Abstract
We tested seven human immunodeficiency virus-infected children for their cytotoxic T-lymphocyte (CTL) activities towards the p24gag QASQEVKNW epitope and its nine variant sequences. Our data confirm that most, but not all, CTL responses are broadly cross-specific. For the first time, we show the high interpatient variability in cross-recognition of mutant CTL epitopes. These interindividual variations in the CTL response to the same epitope should be taken into account in the design and the evaluation of vaccines.
Human immunodeficiency virus (HIV) infection is a major public health problem, in particular in developing countries. Therefore, a vaccine against this virus is urgently needed to reduce the propagation of the epidemic. The contribution of cytotoxic T lymphocytes (CTL) in controlling HIV replication in infected hosts has been well demonstrated, though they do not eradicate the virus (15, 16). Meanwhile, vaccine studies in animal models have shown that preexisting CTL can reduce viral load during primary infection and eventually slow the progression of the disease (15, 18). An important issue for vaccine development is the antigenic diversity among different HIV isolates. In 1992, comparison of genetic sequences from various HIV strains led to the definition of HIV type 1 (HIV-1) subtypes. Since then, the number of known HIV-1 subtypes has been growing (21, 22). In order to confer protection against most field isolates, immunogens able to induce broadly cross-reactive CTL are required. In 1998, we showed that HIV-specific CTL responses from children infected with various HIV subtypes were mostly cross-reactive against two HIV clades (5). At the same time, several other studies on vaccinated or HIV-infected persons reached the same conclusion (1, 8, 11, 12, 19, 20, 28).
These previous studies described the cross-recognition of full-length HIV proteins by bulk CTL from genetically diverse populations exposed to different strains of HIV. They gave an appropriate evaluation of cross-reactive CTL, at the level of the infected patient and at the level of the population infected by various HIV subtypes. This approach is worthy for estimation of the level of CTL cross-reactivity that may be attained in vaccinated populations. However, it may overestimate the cross-recognition at the level of single CTL epitopes for two main reasons. First, the CTL response is multispecific, and distinct CTL populations present in bulk cultures can recognize several CTL epitopes on the same protein (6, 7). Second, some CTL epitopes are conserved between several HIV subtypes. As the use of target cells expressing full-length HIV proteins may have overestimated the cross-recognition capacities of CTL, we studied the capacity of CTL to recognize variant sequences of a single epitope. When cross-recognition of a single epitope has been studied, reactivity from only one CTL population was reported (8, 10, 20). Here, we tested several CTL lines specific for the same epitope in order to establish if cross-recognition of variant epitopes is shared by all CTL or is specific for each CTL population. We chose the HIV p24gag epitope, QASQEVKNW, because it is presented in association with two HLA molecules, HLA-B53 (our results) and HLA-B57 (14), and because several naturally occurring variants have been reported (17). We found that recognition of variant sequences of the CTL epitope was broad, as the five responders recognized two to nine variant sequences. Most importantly, we observed that the CTL responses from each individual differed from the others by the identity of sequences recognized and by the level of lysis of target cells expressing these sequences.
Recognition of HIV Gag protein and CTL epitope QASQEVKNW by PBMCs after nonspecific stimulation.
Seven children included in a longitudinal follow-up study of their CTL activities were selected for their expression of HLA-B53 or -B57, as determined by amplification refractory mutation system PCR (4, 5). Peripheral blood mononuclear cell (PBMC) samples exhibiting significant lysis against the HIV p55gag protein were chosen for this study. The patients were monitored at Hôpital Necker, Paris, France. The legal guardians gave informed consent before the children entered the study. Child EM100 was infected through transfusion of a contaminated blood product. Mother-to-child transmission of HIV was responsible for infection of the other children. Their infecting subtypes were determined by heteroduplex mobility assay or sequencing of the Env gene, as previously reported (5). Patients' clinical and biological characteristics are summarized in Table 1.
TABLE 1.
Characteristics of patients
| Patient | Origin | HIV clade | Age (yr) | CDC stage | Treatment (duration, mo)f | CD4+/mm3 (%)a | CD8+/mm3 (%)a | Plasma viral loadb |
|---|---|---|---|---|---|---|---|---|
| EM3 | Europe | B | 4.1 | A | AZT (41) | 765 (11) | 4,312 (62) | 4.7d |
| EM17 | Africa | D | 7.5 | B | None | 323 (11) | 1,720 (69) | 4.3d |
| 11.6 | B | D4T+3TC+nelfinavir (8) | 782 (24) | 782 (24) | <2.2e | |||
| EM23 | Africa | B | 7.6 | B | AZT (73) | 485 (14) | 2,080 (60) | 5d |
| EM34 | Caribbean | NDc | 2.2 | C | AZT (15) | 1,367 (24) | 1,024 (28) | ND |
| EM47 | Europe | B | 11.1 | A | AZT (84) | 459 (17) | 1,674 (62) | 3.8d |
| 14.5 | A | AZT+DDI (25) | 528 (24) | 1,254 (57) | 3.7e | |||
| EM74 | Haiti | B | 8.9 | B | None | 572 (22) | 910 (35) | <3.3d |
| EM100 | Europe | B | 15.8 | N | None | 768 (24) | 1,248 (39) | 3.6e |
Absolute number per cubic millimeter of blood and the percent among total lymphocytes.
Log HIV RNA copies per milliliter of plasma.
ND, not done.
bDNA assay (Chiron).
Amplicor assay (Roche).
AZT, zidovudine; D4T, stavudine; 3TC, lamivudine; DDI, dideoxyinosine.
Bulk CTL cultures were generated by mitogenic stimulation with phytohemagglutinin (PHA) and expansion in the presence of recombinant interleukin-2 (100 IU/ml; Chiron, Suresnes, France; a generous gift from R. Lebour) as described previously (5). A standard 51Cr release assay was used to measure lysis of target cells expressing p55gag from a clade A and a clade B isolate, encoded by recombinant vaccinia viruses vvTG6026 and vvTG1144 (Transgène, Strasbourg, France [5]). Cells from patients EM47 and EM74 recognized p55gag from both HIV isolates (Table 2). In contrast, cells from patient EM23 recognized only p55gag from the clade A isolate, and CTLs from patient EM3 lysed only target cells expressing HIV clade B p55gag.
TABLE 2.
Recognition of CTL epitope p24gag QASQEVKNW by HIV-infected children carrying HLA-B53 or HLA-B57 molecules
| Cell treatment and patient no. | HLA restricting elementa | % specific lysis ofb:
|
||||||
|---|---|---|---|---|---|---|---|---|
| rVV-infected targetsg
|
Peptide-coated targets
|
|||||||
| WTe | Gag-A | Gag-B | Medium | 25-17B | Pool clade variants | Pool minor variants | ||
| PHA stimulationc | ||||||||
| EM17 | B53 | 1 | NDf | 33 | 6 | 10 | 14 | 11 |
| EM23 | B53 | 4 | 17 | 10 | 8 | 13 | 14 | 2 |
| EM34 | B53 | ND | ND | ND | ND | ND | ND | ND |
| EM47 | B53 | 18 | 45 | 51 | 0 | 38 | 64 | 59 |
| EM74 | B53 | 8 | 26 | 26 | 5 | 5 | 5 | 2 |
| EM3 | B57 | 1 | 8 | 22 | 4 | 4 | 5 | 1 |
| EM100 | B57 | 3 | ND | 18 | ND | ND | ND | ND |
| Peptide stimulationd | ||||||||
| EM17 | B53 | ND | ND | ND | 4 | 7 | 40 | 29 |
| EM23 | B53 | 3 | 23 | 15 | 4 | 47 | 44 | 36 |
| EM34 | B53 | 3 | 22 | 32 | 3 | 28 | 28 | 22 |
| EM47 | B53 | 5 | 44 | 44 | 8 | 62 | 77 | 66 |
| EM74 | B53 | 5 | 37 | 26 | 10 | 14 | 12 | 11 |
| EM3 | B57 | 5 | 37 | 34 | 5 | 26 | 28 | 6 |
| EM100 | B57 | 5 | 11 | 15 | 5 | 4 | 4 | 5 |
HLA genotypes: EM17 (A29, A33, B44, B53, C4, C4); EM23 (A2, A2, B51, B53, C4, C16); EM34 (A1, A26, B3502, B53, C4, C4); EM47 (A30, A68, B53, B53, C4, C4); EM74 (A34, A34, B13, B53, C4, C6); EM3 (A1, A2, B51, B57, C6, C16); EM100 (A3, A11, B44, B57, C5, C6).
Positive results are indicated in bold. Cell lines were tested at three different effector/target (E:T) ratios. Results obtained at an E:T ratio of 60/1 are shown for PHA stimulation. Results obtained at an E:T ratio of 60/1 or 20/1 are shown for peptide stimulation.
Fresh PBMCs were stimulated once with PHA before the CTL assay.
PHA-stimulated PBMCs were restimulated with peptide-coated EBV-B cells before the CTL assay.
WT, wild type.
ND, not done.
rVV, recombinant vaccinia viruses.
Then, we tested the ability of the CTL lines to recognize the variants of the QASQEVKNW epitope using synthetic peptides. Peptides corresponding to the naturally occurring variants recorded in 1997 (17) for the p24(176–184) epitope were synthesized by Neosystem (Strasbourg, France) and obtained through the Agence Nationale de la Recherche sur le SIDA. Peptides corresponding to consensus sequences of major HIV clades A, B, C, D, and F are, respectively, 25-17A (QATQEVKNW), 25-17B (QASQEVKNW), 25-17C (QATQDVKNW), 25-17D (QASQDVKNW), and 25-17F (QATQEVKGW). The five minor variants are 25-17M1 (QATQAVKNW), 25-17M2 (QASQEVKNR), 25-17M3 (QASQEVKGW), 25-17M4 (EATQEVKGW), and 25-17M5 (QATQDVKDN). After PHA stimulation, one out of five CTL cell lines (EM47) recognized peptides corresponding to the QASQEVKNW CTL epitope and its variant sequences. Target cells pulsed with the clade B index peptide as well as with pools of peptides corresponding to the four major clade variants and the five minor variant sequences were lysed by CTL cell line EM47 (Table 2).
Patient-specific patterns of CTL cross-recognition of QASQEVKNW CTL epitope variants after stimulation of PBMCs with peptides.
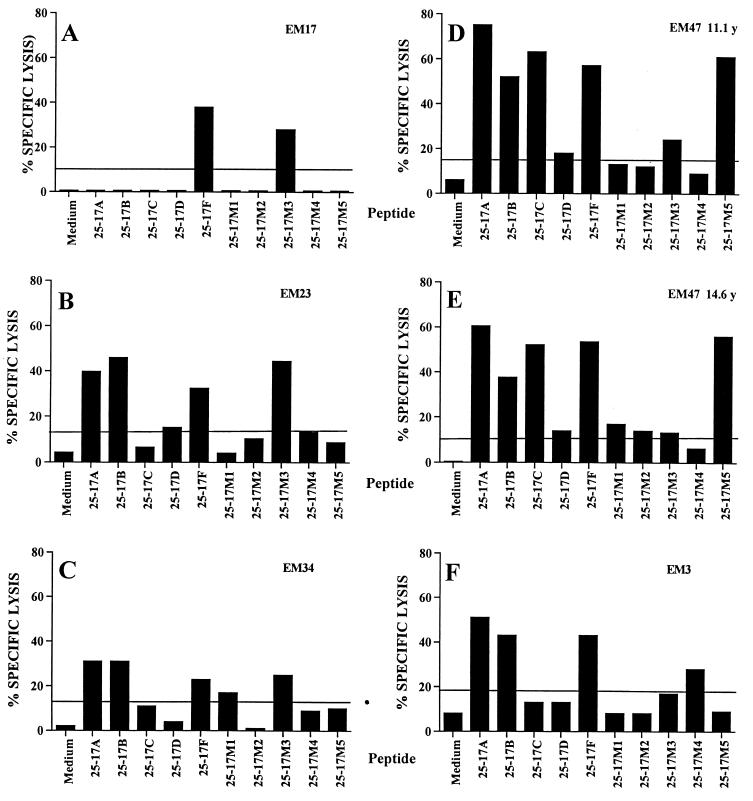
We first used PHA to stimulate CTL because it will expand all CTL independently of their specificity. PHA-stimulated PBMCs recognized full-length HIV p55gag protein, but only one CTL population recognized the p24gag QASQEVKNW epitope (Table 2). Therefore, we performed antigen-specific stimulations of the CTL cell lines in order to increase the frequency of CTL specific for the epitope. We chose to use a mix of the 10 variant peptides corresponding to all sequences studied, to avoid introducing biases in the amplification of CTL specific for a particular sequence. Previously PHA-stimulated PBMCs were restimulated with autologous Epstein-Barr virus-transformed B cells (EBV-B cells) coated with a mix of the 10 peptides, each at a final concentration of 1 μg/ml. Before mixing with responding cells at a ratio of 1:1, EBV-B cells were irradiated at 100 Gy and washed twice to eliminate unbound peptides. This procedure led to the detection of lysis of peptide-pulsed target cells for four of the five HLA-B53-positive patients and one of the two HLA-B57-positive patients (Table 2). Figure 1 shows the recognition of each variant peptide by the responding CTL cell lines. Patient EM17 had the more focused response, as she recognized only peptides 25-17F and 25-17M3 (Fig. 1A). Despite being infected with an HIV-1 clade D isolate, she did not lyse targets pulsed with the consensus clade D sequence of the epitope. A CTL cell line obtained 4 years later from this patient had the same specificity for the 25-17F and 25-17M3 sequences (data not shown). Lysis of HIV-p55gag clade B-expressing target cells (Table 2) was due to immunodominant effectors recognizing the HLA-B44-restricted HIV p24gag SEGATPQDL epitope (data not shown).
FIG. 1.
Recognition of the 10 variant sequences of CTL epitope p24gag QASQEVKNW by CTL cell lines from five responders. Autologous EBV-B cell lines were used as target cells in a 51Cr release assay. Peptides were tested at 1 μg/ml. The line indicates the level of significantly positive lysis. (A) EM17, two peptide stimulations, effector/target ratio (E:T) = 60:1. (B) EM23, one peptide stimulation, E:T = 20:1. (C) EM34, two peptide stimulations, E:T = 20:1. (D) EM47, two peptide stimulations, E:T = 20:1. (E) EM47, PHA stimulation, E:T = 60:1. (F) EM3, three peptide stimulations, E:T = 60:1.
In contrast to patient EM17, the four other responders had broadly cross-reactive responses to the epitope, as they recognized four to nine variant sequences. Patient EM23 is of African origin and has been infected with a clade B isolate. Her CTL efficiently recognized four sequences (25-17A, -B, -F, and -M3; Fig. 1B). The 25-17D peptide was also recognized, albeit with reduced efficiency. Child EM34 is of Caribbean origin. Unfortunately, typing of her HIV type was not performed, due to the lack of available samples. She has been probably infected by a clade B isolate. She had a broad CTL response directed against sequences 25-17A, -B, -F, -M1, and -M3 (Fig. 1). The fourth HLA-B53-positive patient, EM47, had the widest spectrum of cross-clade recognition. His PBMCs, which were isolated in 1995 and 1998, efficiently recognized sequences 25-17A, -B, -C, -F, and -M5 (Fig. 1D and E). Low-level recognition of variants 25-17D, -M1, -M2, or -M3 was observed in one or both samples tested. Of the two HLA-B57-positive patients, only patient EM3 recognized the CTL epitope QASQEVKNW. He had a broad cross-recognition pattern, as four sequences (25-17A, -B, -F, and -M4) were well recognized (Fig. 1F).
The patient-specific response observed might be due either to the viral strains infecting each subject or to host-specific factors and in particular to their T-cell receptor (TCR) repertoire. Three of the four children with broad CTL responses have been infected with a clade B isolate, and the fourth one (EM34) has probably been infected by a clade B strain, as she is of Caribbean origin. As HIV typing is based on the envelope sequence, one cannot exclude the possibility that the patients carry CTL sequences different from the clade B consensus or a recombinant HIV strain (23). Presence of variant sequences of the CTL epitope in vivo may lead to preferential expansion of CTL specific for these variants. However, vaccinees exposed to a single HIV sequence have cross-reactive CTL, suggesting that the viral strain infecting the patient is not the only factor influencing the CTL specificity (12). In addition, two CTL clones derived from the same patient (EM47) differed widely in the variants recognized (F. Buseyne and Y. Rivière, unpublished results). From various studies, it appears that TCR repertoires selected in response to defined peptide-major histocompatibility complex (MHC) complexes can vary from being relatively limited to extremely diverse. What is more, it has been shown that the TCR repertoire specific for a peptide-MHC complex of naïve, effector, or memory CTL differs significantly between individual syngeneic mice (2, 3). These experiments indicate that for the human population that displays high genetic diversity, the interindividual variability in the TCR repertoire will lead to different CTL responses to the same peptide-MHC complex, as was observed in this study.
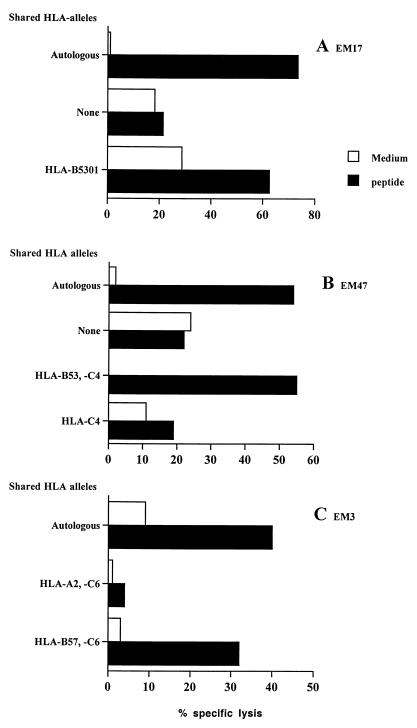
Recognition of endogenously synthesized QASQEVKNW CTL epitope and HLA restriction.
Using a panel of allogeneic target cells partially matched for some HLA molecules, we checked that recognition of the epitope was HLA-B53-restricted for patients EM17 and EM47 (Fig. 2A and B) and HLA-B57-restricted for patient EM3 (Fig. 2C). The four CTL cell lines recognizing peptides 25-17A and -B (EM23, EM34, EM47, and EM3) also lysed target cells expressing full-length p55gag clade A and B, indicating that cross-clade recognition occurred when the epitope was derived from intracellular synthesis and processing of the viral protein (Table 2). Peptide titration experiments showed that CTL had high affinity for the epitope. Peptide concentrations giving 50% of maximal lysis were 0.42 ng of peptide 25-17F/ml for patient EM17, 0.26 ng of peptide 25-17B/ml for patient EM34, and 0.32 ng of peptide 25-17A/ml for patient EM47.
FIG. 2.
CTL epitope p24gag QASQEVKNW is presented by both HLA-B53 and HLA-B57. Autologous and partially HLA-matched EBV-B cells were used as target cells in a 51Cr release assay. (A) EM17, effector/target ratio (E:T) = 10:1, peptide 25-17F at 10 ng/ml. (B) EM47, E:T = 20:1, peptide 25-17A at 1 μg/ml. (C) EM3, E:T = 10:1, peptide 25-17B at 1 μg/ml.
The capacity of a peptide to bind multiple MHC molecules and, consequently, to be immunogenic in the context of individuals of different MHC types has been referred to as degeneracy. The p24gag CTL epitope QASQEVKNW described here is presented in the context of HLA-B57 (14). In the present study, we showed that HLA-B53 is able to present the same peptide. In contrast to previous reports where degenerate epitopes were presented by members of the same HLA supertype (13, 25, 27), the two HLA molecules presenting the CTL epitope QASQEVKNW do not belong to the same HLA supertype. Indeed, HLA-B53 belongs to the B7 supertype, whereas HLA-B57 has been proposed to belong to the distinct HLA-B58 supertype (24). However, degenerate presentation of CTL epitopes from Plasmodium falciparum by both HLA-B53 and HLA-B57 was previously reported (9). Interestingly, both B7 and B58 supertypes accept hydrophobic aliphatic or aromatic residues at the C-terminal position of their peptide ligands (24). B58 supertype has a preference for small aliphatic residues at position 2 (A, S, or T) that is reminiscent of the requirement for amino acid fixation in the small B pocket of HLA-B53 (26). In conclusion, even if HLA-B53 and HLA-B57 belong to different supertypes, their peptide-binding characteristics are consistent with the presentation of the same epitope, as we observed here.
With regard to peptide sequences, three were efficiently recognized by most patients (variants A, B, and F), three were efficiently recognized by a single patient (variants C, M3, and M5), and four were weakly recognized (variants D, M1, M2, and M4). Differences in recognition of efficiently presented variants might be due to a bias in the sequences infecting the patients or in the TCR repertoire, as discussed above. The four poorly recognized peptides may have reduced immunogenicity. However, amino acid changes in variant 25-17D are also present in other variants that are well recognized. In contrast, presence of the positively charged R in position 9 of variant 25-17M2 may decrease binding to the F pocket of HLA-B53, which prefers hydrophobic or aromatic residues. The two other weakly recognized variants have a nonconservative mutation in position 1 (25-17M4) or in position 5 (25-17M1). In conclusion, it is likely that amino acid changes in these three naturally occurring mutants may lead to decreased immunogenicity.
Conclusions.
In the present work, we have looked at the recognition of 10 naturally occurring variants of the HIV p24gag QASQEVKNW CTL epitope. The specificity of our approach was to investigate the cross-recognition of variants of a defined CTL epitope by several patients, whereas former studies tested a single CTL population specific for each epitope (8, 10, 20). Thus, our approach shows that the pattern of cross-recognition is dependent on the patients tested, as some patients had a focused response whereas others were broadly cross-reactive (Fig. 1). Most importantly, each patient recognized a distinct set of variants. This interpatient variability in CTL cross-recognition of variant sequences is described for the first time in HIV-infected patients and should be taken into account as a factor that may influence the response to and the efficacy of an HIV vaccine.
Because of the broad cross-clade reactivity of CTL observed in former studies (1, 5, 8, 11, 12, 19, 20, 28), it is believed that vaccines based on a single immunogen will be sufficient to protect against most field isolates. This is true for the most conserved epitopes and if several epitopes are present in the vaccine. The present results corroborate the broad cross-clade reactivity of CTL at the level of a single epitope for the majority of patients studied, though some patients had a more focused CTL response. The actual trend in vaccine strategies is to incorporate additional HIV genes, which have been shown to encode a significant number of determinants recognized by CTL. It is hoped that each of these additional sequences will result both in an added breadth in the CTL response as well as a greater response rate among vaccinees. The patient-specific pattern of CTL cross-recognition observed in the present study strongly supports the use of vaccine strategies designed to induce a polyclonal CTL response, as the response to a defined CTL epitope might differ significantly among individuals.
Acknowledgments
This work was supported by Institut Pasteur, Agence Nationale de Recherche sur le SIDA, and SIDACTION. Y.R. is an Elisabeth Glaser Scientist.
We thank F. Porrot and B. Corre for the follow-up of CTL activities from children presented in this study and E. Bui for collecting clinical data. Genotyping was performed by F. Gotch at Chelsea and Westminster Hospital, London, United Kingdom.
REFERENCES
- 1.Betts M R, Krowka J, Santamaria C, Balsamo K, Gao F, Mulundu G, Luo C, N'Gandu N, Sheppard H, Hahn B H, Allen S, Frelinger J A. Cross-clade human immunodeficiency virus (HIV)-specific cytotoxic T-lymphocyte responses in HIV-infected Zambians. J Virol. 1997;71:8908–8911. doi: 10.1128/jvi.71.11.8908-8911.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bousso P, Casrouge A, Altman J D, Haury M, Kanellopoulos J, Abastado J P, Kourilsky P. Individual variations in the murine T cell response to a specific peptide reflect variability in naive repertoires. Immunity. 1998;9:169–178. doi: 10.1016/s1074-7613(00)80599-3. [DOI] [PubMed] [Google Scholar]
- 3.Busch D H, Pilip I, Pamer E G. Evolution of a complex T cell receptor repertoire during primary and recall bacterial infection. J Exp Med. 1998;188:61–70. doi: 10.1084/jem.188.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buseyne F, Blanche S, Schmitt D, Griscelli C, Rivière Y. Detection of HIV-specific cell-mediated cytotoxicity in the peripheral blood from infected children. J Immunol. 1993;150:3569–3581. [PubMed] [Google Scholar]
- 5.Buseyne F, Chaix M L, Fleury B, Manigard O, Burgard M, Blanche S, Rouzioux C, Rivière Y. Cross-clade-specific cytotoxic T lymphocytes in HIV-1 infected children. Virology. 1998;250:316–324. doi: 10.1006/viro.1998.9373. [DOI] [PubMed] [Google Scholar]
- 6.Buseyne F, Janvier G, Fleury B, Schmidt D, Rivière Y. Multispecific and heterogeneous recognition of the gag protein by cytotoxic T lymphocytes (CTL) from HIV-infected patients: factors other than the MHC control the epitopic specificities. Clin Exp Immunol. 1994;97:353–360. doi: 10.1111/j.1365-2249.1994.tb06094.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buseyne F, McChesney M, Porrot F, Kovarik S, Guy B, Rivière Y. Gag-specific cytotoxic T lymphocytes from human immunodeficiency virus type 1-infected individuals: Gag epitopes are clustered in three regions of the p24gag protein. J Virol. 1993;67:694–702. doi: 10.1128/jvi.67.2.694-702.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cao H, Kanki P, Sankale J L, Dieng-Sarr A, Mazzara G P, Kalams S A, Korber B, Mboup S, Walker B D. Cytotoxic T-lymphocyte cross-reactivity among different human immunodeficiency virus type 1 clades: implications for vaccine development. J Virol. 1997;71:8615–8623. doi: 10.1128/jvi.71.11.8615-8623.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Doolan D L, Hoffman S L, Southwood S, Wentworth P A, Sidney J, Chesnut R W, Keogh E, Appella E, Nutman T B, Lal A A, Gordon D M, Oloo A, Sette A. Degenerate cytotoxic T cell epitopes from P. falciparum restricted by multiple HLA-A and HLA-B supertype alleles. Immunity. 1997;7:97–112. doi: 10.1016/s1074-7613(00)80513-0. [DOI] [PubMed] [Google Scholar]
- 10.Dorrell L, Dong T, Ogg G S, Lister S, McAdam S, Rostron T, Conlon C, McMichael A J, Rowland-Jones S L. Distinct recognition of non-clade B human immunodeficiency virus type 1 epitopes by cytotoxic T lymphocytes generated from donors infected in Africa. J Virol. 1999;73:1708–1714. doi: 10.1128/jvi.73.2.1708-1714.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Durali D, Morvan J, Letourneur F, Schmitt D, Guegan N, Dalod M, Saragosti S, Sicard D, Levy J P, Gomard E. Cross-reactions between the cytotoxic T-lymphocyte responses of human immunodeficiency virus-infected African and European patients. J Virol. 1998;72:3547–3553. doi: 10.1128/jvi.72.5.3547-3553.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ferrari G, Humphrey W, McElrath M J, Excler J L, Duliege A M, Clements M L, Corey L C, Bolognesi D P, Weinhold K J. Clade B-based HIV-1 vaccines elicit cross-clade cytotoxic T lymphocyte reactivities in uninfected volunteers. Proc Natl Acad Sci USA. 1997;94:1396–1401. doi: 10.1073/pnas.94.4.1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fleischhauer K, Tanzarella S, Wallny H J, Bordignon C, Traversari C. Multiple HLA-A alleles can present an immunodominant peptide of the human melanoma antigen Melan-A/MART-1 to a peptide-specific HLA-A∗0201+ cytotoxic T cell line. J Immunol. 1996;157:787–797. [PubMed] [Google Scholar]
- 14.Goulder P J, Bunce M, Krausa P, McIntyre K, Crowley S, Morgan B, Edwards A, Giangrande P, Phillips R E, McMichael A J. Novel, cross-restricted, conserved, and immunodominant cytotoxic T lymphocyte epitopes in slow progressors in HIV type 1 infection. AIDS Res Hum Retrovir. 1996;12:1691–1698. doi: 10.1089/aid.1996.12.1691. [DOI] [PubMed] [Google Scholar]
- 15.Heeney J L, Bruck C, Goudsmit J, Montagnier L, Schultz A, Tyrrell D, Zolla-Pazner S. Immune correlates of protection from HIV infection and AIDS. Immunol Today. 1997;18:4–8. doi: 10.1016/s0167-5699(97)80005-9. [DOI] [PubMed] [Google Scholar]
- 16.Klein M. AIDS and HIV vaccines. Vaccine. 1999;17:S65–S70. doi: 10.1016/s0264-410x(99)00236-4. [DOI] [PubMed] [Google Scholar]
- 17.Korber B, Moore J, Brander C, Koup R, Heynes B, Walker B. HIV molecular immunology database 1997. Los Alamos, N.Mex: Los Alamos National Laboratory; 1997. [Google Scholar]
- 18.Letvin N L. Progress in the development of an HIV-1 vaccine. Science. 1998;280:1875–1880. doi: 10.1126/science.280.5371.1875. [DOI] [PubMed] [Google Scholar]
- 19.Lynch J A, deSouza R, Robb M D, Markowitz L, Nitayaphan S, Sapan C V, Mann D L, Birx D L, Cox J H. Cross-clade cytotoxic T cell response to human immunodeficiency virus type 1 proteins among HLA disparate North Americans and Thais. J Infect Dis. 1998;178:1040–1046. doi: 10.1086/515652. [DOI] [PubMed] [Google Scholar]
- 20.McAdam S, Kaleebu P, Krausa P, Goulder P, French N, Collin B, Blanchard T, Whitworth J, McMichael A, Gotch F. Cross-clade recognition of p55 by cytotoxic T lymphocytes in HIV-1 infection. AIDS. 1998;12:571–579. doi: 10.1097/00002030-199806000-00005. [DOI] [PubMed] [Google Scholar]
- 21.McCutchan F E. Understanding the genetic diversity of HIV-1. AIDS. 2000;14:S31–S44. [PubMed] [Google Scholar]
- 22.Robertson D L, Anderson J P, Bradac J A, Carr J K, Foley B, Funkhouser R K, Gao F, Hahn B H, Kalish M L, Kuiken C, Learn G H, Leitner T, McCutchan F, Osmanov S, Peeters M, Pieniazek D, Salminen M, Sharp P M, Wolinsky S, Korber B. HIV-1 nomenclature proposal. Science. 2000;288:55–56. doi: 10.1126/science.288.5463.55d. [DOI] [PubMed] [Google Scholar]
- 23.Robertson D L, Sharp P M, McCutchan F E, Hahn B H. Recombination in HIV-1. Nature. 1995;374:124–126. doi: 10.1038/374124b0. [DOI] [PubMed] [Google Scholar]
- 24.Sette A, Sidney J. Nine major HLA class I supertypes account for the vast preponderance of HLA-A and -B polymorphism. Immunogenetics. 1999;50:201–212. doi: 10.1007/s002510050594. [DOI] [PubMed] [Google Scholar]
- 25.Shiga H, Shioda T, Tomiyama H, Takamiya Y, Oka S, Kimura S, Yamaguchi Y, Gojoubori T, Rammensee H G, Miwa K, Takiguchi M. Identification of multiple HIV-1 cytotoxic T-cell epitopes presented by human leukocyte antigen B35 molecules. AIDS. 1996;10:1075–1083. [PubMed] [Google Scholar]
- 26.Smith K J, Reid S W, Harlos K, McMichael A J, Stuart D I, Bell J I, Jones E Y. Bound water structure and polymorphic amino acids act together to allow the binding of different peptides to MHC class I HLA-B53. Immunity. 1996;4:215–228. doi: 10.1016/s1074-7613(00)80430-6. [DOI] [PubMed] [Google Scholar]
- 27.Threlkeld S C, Wentworth P A, Kalams S A, Wilkes B M, Ruhl D J, Keogh E, Sidney J, Southwood S, Walker B D, Sette A. Degenerate and promiscuous recognition by CTL of peptides presented by the MHC class I A3-like superfamily: implications for vaccine development. J Immunol. 1997;159:1648–1657. [PubMed] [Google Scholar]
- 28.Wilson S E, Pedersen S L, Kunich J C, Wilkins V L, Mann D L, Mazzara G P, Tartaglia J, Celum C L, Sheppard H W. Cross-clade envelope glycoprotein 160-specific CD8(+) cytotoxic T lymphocyte responses in early HIV type 1 clade B infection. AIDS Res Hum Retrovir. 1998;14:925–937. doi: 10.1089/aid.1998.14.925. [DOI] [PubMed] [Google Scholar]