Abstract
Lung cancer as a molecularly and histologically high heterogonous disease, there is an urgent need to predict lung cancer patients' responses to anti-cancer treatment, and patient-derived organoids (PDOs) have been recognized as a valuable platform for preclinical drug screening. In this study, we successfully established 26 PDO lines from various subtypes of lung cancers including benign tumor, adenocarcinoma, squamous cell carcinoma, adenosquamous carcinoma, large-cell carcinoma, and small-cell carcinoma. These PDOs were shown to retain the major genomic and histological characteristics of primary tumors and remain stable during long-term culture. With the help of targeted genomic sequencing, we found that lung cancer that harbors METN375S mutation is selectively sensitive to afatinib, and a combination of afatinib and gemcitabine induced synthetic lethality in PDO and mini-PDX models. In summary, our findings demonstrate the potential of PDO in predicting lung cancer drug response, and reveal a promising strategy for METN375S mutant lung cancer treatment.
1. Introduction
Lung cancer is the leading cause of cancer-related death worldwide, with a 5-year overall survival rate of less than 15 % [1]. Due to the high heterogeneity of both its molecular structure and its pathology, the treatment of lung cancer remains a tremendous challenge [2,3]. Benefitting from the development of high-throughput sequencing technology, molecular targeted therapy continues to be developed and shows great progress in individualized treatment and precision cancer medicine [4]. However, certain genomic mutations do not also respond to drug treatments, and it is therefore urgent to uncover a new preclinical model for drug efficacy screening.
In recent years, patient-derived tumor Organoids (PDOs) produced by three-dimensional culture of primary patient material have emerged as robust preclinical model for drug testing [[5], [6], [7], [8]]. PDOs require less time yet have a higher success rate than patient-derived xenograft (PDX), which usually takes 4–8 months. Importantly, PDOs have demonstrated stable morphological and genetic features even after long-term expansion, thus making them a suitable model to guide clinical treatment [8,9]. PDOs have already been successfully used for drug screening in several tumors including colorectal cancer [10,11], breast cancer [12], and bladder cancer [13]. Specifically, in lung cancer, a PDO model coupled with a microwell device has been reported to provide a technically feasible means for predicting patient-specific drug responses in clinical settings [[14], [15], [16]].
Lung organoid models of tracheobronchial and alveolar tissue have been developed from epithelial stem and progenitor cells [17]. Within the mammalian lung, a large number of epithelial cells are distributed along the tracheal and alveolar zones of the lung axis, maintained by regional epithelial stem and progenitor cells with the potential to generate lung organoids [18]. 2D air-liquid interface (ALI) culture mimics a realistic lung environment and drives airway epithelial cells to proliferate and differentiate in vitro [19]. However, the concomitant lack of tissue architecture with multi-cellular differentiation necessitated in the development of 3D culture system. Here, tissue is embedded in a basement membrane mixture to mimic the physical framework of in vivo tissue. Differentiation-inducing growth factors are added to complete the 3D culture [20]. At present, previous studies have utilized PDO for targeted therapy in NSCLC including lung squamous cell carcinoma [21], lung adenocarcinoma [22,23], small-cell carcinoma [24], and large-cell carcinoma [[25], [26], [27]]. Lung organoid models of tracheobronchial and alveolar tissue have also been developed from adult stem cells and pluripotent stem cells 25–27 and have both grown in vitro epithelial of both these two different tissue types [14,26,28].
In this study, we successfully established and characterized PDOs from 6 subtypes of lung cancers including benign tumor, adenocarcinoma, squamous cell carcinoma, adenosquamous carcinoma, large-cell carcinoma, and small-cell carcinoma. Using these PDO lines and a PDO-derived mini-PDX model, we found that the combination of afatinib and gemcitabine induced synthetic lethality in METN375S mutant lung cancer. Our study demonstrates that PDOs combined with targeted genomic sequencing can assist providers in making clinical decision.
2. Materials and methods
2.1. Human specimens
Lung cancer tissue (about 1–3 cm3) was taken from surgically resected lung cancer specimens for the cultivation of lung cancer organoids at the Department of Thoracic Surgery of Thoracic Surgery Department. The samples were collected in accordance with the relevant regulations on the management of human genetic resources in China after the study was approved by the Ethical Examination and approval Committee of Xi ‘an Jiaotong University. Informed consent was obtained from each patient/family member.
2.2. Tissue preparation
The tissue samples were placed in serum-free RPMI medium 1640 with 2 % antibiotics and transported to the laboratory within 1–2 h of harvest. Samples were washed three times with cold PBS containing 2 % antibiotics and sectioned with sterile scissors and tweezers to1-2 mm3. A small piece of tissue was fixed in formalin for hematoxylin-eosin staining (HE)/immunohistochemistry (IHC), another piece of tissue was taken into a cryovial to be ready for sequencing, and the rest of the tissue was used in the PDO model.
2.3. Organoid culture
For the PDOs, the patient's tissue was placed in a 15 ml centrifuge tube to which 4 ml serum-free Advanced DMEM/F12 medium (Gibco) with 2 % antibiotics and 1 ml Liberase TH (5 mg/ml) was added and kept at 37 °C for 30 min with intermittent agitation [28]. After sufficient dissociation, the tissue suspension was repeatedly blown with a sterile pipette to isolate the cells. After waiting 2–3 min for the tissue deposition to the bottom of the centrifuge tube each time, the cell suspension was passed with the medium through a 70-μm cell filter, all of which was repeated 3–5 times. The filtered cell suspension was then centrifuged at 4 °C, 500g for 5 min, and the pellet was resuspended in 3 ml PBS(2 % antibiotics) for cleaning. Next, the cells were centrifuged again (4 °C, 500g, 5 min), and 200 μl serum-free Advanced DMEM/F12 medium was used to resuspend them. Even though the red blood cells died after 4–5 days, a large number of red blood cells and subsequent dead red blood cell fragments could still have potentially affected the initial culture environment of the organoid. We therefore used red blood cell lysis buffer if there was any red precipitation. Matrigel (400ul) was added to the 200 μl suspension to establish the PDOs, and the resulting cell(3^104/80ul/drop) suspension was allowed to solidify in pre-warmed 24-well culture plates at 37 °C for 20 min. After gelation, 750 μl M26 (Table S1) complete medium was added to the wells. We also added Nutlin-3 [29,30], which can enrich tumor organoids, on the basis of the patient tissue [26]. New M26 medium was used that was subsequently changed every 3–5 days, and the organoids were passaged after 1–3 weeks.
For passaging, the original medium was aspirated, and each well received 1 ml TrypLE Express (Gibco) using a pipettor to resuspend the coagulated Matrigel. The samples were then incubated for 10–15 min at 37 °C for dissociation, and the organoid state was observed under a microscope every 5 min and repeatedly blown with a pipettor until the samples dissociated into single cells. Then, the organoids were centrifuged at 500g for 5 min, washed once with cold PBS and centrifuged again. The pellets were washed with cold PBS and centrifuged at 500g for 5 min as well before being resuspended in serum-free Advanced DMED/F12 medium. Matrigel (1:2) was inoculated into the new 24-well plate in a 1:3 ratio for passaging.
For freezing, the original medium was aspirated and combined with 1 ml cell freezing solution for each well. Using a pipettor to resuspend the organoids, the organoids was transferred to a 2 ml cryotube and stored in a gradient cooling box at −80 °C. The next day, the cryotube was stored in a gaseous nitrogen tank.
2.4. Organoid drug screening
Organoids were cultured in 24-well plates over 2–3 weeks and then were harvested and dissociated using TrypLE Express after passaging and resuspended in M26 medium: Matrigel (1:3 ratio). They were then seeded into 96-well (2 × 103 cells per well) plates. After gelation, 100 μl M26 medium was added to each well, and drugs (all drugs were sourced from Selleckchem, TX, USA) were dispensed 24 h after plating. After 3 days of drug incubation, cell viability was assayed using Cell Titer Glo 3D (Promega) according to the manufacturer's instructions. The samples were then incubated room temperature for 25 min immediately prior to luminescence reading. The determination of IC50 values was conducted using Graph Pad version 6.
2.5. Histology and immunostaining
For IHC staining, 4-mm paraffin-embedded tumor tissue and PDO sections were deparaffinized and rehydrated. Slices were then incubated overnight with primary antibody at 4 °C, and subsequently incubated with secondary antibodies. Stained slides were visualized using a Nikon microscope.
2.6. Immunofluorescence
Harvested cultured PDOs were fixed in 4 % paraformaldehyde for 15 min, and the fixed cells were blocked and permeabilized with 2 % BSA, 0.1 % Triton X-100 in TBS. Immunostaining was then performed using PanCK, P63, MUC1, CC10, KRT7, KRT5, ARL13B, Epcam in an Ac-Tub overnight at 4 °C, followed by incubation with Alexa Fluor-488 or-Cy3 secondary antibodies for 2 h at 25 °C. The samples were counterstained with DAPI for 8–10 min by nuclei, and imaging was performed on a digital scanner for tissue sections (Pannoramic MIDI, 3DHISTECH).
2.7. Western blotting
For western blotting Matrigel was dissociated with TrypLE Express to obtain organoid suspensions. Briefly, cells were lysed in RIPA buffer containing protease inhibitors, and protein concentration was measured by BCA Protein Assay Kit (Pierce Biotechnology, USA). The protein was then denatured at high temperature and transferred to a nitrocellulose membrane (Bio-rad) using SDS-PEG electrophoresis. After blocking in 5 % skim milk for 1 h, membranes were incubated with primary antibodies (1:1000) overnight at 4 °C and secondary antibody (1:5000) for 1h at room temperature (Table S2). Finally, protein bands were visualized by ECL imaging (GE Healthcare, Chicago, IL, USA).
2.8. Mini-PDX model
Mini-PDX was performed as previously reported [31,32]. 6-8 week-old immunodeficient BALB/c-Nude mice (BALB/cNj-Foxn1nu/Gpt) were purchased from GemPharmatech (JiangsuProvince), and fed in the Animal Center of Xi'an Jiaotong University under specified pathogen-free (SPF) conditions. All animal experiments were approved by the Institutional Animal Care and Use Committee Of Xi'an Jiaotong University.
The fifth passage organoids were digested with TrypLE digested solution for 15 min to generate single cells, and the organoid suspension was transferred into PBS-washed capsules and transplanted into the BALB/c nude mice. Each mouse received 2 capsules. Drug sensitivity was examined using the OncoVee™-MiniPDX assay (LIDE Biotech, Shanghai, China). Gemcitabine (50 mg/kg, IP, biw) and Afatinib (20 mg/kg, po, Daily) were administered for 7 days each, and normal saline was used as the control During this period. Relative tumor growth was detected using a CellTiter-Glo® Luminescent Cell Viability Assay (Promega, Madison, WI, USA).
2.9. Targeted genomic sequencing
Genomic DNA was extracted following the manufacturer's instructions (QIAGEN, Valencia, CA, USA). For sequencing, 1 μg of fragmented genomic DNA underwent end-repairing, A-tailing and ligation with indexed adapters sequentially, followed by size selection using Agencourt AMPure XP beads (Beckman Coulter). Hybridization-based target enrichment was then carried out with a GeneseeqOne™ pancancer gene panel (425-cancer-relevant genes, Geneseeq Technology Inc.), and xGen Lockdown Hybridization and Wash Reagents Kit (Integrated DNA Technologies). Captured libraries by Dynabeads M − 270 (Life Technologies) were amplified in KAPA HiFi HotStart ReadyMix (KAPA Biosystems) and quantified by qPCR using the KAPA Library Quantification kit (KAPA Biosystems), and target enriched libraries were sequenced on the HiSeq 4000 platform (Illumina) with 2 × 150 bp pair-end reads. Next, sequencing data were demultiplexed by bcl2 fastq (v2.19), analyzed by Trimmomatic [33] to remove low-quality (quality<15) or N bases, and mapped to the reference hg19 genome (Human Genome version 19) using a Burrows-Wheeler Aligner [34]. The Genome Analysis Toolkit (GATK) [35] was used to perform local realignments around indels and base quality reassurance. SNPs and indels were called by VarScan2 [36] and HaplotypeCaller/UnifiedGenotyper in GATK, with the mutant allele frequency (MAF) cutoff set to 0.5 % for tissue samples and a minimum of three unique mutant reads.
Common variants were removed using dbSNP and the 1000 Genomes project. Germline mutations were screened by whole blood controls with patients, and the resulting somatic variants were further filtered through an in-house list of recurrent sequencing errors that was generated from over 10,000 normal control samples on the same sequencing platform. Gene fusions were identified by FACTERA [37], and copy number variations (CNVs) were analyzed with ADTEx [38]. The log2 ratio cut-off for copy number gain was defined as 2.0 for tissue samples. A log2 ratio cut-off of 0.67 was used for copy number loss detection in all sample types. The thresholds were determined from previous assay validation using the absolute CNVs detected by droplet digital PCR (ddPCR). Next, allele-specific CNVs were analyzed by FACETS [39] with a 0.2 drift cut-off for unstable joint segments. Finally, the proportion of chromosomal instability (CIN) was calculated by dividing the size of drifted segments by the total segment size.
2.10. Statistical analysis
All data were expressed as means ± SEM prior to being analyzed by two-tailed paired Student's t-test, unless otherwise indicated. The threshold for statistically significant results was set to p < 0.05. All statistical analysis was carried out in GraphPad Prism 6, and gene mutation patterns were visualized using GenVisR in R (version = 4.3.0). SynergyFinder—Bliss synergy scores were calculated using the SynergyFinder (version 1.6.1) R (version 3.5.1) package as described in “Proliferation Assays” [40].
3. Results
3.1. Establishment of PDOs from different subtypes of lung cancer
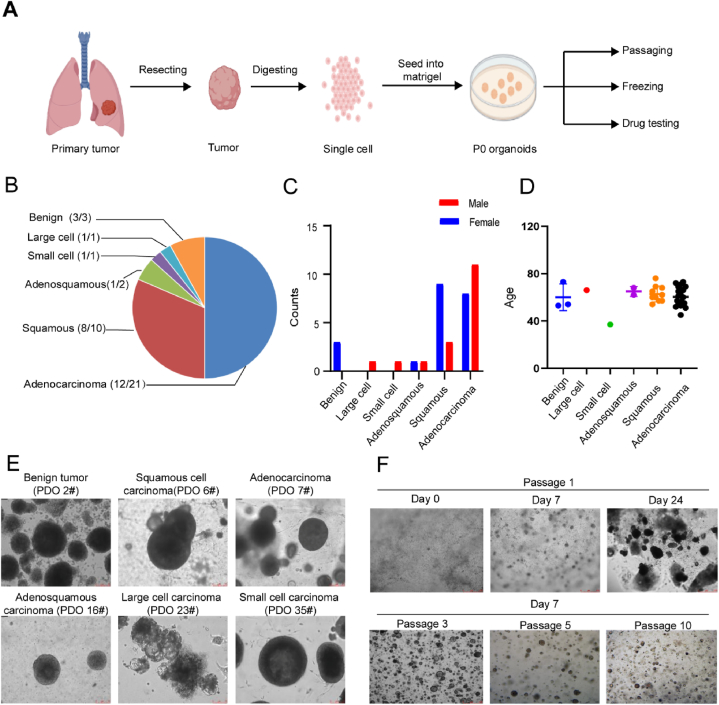
To establish PDOs from the various types of lung cancers, resected tumor tissues were transported directly from the operating room to the laboratory for processing, where they were divided into <2 mm pieces and treated with collagenase to generate single cells or cell clusters. Afterwards, the cells were seeded in Matrigel and cultured in M26 medium (Fig. 1A). Separation from normal organoid was achieved by manual separation and in the case of TP53 mutations by the addition of 5 μM Nutlin-3a in the culture medium. Organoids that were successfully maintained for 5 passages were considered to be successfully established. Using this protocol, we successfully established 26 human lung cancer-derived organoids from 38 patients with an average success rate of 66 % (Fig. 1B, Supplementary Fig. 3, and Table 1). The age and gender distribution of the patients are shown in Fig. 1C–D. Our established lung cancer organoid lines consisted of six subtypes including benign tumor, adenocarcinoma, squamous cell carcinoma, adenosquamous carcinoma, large-cell carcinoma, and small-cell carcinoma (Fig. 1B and E). The organoids were usually formed within 1 week and were passaged in a 1:2 or 1:3 ratio after 2–3 weeks (Fig. 1D). Additionally, as shown in Fig. 1E, long-term expansion (>3 months, >10 passages) had a minimal effect on the spherical morphology of the organoids. These findings suggested that we successfully generated PDOs that could be stably passaged for subsequent applications.
Fig. 1.
Establishment of PDOs from different subtypes of lung cancer A. Diagram of the process of establishing PDOs from patient tumors for the subsequent passaging, freezing and drug testing; B. Pie chart showing the subtypes of established 38 PDOs for the lung cancer, also see Table 1. C. The ratio of male to female patients in each subtype D. The scatter plot shows the distribution of patient age among lung cancer subtypes. E. Representive bright-field microscopy images of six subtypes PDOs cultured for day 24. Scale bar, 75 μm. F. Representative bright-field pictures showing growth status of PDO at passage 1, days 0, 7 and 24, the image was obtained from PDO-3; And, representative bright-field pictures showing growth status of PDO at passage 3, 5 and 10; the image was obtained from PDO-3 on day 7 after passage.
Table 1.
PDOs established in this study.
| ID | Sex | Age | Lung cancer type | Degree of tissue differentiation | Success /Failure |
|---|---|---|---|---|---|
| PDO-1 | F | 69 | Adenocarcinoma | S | |
| PDO-2 | F | 53 | Benign tumor | S | |
| PDO-3 | F | 54 | Squamous cell carcinoma | Poor | S |
| PDO-4 | M | 53 | Adenocarcinoma | F | |
| PDO-5 | F | 69 | Squamous cell carcinoma | Poor | S |
| PDO-6 | F | 62 | Squamous cell carcinoma | Moderate | S |
| PDO-7 | F | 54 | Adenocarcinoma | S | |
| PDO-8 | F | 61 | Adenocarcinoma | Poor | S |
| PDO-9 | M | 62 | Adenocarcinoma | F | |
| PDO-10 | M | 66 | Adenocarcinoma | F | |
| PDO-11 | M | 56 | Adenocarcinoma | Well | S |
| PDO-12 | M | 52 | Adenocarcinoma | Moderate | F |
| PDO-13 | F | 68 | Squamous cell carcinoma | Moderate | S |
| PDO-14 | F | 76 | Squamous cell carcinoma | Moderate | F |
| PDO-15 | F | 57 | Squamous cell carcinoma | Moderate | S |
| PDO-16 | M | 68 | Adenosquamous | Moderate | S |
| PDO-17 | M | 60 | Adenocarcinoma | S | |
| PDO-18 | M | 60 | Squamous cell carcinoma | F | |
| PDO-19 | F | 54 | Benign tumor | S | |
| PDO-20 | M | 57 | Adenocarcinoma | S | |
| PDO-21 | F | 60 | Squamous cell carcinoma | Moderate | S |
| PDO-22 | M | 69 | Adenocarcinoma | S | |
| PDO-23 | M | 66 | Large cell carcinoma | S | |
| PDO-24 | F | 55 | Adenocarcinoma | Poor | F |
| PDO-25 | F | 45 | Adenocarcinoma | Poor | S |
| PDO-26 | F | 72 | Adenocarcinoma | Moderate | F |
| PDO-27 | F | 65 | Squamous cell carcinoma | Moderate | S |
| PDO-28 | M | 73 | Adenocarcinoma | F | |
| PDO-29 | F | 62 | Adenosquamous | F | |
| PDO-30 | F | 65 | Adenocarcinoma | S | |
| PDO-31 | M | 56 | Adenocarcinoma | S | |
| PDO-32 | F | 73 | Benign tumor | S | |
| PDO-33 | F | 66 | Adenocarcinoma | S | |
| PDO-34 | M | 71 | Adenocarcinoma | Poor | F |
| PDO-35 | M | 37 | Small cell carcinoma | Moderate | S |
| PDO-36 | F | 57 | Squamous cell carcinoma | Poor | S |
| PDO-37 | M | 51 | Adenocarcinoma | Well | S |
| PDO-38 | M | 54 | Adenocarcinoma | F |
3.2. PDO retention of histological and genetic characteristics of their original tumors
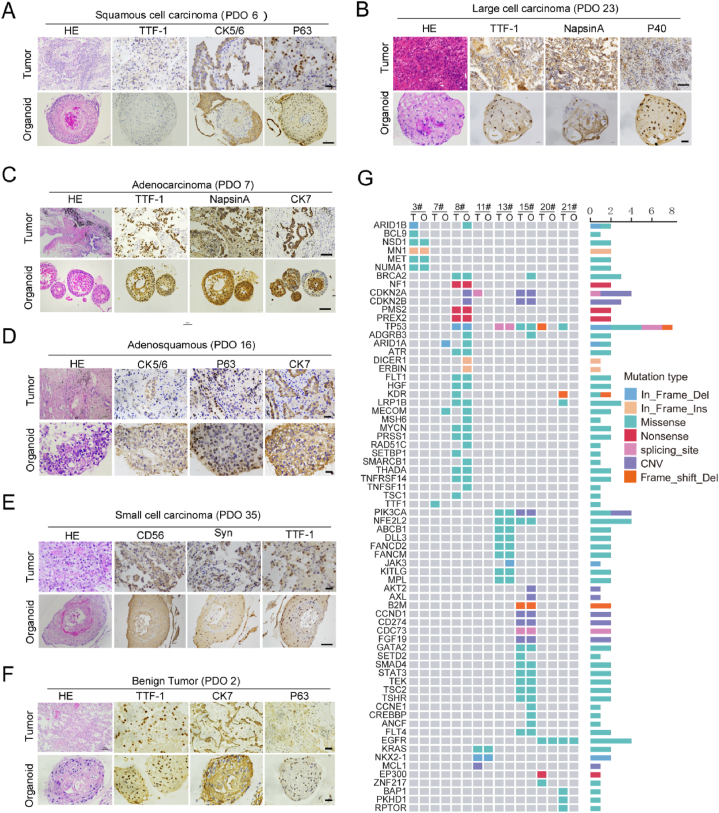
To test whether PDOs retained characteristics of their original tumors, we compared the tissue pathology of the tumors and PDOs using immunohistochemistry. Based on HE staining, there was a high similarity in cellular and nuclear atypia, indicating that organoids retained the malignant characteristics of their original tumors (Fig. 2A–F). Next, we analyzed the expression of lung cancer biomarkers such as Thyroid Transcription Factor 1 (TTF-1), Cytokeratin 5/6/7 (CK5/6/7), Napsin-A, CD56, Synaptophysin and P63, and found that these biomarkers were expressed consistently in five subtypes of malignant cancers and paired organoids (Fig. 2A–E). Interestingly, in organoids derived from benign tumors, CK7 and P63 expression seemed to have been activated (Fig. 2F).
Fig. 2.
PDO retention of histological and genetic characteristics of their original tumors. A-F. Representative image of HE staining and IHC staining of six subtypes of lung cancers and paired PDOs using indicated antibodies. G. Targeted genomic sequencing of 425 cancer-related genes was performed using 8 paired primary tumor and organiods, and 70 gene mutations were shown.
Gene mutation in cancer may guide the clinical treatment, so to determine whether genetic mutations were preserved in the organoids, genomic profiling was performed on 8 surgical samples and paired organoids using targeted next-generation sequencing of 425 cancer-related genes. In total, 70 mutant genes were detected, including some common driver gene mutations in lung cancer, such as TP53 and EGFR [41], among which the TP53 mutation occurred with the highest frequency (Fig. 2G). We also compared the gene mutations in original cancer tissues and paired organoids, as shown in Fig. 2G, and as expected there was some heterogeneity in the gene mutations. However, most of the somatic mutations in the original tissues were maintained in the PDOs. These results showed that the cultured PDOs in vitro retained both histological and genetic characteristics of their parental tumors.
3.3. Characterization of organoids
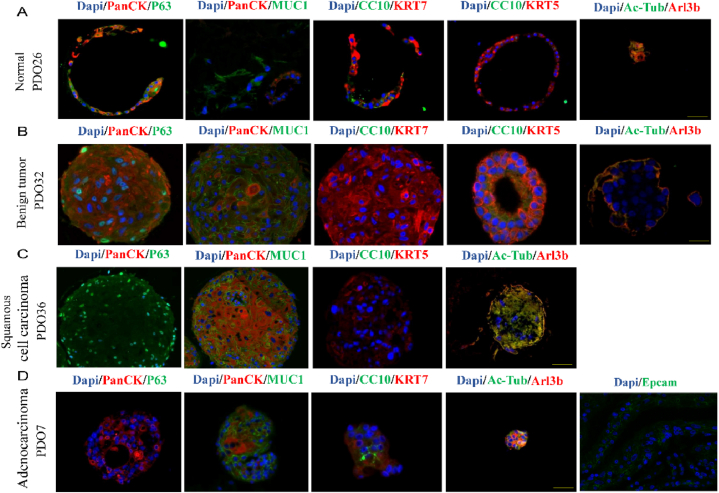
To identify the cell types in the PDOs, we performed immunofluorescence staining and observed normal and benign organoids with lung epithelial cells, basal cells, goblet cells (MUC1), club cell, (CC10), cilia (Arl13b), and epithelial cell (Epcam) (Fig. 3A and B). P63-expressing basal cells were found only in lung squamous cell carcinoma organoids (Fig. 3C), as adenocarcinoma organoids consisting of PanCK-positive cells with ciliated cells (Fig. 3D). Additionally, Epcam showed positive expression in lung adenocarcinoma organoids (Fig. 3D).
Fig. 3.
Identification of PDOs cell types. A-D. Immunofluorescence images showing the expression of PanCK, p63, MUC1, KRT5, KRT7, CC10, Ac-Tub, Arl13b, Epcam. The nuclei were stained with DAPI(Blue). Scale bar, 50 μm. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.4. Drug response of organoid lines
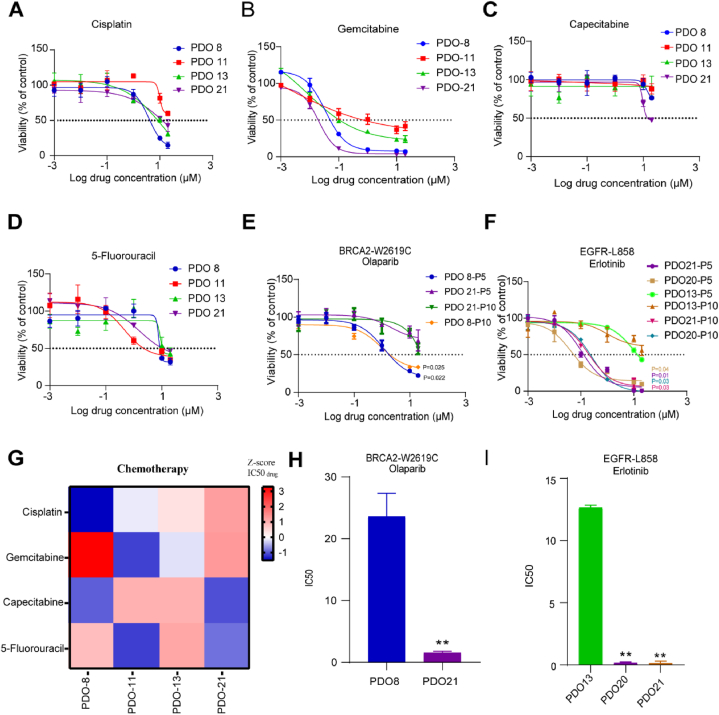
To explore the potential of the organoid lines to be used as preclinical models for evaluation of drug responses, we treated the organoid lines with 4 first-line chemotherapy drugs including cisplatin, gemcitabine, capecitabine, and 5-Fu. Each organoid line was tested between passages 5 and 10, and the organoids were found to be stable for passage and sufficient for drug screening. Similar to the drug responses of tumor patients, the organoid lines showed a marked difference in drug sensitivity (Fig. 4A–D). We found several relative IC50 values, indicating the presence of differentially susceptible organoids (Fig. 4G).
Fig. 4.
Drug response of organoid lines A-D. The PDO lines were seeded in 96 well plate, and treated with different doses of Cisplatin (A), Gemcitabine (B), Capecitabine (C) and 5-Fluorouracil (D) for 72 h, the PDO viability was measured by CELL TITER-GLO (CTG); E-F. The PDO lines (Passage5, Passage10) were treated with various doses of Olaparib (E) and Erlotinib (F) for 72 h, the PDO viability was measured using CTG. G. Heat-map of IC50 values were the four chemotherapeutic drugs treated in the four organoid lines. Where organoids showed multiple relative IC50 values. As expected, organoids varied significantly in their sensitivity to chemotherapy drugs. H-I. IC50 values are the average ± SD of each condition analyzed in triplicate. Data analysis using two-tailed paired student's t-test. Error bars represent SEM of two to three independent experiments. ∗∗p < 0.05.
Further, we tested the corresponding targeted drugs with 6 organoids based on the sequencing data. Breast-cancer susceptibility gene 2 (BRCA2) is an important tumor suppressor, and its mutation can cause deficiencies of homologous recombination repair and sensitivity to the PARP inhibitor Olaparib [42,43]. As expected, organoids with BRCA2 mutation (PDO8-P5) were more sensitive to Olaparib compared to BRCA2 wildtype organoids (PDO13-P5) (Fig. 4E). EGFRL858R is a classic activating mutation in lung cancer and is associated with sensitivity to EGFR tyrosine kinase inhibitors (TKIs) [44]. Consistently, PDOs (PDO20-P5 and PDO21-P5) that harbored an EGFRL858R mutation were more sensitive to the EGFR inhibitor erlotinib (Fig. 4F). In addition, we performed the same validation on the ninth passage organoids (Fig. 4F) and counted the IC50 values (Fig. 4H and I). These results suggest that PDOs such as those in our experiment can be used to predict patient-specific drug responses, as well as gene-based targeted drug responses.
3.5. The combination of afatinib and gemcitabine induced synthetic lethality in METN375S mutant lung cancer
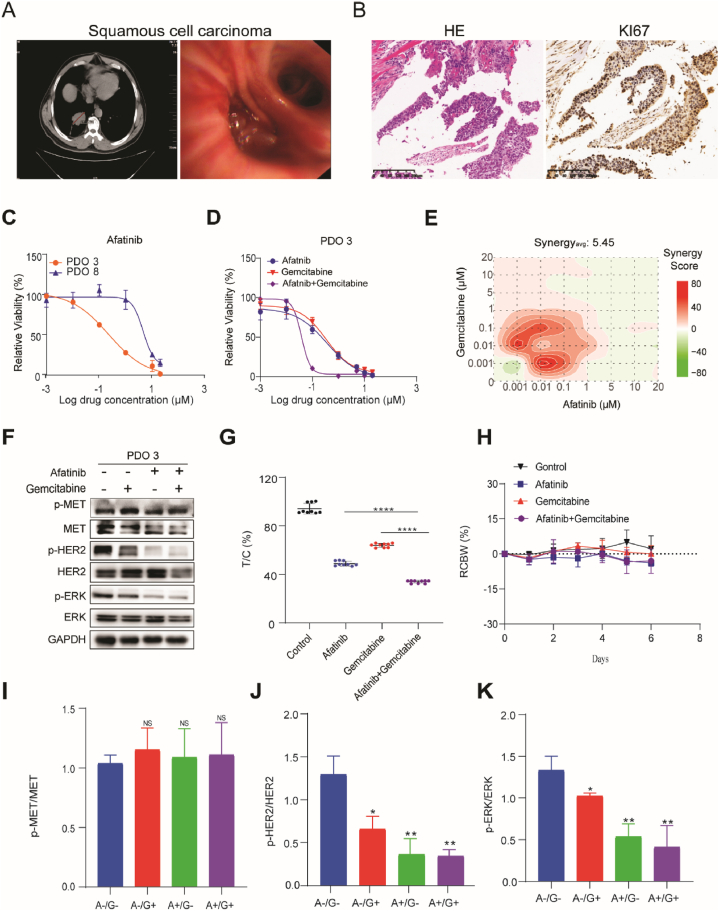
To explore how PDOs can be used for predicting drug response even further, we selected a 54-year-old man with a highly aggressive squamous cell carcinoma of the lung (PT-3) as a research subject. CT scan showed that the tumor size was 46.5 mm, and the tumor mass was visible under bronchoscopy (Fig. 5A). HE and Ki67 staining also confirmed that it was highly malignant (Fig. 5B). Based on next-generation sequencing, we detected METN375Smutation in the primary tumor as well as the PDO (PDO-3). Oncogenic MET has been recognized as a promising target in cancer therapy [45], and interestingly METN375S, the most frequent mutation of MET, has recently been reported to suggest a more aggressive oncogenic phenotype in lung cancer [46]. However, exactly which pathways may represent a potential therapeutic weakness in METN375S mutant lung cancers remain unknown.
Fig. 5.
The combination of afatinib and gemcitabine induced synthetic lethality in METN375S mutant lung cancer A. left: CT scan image showing the size of the patient's originating tumor (red line), right: the bronchial placeholder image of the tumor under thoracoscopy; B. Representative staining of HE and Ki67 in primary tissues (patient-3); C. PDO-3 (MET N375S) and PDO-8 (MET WT) were treated with various concentration of Afatinib for 72 h, and the PDOs viability were detected using CTG; D. PDO-3 was treated with a single drug or combination of Afatinib and Gemcitibabine for 72 h, the PDOs viability were detected using CTG; E. Representative bliss synergy score heatmap for three independent experiments is shown. Red, synergy; green, antagonism; white, no effect. Synergyavg = average bliss synergy score. F. PDO-3 was treated with Afatinib (1 μM) and/or gemcitabine (1 μM) for 72 h, indicated proteins expression was analyzed using immunoblot analysis, GAPDH was used as loading control. Full-length blot is shown in the supplementary information, and the gels have been run under the same experimental conditions. G. Cytotoxicity analysis of single drug or combination of Afatinib and Gemcitibabine in mini-PDX model, relative tumor organoid growth was shown; H. The body weight of mice used in G during drug treatment. I-K. Expression of p-MET, p-HER2 and P-ERK were quantified and expressed as mean of total MET, HER2 and ERK ± SD (n = 3). Two-tailed Student's t-test; ∗P < 0.05. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
We treated the PDOs with several small molecule inhibitors including Savolitinib, JNJ-388877618, Gefitinib, Crizotinib, Trametinib, and Afatinib, which are widely used in basic and clinical research of lung cancer. Interestingly, PDO-3(METN375S) showed no significant difference in MET inhibitor sensitivity compared to PDO-21(METWT) (Supplementary Figs. 1A and 1B) but did show a specifically increased sensitivity to Afatinib, an irreversible ErbB family blocker that is a specific and potent inhibitor of HER2, EGFR, and HER4 and blocks signaling from all homo- and heterodimers formed by members of ErbB family [47,48] (Fig. 5C and Supplementary Figs. 1C–1E). Since the use of targeted drugs is prone to drug resistance, we also introduced two first-line lung cancer chemotherapy drugs, cisplatin and gemcitabine, combined with Afatinib. We found that the combination of afatinib and gemcitabine, but not cisplatin, induced a significant synthetic lethality in PDO-3 (Fig. 5D Supplementary Fig. 2A). Furthermore, bliss analysis was performed to determine whether the inhibitory activities of each combination was synergistic, where bliss scores greater than 1.0 indicate synergy [49]. The combination of Afatinib and gemcitabine (synergyavg = 5.45) but not cisplatin (synergyavg = −11.03) mechanically reduced the phosphorylation level of HER2 and ERK (Fig. 5E, F, 5I-5K). In order to demonstrate the combinational effect in vivo, we used the MINI-PDX model to evaluate the efficacy of afatinib both alone and in combination with gemcitabine. Drug-sensitivity analysis (as T/C%) revealed that the combination was significantly better than the single drug but did not affect the mice's bodyweight (Fig. 5G and H). Thus, these results indicate that the combination of afatinib and gemcitabine may be a promising therapeutic strategy for METN375S mutant lung cancers.
4. Discussion
As a highly heterogeneous disease, there is an urgent need for personalized medicine in lung cancer treatment, and several studies have already shown that PDOs are the ideal research model for effective drug screening of lung cancer [16,23,24,50]. In this study, we successfully established 26 PDO lines from 38 primary lung cancers including 5 common tumor subtypes and benign tumors, with an overall success rate of 66 %, and confirmed the consistency of histological and genomic characteristics between primary tumors and paired PDOs. CK7 is expressed in glandular epithelial cells, and P63 is a myoepithelial cell marker [17]. Both of these cells are developed from progenitor cells. Therefore, we suspect that the detection of CK7 and P63 activation is a signal for the transition from benign to malignant tumor. However, this speculation needs a great deal of empirical evidence to support it.
In the era of individualized cancer drugs, preclinical drug reuse trials and drug combination trials are becoming more and more important [51]. The results of this paper show that PDOs can be used for anticancer drug screening or in vitro testing to predict drug response in individual patients. Just like their primary lung cancer, the PDOs in this study showed significant heterogeneity in response to chemotherapy and targeted therapy drugs, implying that PDOs retain the specific drug sensitivity of their primary cancers. These findings, along with those of previous studies, strongly indicate that PDOs are the most realistic platform for specific drug screening in personalized cancer treatment. Genetic alterations in lung cancer are extraordinarily diverse [52]. Gene sequencing is necessary to guide drug therapy. For example, organoids PDO-20 and PDO-21 containing the EGFR-L858 mutation show increased sensitivity relative to wild-type PDO-13 and also show the same effect across passages.
METN375S, the most frequent mutation of MET, occurs in 13 % of East Asians compared to 0 % of African Americans [50], and overexpression of this mutant in COS-7, an SV40 transformed African green monkey kidney cell line, has been reported to induce resistance to MET inhibitor cytotoxicity [50]. However, the potential role of METN375S mutant in cancer remains largely unknown, although it causes conformational changes at the ligand-binding site [53]. In this study, in order to examine whether the MET-driven signal of the N375S variant is a predictive biomarker for therapeutic intervention, the viability of isocells after organoid lysis was measured after treatment with known MET inhibitors Trametinib, JNJ-38877618, Savolitinib, Gefitinb, and crizotinib. We found that METN375S organoids did not exhibit differential half-maximal inhibitory concentration (IC50) values compared to METwt organoids, but organoids showed better sensitivity to afatinib. Therefore, we determined that lung cancer that harbors METN375S mutant is more sensitive to the tyrosine kinase inhibitor (TKI) afatinib (an inhibitor of HER2 and EGFR) in the PDOs and PDO-derived mini-PDX model. Moreover, a combination of afatinib and gemcitabine induced synthetic lethality in METN375S mutant lung cancer. Our findings thus strongly indicate that phosphorylated METN375S leads to constitutively active HER2 that mediates signaling regardless of MET inhibition. Consistent with our study, METN375S mutant squamous cell carcinoma cell lines have recently been reported to have a unique affinity for HER2 blockade therapy [46].
In conclusion, we successfully established and characterized PDOs from several different lung cancers, and revealed a novel potential therapeutic strategy for METN375S mutant lung cancers using these PDOs.
5. Limitations and future work
A key limitation of organoid models is the lack of a cancer microenvironment including stromal and immune cells. The spatial structure of cancer organoids is made up of different types of epithelial cells, and thus in order to study the interaction between cancer cells and the microenvironment we intend to use a co-culture system of tumor-infiltrating lymphocytes and PDOs to study a combination of immune and targeted therapy.
Ethics declarations
The study complies with all ethics regulations.
-
•
This study were approved by the Ethical Examination and approval Committee of Xi ‘an Jiaotong University (No.2021-334), and Animal experiments were approved by the Institutional Animal Care and Use Committee(XJTUAE2023-1581).
-
•
All participants provided written informed consent to participate in the study
Funding
This work was supported by grants from the Innovation Capability Support Program of Shaanxi (No.2021 PT-050).
Data availability
Data will be made available on request.
CRediT authorship contribution statement
Meng Jiang: Writing – original draft, Conceptualization. Rongfu Tu: Writing – original draft. Yiwen Pan: Validation. Yuxin Cui: Data curation. Xin Qi: Software. Hongyu Qin: Validation. Lijuan Liu: Investigation. Xiaorui Wang: Formal analysis. Ying Xue: Funding acquisition. Yao Xu: Project administration. Ziyang Peng: Resources. Chengsheng Zhang: Visualization, Methodology. Jin Yang: Writing – review & editing, Project administration.
Declaration of competing interest
We declare that we have no conflicts of interest.
Acknowledgments
The authors thank AiMi Academic Services (www.aimieditor.com) for English language editing and review services.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2024.e37884.
Contributor Information
Chengsheng Zhang, Email: cszhang99@126.com.
Jin Yang, Email: yangjin@xjtu.edu.cn.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Siegel R.L., et al. Cancer statistics. CA A Cancer J. Clin. 2021;71(1) doi: 10.3322/caac.21654. 2021. [DOI] [PubMed] [Google Scholar]
- 2.Zito Marino F., et al. Molecular heterogeneity in lung cancer: from mechanisms of origin to clinical implications. Int. J. Med. Sci. 2019;16(7):981–989. doi: 10.7150/ijms.34739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Leitao, et al. Pathobiology Journal of Immunopathology Molecular & Cellular Biology; 2018. Heterogeneity in Lung Cancer. [Google Scholar]
- 4.Bedard P., et al. Small molecules, big impact: 20 years of targeted therapy in oncology. Lancet (London, England) 2020;395(10229):1078–1088. doi: 10.1016/S0140-6736(20)30164-1. [DOI] [PubMed] [Google Scholar]
- 5.Patient-derived organoids model treatment response of metastatic gastrointestinal cancers. Science. 2018;359(6378):920–926. doi: 10.1126/science.aao2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Driehuis E., Kretzschmar K., Clevers H. Establishment of patient-derived cancer organoids for drug-screening applications (vol 63, pg 891, 2020) Nature protocols erecipes for researchers. 2021;(12):16. doi: 10.1038/s41596-021-00494-5. [DOI] [PubMed] [Google Scholar]
- 7.Ebisudani T., et al. Genotype-phenotype mapping of a patient-derived lung cancer organoid biobank identifies NKX2-1-defined Wnt dependency in lung adenocarcinoma. Cell Rep. 2023;42(3) doi: 10.1016/j.celrep.2023.112212. [DOI] [PubMed] [Google Scholar]
- 8.Li Y.F., et al. Patient-derived organoids of non-small cells lung cancer and their application for drug screening. Neoplasma. 2020;67(2) doi: 10.4149/neo_2020_190417N346. [DOI] [PubMed] [Google Scholar]
- 9.Pauli C., et al. Personalized in vitro and in vivo cancer models to guide precision medicine. Cancer Discov. 2017;7(5):462–477. doi: 10.1158/2159-8290.CD-16-1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fujii M., et al. A colorectal tumor organoid library demonstrates progressive loss of niche factor requirements during tumorigenesis. Cell Stem Cell. 2016;18(6):827–838. doi: 10.1016/j.stem.2016.04.003. [DOI] [PubMed] [Google Scholar]
- 11.Sato T., et al. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett's epithelium. Gastroenterology. 2011;141(5):1762–1772. doi: 10.1053/j.gastro.2011.07.050. [DOI] [PubMed] [Google Scholar]
- 12.Sachs N., et al. A living biobank of breast cancer organoids captures disease heterogeneity. Cell. 2018;172(1–2):373–386.e10. doi: 10.1016/j.cell.2017.11.010. [DOI] [PubMed] [Google Scholar]
- 13.Lee S.H., et al. Tumor evolution and drug response in patient-derived organoid models of bladder cancer. Cell. 2018;173(2):515–528.e17. doi: 10.1016/j.cell.2018.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hu Y., et al. Lung cancer organoids analyzed on microwell arrays predict drug responses of patients within a week. Nat. Commun. 2021;12(1):2581. doi: 10.1038/s41467-021-22676-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koga T., et al. Clinical relevance of patient-derived organoid of surgically resected lung cancer as an in vitro model for biomarker and drug testing. JTO Clin Res Rep. 2023;4(9) doi: 10.1016/j.jtocrr.2023.100554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pan Y., Cui H., Song Y. Organoid drug screening report for a non-small cell lung cancer patient with EGFR gene mutation negativity: a case report and review of the literature. Front. Oncol. 2023;13 doi: 10.3389/fonc.2023.1109274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Choi J., Iich E., Lee J.H. Organogenesis of adult lung in a dish: differentiation, disease and therapy. Dev. Biol. 2016:278. doi: 10.1016/j.ydbio.2016.10.002. [DOI] [PubMed] [Google Scholar]
- 18.Deuse T., Schrepfer S. Distal airway stem cells are essential for lung regeneration. Transplantation. 2015;99(8):1540–1541. doi: 10.1097/TP.0000000000000794. [DOI] [PubMed] [Google Scholar]
- 19.A three-dimensional model of differentiation of immortalized human bronchial epithelial cells. Differentiation. 2006;74(4):141–148. doi: 10.1111/j.1432-0436.2006.00069.x. [DOI] [PubMed] [Google Scholar]
- 20.Wong, A.P., et al., Directed differentiation of human pluripotent stem cells into mature airway epithelia expressing functional CFTR protein. Nat. Biotechnol. [DOI] [PMC free article] [PubMed]
- 21.Wang H.M., et al. Using patient-derived organoids to predict locally advanced or metastatic lung cancer tumor response: a real-world study. Cell Rep Med. 2023;4(2) doi: 10.1016/j.xcrm.2022.100911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang J., Li X., Chen H. Organoid models in lung regeneration and cancer. Cancer Lett. 2020;475:129–135. doi: 10.1016/j.canlet.2020.01.030. [DOI] [PubMed] [Google Scholar]
- 23.Kim S.Y., et al. Modeling clinical responses to targeted therapies by patient-derived organoids of advanced lung adenocarcinoma. Clin. Cancer Res. 2021;27(15):4397–4409. doi: 10.1158/1078-0432.CCR-20-5026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sen C., et al. Development of a small cell lung cancer organoid model to study cellular interactions and survival after chemotherapy. Front. Pharmacol. 2023;14 doi: 10.3389/fphar.2023.1211026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dijkstra K.K., et al. Challenges in establishing pure lung cancer organoids limit their utility for personalized medicine. Cell Rep. 2020;31(5) doi: 10.1016/j.celrep.2020.107588. [DOI] [PubMed] [Google Scholar]
- 26.Kim M., et al. Patient-derived lung cancer organoids as in vitro cancer models for therapeutic screening. Nat. Commun. 2019;10(1):3991. doi: 10.1038/s41467-019-11867-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shi R., et al. Organoid cultures as preclinical models of non-small cell lung cancer. Clin. Cancer Res. 2020;26(5):1162–1174. doi: 10.1158/1078-0432.CCR-19-1376. [DOI] [PubMed] [Google Scholar]
- 28.Li Z., et al. Protocol for generation of lung adenocarcinoma organoids from clinical samples. STAR Protoc. 2021;2(1) doi: 10.1016/j.xpro.2020.100239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sachs N., et al. Long-term expanding human airway organoids for disease modeling. EMBO J. 2019;38(4) doi: 10.15252/embj.2018100300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yan H.H.N., et al. A comprehensive human gastric cancer organoid biobank captures tumor subtype heterogeneity and enables therapeutic screening. Cell Stem Cell. 2018;23(6):882–897. doi: 10.1016/j.stem.2018.09.016. [DOI] [PubMed] [Google Scholar]
- 31.Huang Y., et al. A novel, personalized drug-screening system for platinum-resistant ovarian cancer patients: a preliminary clinical report. Cancer Manag. Res. 2021;13:2849–2867. doi: 10.2147/CMAR.S276799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhan M., et al. Guided chemotherapy based on patient-derived mini-xenograft models improves survival of gallbladder carcinoma patients. Cancer Commun. 2018;38(1):48. doi: 10.1186/s40880-018-0318-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bolger A.M., Lohse M., Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.DePristo M.A., et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 2011;43(5):491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Koboldt D.C., et al. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 2012;22(3):568–576. doi: 10.1101/gr.129684.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Newman A.M., et al. FACTERA: a practical method for the discovery of genomic rearrangements at breakpoint resolution. Bioinformatics. 2014;30(23):3390–3393. doi: 10.1093/bioinformatics/btu549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Amarasinghe K.C., Li J., Halgamuge S.K. CoNVEX: copy number variation estimation in exome sequencing data using HMM. BMC Bioinf. 2013;14(Suppl 2):S2. doi: 10.1186/1471-2105-14-S2-S2. Suppl 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shen R., Seshan V.E. FACETS: allele-specific copy number and clonal heterogeneity analysis tool for high-throughput DNA sequencing. Nucleic Acids Res. 2016;44(16):e131. doi: 10.1093/nar/gkw520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ozkan-Dagliyan I., et al. Low-dose vertical inhibition of the RAF-MEK-ERK cascade causes apoptotic death of KRAS mutant cancers. Cell Rep. 2020;31(11) doi: 10.1016/j.celrep.2020.107764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li H., et al. Disparate genomic characteristics of patients with early-stage lung adenocarcinoma manifesting as radiological subsolid or solid lesions. Lung Cancer. 2022;166:178–188. doi: 10.1016/j.lungcan.2022.02.012. [DOI] [PubMed] [Google Scholar]
- 42.Helleday T. The underlying mechanism for the PARP and BRCA synthetic lethality: clearing up the misunderstandings. Mol. Oncol. 2011;5(4):387–393. doi: 10.1016/j.molonc.2011.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dziadkowiec K.N., et al. PARP inhibitors: review of mechanisms of action and BRCA1/2 mutation targeting. Prz Menopauzalny. 2016;15(4):215–219. doi: 10.5114/pm.2016.65667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yasuda H., Kobayashi S., Costa D.B. EGFR exon 20 insertion mutations in non-small-cell lung cancer: preclinical data and clinical implications. Lancet Oncol. 2012;13(1):e23–e31. doi: 10.1016/S1470-2045(11)70129-2. [DOI] [PubMed] [Google Scholar]
- 45.Paik P.K., et al. Response to MET inhibitors in patients with stage IV lung adenocarcinomas harboring MET mutations causing exon 14 skipping. Cancer Discov. 2015;5(8):842–849. doi: 10.1158/2159-8290.CD-14-1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kong L.R., et al. A common MET polymorphism harnesses HER2 signaling to drive aggressive squamous cell carcinoma. Nat. Commun. 2020;11(1):1556. doi: 10.1038/s41467-020-15318-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Solca F., et al. Target binding properties and cellular activity of afatinib (BIBW 2992), an irreversible ErbB family blocker. J. Pharmacol. Exp. Therapeut. 2012;343(2):342–350. doi: 10.1124/jpet.112.197756. [DOI] [PubMed] [Google Scholar]
- 48.Wang J., Li X., Chen H. Organoid models in lung regeneration and cancer. Cancer Lett. 2020;475:129–135. doi: 10.1016/j.canlet.2020.01.030. [DOI] [PubMed] [Google Scholar]
- 49.Ozkan-Dagliyan I., et al. Low-dose vertical inhibition of the RAF-MEK-ERK cascade causes apoptotic death of KRAS mutant cancers. Cell Rep. 2020;31(11) doi: 10.1016/j.celrep.2020.107764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Krishnaswamy S., et al. Ethnic differences and functional analysis of MET mutations in lung cancer. Clin. Cancer Res. 2009;15(18):5714–5723. doi: 10.1158/1078-0432.CCR-09-0070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shamir E.R., Ewald A.J. Three-dimensional organotypic culture: experimental models of mammalian biology and disease. Nat. Rev. Mol. Cell Biol. 2014;15(10):647–664. doi: 10.1038/nrm3873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pikor L.A., et al. Genetic alterations defining NSCLC subtypes and their therapeutic implications. Lung Cancer. 2013;82(2):179–189. doi: 10.1016/j.lungcan.2013.07.025. [DOI] [PubMed] [Google Scholar]
- 53.Yeh Y.M., et al. MET mutation is a potential therapeutic target for advanced endometrial cancer. Cancers. 2021;13(16) doi: 10.3390/cancers13164231. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.