Abstract
Proteins can become modified by a large number of reactions involving reactive oxygen species. Among these reactions, carbonylation has attracted a great deal of attention due to its irreversible and unrepairable nature. Carbonylated proteins are marked for proteolysis by the proteasome and the Lon protease but can escape degradation and form high-molecular-weight aggregates that accumulate with age. Such carbonylated aggregates can become cytotoxic and have been associated with a large number of age-related disorders, including Parkinson's disease, Alzheimer's disease, and cancer. This review focuses on the generation of and defence against protein carbonyls and speculates on the potential role of carbonylation in protein quality control, cellular deterioration, and senescence.
Keywords: damage segregation, mistranslation, protein carbonylation, proteolysis, senescence
Introduction
The widespread use of carbonylation assays for the detection of oxidative damage to proteins has its origin in studies on bacterial glutamine synthetase activity (Stadtman, 2001). Upon ammonium starvation, this enzyme undergoes a modification that marks it for proteolytic degradation and this marker reaction requires the ingredients of a mixed-function oxidation reaction, that is, oxygen, iron, and reducing equivalents (Levine et al, 1981; Roseman and Levine, 1987; Rivett and Levine, 1990; Stadtman, 2001). Biochemical analysis revealed that carbonyl groups introduced into the side chains of specific amino acids in the active center of the protein trigger the initial steps in the degradation of the enzyme (Levine, 1983, 2002). Parallel studies on aging animals have similarly identified oxidative carbonylation as one important factor in protein function and removal (e.g. Oliver et al, 1987; Stadtman, 1992; Levine, 2002).
A large number of studies have shown that protein carbonylation increases with the age of cells, organelles, and tissues of varied species and, in some cases, carbonylation has been linked to age-dependent wear and tear of specific enzymes, such as aconitase and the nucleotide translocator ANT (e.g. Yan et al, 1997; Yan and Sohal, 1998). Diseases associated with increased carbonylation include Parkinson's disease, Alzheimer's disease, cancer, cataractogenesis, diabetes, and sepsis (Levine, 2002; Dalle-Donne et al, 2003). In addition, manipulations leading to prolonged lifespan, that is, caloric restriction, decrease carbonylation levels in mouse mitochondria (Lass et al, 1998). In some cases, the levels of carbonylated-damaged proteins have been shown to be associated with the physiological age or life expectancy of an organism rather than with its chronological age. For example, carbonyl levels are higher in crawlers (low life expectancy) than fliers in a cohort of houseflies of the same chronological age (Sohal et al, 1993). Similarly, using in situ detection of protein oxidation in single Escherichia coli cells and a technique to separate culturable and nonculturable cells of the same chronological age, it was demonstrated that proteins of the nonculturable cell population exhibited markedly higher levels of irreversible carbonylation (Desnues et al, 2003). However, it is still unclear to what extent carbonylation may be a cause of cellular senescence and aging or yet another, albeit highly useful, diagnostic marker of age-related deterioration.
Generation of protein carbonyls
Carbonyl derivatives are formed by a direct metal catalyzed oxidative (MCO) attack on the amino-acid side chains of proline, arginine, lysine, and threonine. In addition, carbonyl derivatives on lysine, cysteine, and histidine can be formed by secondary reactions with reactive carbonyl compounds on carbohydrates (glycoxidation products), lipids, and advanced glycation/lipoxidation end products. The quantitatively most important products of the carbonylation reaction are glutamic semialdehyde from arginine (Figure 1) and proline, and aminoadipic semialdehyde from lysine (Requena et al, 2003). Compared to other oxidative modifications, carbonyls are relatively difficult to induce and in contrast to, for example, methionine sulfoxide and cysteine disulfide bond formation, carbonylation is an irreversible oxidative process (Dalle-Donne et al, 2003). Thus, a cell must rid itself of carbonylated proteins by degrading them. Different sensitive methods have been developed for the detection and quantification of protein carbonyl groups and most of these involve derivatization of the carbonyl group with 2,4-dinitrophenol hydrazine and subsequent immunodetection of the resulting hydrazone (Figure 1) using monoclonal or polyclonal antibodies (Levine, 2002).
Figure 1.
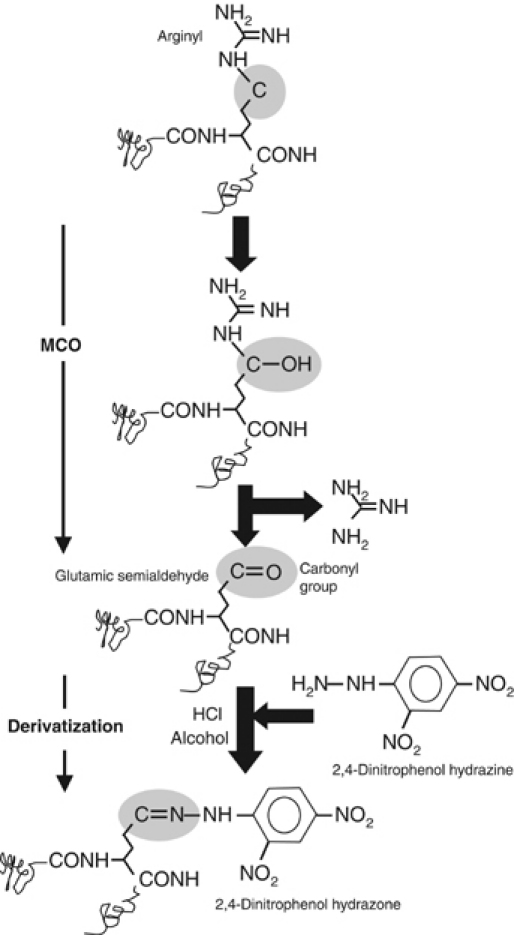
Carbonylation and derivatization of a protein amino-acid side chain. A scheme for the formation of glutamic semialdehyde from an arginyl residue is depicted as a consequence of an MCO. For detection, the carbonyl group, in this case glutamic semialdehyde, is subsequently derivatized by 2,4-dinitrophenol hydrazine. The resulting protein 2,4-dinitrophenol hydrazone can be detected by specific monoclonal or polyclonal antibodies (see Requena et al, 2001).
Aging-, starvation-, and stress-induced carbonylation does not affect the proteome uniformly (Levine, 2002). In E. coli, the targeted proteins include the Hsp70 and Hsp60 chaperones, the histone-like protein, H-NS, elongation factors, EF-Tu and EF-G, glutamine synthetase, glutamate synthase, aconitase, malate dehydrogenase, and pyruvate kinase (Dukan and Nyström, 1998, 1999; Tamarit et al, 1998). Some of these proteins have been demonstrated to be specifically carbonylated also in oxidation stressed yeast cells (Cabiscol et al, 2000), aging flies (Yan et al, 1997; Sohal, 2002), plants (Johansson et al, 2004; Kristensen et al, 2004), and in Alzheimer's diseased brain (Castegna et al, 2002). Thus, there is a general pattern of carbonylation specificity in distantly related organisms. The molecular basis for the apparent sensitivity of some proteins to carbonylation is not understood, but it is likely that MCO is an intrinsic problem for proteins containing transition metals (however, only a few of the carbonylated proteins are known to bind metals). In addition, some proteins (e.g. enzymes of the Kreb's cycle and electron transport chain) may have been classified as ‘oxidation-sensitive' mainly because they are located in the proximity of sites generating reactive oxygen species (ROS).
Different origins of protein carbonylation
The task of identifying the causal factors behind age-dependent increased carbonylation has proven difficult. Several principal possibilities can be envisioned, including (I) a decline in the antioxidant defence system, (II) an increased production of ROS, (III) a diminished capacity for removal of oxidized proteins, or (IV) an increased susceptibility of proteins to oxidative attack. Of course, these possibilities are not mutually exclusive, and evidence both for and against all these possibilities can be found in the literature depending on the cells, tissues, or organisms studied (e.g. Ji et al, 1990; Sohal and Weindruch, 1996; Dukan and Nyström, 1998, 1999; Dukan et al, 2000; Nyström, 2002; Sohal, 2002). For example, catalases have been demonstrated to decrease with age in some tissues, and it is possible that carbonylation levels increase as a direct consequence of a diminished activity of the primary antioxidant defence systems (Ji et al, 1990; Sohal et al, 1995). It appears that the classical enzymes involved in ROS detoxification, that is, superoxide dismutases, catalases, and peroxidases, are key members of the cellular defence also against protein carbonylation. But, in contrast to its essential role in attenuating illegitimate disulfide bond formation, the glutathione reductase system appears less important, at least in E. coli (Dukan and Nyström, 1998, 1999). Another important factor in the generation of ROS and protein carbonyls during aging and oxidative stress is the intracellular availability of free iron (Stadtman, 1992; Stadtman and Levine, 2000). For example, carbonylation levels are markedly higher in yeast mutants lacking the iron storage protein YFH1p, a homolog to the human frataxin (Desmyter et al, 2004). Interestingly, the human ferritin L gene can partly compensate for the lack of functional YFH1p, that is, it counteracts elevated carbonylation, and also prolongs the replicative lifespan of the yeast cells (Desmyter et al, 2004).
In yeast, increased carbonylation has also been linked to an increased tendency of the aging mitochondria to produce ROS rather than a diminished activity (or abundance) of antioxidant defence systems (Aguilaniu et al, 2001; see also Jazwinski, 2004). During carbon or nitrogen starvation, the respiratory state of the yeast mitochondria was found to shift from a coupled state-3 to a state-4 type respiration, which is characterized by a low activity of the ATP synthase, an elevated membrane potential, and an increased production of superoxide (Aguilaniu et al, 2001). This propensity of the mitochondria to produce ROS is subjected to control by the RAS signaling pathway (Hlavata et al, 2003) such that overactive RAS signaling elevates ROS production (and shortens lifespan) (Hlavata et al, 2003).
The third possibility listed above as a possible cause of increased carbonylation during aging relates to the activity of the cellular protease systems. It has been shown that the function of the proteasome decreases during aging in several human tissues as well as in senescent primary cultures, which may result in the accumulation of damaged proteins (Friguet et al, 2000; Sitte et al, 2000; Shringarpure and Davies, 2002). It has been suggested that this decreased proteolysis is a consequence of the accumulation of protease-resistant aggregates, ‘aggresomes', which bind to the proteasome (Grune et al, 2004). In other words, the aggresomes are argued to clog up the proteasomes and as a consequence, damaged (e.g. carbonylated) and potentially protease-susceptible substrates accumulate with time (Grune et al, 2004).
Recent data indicate that carbonylation of proteins may occur also in the absence of an increased ROS production, diminished ROS defence, or reduced protease activity. This pathway of carbonylation is rather linked to an increased production of substrates available for oxidative attack (Dukan et al, 2000; Ballesteros et al, 2001). This carbonylation is less specific than the one observed in aging organisms and long-term starved microbes and highlights an until now unknown link between ribosomal proofreading and protein oxidation. Proteomics demonstrated that this carbonylation is strongly associated with the production of aberrant protein isoforms (Ballesteros et al, 2001). In addition, the level of protein carbonylation was found to increase upon treatment of cells with antibiotics, for example, streptomycin and puromycin, and introduction of mutations causing increased mistranslation (Dukan et al, 2000). During these treatments, the rate of superoxide production and the activity of the superoxide dismutases and catalases were unchanged and the expression of oxidative stress defense genes did not increase (Dukan et al, 2000). In addition, it was demonstrated that stasis-induced carbonylation is drastically attenuated in mutants harboring intrinsically hyperaccurate ribosomes (Ballesteros et al, 2001). In other words, protein carbonylation of aberrant proteins does not require increased generation of ROS. Instead, the elevated oxidation of proteins in growth-arrested cells may be due to aberrant proteins being more susceptible to oxidative attack. The pool of aberrant proteins is elevated during bacterial stasis due to a reduced fidelity of the translational apparatus (O'Farrell, 1978; Barak et al, 1996; Wenthzel et al, 1998; Ballesteros et al, 2001).
Possible role of carbonyls in protein quality control
The rapid carbonylation of mistranslated or otherwise aberrant proteins points to an important physiological role of carbonylation in protein quality control. Since carbonylated proteins are more susceptible to proteolytic degradation than their nonoxidized counterparts (Dukan et al, 2000; Bota and Davies, 2002; Grune et al, 2003, 2004), the rapid carbonylation of an erroneous protein may ensure that such a polypeptide is directed to the proteolysis apparatus. Thus, carbonylation may act as a signal ensuring that damaged proteins enter the degradation pathway rather than the chaperone/repair pathway since carbonylation is an irreversible/unrepairable modification. This could effectively reduce incorporation of mistranslated proteins into mature machines (e.g. ribosomes and RNA and DNA polymerases) involved in information transfer. In line with this notion, carbonylated proteins generated as a result of increased mistranslation were found to be less stable than the average bulk protein (Dukan et al, 2000) and the degree of carbonylated proteins in mature ribosomes is small in healthy cells of E. coli (Desnues et al, 2003).
Another potentially important role of carbonylation is its involvement in autophagy-like mechanisms. In this context, carbonylation may act as a mechanism providing amino acids for de novo protein synthesis by targeting proteins that are no longer required, or have become damaged/aberrant, for degradation. It is not clear whether carbonylation and ubiquitinylation operate independently or in concert in tagging a protein for degradation in eukaryotic cells. However, the fact that carbonylation of a protein isolated from E. coli (lacking ubiquitin tagging systems) is recognized by the mammalian proteasome in vitro (Roseman and Levine, 1987; Rivett and Levine, 1990) suggests that carbonylation may bypass the need for ubiquitinylation, at least in this specific case (see also Shringarpure et al, 2003).
The above-mentioned carbonylation of aberrant proteins is rather unspecific and one may ask how the cell can achieve specificity using carbonylation as a marker for protein degradation. In other words, how can, for example, specific degradation of glutamine synthetase be achieved during nitrogen starvation by a carbonylation reaction? As mentioned, introduction of carbonyl groups in this enzyme marks it for subsequent degradation upon nitrogen (ammonium) starvation. One possibility for such a specific carbonylation of the enzyme during ammonium starvation is that the idle enzyme, like aberrant proteins, is more susceptible for oxidative carbonylation. Enzymes are frequently found to be protected by their substrates from proteolysis (Miller, 1987). Thus, certain enzymes become more susceptible to degradation during conditions where their substrates are present at diminishing levels (Miller, 1987). This is conceivably due to subtle conformational differences in the working and unoccupied forms of the enzyme. If the idle form of glutamine synthetase is more susceptible to oxidative carbonylation, this may impart an apparent specificity and regulation of carbonylation-dependent tagging for protein degradation during ammonium starvation. A summary of possible mechanisms rendering a protein more susceptible to carbonylation, and therefore degradation, is illustrated in Figure 2. It is conceivable that any condition, such as elevated temperatures, exposure to denaturing agents, and limitations or defects in the molecular chaperone systems, could cause an elevated concentration of aberrant proteins and therefore increased carbonylation (Figure 2). However, so far, the only means of generating aberrant proteins that have been shown experimentally to elevate carbonylation are those affecting the proofreading capacity of the ribosome (Nyström, 2002, 2004).
Figure 2.
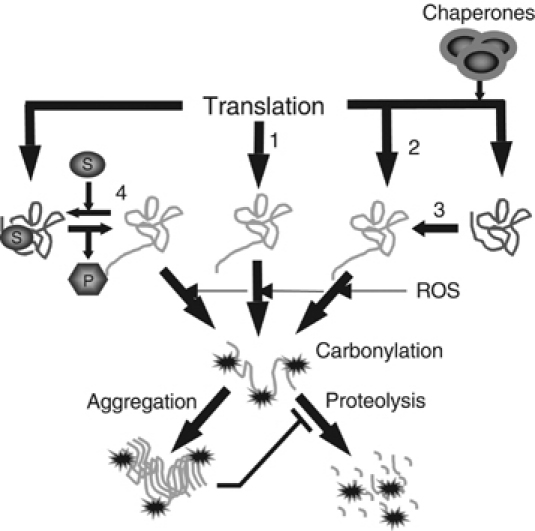
Schematic representation of pathways that can generate carbonylation-prone protein species and the fate of such proteins. Pathway 1 portrays the generation of aberrant proteins by mistranslation, for example, missense incorporation, frame-shifting, and stop-codon read-through. Pathway 2 signifies the formation of aberrant folding structures during chaperone deficiency, whereas pathway 3 is the formation of similar aberrant structures during specific stresses such as heat and exposure to denaturing agents. Pathway 4 illustrates the hypothetical carbonylation of an idle enzyme structure, which prevails during substrate deficiency. As long as the substrate is present and the enzyme working, it may be safeguarded from carbonylation and less prone to be attacked by proteases. Soluble species of carbonylated proteins appear ordained for proteolysis by, for example, the proteasome or Lon protease. However, highly carbonylated proteins form high-molecular-weight aggregates that are proteolysis-resistant. Such aggregates appear to inhibit protease functions.
Carbonylation—friend or foe?
It has been argued that oxidative modification of a protein makes it more susceptible to proteolysis largely due to unfolding of the targeted protein domains (Grune et al, 2004). Unfolding results in an increased exposure of hydrophobic residues that are normally hidden in the interior of soluble proteins and such hydrophobic patches are known to favor recognition and degradation by the proteasome and the Lon protease (Grune et al, 2003, 2004). A partly aberrant unfolded protein exposes amino-acid residues normally hidden in the protein structure, and introduction of carbonyl groups on those amino acids may simply result in further loss of the protein's integrity (Davies and Delsignore, 1987; Davies et al, 1987). In this scenario, carbonylation is just one of many possible oxidative modifications that may render a protein more prone for degradation. Since carbonylation is unrepairable, this modification may, however, be of special importance in directing the affected protein to the path toward degradation.
The described functions of carbonylation point to a beneficial role in protein quality control and protein metabolism. However, it turns out that carbonylation may be a mixed blessing. For example, while a mildly carbonylated aconitase is marked for proteolysis, a heavily oxidized form of the enzyme tends to form high-molecular-weight aggregates that escape degradation by the mitochondrial Lon protease (Bota and Davies, 2002). In addition, aggregates of severely oxidized proteins are poor substrates also for the proteasome and such aggregates can inhibit proteasome activity (e.g. Grune et al, 2004; Figure 2). It has been suggested that the decline in proteosomal activity during aging (e.g. Agarwal and Sohal, 1994; Bota et al, 2002) may, in fact, be closely connected to a gradual accumulation of proteolysis-resistant aggregates of oxidized proteins that bind and inhibit proteosomal function (Grune et al, 2004). Further, it is argued that this mechanism of inhibition of the proteasome is more dramatic in postmitotic cells, for example, neurons, than dividing cells since the latter can effectively dilute damaged proteins and aggregates by growth and proliferation (Grune et al, 2004).
Protein carbonylation is not an inevitable consequence of tissue aging
It has been demonstrated recently that the quantitative and qualitative pattern of protein carbonylation during progression through the life cycle of the plant Arabidopsis thaliana is distinct from that of the yeast, fly, nematode, bird, and mammalian aging systems. Carbonylation first increases with the age of the plant, similar to animals, but drops abruptly prior to the vegetative to reproductive transition (Johansson et al, 2004). Proteomic identification of the major targets for protein carbonylation indicates that proteins directly and indirectly linked to chloroplast activities exhibit special problems with respect to oxidative damage (Johansson et al, 2004; Kristensen et al, 2004). At first glance, this suggests that the progression of oxidative damage in the life cycle of animals and plants is fundamentally different. Indeed, it clearly demonstrates that increased protein carbonylation is not universally and inevitably related to the age of a biological tissue. However, one aspect of protein carbonylation is similar in animals and plants; that is, production of offspring occurs at a time at which the overall oxidative damage in the organism is low. In animals, this coincides with the early to middle stages of the organism's life cycle, whereas in A. thaliana, it signifies the end point of its developmental progression. This begs the question of the physiological significance of the drastic drop in protein oxidation preceding flower development and to what extent this may affect the fitness of the offspring. In addition, what is the temporal and molecular nature of the signals triggering the removal of damaged proteins and is the elimination of such damage (or the peak in damage preceding this removal) a prerequisite for the transition to flowering?
Segregation of carbonylated proteins—a novel defence mechanism of asymmetrically dividing cells
The yeast Saccharomyces cerevisiae, like A. thaliana, appears to have evolved the means of keeping the offspring free of carbonylated proteins but the mechanism is different. Yeast belong to the exclusive club of organisms exhibiting replicative aging. Most laboratory strains of S. cerevisiae can complete, on average, 20–30 divisions followed by death. During these progressive divisions, the cells undergo age-related changes, including an increased generation time, increase in size, decline in mating ability, and accumulation of extrachromosomal rDNA circles (ERCs). Thus, with each division, the mother cell becomes phenotypically older and more deteriorated but generates offspring exhibiting a full replicative potential. In other words, the acquired aging phenotype of the mother cell is somehow expunged in the progeny during the process of asymmetrical division (Jazwinski, 2004). This argues for the existence of a senescence factor that accumulates in the mother cells but is prevented from transmission to the offspring (Egilmez and Jazwinski, 1989). ERCs (Sinclair and Guarente, 1997) and dysfunctional mitochondria (Jazwinski, 2004) have been suggested to be such senescence factors, but conclusive evidence for one primary senescence factor in yeast is still lacking. In addition, an unbiased screen for age asymmetry mutants identified carbonylated proteins as one such potential senescence factor. Specifically, it was demonstrated that stable protein carbonyls are not inherited by the daughter cells during cytokinesis (Figure 3; Aguilaniu et al, 2003). The ability of mother cells to retain carbonylation-damaged proteins is diminished with the replicative age of the mother. Moreover, a screen for mutants with an abrogated ability to retain carbonylated proteins in the mother cell identified the sirtuin, Sir2p, as an essential component in segregating oxidative damage during mitosis (Aguilaniu et al, 2003). Intriguingly, Sir2p has previously been shown to be an aging determinant in organisms ranging from yeast, worms, and flies (Guarente, 2000; Tissenbaum and Guarente, 2001; Sinclair, 2002; Rogina and Helfand, 2004). Whether the aging phenotype of organisms lacking functional Sir proteins is somehow linked to management of oxidative damage is presently not known.
Figure 3.
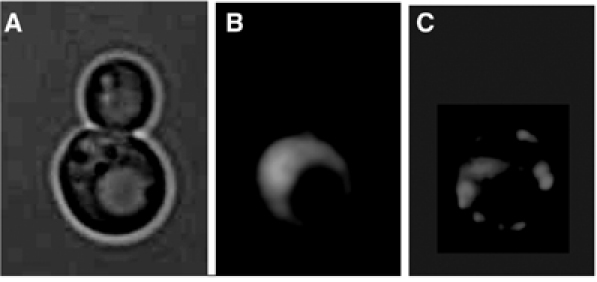
Segregation of carbonylated proteins during cytokinesis of the yeast S. cerevisiae. (A) Transmission electrograph of a yeast mother cell in the process of generating a daughter cell. (B) Levels of superoxide as determined by DHE staining in cells shown in panel A. (C) In situ detection of carbonylated proteins (see Aguilaniu et al, 2003) of cells shown in panel A.
Hints toward a mechanistic explanation for damage segregation in yeast come form experiments using drugs that effect proper arrangement of the cytoskeleton. Specifically, drugs that debilitate actin filament assembly inhibit damage segregation during cytokinesis (i.e. gives a phenocopy of sir2; Aguilaniu et al, 2003). In other words, it appears as if the cytoskeleton has a dual function in ensuring that daughters inherit a full complement of genomic material and active mitochondria while preventing inheritance of old and damaged macromolecules. Moreover, actin staining indicates that sir2 mutants are somewhat defective in actin filament organization during cytokinesis (Aguilaniu et al, 2003).
Future analysis may clarify whether damage segregation is restricted to asymmetrically dividing microbes or if this phenomenon is a common feature also in eukaryotic development, for example, during stem cell renewal or generation of the germ cell line.
Conclusion
From being first recognized as an oxidative modification tagging enzymes for proteolytic degradation, protein carbonylation has become a widespread indicator of severe oxidative damage and disease-derived protein dysfunction. The increase in protein carbonyls during aging in animals is quite substantial (Levine, 2002) eventually reaching a level of one out of every third protein molecule carrying this modification (Stadtman and Levine, 2000). In view of the fact that carbonylation is reducing, or totally abrogating, the targeted proteins catalytic functions and may trigger formation of high–molecular-weight, potentially cytotoxic, aggregates, this modification is likely to play havoc with cellular/tissue functions in the aging organism.
One might wonder why evolution has failed to provide the soma with more proficient defence systems that can more efficiently combat this damage throughout the organism's lifespan. This problem is adequately explained by the evolutionary theory of aging, which states that there is no need for such a defence. Aging is argued to be a consequence of the fact that natural selection's power to influence the fate of genes gradually wanes as the age at which those genes have their effects increases. In line with this concept, the carbonyl content increases at a drastically accelerated rate in the last third of an animal's lifespan (Levine, 2002); that is, after termination of the reproductive period. In other words, the carbonylation load of young individuals is sufficiently low not to affect the fitness of the offspring and the increased load of damage of the soma in late life is impervious to natural selection. However, these arguments are only applicable to organisms whose life history entails early reproduction. If a high carbonyl load is a genuine hazard to fitness, organisms producing reproductive organs at the closing stages of their development should have evolved means of keeping protein damage low throughout their lifespan or, alternatively, be equipped with systems that can clear out damage prior to reproduction. Interestingly, it appears that the plant A. thaliana has evolved defence systems to do just that (Johansson et al, 2004). In addition, some unicellular organisms display previously unrecognized means of keeping the young free of carbonylation damage. The asymmetrically dividing yeast S. cerevisiae has evolved a Sir2p-dependent system that retains carbonylated proteins in the mother cell compartment during mitotic cytokinesis (Aguilaniu et al, 2003). Thus, the progeny, exhibiting a full reproductive potential in contrast to the mother cell, starts out with a markedly reduced load of damage compared to the ancestor cell. Prokaryotic cell cultures, that is, of E. coli, subjected to starvation break up into two populations exhibiting markedly different loads of carbonylated proteins (Desnues et al, 2003). The cells displaying low carbonyl levels remain reproductively competent, whereas cells with a high carbonyl load become genetically dead (nonculturable). Whether this starvation-induced heterogeneity in carbonylation and fitness is programmed or stochastic remains to be elucidated.
The role of carbonylation may change with the age of the organisms. From being an important part of protein quality control and a reaction that labels aberrant, damaged, or idle enzymes for degradation, it may later become a menace and negatively influence cellular functions, including the function of the proteasomes. Future analysis may illustrate whether an organism's ability to retain the former function of carbonylation modifications as long as possible also affects its longevity or fitness.
Acknowledgments
Past and present colleagues of the ‘Nyström lab' are greatly acknowledged for contributing to the work described in this review. I thank The Swedish Research Council, VR, for long-term support of the cellular senescence project and the Göran Gustafsson Foundation for an award in Molecular Biology.
References
- Agarwal S, Sohal RS (1994) Aging and proteolysis of oxidized proteins. Arch Biochem Biophys 309: 24–28 [DOI] [PubMed] [Google Scholar]
- Aguilaniu H, Gustafsson L, Rigoulet M, Nyström T (2001) Protein oxidation in G0 cells of Saccharomyces cerevisiae depends on the state rather than rate of respiration and is enhanced in pos9 but not yap1 mutants. J Biol Chem 276: 35396–35404 [DOI] [PubMed] [Google Scholar]
- Aguilaniu H, Gustafsson L, Rigoulet M, Nyström T (2003) Asymmetric inheritance of oxidatively damaged proteins during cytokinesis in Saccharomyces cerevisiae: a Sir2p dependent mechanism. Science 299: 1751–1753 [DOI] [PubMed] [Google Scholar]
- Ballesteros M, Fredriksson A, Henriksson J, Nyström T (2001) Bacterial senescence: protein oxidation in non-proliferating cells is dictated by the accuracy of the ribosomes. EMBO J 20: 5280–5289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barak Z, Gallant J, Lindsley D, Kwieciszewki B, Heidel D (1996) Enhanced ribosome frameshifting in stationary phase cells. J Mol Biol 263: 140–148 [DOI] [PubMed] [Google Scholar]
- Bota DA, Davies KJ (2002) Lon protease preferentially degrades oxidized mitochondrial aconitase by an ATP-stimulated mechanism. Nat Cell Biol 4: 674–680 [DOI] [PubMed] [Google Scholar]
- Bota DA, Van Remmen H, Davies KJ (2002) Modulation of Lon protease activity and aconitase turnover during aging and oxidative stress. FEBS Lett 532: 103–106 [DOI] [PubMed] [Google Scholar]
- Cabiscol E, Piulats E, Echave P, Herrero E, Ros J (2000) Oxidative stress promotes specific protein damage in Saccharomyces cerevisiae. J Biol Chem 275: 27393–27398 [DOI] [PubMed] [Google Scholar]
- Castegna A, Aksenov M, Aksenova M, Thongboonkerd V, Klein JB, Pierce WM, Booze R, Markesbery WR, Butterfield DA (2002) Proteomic identification of oxidatively modified proteins in Alzheimer's disease brain. Part I: creatine kinase BB, glutamine synthase, and ubiquitin carboxy-terminal hydrolase L-1. Free Radic Biol Med 33: 562–571 [DOI] [PubMed] [Google Scholar]
- Dalle-Donne I, Giustarini D, Colombo R, Rossi R, Milzani A (2003) Protein carbonylation in human diseases. Trends Mol Med 9: 169–176 [DOI] [PubMed] [Google Scholar]
- Davies KJ, Delsignore ME (1987) Protein damage and degradation by oxygen radicals. III. Modification of secondary and tertiary structure. J Biol Chem 262: 9908–9913 [PubMed] [Google Scholar]
- Davies KJ, Delsignore ME, Lin SW (1987) Protein damage and degradation by oxygen radicals. II. Modification of amino acids. J Biol Chem 262: 9902–9907 [PubMed] [Google Scholar]
- Desmyter L, Dewaele S, Verstraelen J, Nyström T, Contreras R, Chen C (2004) Expression of the human ferritin light chain in a budding yeast frataxin mutant affects aging and cell death. Exp Gerontol 39: 707–715 [DOI] [PubMed] [Google Scholar]
- Desnues B, Gregori G, Dukan S, Aguilaniu H, Nyström T (2003) Differential oxidative damage and expression of stress regulons in culturable and nonculturable cells of Escherichia coli. EMBO Rep 4: 400–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dukan S, Farewell A, Ballesteros M, Taddei F, Radman M, Nyström T (2000) Protein oxidation in response to increased transcriptional or translational errors. Proc Natl Acad Sci USA 97: 5746–5749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dukan S, Nyström T (1998) Bacterial senescence: stasis results in increased and differential oxidation of cytoplasmic proteins leading to developmental induction of the heat shock regulon. Genes Dev 12: 3431–3441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dukan S, Nyström T (1999) Oxidative stress defense and deterioration of growth-arrested Escherichia coli cells. J Biol Chem 274: 26027–26032 [DOI] [PubMed] [Google Scholar]
- Egilmez NK, Jazwinski SH (1989) Evidence for the involvement of a cytoplasmic factor in the aging of the yeast Saccharomyces cerevisiae. J Bacteriol 171: 37–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friguet B, Bulteau AL, Chondrogianni N, Conconi M, Petropoulos I (2000) Protein degradation by the proteasome and its implications in aging. Ann NY Acad Sci 908: 143–154 [DOI] [PubMed] [Google Scholar]
- Grune T, Jung T, Merker K, Davies KJA (2004) Decreased proteolysis caused by protein aggregates, inclusion bodies, plaques, lipofuscin, ceroid, and ‘aggresomes' during oxidative stress, aging, and disease. Int J Biochem Cell Biol 36: 2519–2530 [DOI] [PubMed] [Google Scholar]
- Grune T, Merker K, Sandig G, Davies KJ (2003) Selective degradation of oxidatively modified protein substrates by the proteasome. Biochem Biophys Res Commun 305: 709–718 [DOI] [PubMed] [Google Scholar]
- Guarente L (2000) Sir2 links chromatin silencing, metabolism, and aging. Genes Dev 14: 1021–1026 [PubMed] [Google Scholar]
- Hlavata L, Aguilaniu H, Pichova A, Nystrom T (2003) The oncogenic RAS2(val19) mutation locks respiration, independently of PKA, in a mode prone to generate ROS. EMBO J 22: 3337–3345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jazwinski SM (2004) Yeast replicative life span—the mitochondrial connection. FEMS Yeast Res 5: 119–125 [DOI] [PubMed] [Google Scholar]
- Ji LL, Dillon D, Wu E (1990) Alteration of antioxidant enzymes with aging in rat skeletal muscle and liver. Am J Physiol 258: 918–923 [DOI] [PubMed] [Google Scholar]
- Johansson EC, Olsson O, Nystrom T (2004) Progression and specificity of protein oxidation in the life cycle of Arabidopsis thaliana. J Biol Chem 279: 22204–22208 [DOI] [PubMed] [Google Scholar]
- Kristensen BK, Askerlund P, Bykova NV, Egsgaard H, Moller IM (2004) Identification of oxidised proteins in the matrix of rice leaf mitochondria by immunoprecipitation and two-dimensional liquid chromatography–tandem mass spectrometry. Phytochemistry 65: 1839–1851 [DOI] [PubMed] [Google Scholar]
- Lass A, Sohal BH, Weindruch R, Forster MJ, Sohal RS (1998) Caloric restriction prevents age-associated accrual of oxidative damage to mouse skeletal muscle mitochondria. Free Radic Biol Med 25: 1089–1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine RL (1983) Oxidative modification of glutamine synthetase. I. Inactivation is due to loss of one histidine residue. J Biol Chem 258: 11823–11827 [PubMed] [Google Scholar]
- Levine RL (2002) Carbonyl modified proteins in cellular regulation, aging, and disease. Free Radic Biol Med 32: 790–796 [DOI] [PubMed] [Google Scholar]
- Levine RL, Oliver CN, Fulks RM, Stadtman ER (1981) Turnover of bacterial glutamine synthetase: oxidative inactivation precedes proteolysis. Proc Natl Acad Sci USA 78: 2120–2124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller CG (1987) Protein degradation and proteolytic modification. In Escherichia coli and Salmonella typhimurium, Cellular and Molecular Biology, Neidhardt FC (ed) pp 680–691. Washington, DC: ASM press [Google Scholar]
- Nyström T (2002) Translational fidelity, protein oxidation, and senescence: lessons from bacteria. Aging Res Rev 1: 693–703 [DOI] [PubMed] [Google Scholar]
- Nyström T (2004) Stationary phase physiology. Ann Rev Microbiol 58: 161–181 [DOI] [PubMed] [Google Scholar]
- O'Farrell PH (1978) The suppression of defective translation by ppGpp and its role in the stringent response. Cell 14: 545–557 [DOI] [PubMed] [Google Scholar]
- Oliver CN, Ahn BW, Moerman EJ, Goldstein S, Stadtman ER (1987) Age-related changes in oxidized proteins. J Biol Chem 262: 5488–5491 [PubMed] [Google Scholar]
- Requena JR, Chao C-C, Levine RL, Stadtman ER (2001) Glutamic and aminoadipic semialdehydes are the main carbonyl products of metal-catalyzed oxidation of proteins. Proc Natl Acad Sci USA 98: 69–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Requena JR, Levine RL, Stadtman ER (2003) Recent advances in the analysis of oxidized proteins. Amino Acids 25: 221–226 [DOI] [PubMed] [Google Scholar]
- Rivett AJ, Levine RL (1990) Metal-catalyzed oxidation of Escherichia coli glutamine synthetase: correlation of structural and functional changes. Arch Biochem Biophys 278: 26–34 [DOI] [PubMed] [Google Scholar]
- Rogina B, Helfand SL (2004) Sir2 mediates longevity in the fly through a pathway related to calorie restriction. Proc Natl Acad Sci USA 101: 15998–16003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roseman JE, Levine RL (1987) Purification of a protease from Escherichia coli with specificity for oxidized glutamine synthetase. J Biol Chem 262: 2101–2110 [PubMed] [Google Scholar]
- Shringarpure R, Davies KJ (2002) Protein turnover by the proteasome in aging and disease. Free Radic Biol Med 32: 1084–1089 [DOI] [PubMed] [Google Scholar]
- Shringarpure R, Grune T, Mehlhase J, Davies KJ (2003) Ubiquitin conjugation is not required for the degradation of oxidized proteins by proteasome. J Biol Chem 278: 311–318 [DOI] [PubMed] [Google Scholar]
- Sinclair DA (2002) Paradigms and pitfalls of yeast longevity research. Mech Ageing Dev 123: 857–867 [DOI] [PubMed] [Google Scholar]
- Sinclair DA, Guarente L (1997) Extrachromosomal rDNA circles—a cause of aging in yeast. Cell 91: 1033–1042 [DOI] [PubMed] [Google Scholar]
- Sitte N, Merker K, von Zglinicki T, Grune T, Davies KJA (2000) Protein oxidation and degradation during cellular senescence of human BJ fibroblasts: part I—effects of proliferative senescence. FASEB J 14: 2495–2502 [DOI] [PubMed] [Google Scholar]
- Stadtman ER (1992) Protein oxidation and aging. Science 257: 1220–1224 [DOI] [PubMed] [Google Scholar]
- Stadtman ER (2001) The story of glutamine synthetase regulation. J Biol Chem 276: 44357–44364 [DOI] [PubMed] [Google Scholar]
- Stadtman ER, Levine RL (2000) Protein oxidation. Ann NY Acad Sci 899: 191–208 [DOI] [PubMed] [Google Scholar]
- Sohal RS (2002) Role of oxidative stress and protein oxidation in the aging process. Free Radic Biol Med 33: 37–44 [DOI] [PubMed] [Google Scholar]
- Sohal RS, Agarwal S, Dubey A, Orr WC (1993) Protein oxidative damage is associated with life expectancy of houseflies. Proc Natl Acad Sci USA 90: 7255–7259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohal RS, Agarwal S, Sohal BH (1995) Oxidative stress and aging in the Mongolian gerbil (Meriones unguiculatus). Mech Ageing Dev 81: 15–25 [DOI] [PubMed] [Google Scholar]
- Sohal RS, Weindruch R (1996) Oxidative stress, caloric restriction, and aging. Science 273: 59–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamarit J, Cabiscol E, Ros J (1998) Identification of the major oxidatively damaged proteins in Escherichia coli cells exposed to oxidative stress. J Biol Chem 273: 3027–3032 [DOI] [PubMed] [Google Scholar]
- Tissenbaum HA, Guarente L (2001) Increased dosage of a sir-2 gene extends lifespan in Caenorhabditis elegans. Nature 410: 227–230 [DOI] [PubMed] [Google Scholar]
- Yan LJ, Levine RL, Sohal RS (1997) Oxidative damage during aging targets mitochondrial aconitase. Proc Natl Acad Sci USA 94: 11168–11172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan LJ, Sohal RS (1998) Mitochondrial adenine nucleotide translocase is modified oxidatively during aging. Proc Natl Acad Sci USA 95: 12896–12901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenthzel AM, Stancek M, Isaksson LA (1998) Growth phase dependent stop codon readthrough and shift of translation reading frame in Escherichia coli. FEBS Lett 421: 237–242 [DOI] [PubMed] [Google Scholar]


