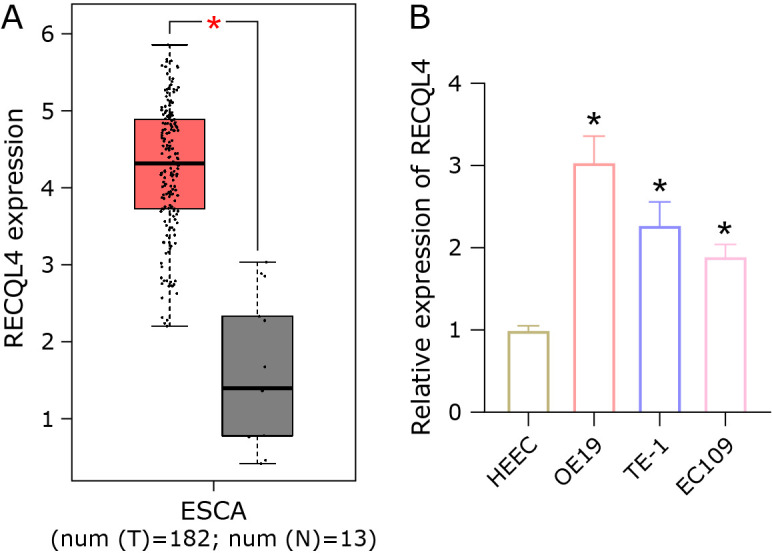
RECQL4 is highly expressed in esophageal cancer. (A) Expression of RECQL4 in normal tissues and esophageal cancer tissues in TCGA database. (B) Expression of RECQL4 in HEEC and esophageal cancer cells (OE19, TE-1, EC109) determined by qRT-PCR. * represents p<0.05.
The role of RECQL4 in esophageal cancer metastasis
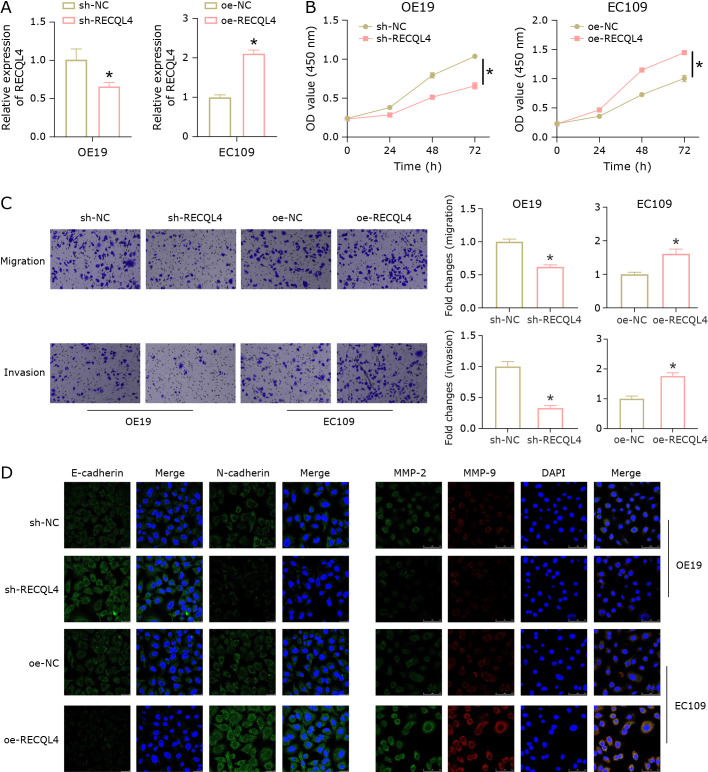
To investigate the role of RECQL4 in the metastasis of esophageal cancer, we constructed sh-NC/sh-RECQL4 cell lines based on OE19 cells with high RECQL4 expression and oe-NC/oe-RECQL4 cell lines based on EC109 cells with low RECQL4 expression. The qRT-PCR analysis showed that RECQL4 expression was decreased by sh-RECQL4 and increased by oe-RECQL4, suggesting a good transfection efficiency (Fig. 2A). The viability of cells was evaluated using CCK-8. Knockdown of RECQL4 reduced cell viability, while overexpression of RECQL4 increased it when compared to the control group (Fig. 2B). Next, Transwell assays were conducted to investigate changes in cell migration and invasion. The results showed that oe-RECQL4 facilitated the migration and invasion of EC109 cells, while sh-RECQL4 inhibited the migration and invasion of OE19 cells (Fig. 2C). Immunofluorescence detection results showed that silencing RECQL4 increased the E-cadherin expression and decreased the N-cadherin expression in OE19 cells, as well as reduced the expression of metastasis-related proteins MMP-2 and MMP-9. Comparing EC109 cells overexpressing RECQL4 to cells with RECQL4 silenced, the expression levels of these proteins displayed entirely different changes (Fig. 2D). These findings suggested that RECQL4 could promote the metastasis of esophageal cancer.
Fig. 2.
The role of RECQL4 in esophageal cancer metastasis. (A) qRT-PCR was used to determine the RECQL4 expression. (B) CCK-8 assay was used to measure cell viability. (C) Transwell assays were used to measure cell migration and invasion. (D) Immunofluorescence was used to detect the expression of EMT-related proteins (E-cadherin and N-cadherin) and metastasis-related proteins (MMP-2 and MMP-9). * represents p<0.05.
RECQL4 promotes esophageal cancer metastasis by mitophagy regulation
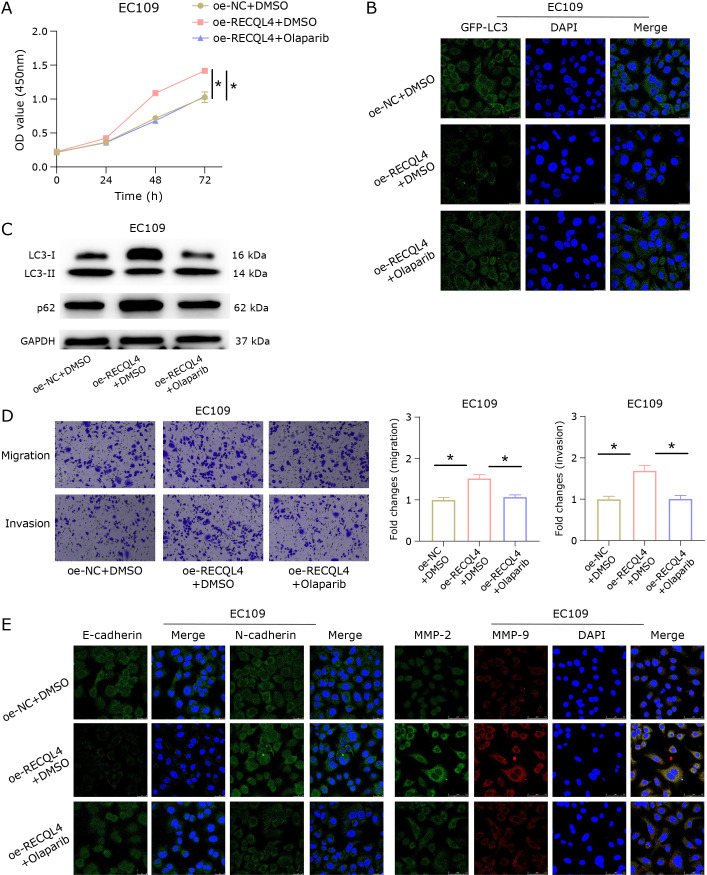
Previous studies have reported that RECQL4 is related to regulating mitophagy in osteosarcoma cells, and the loss of RECQL4 induces mitophagy in U2OS cells.(15) Therefore, we speculated that RECQL4 may affect esophageal cancer cell metastasis by regulating mitophagy. To verify this hypothesis, we first constructed the following groups based on EC109 cells: oe-NC + DMSO, oe-RECQL4 + DMSO, oe-RECQL4 + Olaparib (mitophagy inducer). The CCK-8 results indicated that the oe-RECQL4 + DMSO group had much higher cell viability in EC109 cells when compared to the oe-NC + DMSO group. However, after adding Olaparib to induce mitophagy, the cell viability in the oe-RECQL4 group returned to the control group level (Fig. 3A). To investigate how RECQL4 affects the activation of mitophagy, we used GFP-LC3 to monitor the LC3 protein. Through fluorescence microscopy, we observed that GFP-LC3 aggregation (green fluorescence) in EC109 cells of the oe-RECQL4 + DMSO group was reduced, while oe-GFP-LC3 aggregation of the RECQL4 + Olaparib group returned to the control group level (Fig. 3B). The WB results suggested that overexpression of RECQL4 resulted in a significant decrease in the level of autophagy-related protein LC3II, an increase in the expression of p62, and a decrease in the LC3II/LC3I ratio. Following the addition of Olaparib, the p62 expression trended downward and eventually reached the level of the control group (Fig. 3C). Transwell assays were used to measure cell invasion and migration. The results demonstrated that the capacity of EC109 cells for migration and invasion was increased in the oe-RECQL4 + DMSO group, whereas oe-RECQL4 + Olaparib attenuated the effect of RECQL4 overexpression on these processes (Fig. 3D). According to immunofluorescence results, overexpression of RECQL4 in EC109 cells resulted in decreased expression of the EMT-related protein E-cadherin, increased expression of N-cadherin, and elevated expression of the proteins MMP-2 and MMP-9 related to metastasis. However, after induction of mitophagy with Olaparib, the expression of these proteins returned to the control group level (Fig. 3E). The above results indicated that RECQL4 could inhibit mitophagy and promote the metastasis of esophageal cancer cells.
Fig. 3.
RECQL4 regulates mitophagy and affects the metastasis of esophageal cancer cells. (A) CCK-8 detected the viability of EC109 cells in the oe-NC + DMSO, oe-RECQL4 + DMSO, and oe-RECQL4 + Olaparib groups. (B) Immunofluorescence results of GFP-LC3. (C) WB detected the expression of LC3I, LC3II, and p62. (D) Transwell determined cell migration and invasion. (E) Immunofluorescence detected the expression of EMT-related proteins (E-cadherin and N-cadherin) and metastasis-related proteins (MMP-2 and MMP-9). * represents p<0.05.
CREB1 regulates the RECQL4 expression
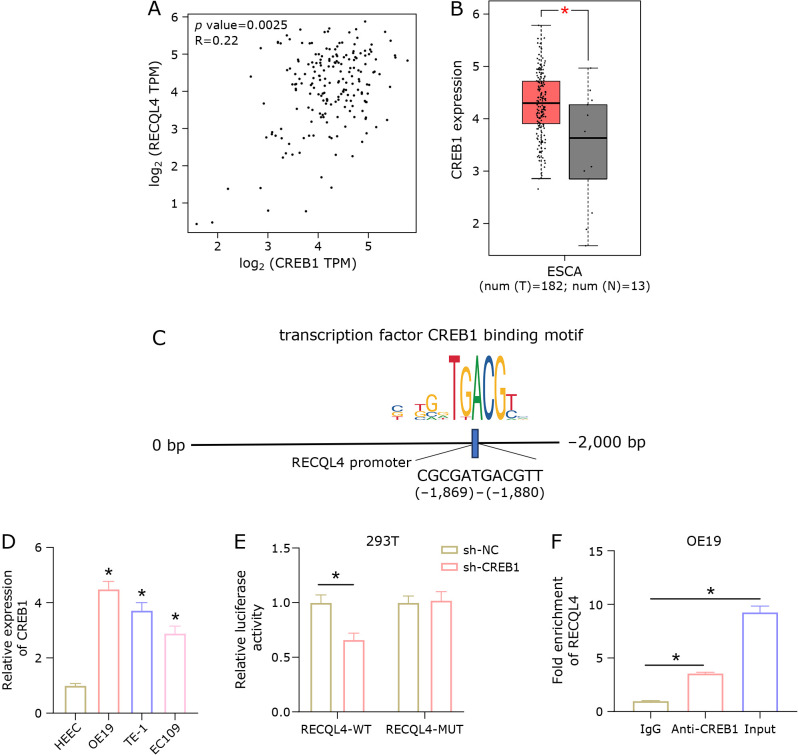
The possible transcription factor CREB1 upstream of RECQL4 in esophageal cancer was predicted using the KnockTF database to investigate the upstream regulatory factors of RECQL4. A positive correlation was found between CREB1 and RECQL4 (Fig. 4A). Bioinformatics analysis found that esophageal cancer tissues exhibit high expression of CREB1 (Fig. 4B). Afterward, motif binding sites within 2,000 bp upstream of RECQL4 were predicted using JASPAR (Fig. 4C). Furthermore, qRT-PCR analysis revealed that CREB1 was significantly upregulated in esophageal cancer cell lines (OE19, TE-1, EC109) (Fig. 4D). The binding relationship between CREB1 and RECQL4 was validated through dual-luciferase and ChIP experiments. The results showed that sh-CREB1 notably reduced the luciferase activity of the RECQL4-WT group, but had no evident effect on the RECQL4-MUT group (Fig. 4E), and RECQL4 was markedly enriched by Anti-CREB1 (Fig. 4F). The above results indicated that CREB1 transcriptionally regulated the expression of RECQL4.
Fig. 4.
CREB1 transcriptionally regulates RECQL4 expression. (A) Pearson correlation analysis with CREB1 and RECQL4. (B) Expression of CREB1 in esophageal cancer tissues. (C) Targeted binding sites of CREB1 and RECQL4. (D) Expression of CREB1 in esophageal cancer cells. (E, F) The binding relationship between CREB1 and RECQL4 was confirmed by dual luciferase and ChIP experiments. * represents p<0.05.
CREB1 eregulates RECQL4 to control mitophagy esophageal cancer cells metastasis
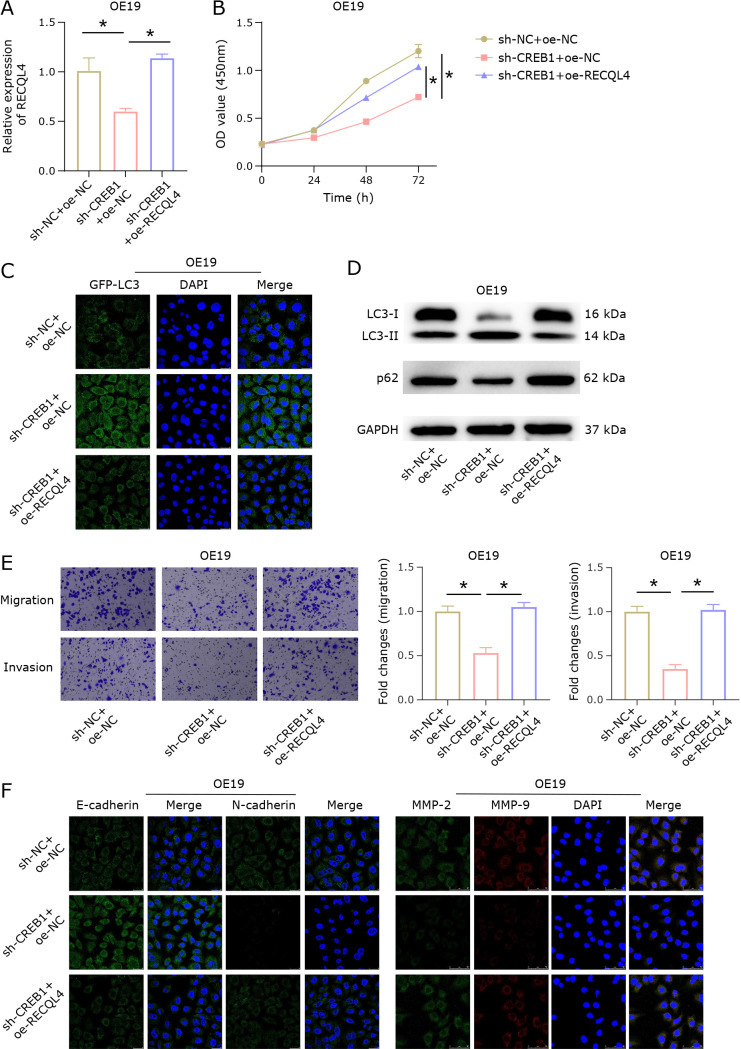
Our study demonstrated that CREB1 could transcriptionally regulate the expression of RECQL4, and that RECQL4 was associated with mitophagy. Based on this, we speculated that CREB1 could regulate RECQL4 to control mitophagy and affect the metastasis of esophageal cancer cells. Therefore, we constructed the following groups based on OE19 cells: sh-NC + oe-NC, sh-CREB1 + oe-NC, sh-CREB1 + oe-RECQL4. First, RECQL4 expression was detected by qRT-PCR. The results demonstrated that compared to the control group, RECQL4 expression was significantly decreased in sh-CREB1 + oe-NC cells, and RECQL4 expression in sh-CREB1 + oe-RECQL4 cells was restored to the level of the control group (Fig. 5A). Sh-CREB1 decreased OE19 cell viability, according to CCK-8 results, but concurrent oe-RECQL4 treatment lessened the inhibitory effect of sh-CREB1 on cell viability (Fig. 5B). Next, the level of mitophagy was evaluated. It was observed under a fluorescence microscope that GFP-LC3 aggregation increased in OE19 cells treated with sh-CREB1 + oe-NC, while GFP-LC3 aggregation in OE19 cells treated with sh-CREB1 + oe-RECQL4 was close to the control group level (Fig. 5C). To study mitophagy, the conversion of LC3-I to LC3-II was further analyzed. The results showed that the ratio of LC3-II/LC3-I increased and the expression of p62 reduced in the sh-CREB1 + oe-NC group; while in the sh-CREB1 + oe-NC group, these indicators returned to the control group level (Fig. 5D). According to the Transwell results, OE19 cells’ capacity for migration and invasion was reduced by sh-CREB1 + oe-NC, whereas this capacity was increased by sh-CREB1 + oe-RECQL4 treatment, approaching the level of the control group (Fig. 5E). Finally, immunofluorescence detection of EMT-related proteins (E-cadherin and N-cadherin) and metastasis-related proteins (MMP-2 and MMP-9) was used to evaluate the cells’ metastatic ability. The findings demonstrated that while N-cadherin, MMP-2, and MMP-9 expression was reduced in OE19 cells treated with sh-CREB1 + oe-RECQL4, the expression of these proteins was increased in OE19 cells treated with sh-CREB1 and returned to the level of the control group (Fig. 5F). In conclusion, CREB1 could activate RECQL4 to inhibit mitophagy and promote esophageal cancer cell metastasis.
Fig. 5.
CREB1 regulates RECQL4 to control mitophagy and affect esophageal cancer cell metastasis. (A) qRT-PCR was used to detect the RECQL4 expression in OE19 cells of sh-NC + oe-NC, sh-CREB1 + oe-NC, and sh-CREB1 + oe-RECQL4 groups. (B) CCK-8 assay was performed to measure the cell viability of OE19 cells in sh-NC + oe-NC, sh-CREB1 + oe-NC, and sh-CREB1 + oe-RECQL4 groups. (C) Immunofluorescence results of GFP-LC3. (D) WB was conducted to detect the expression of LC3I, LC3II, and p62. (E) Transwell assays were used to measure cell migration and invasion. (F) Immunofluorescence was performed to detect the expression of EMT-related proteins (E-cadherin and N-cadherin) and metastasis-related proteins (MMP-2 and MMP-9). * represents p<0.05.
Discussion
In this research, we discovered that RECQL4 was upregulated in esophageal cancer and may facilitate the metastasis of esophageal cancer by affecting mitophagy. We found that CREB1 was an upstream regulatory factor of RECQL4 and activated RECQL4 transcription, which in turn promoted the metastasis of esophageal cancer through bioinformatics analysis. In conclusion, this study elucidates the specific mechanism of CREB1/RECQL4 axis in regulating esophageal cancer metastasis.
Mitophagy is a type of selective autophagy that helps the body to maintain homeostasis by breaking down malfunctioning mitochondria.(16) Prior research has revealed that mitophagy is a major factor in the development of various human diseases, including cancer.(17) Existing research has demonstrated that mitophagy functions as a “double-edged sword” in tumor development, either accelerating or preventing the cancer progression.(18,19) In this study, we examined mitophagy by transfecting esophageal cancer cells with GFP-LC3 plasmid. Our findings indicated that RECQL4 overexpression suppressed mitophagy and enhanced the metastasis of esophageal cancer cells. The addition of Olaparib induced mitophagy and inhibited the metastatic ability of esophageal cancer cells. Xie et al.(20) found that the antimalarial medication mefloquine (MQ) can suppress the tumor growth of esophageal squamous cell carcinoma by inducing mitophagy, which is consistent with our results that demonstrated the inhibitory effect of induced mitophagy on cancer metastasis. Similar phenomena have also been observed in triple-negative breast cancer (TNBC), where mitochondrial protein UCP1 can prevent the metastasis of TNBC by activating mitophagy and apoptosis, While UCP1’s inhibition of TNBC metastasis is lessened by damaged mitophagy activation.(21) Therefore, unraveling the regulatory mechanism that affects mitophagy may provide promising targets for future anti-tumor therapy.
In addition, beyond its impact on mitophagy, RECQL4 is implicated in numerous biological processes, and the underlying mechanisms of its anti-tumor properties can be intricate. Elevated RECQL4 expression has been found in many human cancers. For example, in breast cancer, increased RECQL4 mRNA expression is associated to a reduced overall survival rate of patients.(22) Similarly, the expression level of RECQL4 mRNA in gastric cancer is much higher compared to normal gastric tissues.(23) Meanwhile, previous studies have demonstrated a close correlation between abnormal expression of RECQL4 and cancer progression. For example, Ye et al.(24) investigated the impact of RECQL4 on the progression of hepatocellular carcinoma, and found that knocking down RECQL4 inhibits the progression and EMT of hepatocellular carcinoma cells. A similar conclusion was also reached in a study on esophageal squamous cell carcinoma, which showed that EMT and proliferation of the cancer are greatly decreased by knocking down RECQL4.(13) Consistently, our results also indicated that knocking down RECQL4 could reduce the vitality, migration, invasion, and EMT of esophageal cancer cells. Additionally, research has also pointed out that RECQL4 has the opposite function, as evidenced by the higher risk of osteosarcoma or lymphoma in individuals with RTS due to RECQL4 deficiency.(11) This may be due to tumor heterogeneity between different tumors. The results amply demonstrated the critical role of RECQL4 in the advancement of esophageal cancer.
We eliminated the upstream transcription factor CREB1 of RECQL4 by bioinformatics analysis and verified their binding relationship to delve deeper into the molecular mechanism of RECQL4 in controlling esophageal cancer metastasis. CREB1 is a transcription factor in the nucleus of eukaryotic cells, which can be activated by signaling pathways like cAMP to regulate the expression of downstream target genes.(25,26) Recent studies have shown that CREB1 abnormally initiates the transcription of genes related to cancer, which is vital in the development and occurrence of tumors. CREB1 expression levels are elevated in a variety of tumors, including colorectal cancer,(27) ovarian cancer,(28) and gastric cancer.(29) Similarly, our results also indicate that CREB1 is upregulated in esophageal cancer. Furthermore, in this research, knocking down CREB1 suppressed the EMT process, migration and invasion of esophageal cancer cells, while overexpression of RECQL4 restored these indicators. These results demonstrated the promoting effect of CREB1 transcription activation of RECQL4 on esophageal cancer metastasis, enriching the study on CREB1 in esophageal cancer, and suggesting that the CREB1/RECQL4 axis may be an important molecular target for inhibiting esophageal cancer metastasis.
As this study elucidates, CREB1 activates RECQL4 to inhibit mitochondrial autophagy and promote esophageal cancer metastasis. This study can offer a fresh perspective for the esophageal cancer treatment, suggesting that regulating mitochondrial autophagy or targeting the CREB1/RECQL4 axis can be a novel approach for esophageal cancer therapy. Nevertheless, this study has certain limitations that require additional validation, such as the absence of animal-level studies on the impact of the CREB1/RECQL4 axis on esophageal cancer metastasis.
Author Contributions
(I) Conception and design: SZ
(II) Provision of study materials or patients: YZ
(III) Collection and assembly of data: XG
(IV) Data analysis and interpretation: ZT
(V) Manuscript writing: JC
(VI) Final approval of manuscript: All authors
Data Availability Statement
The data and materials in the current study are available from the corresponding author on reasonable request.
Funding
This research received no specific grant from any funding agency in the public, commercial, or not for profit sectors.
Conflict of Interest
No potential conflicts of interest were disclosed.
References
- 1.Morgan E, Soerjomataram I, Rumgay H, et al. The global landscape of esophageal squamous cell carcinoma and esophageal adenocarcinoma incidence and mortality in 2020 and projections to 2040: new estimates from GLOBOCAN 2020. Gastroenterology 2022; 163: 649–658.e2. [DOI] [PubMed] [Google Scholar]
- 2.Harada K, Rogers JE, Iwatsuki M, Yamashita K, Baba H, Ajani JA. Recent advances in treating oesophageal cancer. F1000Res 2020; 9: F1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Song M, Zhou Y, Fan X. Mitochondrial quality and quantity control: mitophagy is a potential therapeutic target for ischemic stroke. Mol Neurobiol 2022; 59: 3110–3123. [DOI] [PubMed] [Google Scholar]
- 4.Mrakovcic M, Kleinheinz J, Fröhlich LF. p53 at the crossroads between different types of HDAC inhibitor-mediated cancer cell death. Int J Mol Sci 2019; 20: 2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zheng Y, Huang C, Lu L, et al. STOML2 potentiates metastasis of hepatocellular carcinoma by promoting PINK1-mediated mitophagy and regulates sensitivity to lenvatinib. J Hematol Oncol 2021; 14: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhen Y, Yuan Z, Zhang J, et al. Flubendazole induces mitochondrial dysfunction and DRP1-mediated mitophagy by targeting EVA1A in breast cancer. Cell Death Dis 2022; 13: 375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang SQ, Zhou KR, Shi XL, et al. Steroidal dimer by001 inhibits proliferation and migration of esophageal cancer cells via multiple mechanisms. Cancer Chemother Pharmacol 2019; 83: 179–189. [DOI] [PubMed] [Google Scholar]
- 8.Chang JY, Yi HS, Kim HW, Shong M. Dysregulation of mitophagy in carcinogenesis and tumor progression. Biochim Biophys Acta Bioenerg 2017; 1858: 633–640. [DOI] [PubMed] [Google Scholar]
- 9.Petkovic M, Dietschy T, Freire R, Jiao R, Stagljar I. The human Rothmund–Thomson syndrome gene product, RECQL4, localizes to distinct nuclear foci that coincide with proteins involved in the maintenance of genome stability. J Cell Sci 2005; 118 (Pt 18): 4261–4269. [DOI] [PubMed] [Google Scholar]
- 10.Matsuno K, Kumano M, Kubota Y, Hashimoto Y, Takisawa H. The N-terminal noncatalytic region of Xenopus RecQ4 is required for chromatin binding of DNA polymerase alpha in the initiation of DNA replication. Mol Cell Biol 2006; 26: 4843–4852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lu L, Jin W, Wang LL. RECQ DNA helicases and osteosarcoma. Adv Exp Med Biol 2020; 1258: 37–54. [DOI] [PubMed] [Google Scholar]
- 12.Guo L, Li Y, Zhao C, et al. RECQL4, negatively regulated by miR-10a-5p, facilitates cell proliferation and invasion via MAFB in ovarian cancer. Front Oncol 2020; 10: 524128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lyu G, Su P, Hao X, et al. RECQL4 regulates DNA damage response and redox homeostasis in esophageal cancer. Cancer Biol Med 2021; 18: 120–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Santiago-O'Farrill JM, Weroha SJ, Hou X, et al. Poly(adenosine diphosphate ribose) polymerase inhibitors induce autophagy-mediated drug resistance in ovarian cancer cells, xenografts, and patient-derived xenograft models. Cancer 2020; 126: 894–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Duan Y, Fang H. RecQL4 regulates autophagy and apoptosis in U2OS cells. Biochem Cell Biol 2016; 94: 551–559. [DOI] [PubMed] [Google Scholar]
- 16.Yao J, Wang J, Xu Y, et al. CDK9 inhibition blocks the initiation of PINK1-PRKN-mediated mitophagy by regulating the SIRT1-FOXO3-BNIP3 axis and enhances the therapeutic effects involving mitochondrial dysfunction in hepatocellular carcinoma. Autophagy 2022; 18: 1879–1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ferro F, Servais S, Besson P, Roger S, Dumas JF, Brisson L. Autophagy and mitophagy in cancer metabolic remodelling. Semin Cell Dev Biol 2020; 98: 129–138. [DOI] [PubMed] [Google Scholar]
- 18.Chourasia AH, Boland ML, Macleod KF. Mitophagy and cancer. Cancer Metab 2015; 3: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Drake LE, Springer MZ, Poole LP, Kim CJ, Macleod KF. Expanding perspectives on the significance of mitophagy in cancer. Semin Cancer Biol 2017; 47: 110–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xie Y, Zhang J, Lu B, et al. Mefloquine inhibits esophageal squamous cell carcinoma tumor growth by inducing mitochondrial autophagy. Front Oncol 2020; 10: 1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xia J, Chu C, Li W, et al. Mitochondrial protein UCP1 inhibits the malignant behaviors of triple-negative breast cancer through activation of mitophagy and pyroptosis. Int J Biol Sci 2022; 18: 2949–2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhu X, Chen H, Yang Y, et al. Distinct prognosis of mRNA expression of the five RecQ DNA-helicase family members - RECQL, BLM, WRN, RECQL4, and RECQL5 - in patients with breast cancer. Cancer Manag Res 2018; 10: 6649–6668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen H, Yuan K, Wang X, et al. Overexpression of RECQL4 is associated with poor prognosis in patients with gastric cancer. Oncol Lett 2018; 16: 5419–5425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ye Y, Yu F, Li Z, Xie Y, Yu X. RNA binding protein serine/arginine splicing factor 1 promotes the proliferation, migration and invasion of hepatocellular carcinoma by interacting with RecQ protein-like 4 mRNA. Bioengineered 2021; 12: 6144–6154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mayr B, Montminy M. Transcriptional regulation by the phosphorylation-dependent factor CREB. Nat Rev Mol Cell Biol 2001; 2: 599–609. [DOI] [PubMed] [Google Scholar]
- 26.Montminy MR, Sevarino KA, Wagner JA, Mandel G, Goodman RH. Identification of a cyclic-AMP-responsive element within the rat somatostatin gene. Proc Natl Acad Sci U S A 1986; 83: 6682–6686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yan L, You WQ, Sheng NQ, et al. A CREB1/miR-433 reciprocal feedback loop modulates proliferation and metastasis in colorectal cancer. Aging (Albany NY) 2018; 10: 3774–3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li CJ, Lin LT, Chu PY, et al. Identification of novel biomarkers and candidate drug in ovarian cancer. J Pers Med 2021; 11: 316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang X, Liu F, Jiang Z, Guan H, Jia Q. CREB1 suppresses transcription of microRNA-186 to promote growth, invasion and epithelial-mesenchymal transition of gastric cancer cells through the KRT8/HIF-1α axis. Cancer Manag Res 2020; 12: 9097–9111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data and materials in the current study are available from the corresponding author on reasonable request.