Abstract
The gp120 V3-encoding region of human immunodeficiency virus type 1 (HIV-1) RNA derived from the saliva and blood plasma of 11 individuals was characterized by heteroduplex tracking assay and sequence analyses. R5-like viral variants were identified in both fluids of all subjects. X4-like variants were detected in the plasma and/or saliva of three subjects, indicating that X4-like variants are not excluded from the saliva compartment. Viral subpopulations were similar in both fluids of most subjects, suggesting that HIV-1 in oral fluids and blood may stem from a common source. These findings raise the possibility of using saliva as a noninvasive fluid for evaluating and monitoring viral evolution in infected persons.
Body compartments (e.g., the genital tract, central nervous system, and breast) are reservoirs of human immunodeficiency virus type 1 (HIV-1) infection. These sites serve as potential sources of transmission and rebound virus in individuals whose viral loads (HIV-1 RNA copies per milliliter) in blood plasma have been reduced by potent antiretroviral therapy (ART) (28). Monitoring the HIV-1 burden in viral compartments is important for assessing transmission risk, disease progression, and responses to ART.
Mounting evidence suggests that the oral cavity may be a previously unrecognized viral reservoir. HIV-1 infection can be acquired through the oral cavity during breast-feeding (19, 31), oral-genital sex (26), and direct deposition in macaques (27), despite the low transmission risk associated with saliva (35). Viral loads exceeding those in matched plasma by up to 102 copies/ml in saliva (33, 34), 103 copies/ml in the tonsils (13), and 107 copies/ml in lymphoepithelial parotid cyst aspirates (38) have been recently reported. Discordant viral loads argue for a distinct oral compartment in which the virus may evolve independently from blood or other distal compartments.
The heteroduplex tracking assay (HTA) (7) is a powerful tool for characterizing sequence divergence of viral subpopulations. V3-specific HTA (V3-HTA) rapidly identifies R5-like and X4-like HIV-1 variants in blood plasma (22) and seminal plasma (24) based on sequence variability within the gp120 V3-encoding region of env (30). R5 variants utilize CD4 and the CCR5 chemokine coreceptor for viral entry (1, 6, 8, 9, 10), typically exhibit a non-syncytium-inducing phenotype in the MT-2 assay (2), and predominate early in the disease course (29, 37). X4 variants use the CXCR4 chemokine coreceptor for virus entry (14), are often syncytium inducing (2), and may evolve from R5 variants with disease progression (29, 37). R5X4 variants can use either coreceptor (3). Determinants of the R5/X4 and non-syncytium-inducing/syncytium-inducing phenotypes largely reside in the 35-amino-acid V3 domain of gp120 (21).
We adapted the V3-HTA to characterize the V3-encoding sequences of saliva-derived HIV-1. Results obtained from V3-HTA and genotypic analyses of saliva and blood plasma were compared both within and between infected persons as a first step in investigating the potential of the oral cavity to serve as a discrete viral reservoir.
V3-HTA analysis.
Blood plasma and whole saliva were collected from 11 adults with clade B HIV-1 infection. Subjects underwent an oral examination to detect HIV-associated mucosal lesions (11), and medical records were reviewed for immunological, viral, and ART data (Table 1). HIV-1 RNA in saliva was quantitated by Nuclisens assay (Organon Teknika, Durham, N.C.) (32). Following RNA extraction of patient samples (4), V3-encoding env regions were amplified by reverse transcription-PCR (RT-PCR) as previously described (22). RT-PCR products (from 1 μl of plasma or 8 μl of saliva) were annealed to a radiolabeled probe containing the HIV-1 JR-FL V3 sequence, which is nearly identical to the clade B consensus sequence (18), and heteroduplexes were separated by nondenaturing polyacrylamide gel electrophoresis (22). RT-PCR of salivary RNA is typically less efficient than that of blood plasma (data not shown), and viral loads tend to be lower in saliva than in plasma (33). Therefore, a larger volume of saliva-derived amplification products was used in the probe-annealing step in order to obtain HTA bands of comparable intensities from paired saliva and blood samples. Mobility ratios were calculated for each visualized heteroduplex band and used to infer the presence of X4-like (mobility ratio of <0.91) and R5-like (mobility ratio of ≥0.91) genotypes (22).
TABLE 1.
Characteristics of study participantsa
| Patient | Age (yr) | Gender | Race | Lag timeb | Clinical disease stagec | CD4+ cells/μl | ART | Oral mucosal disease | Log10 HIV-1 RNA copies/ml
|
Hemoglobin content in salivad (μg/dl) | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Plasma | Saliva | ||||||||||
| 3040 | 45 | M | W | 8 | Asymptomatic | 205 | None | None | 5.20 | 4.23 | <10 |
| 3004 | 31 | M | W | 70 | Asymptomatic | 358 | 3TC | None | 4.10 | 3.59 | 10–50 |
| 3090 | 39 | M | W | 0 | Symptomatic | 270 | ZDU | HLP | 4.67 | 3.49 | >200 |
| 1341 | 40 | M | W | 9 | AIDS | 288 | ZDU, 3TC | None | 4.14 | 3.85 | 50–200 |
| 9208 | 31 | F | B | 0 | AIDS | 308 | ZDU, 3TC, IND | Candidiasis, SGD | 4.31 | 3.45 | >200 |
| 3051 | 31 | F | W | 48 | Symptomatic | 400 | None | None | 4.91 | 5.90 | ND |
| 1351 | 27 | M | W | 87 | Asymptomatic | 576 | None | None | 4.78 | 5.88 | >200 |
| 9371 | 35 | M | W | 16 | Asymptomatic | 351 | None | None | 4.99 | 5.76 | 10–50 |
| 3058 | 34 | M | B | 0 | AIDS | 169 | ZDU, 3TC | None | 5.12 | 4.87 | 50–200 |
| 9411 | 50 | M | B | 87 | AIDS | 4 | 3TC, NEV, IND | None | 4.00 | 4.26 | 50–200 |
| 1460 | 29 | M | B | 0 | Asymptomatic | 229 | None | None | 4.44 | 3.83 | >200 |
Abbreviations: M, male; F, female; W, Caucasian (non-Hispanic); B, African-American; 3TC, lamivudine; ZDU, zidovudine; IND, indinavir; NEV, nevirapine; SGD, HIV-associated salivary gland disease; HLP, hairy leukoplakia; ND, not done.
Number of days between plasma and saliva collections.
Based on the Centers for Disease Control's revised classification (5).
A 10-μg/dl hemoglobin level corresponds to approximately 60 red blood cells.
All subjects had R5-like variants in both body fluids (Fig. 1). Eight subjects (3004, 3090, 1341, 1351, 9371, 3058, 9411, and 1460) demonstrated only R5-like variants. Heteroduplex migration patterns in plasma and saliva were similar for these subjects, indicating identical or highly related viral subpopulations within both compartments.
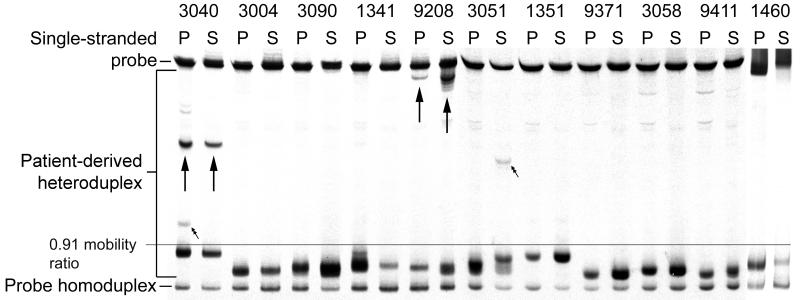
FIG. 1.
V3-HTA analysis of the HIV-1 V3 region in plasma and saliva. Viral RNA was isolated from the plasma (P) and saliva (S) of 11 infected individuals, and the gp120 V3-encoding region was amplified by RT-PCR. Amplification products were annealed to a radiolabeled clade B consensus R5-like sequence probe. Heteroduplexes were separated on a 12% polyacrylamide gel and analyzed by autoradiography. X4-like bands are indicated by arrows. Bands marked by double arrows are unique to a body fluid.
X4-like variants (Fig. 1) were identified in RT-PCR products from subjects 3040, 9208, and 3051. Subject 9208 exhibited similar HTA patterns in plasma and saliva, suggestive of related viral subpopulations in the fluids. Subjects 3040 and 3051 had discordant HTA patterns; bands present in either plasma (3040) or saliva (3051) were not detected in the other fluid sample (Fig. 1), indicating the existence of distinct viral subpopulations in these fluids. To obtain HTA bands of equal intensity, eight times as much of the PCR products derived from saliva samples was loaded onto HTA gels compared to PCR products derived from blood samples. It is possible that the unique band in the blood of subject 3040 was present in the saliva but was not amplified to a detectable level due to fewer viral RNA copies in the saliva than in the blood (4.23 log10 versus 5.20 log10 [Table 1]) or less efficient RT of saliva-derived RNA (data not shown). By the same token, however, the X4-like band in the saliva of subject 3051 was not detected in the blood despite the probable sampling of more viral templates in blood. Because this unique heteroduplex band was detected consistently from two independent PCRs, we are confident of its presence in the saliva of subject 3051. The faint, slowly migrating bands in plasma of patients 3058 and 9411 are primer artifacts that are present in all lanes upon darker exposure of the autoradiogram (data not shown).
Differences in ART responses may contribute to the dynamics of viral replication within the blood and the oral cavity, as has been shown in blood and the genital tract (12, 16, 39). Thus, discordant V3-HTA banding patterns observed in samples from subjects 3040 (plasma) and 3051 (saliva) may be due to sample lag time, i.e., the number of days between blood and saliva collections (Table 1). These unique X4-like bands may have arisen from rapid viral evolution in the compartments during the 8 and 48 days, respectively, between sample collections. HTA patterns for all samples were consistent upon repeat PCR amplification and HTA analysis (data not shown).
Genotypic analysis.
DNA sequences of visualized V3-HTA heteroduplexes were determined by direct sequencing of RT-PCR products (single discrete bands) or cloning prior to sequencing (single wide bands and multiple discrete bands). The predicted amino acid sequences were aligned with the probe V3 sequence and classified as X4-like or R5-like using amino acid sequence criteria (20) (Table 2). With three exceptions, all X4-like genotypes corresponded to a mobility ratio of <0.91 whereas a mobility ratio of ≥0.91 was consistent with an R5-like genotype, verifying previous findings (22). The slowly migrating heteroduplex band identified in saliva-derived products of subject 3051 (Fig. 1) contained a leucine insertion following amino acid position 15 in the V3 loop (Table 2). Although the sequence was R5-like based on genotypic criteria (20), the insertion shifted the band above the X4-like-defining mobility ratio, as has been shown with other patient-derived sequences containing insertions and deletions (22). Sequence data representing X4-like heteroduplex bands from plasma and saliva (0.69 mobility ratio) of patient 3040 indicate an arginine-to-tryptophan substitution in 2 of the 17 sequences (Table 2). Because the change does not involve a basic substitution at position 24 that defines X4-like viruses (20), these sequences are therefore classified as R5-like. The nucleotide sequence of the unique plasma-derived HTA band (0.88 mobility ratio) from patient 3040 differed by only a single noncoding change compared to the sequence of the most closely related R5-like product (0.93 mobility ratio) from the plasma of that patient (Table 2). Compared to the probe sequence, this single mutation clustered with other single mutations to produce a more slowly migrating heteroduplex that is likely a member of the same R5 population rather than a unique viral variant (Table 2).
TABLE 2.
Analysis of patient-derived V3 sequences
| Patient | No. of clonesa | Peptide sequence | Genotypeb | Mobility ratio |
|---|---|---|---|---|
| 111 13 24 25 35 | ||||
| None (probec) | C T R P N N N T R K S I H I G . P G R A F Y T T G E I I G D I R Q A H C | R5 | ||
| 1341 | 6 P | – – – – G – – – – – – – P M – . – – K – V – A – – D – – – – – – – – – – | R5-like | 0.96 |
| 1 P | – – – – G – – – – – – – P M – . – – K – – – A – – D – – – – – – – – – – | R5-like | 0.96 | |
| 1 P | – – – – G – – – – – – – – M – . – – K – – – A – – D – – – – – – – – – – | R5-like | 0.96 | |
| 1 P | – – – – G – – – – R – – – M – . – – K – – – A – – D – – – – – – – – – – | R5-like | 0.96 | |
| 1 P | – – – – G – – – – R – – – M – . – – – – – – A – – D – – – – – – – – – – | R5-like | 0.96 | |
| 1 P | – – – – G – – – – R – – – M – . – – – – – – A – – D – – – – – K – – – – | R5-like | 0.96 | |
| 4 P, 2 S | – – – – G – – – – – – – – – – . – – K – – – A – – D – – – – – – – – – – | R5-like | 0.96 | |
| 3040 | 1 S | – – – – – – – – – R – – – M – . – – K – – – – – – D – – – – – – – – – – | R5-like | 0.93 |
| 1 S | – – – – – – – – – R – – P M – . – – K – – – A – – D – – – N – – – – – – | R5-like | 0.93 | |
| 2 P | – – – – – – – – – – G – N – – . – – – – W – A – – D – – – – – – – – – – | R5-like | 0.93 | |
| 1 P | – – – – – – – – – – G – N – – . – – – – C – A – S D – – – – – – – – – – | R5-like | 0.93 | |
| 6 P, 1 S | – – – – – – – – – – G – N – – . – – – – W – A – S D – – – – – – – – – – | R5-like | 0.93 | |
| 1 P | – – – – – I – – – – – – N – – . – – K – L – A – S D – – – – – – – – – – | R5-like | 0.93 | |
| 1 P | – – – – – – – – – – – – N – – . – – – – W – A – – D – – – – – – – – – – | R5-like | 0.93 | |
| 1 P | – – – – – – – – – – G – N – – . – – – – W – A – S D – – – – – – – – – – | R5-like | 0.88 | |
| 1 P, 1 S | – – – – – – – – – R – – Q – – . – – – – W H – – W . – – – – – – – – – – | R5-like | 0.69 | |
| 7 P | – – – – – – – – – R – – Q – – . – – – – W H – – R . – – – – – – – – – – | X4-like | 0.69 | |
| 8 P | – – – – – – – – – R – – Q – – . – – – – W H – – R . T – – – – – – – – – | X4-like | 0.69 | |
| 9208 | 1 P, 1 S | – – – – – – – – – R – – – M – . – – K – – – – – – D – – – – – – – – – – | R5-like | 0.95 |
| 1 S | – – – – – – – – – R – – – M – . – – K – – – – – – N – – – – – – – – – – | R5-like | 0.95 | |
| 1 S | – – – – – – – – – G – – P M – . – – K – – – A S – D – – – N – V – – – – | R5-like | 0.95 | |
| 1 P, 1 S | – – – – Y – H – – – – – R – – . – – – – – – – – R N V – – – – – – – – – | X4-like | 0.54 | |
| 1 P, 1 S | – – – – Y – H – – – – – – – – . – – – – – – – – R N V – – – – – – – – – | X4-like | 0.54 | |
| 1 S | – – – – – – H – – – – – – – – . – – – – – – – – R N V – – – – – – – – – | X4-like | 0.54 | |
| 1 S | – – – – – – H – T R R – – – – . – – – – – – – – – N V – – – – – – – – – | X4-like | 0.54 | |
| 1351 | 2 P | – – – – – – – – – – – – – – – . – – – T – – A – – D – – – N – – – – – – | R5-like | 0.94 |
| 2 S | – – – – – – – – – – – – – – – . – – – T – – A – – D – – – – – – – – – – | R5-like | 0.94 | |
| 1 S | – – – – H R T – – – – – – – – . – – – T – – A – – D – L – – – – – – – – | R5-like | 0.94 | |
| 3051 | 2 P, 2 S | – – – – – – – – – R – L N – – . – – – – – – – – – Q – – – – – – – – – – | R5-like | 0.97 |
| 1 S | – – – – G – – – – R – – N – – L – – G – – – – – – Q – – – – – – – – – – | R5-like | 0.73 | |
| 3058 | 2 P | – – – – – – – – – R – – S – – . – – – – – F – – – – V – – N – – – – – – | R5-like | 0.97 |
| 1 S | – – – – S – – – – R – – S – – . – – – – – F – – – – V – – N – – – – – – | R5-like | 0.97 | |
| 3090 | 2 P | – – – – – – – – – – – – – – – . – – S – – – – – – – – – – – – – – – – – | R5-like | 0.97 |
| 6 S | – – – – – – – – – – – – – – – . – – S – – – – – – A – – – – – – – – – – | R5-like | 0.97 | |
| 1460 | 2 P, 2 S | – – – – – – – – – – G – – – – . – – – – – – – – – Q – – – – – – – – – – | R5-like | 0.97 |
| 3004 | 2 P, 2 S | – – – – F – H – – – – – N – – . – – – – – – A – – – – – – – – – – – – – | R5-like | 0.96 |
| 9371 | 2 P, 2 S | – – – – – – – – – – – – – – – . – – – – – – A – – – – – – – – – – – – – | R5-like | 0.98 |
| 9411 | 2 P, 2 S | – – – – – – – – – – G – P – – . – – – – – – A – – – – – – – – – – – – – | R5-like | 0.98 |
Number of clones from plasma (P) or saliva (S) with the predicted amino acid sequence.
R5-like or X4-like genotype determined by presence of basic amino acids at the bold positions.
Residues overlapping PCR amplification primers are underlined in the probe amino acid sequence.
The nucleotide sequence of this unique band from patient 3040 was identified only once in a screening of over 100 clones from multiple RT-PCR products. Also, screening of more than 200 clones from several RT-PCR products yielded a single sequence representing the X4-like heteroduplex band derived from the saliva of this patient (Table 2). Therefore, viral variants corresponding to these bands likely represent a very small proportion of the total viral populations in this patient.
Comparison of patient-derived sequences revealed that no sequence was represented in more than one patient, and in a neighbor-joining tree, sequences clustered within but not between patients (data not shown). Based on concordance between V3-HTA and DNA sequence data, most subjects harbored genetically similar virus populations in both fluids.
Contaminating blood in saliva.
HIV-1 may arise from bleeding oral tissues. To determine whether contaminating blood influenced study findings, hemoglobin content was estimated in saliva using a quantitative test strip method (17, 33). Neither the presence nor the quantity of blood in saliva was predictive of an R5-like/X4-like genotype, as R5-like variants were identified in the saliva of all patients despite highly variable hemoglobin levels and X4-like variants were present in subjects with unquantifiable (3040) as well as high (9208) hemoglobin levels (Table 1). Also, a high HIV-1 RNA level in hemoglobin-free saliva (subject 3040) suggests that a nonblood source of virus exists in the oral cavity of this subject. Potential oral sources of the virus include the tonsils (13, 15, 25), mononuclear cells trafficking into the oral cavity (23), the salivary glands (38, 40), and gingival crevicular fluid (serum transudate bathing gingival tissues) (36). Additional study is needed to assess the contributions of these sources to oral viral shedding and the potential for the oral cavity to serve as a reservoir of residual infection following ART.
Conclusions.
To develop effective ART regimens, all anatomical sources of HIV-1 infection must be identified and techniques must be developed to monitor viral loads within the affected sites. This study identified oral secretions as a substantial source of viral RNA and raises the possibility of using saliva as a noninvasive body fluid for evaluating and monitoring viral evolution and ART intervention. We also documented one case of discordant HIV-1 subpopulations in peripheral blood and the oral cavity (subject 3051). The latter finding suggests that selected individuals (e.g., subject 3051) may harbor different viral populations in the blood and oral fluids, as has been widely documented for blood and other body fluids (28), and warrants further analysis of other viral gene-encoding regions in paired fluids from a larger cohort.
Nucleotide sequence accession numbers.
Sequences have been submitted to GenBank and have been given accession numbers AF362846 through AF362885.
Acknowledgments
We thank Dawn Rogers for patient recruitment, Jody Shock and Ada Cachafeiro for viral load determinations, and Patrick Garrison for technique assistance.
This study was supported by the National Institutes of Health (R01-DE12162, R29-DE11369, and R01-AI44667), the National Institute of Dental Research (1-P60-DE13079), and the UNC Center for AIDS Research (NIH P30-HD37260). J.A.E.N. was supported by grant NIH NRSA F32-AI09749.
REFERENCES
- 1.Alkhatib G, Combadiere C, Broder C C, Feng Y, Kennedy P E, Murphy P M, Berger E A. CC CKR5: a RANTES, MIP-1α, MIP-1β receptor as a fusion cofactor for macrophage-tropic HIV-1. Science. 1996;272:1955–1958. doi: 10.1126/science.272.5270.1955. [DOI] [PubMed] [Google Scholar]
- 2.Asjö B, Morfeldt-Månson L, Albert J, Biberfeld G, Karlsson A, Lidman K, Fenyö E M. Replicative capacity of human immunodeficiency virus from patients with varying severity of HIV infection. Lancet. 1986;ii:660–662. [PubMed] [Google Scholar]
- 3.Berger E A, Doms R W, Fenyö E M, Korber B T M, Littman D R, Moore J P, Sattentau Q J, Schuitemaker H, Sodroski J, Weiss R A. A new classification for HIV-1. Nature. 1998;391:240. doi: 10.1038/34571. [DOI] [PubMed] [Google Scholar]
- 4.Boom R, Sol C J A, Salimans M M M, Jansen C L, Wertheim-van Dillen P M E, van der Nordaa J. Rapid and simple method for purification of nucleic acids. J Clin Microbiol. 1990;28:495–503. doi: 10.1128/jcm.28.3.495-503.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Centers for Disease Control. 1993 Revised classification system for HIV infection and expanded surveillance case definition for AIDS among adolescents and adults. Morb Mortal Weekly Rep. 1992;41:1–19. [PubMed] [Google Scholar]
- 6.Choe H, Farzan M, Sun Y, Sullivan N, Rollins B, Ponath P D, Wu L, Mackay C R, LaRosa G, Newman W, Gerard N, Gerard C, Sodroski J. The β-chemokine receptors CCR3 and CCR5 facilitate infection by primary HIV-1 isolates. Cell. 1996;85:1135–1148. doi: 10.1016/s0092-8674(00)81313-6. [DOI] [PubMed] [Google Scholar]
- 7.Delwart E L, Shpaer E G, Louwagie J, McCutchan F E, Grez M, Rubsamen-Waigmann H, Mullins J I. Genetic relationships determined by a DNA heteroduplex mobility assay: analysis of HIV-1 env genes. Science. 1993;262:1257–1261. doi: 10.1126/science.8235655. [DOI] [PubMed] [Google Scholar]
- 8.Deng H, Liu R, Ellmeier W, Choe S, Unutmaz D, Burkhart M, Di Marzio P, Marmon S, Sutton R E, Hill C M, Davis C B, Peiper S C, Schall T J, Littman D R, Landau N R. Identification of a major co-receptor for primary isolates of HIV-1. Nature. 1996;381:661–666. doi: 10.1038/381661a0. [DOI] [PubMed] [Google Scholar]
- 9.Doranz B J, Rucker J, Yi Y, Smyth R J, Samson M, Peiper S C, Parmentier M, Collman R G, Doms R W. A dual-tropic, primary HIV-1 isolate that uses fusin and the β-chemokine receptors CKR-5, CKR-3, and CKR-2b as fusion cofactors. Cell. 1996;85:1149–1158. doi: 10.1016/s0092-8674(00)81314-8. [DOI] [PubMed] [Google Scholar]
- 10.Dragic T, Litwin V, Allaway G P, Martin S R, Huang Y, Nagashima K A, Cayanan C, Maddon P J, Koup R A, Moore J P, Paxton W A. HIV-1 entry into CD4+ cells is mediated by the chemokine receptor CC-CKR-5. Nature. 1996;381:667–673. doi: 10.1038/381667a0. [DOI] [PubMed] [Google Scholar]
- 11.EC-Clearinghouse on Oral Problems Related to HIV Infection; WHO Collaborating Center on Oral Manifestations of the Immunodeficiency Virus. Classification and diagnostic criteria for oral lesions in HIV infection. J Oral Pathol Med. 1993;22:289–291. [PubMed] [Google Scholar]
- 12.Eron J J, Vernazza P L, Johnston D M, Seillier-Moiseiwitsch F, Alcorn T M, Fiscus S A, Cohen M S. Resistance of HIV-1 to antiretroviral agents in blood and seminal plasma: implications for transmission. AIDS. 1998;12:F181–F189. doi: 10.1097/00002030-199815000-00003. [DOI] [PubMed] [Google Scholar]
- 13.Faust R A, Henry K, Dailey P, Melroe H, Sullivan C, Erice A, Haase A T, Boie L R., Jr Outpatient biopsies of the palatine tonsil: access to lymphoid tissue for assessment of human immunodeficiency virus RNA titers. Otolaryngol Head Neck Surg. 1996;114:593–598. doi: 10.1016/S0194-59989670252-8. [DOI] [PubMed] [Google Scholar]
- 14.Feng Y, Broder C C, Kennedy P E, Berger E A. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane G protein-coupled receptor. Science. 1996;272:872–877. doi: 10.1126/science.272.5263.872. [DOI] [PubMed] [Google Scholar]
- 15.Frankel S S, Wenig B M, Burke A P, Mannan P, Thompson L D, Abbondanzo S L, Nelson A M, Pope M, Steinman R M. Replication of HIV-1 in dendritic cell-derived syncytia at the mucosal surface of the adenoid. Science. 1996;272:115–117. doi: 10.1126/science.272.5258.115. [DOI] [PubMed] [Google Scholar]
- 16.Hamed K A, Winters M A, Holodniy M, Katzenstein D A, Merigan T C. Detection of human immunodeficiency virus type 1 in semen: effects of disease stage and nucleoside therapy. J Infect Dis. 1993;167:798–802. doi: 10.1093/infdis/167.4.798. [DOI] [PubMed] [Google Scholar]
- 17.Hart C E, Lennox J L, Pratt-Palmore M, Wright T C, Schinazi R F, Evans-Strickfaden T, Bush T J, Schnell C, Conley L J, Clancy K A, Ellerbrock T V. Correlation of human immunodeficiency virus type 1 RNA levels in blood and the female genital tract. J Infect Dis. 1999;179:871–882. doi: 10.1086/314656. [DOI] [PubMed] [Google Scholar]
- 18.Koyanagi Y, Miles S, Mitsuyasu R T, Merrill J E, Vinters H V, Chen I S Y. Dual infection of the central nervous system by AIDS viruses with distinct cellular tropisms. Science. 1987;236:819–822. doi: 10.1126/science.3646751. [DOI] [PubMed] [Google Scholar]
- 19.Lewis P, Nduati R, Kreiss J K, John G C, Richardson B A, Mbori-Ngacha D, Ndinya-Achola J, Overbaugh J. Cell-free human immunodeficiency virus type 1 in breast milk. J Infect Dis. 1998;177:34–39. doi: 10.1086/513816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Milich L, Margolin B H, Swanstrom R. Patterns of amino acid variability in NSI-like and SI-like V3 sequences and a linked change in the CD4-binding domain of the HIV-1 Env protein. Virology. 1997;239:108–118. doi: 10.1006/viro.1997.8821. [DOI] [PubMed] [Google Scholar]
- 21.Moore J P, Nara P L. The role of the V3 loop of gp120 in HIV infection. AIDS. 1991;5(Suppl.2):S21–S33. doi: 10.1097/00002030-199101001-00004. [DOI] [PubMed] [Google Scholar]
- 22.Nelson J A E, Fiscus S A, Swanstrom R. Evolutionary variants of the human immunodeficiency virus type 1 V3 region characterized by using a heteroduplex tracking assay. J Virol. 1997;71:8750–8758. doi: 10.1128/jvi.71.11.8750-8758.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Odden K, Schenck K, Hurlen B. High numbers of T cells in gingiva from patients with human immunodeficiency virus (HIV) infection. J Oral Pathol Med. 1995;24:413–419. doi: 10.1111/j.1600-0714.1995.tb01211.x. [DOI] [PubMed] [Google Scholar]
- 24.Ping L-H, Cohen M S, Hoffman I, Vernazza P, Seillier-Moiseiwitsch F, Chakraborty H, Kazembe P, Zimba D, Maida M, Fiscus S A, Eron J J, Swanstrom R, Nelson J A E. Effects of genital tract inflammation on human immunodeficiency virus type 1 V3 populations in blood and semen. J Virol. 2000;74:8946–8952. doi: 10.1128/jvi.74.19.8946-8952.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rinfret A, Latendresse H, Lefebvre R, St-Louis G, Jolicoeur P, Lamarre L. Human immunodeficiency virus-infected multinucleated histiocytes in oropharyngeal lymphoid tissues from two asymptomatic patients. Am J Pathol. 1991;138:421–426. [PMC free article] [PubMed] [Google Scholar]
- 26.Robinson E K, Evans B G. Oral sex and HIV transmission. AIDS. 1999;13:737–738. doi: 10.1097/00002030-199904160-00021. [DOI] [PubMed] [Google Scholar]
- 27.Ruprecht R M, Baba T W, Liska V, Ray N B, Martin L N, Murphey-Corb M, Rizvi T A, Bernacky B J, Keeling M E, McClure H M, Andersen J. Oral transmission of primate lentiviruses. J Infect Dis. 1999;179(Suppl. 3):S408–S412. doi: 10.1086/314794. [DOI] [PubMed] [Google Scholar]
- 28.Schrager L K, D'Souza M P. Cellular and anatomical reservoirs of HIV-1 in patients receiving potent antiretroviral combination therapy. JAMA. 1998;280:67–71. doi: 10.1001/jama.280.1.67. [DOI] [PubMed] [Google Scholar]
- 29.Schuitemaker H, Koot M, Kootstra N A, Dercksen M W, de Goede R E Y, van Steenwijk R P, Lange J M A, Eeftinck Schattenkerk J K M, Miedema F, Tersmette M. Biological phenotype of human immunodeficiency virus type 1 clones at different stages of infection: progression of disease is associated with a shift from monocytotropic to T-cell-tropic virus populations. J Virol. 1992;66:1354–1360. doi: 10.1128/jvi.66.3.1354-1360.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Seillier-Moiseiwitsch F, Margolin B H, Swanstrom R. Genetic variability of the human immunodeficiency virus: statistical and biological issues. Annu Rev Genet. 1994;28:559–596. doi: 10.1146/annurev.ge.28.120194.003015. [DOI] [PubMed] [Google Scholar]
- 31.Semba R D, Kumwenda N, Hoover D R, Taha T E, Quinn T C, Mtimavalye L, Biggar R J, Broadhead R, Miotti P G, Sokoll L J, van der Hoeven L, Chiphangwi J D. Human immunodeficiency virus load in breast milk, mastitis, and mother-to-child transmission of human immunodeficiency virus type 1. J Infect Dis. 1999;180:93–98. doi: 10.1086/314854. [DOI] [PubMed] [Google Scholar]
- 32.Shepard R N, Schock J, Robertson K, Shugars D C, Dyer J, Vernazza P, Hall C, Cohen M S, Fiscus S A. Quantitation of human immunodeficiency virus type 1 RNA from different biological compartments. J Clin Microbiol. 2000;38:1414–1418. doi: 10.1128/jcm.38.4.1414-1418.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shugars D C, Slade G D, Patton L L, Fiscus S A. Oral and systemic factors associated with increased levels of human immunodeficiency virus type 1 RNA in saliva. Oral Surg Oral Med Oral Pathol Oral Radiol Endod. 2000;89:432–440. doi: 10.1016/s1079-2104(00)70124-7. [DOI] [PubMed] [Google Scholar]
- 34.Shugars, D. C., L. L. Patton, S. A. Freel, L. R. Gray, J. J. Eron, Jr., and S. A. Fiscus. Hyper-excretion of human immunodeficiency virus type 1 RNA in saliva. J. Dent. Res., in press. [DOI] [PubMed]
- 35.Shugars D C, Wahl S M. The role of the oral environment in HIV-1 transmission. J Am Dent Assoc. 1998;129:851–858. doi: 10.14219/jada.archive.1998.0349. [DOI] [PubMed] [Google Scholar]
- 36.Suzuki T, Tai H, Yoshie H, Jeannel D, Fournier S, Dupont B, De The G, Hara K. Characterization of HIV-related periodontitis in AIDS patients: HIV-infected macrophage exudate in gingival crevicular fluid as a hallmark of distinctive etiology. Clin Exp Immunol. 1997;108:254–259. doi: 10.1046/j.1365-2249.1997.d01-997.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tersmette M, de Goede R E Y, Al B J M, Winkel I N, Gruters R A, Cuypers H T, Huisman H G, Miedema F. Differential syncytium-inducing capacity of human immunodeficiency virus isolates: frequent detection of syncytium-inducing isolates in patients with acquired immunodeficiency syndrome (AIDS) and AIDS-related complex. J Virol. 1988;62:2026–2032. doi: 10.1128/jvi.62.6.2026-2032.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Uccini S, Riva E, Antonelli G, D'Offizi G, Prozzo A, Angelici A, Faggioni A, Angeloni A, Torrisi M R, Gentile M, Baroni C D, Ruco L P. The benign cystic lymphoepithelial lesion of the parotid gland is a viral reservoir in HIV type 1-infected patients. AIDS Res Hum Retrovir. 1999;15:1339–1344. doi: 10.1089/088922299310043. [DOI] [PubMed] [Google Scholar]
- 39.Vernazza P L, Gilliam B L, Dyer J, Fiscus S A, Eron J J, Frank A C, Cohen M S. Quantification of HIV in semen: correlation with antiviral treatment and immune status. AIDS. 1997;11:987–993. [PubMed] [Google Scholar]
- 40.Wahl S M, Worley P, Jin W, McNeely T B, Eisenberg S, Fasching C, Orenstein J M, Janoff E N. Anatomic dissociation between HIV-1 and its endogenous inhibitor in mucosal tissues. Am J Pathol. 1997;150:1275–1284. [PMC free article] [PubMed] [Google Scholar]